Abstract
The ORM1 (Saccharomyces cerevisiae)-like proteins (ORMDLs) and their yeast orthologs, the Orms, are negative homeostatic regulators of the initiating enzyme in sphingolipid biosynthesis, serine palmitoyltransferase (SPT). Genome-wide association studies have established a strong correlation between elevated expression of the endoplasmic reticulum protein ORMDL3 and risk for childhood asthma. Here we test the notion that elevated levels of ORMDL3 decrease sphingolipid biosynthesis. This was tested in cultured human bronchial epithelial cells (HBECs) (an immortalized, but untransformed, airway epithelial cell line) and in HeLa cells (a cervical adenocarcinoma cell line). Surprisingly, elevated ORMDL3 expression did not suppress de novo biosynthesis of sphingolipids. We determined that ORMDL is expressed in functional excess relative to SPT at normal levels of expression. ORMDLs and SPT form stable complexes that are not increased by elevated ORMDL3 expression. Although sphingolipid biosynthesis was not decreased by elevated ORMDL3 expression, the steady state mass levels of all major sphingolipids were marginally decreased by low level ORMDL3 over-expression in HBECs. These data indicate that the contribution of ORMDL3 to asthma risk may involve changes in sphingolipid metabolism, but that the connection is complex.
Keywords: ceramides, enzymology/enzyme regulation, lung, sphingosine phosphate, endoplasmic reticulum, Orm, asthma, ORM1 (Saccharomyces cerevisiae)-like protein
The ORM1 (Saccharomyces cerevisiae)-like proteins (ORMDLs) and their yeast orthologs, the ORMs, regulate the initiating and rate-limiting enzyme in sphingolipid biosynthesis, serine palmitoyltransferase (SPT) (1–4). The three ORMDL isoforms (ORMDL1–3) are small (17.5 kDa) hydrophobic membrane proteins that are situated in the endoplasmic reticulum along with SPT. The three isoforms are highly homologous and their functions appear to be redundant at the cellular level (3, 4). The ORMDLs are negative regulators of SPT and mediate the homeostatic regulation of SPT in response to cellular sphingolipid levels. siRNA depletion of the ORMDLs results in elevated sphingolipid synthesis under control conditions and reverses the homeostatic inhibition of sphingolipid synthesis resulting from elevated cellular sphingolipid content (1, 3, 4). The Orms and ORMDLs form stable physical complexes with SPT (1), even under conditions in which the ORMDLs are minimally inhibiting SPT. In yeast, the Orm regulatory activity is controlled by Orm phosphorylation. This may not be the case for the mammalian ORMDLs, which lack the sequences that are phosphorylated in the yeast Orms. Clinical interest in ORMDL function has been sparked by the observation that SNPs adjacent to the ORMDL3 gene are highly correlated with risk for childhood asthma (5, 6). Cookson and colleagues determined that the risk allele correlates with elevated ORMDL3 mRNA expression (5, 7). It has been suggested that elevated ORMDL3 increases the risk for asthma by depressing sphingolipid biosynthesis (8). Here we directly test whether ORMDL3 overexpression inhibits de novo sphingolipid biosynthesis in cells derived from one of the tissues important for the asthmatic phenotype, the airway epithelium. We find, surprisingly, that elevated ORMDL3 does not suppress de novo sphingolipid biosynthesis and present evidence that this is because ORMDLs are normally expressed in functional excess relative to SPT.
MATERIALS AND METHODS
Materials
Silencer® Select siRNA oligonucleotides for human ORMDL1–3 (catalog numbers s41258, s26474, and s41260, respectively) were from Ambion® (part of Life Technologies, Grand Island, NY). The gene-specific TaqMan® probes for quantitative real-time PCR, TaqMan® Gene Expression Master Mix, high-capacity reverse transcription kit, and MicroAmp® Fast Optical 96-well PCR plates were all from Applied Biosystems® (part of Life Technologies). The Lipofectamine® RNAiMax for siRNA delivery, Lipofectamine® 2000 transfection reagent, and TRIzol® were from Invitrogen (Life Technologies). The In-Fusion® HD Cloning Plus kit and CloneAmp™ HiFi PCR Premix were both from Clontech Laboratories (Mountain View, CA). L-[3H(G)]-serine, scintillation vials, and scintillation fluid were from Perkin Elmer (Waltham, MA). C16-ceramide standard was from Avanti Polar Lipids (Alabaster, AL). Whatman TLC plates were from VWR (Radnor, PA). HeLa cells were from ATCC (Manassas, VA). A549 lung adenocarcinoma cells were from ATCC. Human bronchial epithelial cells (HBECs) (9) were a generous gift of Dr. Jerry Shay, University of Texas, Southwestern, Department of Cell Biology. Keratinocyte serum-free medium for culturing HBECs was from Invitrogen (Life Technologies). DMEM and all other cell culture supplies were from VWR. Anti-SPT long chain base subunit 1 (SPTLC1) antibody was from Santa Cruz Biotechnology, Inc. (Dallas, TX). Anti-ORMDL antibody was from EMD Millipore (Billerica, MA). Anti-beta actin and secondary antibodies were all from Pierce (part of Thermo Scientific, Rockford, IL). Organic solvents were from Thermo Fisher Scientific (Pittsburgh, PA). Phosphorous standard and all other chemicals used were from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated.
Methods
Measurement of de novo ceramide biosynthesis.
Measurement of de novo ceramide biosynthesis in intact cells was accomplished by incorporation of 3H-serine into ceramide during a 60 minute incubation, as previously described (3). Briefly, HeLa cells and HBECs were plated in 12-well plates at various densities depending on the protocol in complete medium overnight. The next day, the HeLa cells were transfected as described below and the stable HBECs were cultured until desired confluence was reached. Prior to labeling, cells were treated with 10 μM ceramide-N-hexanoyl-D-erythro-sphingosine (C6)-ceramide for 60 minutes, where indicated. Cells were labeled in serine-depleted medium (MEM containing 1% dialyzed FBS and 10 μCi/ml 3H-serine) using 1 ml/well for 60 minutes. Total lipids were extracted by a modification of the Bligh-Dyer extraction (3) and the organic phase was dried under nitrogen. The entire organic phase was then subjected to TLC using a solvent system of chloroform:acetic acid:methanol (90:10:2, v/v) and C16-ceramide was spotted in each lane as a standard. After visualizing the ceramide band by exposure in an iodine tank, the ceramide bands were scraped and incorporated 3H-serine was measured by liquid scintillation counting.
Measurement of SPT activity in permeabilized cells.
HBECs were plated into collagen-coated 24-well plates at 0.3 × 106 cells/well overnight. Cells were then washed with PBS and permeabilized by treatment with 200 μg/ml digitonin for 3 minutes. SPT activity was measured in a buffer containing 50 mM HEPES (pH 8.0), 1 mM MgCl2, 1 mM ATP, 20 μM 5′ pyridoxyl phosphate, 50 μM palmitoyl CoA, 1 mM serine, and 3H-serine at 10 μCi/ml in a volume of 400 μl. Incubations were performed for 60 minutes at 37°C. Following incubation, cells were washed with PBS and 400 μl PBS was added to each well. Alkaline methanol (400 μl) (7 g KOH/l) was added and cells were harvested into microcentrifuge tubes. CHCl3 (100 μl) was added and extracts were vortexed. Phases were broken by the addition of 500 μl CHCl3, 500 μl alkaline water (100 μl 2 N NH4OH/100 ml water), and 100 μl 2 N NH4OH. The top aqueous layer was aspirated and the remaining organic layer was washed two times with 1 ml alkaline water. Three hundred and fifty microliters of the organic layer was dried under N2 in a scintillation vial and incorporated 3H-serine was measured by liquid scintillation counting.
siRNA depletion of ORMDL1–3 in HeLa cells.
HeLa cells were plated at 8 × 104 cells/well in collagen-coated 12-well plates overnight in complete medium. siRNA oligonucleotide (5 nmol) for each ORMDL isoform targeted was combined with Lipofectamine® RNAiMax transfection reagent per the manufacturer’s recommendations, added to medium, and cells were incubated 24–48 hours before measurement of de novo ceramide biosynthesis, as described above. The effect of siRNA treatment of expression of ORMDL1–3 was determined by quantitative real-time PCR using gene-specific TaqMan® probes, as previously described (3) and detailed below.
Overexpression of recombinant ORMDL3 and SPT in HeLa cells.
Experiments illustrated in Figs. 2, 3, 5, and 6 utilized mouse ORMDL3 (not epitope tagged), subcloned into the XbaI site of pCMV6-XL5 (Origene, Rockville, MD) by In-Fusion® cloning (Clontech) according to the manufacturer’s instructions using the following primers: forward, 5’-GCT TGT CGA CTC TAG ACC ACC ATG AAT GTG GGC A-3’ reverse, 5’-GCG GCC GCA ATC TAG ATC AGT ACT TAT TGA TTC CAA AG-3’. The immunoprecipitation (IP) experiments illustrated in Fig. 5 utilized human ORMDL3 in pCMV6 (epitope-tagged with Myc and FLAG, both at the carboxy-terminus) purchased from Origene (catalog number RC202279). Note that the amino acid sequences of the mouse (NCBI reference sequence NP_079937.1) and human (NCBI reference sequence ID AAM43507.1) ORMDL3s are 96% identical and of the exact same length. Single-chain SPT (scSPT) consists of a fusion of SPT long chain base subunit 2 (SPTLC2), SPT small subunit a (ssSPTa), and SPTLC1, in that order as described (10), subcloned into the Xba1 site of pCMV6-XL5 (Origene) by In-Fusion® cloning (Clontech) according to the manufacturer’s instructions using the following primers: forward, 5′-GCT TGT CGA CTC TAG ACC ACC ATG GCT AGG CGG CCG GAG C-3′ reverse, 5′-GCG GCC GCA ATC TAG ATC AGA GCA GGA CGG CCT GGG-3′. HeLa cells were plated at 1 × 105 cells/well in collagen-coated 12-well plates overnight in complete medium. The next day, medium was replaced with a transfection mix containing 1.6 μg of total plasmid and 4 μl of Lipofectamine 2000® (Invitrogen) according to the manufacturer’s instructions.
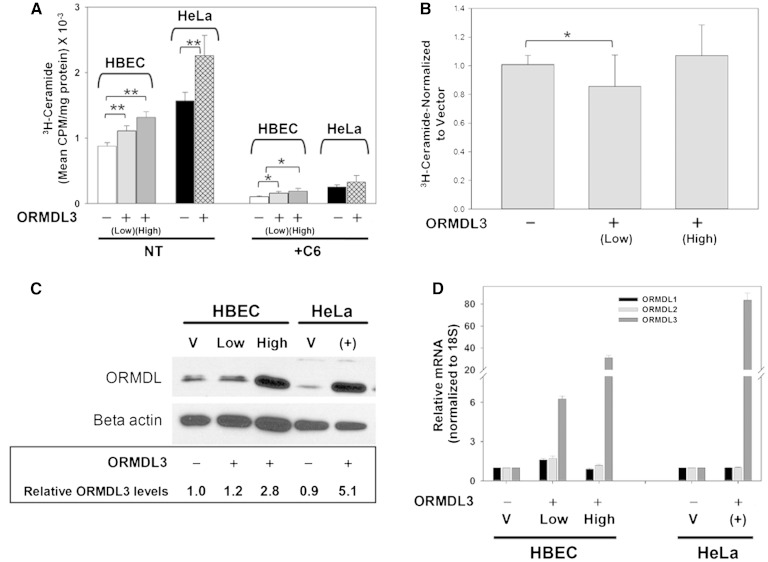
Fig. 2.
Overexpression of ORMDL3, both transiently and in stably transfected cells, does not depress de novo ceramide biosynthesis. A: Stable cell lines overexpressing either empty vector or human ORMDL3 (not epitope tagged) were generated in HBECs using lentiviral constructs as described in the Materials and Methods. Stable lines were chosen based on levels of ORMDL3 protein expression and data is reported for low (light gray bar) and high (dark gray bar) expressing lines. Cell monolayers were incubated for 60 min in the presence (+C6) or absence (NT) of 10 μM C6-ceramide after which they were labeled for 60 min with 3H-serine and incorporation of 3H-serine into ceramide was determined as described in the Materials and Methods. HeLa cells were transiently transfected with either empty vector or mouse ORMDL3 (not epitope tagged) for 24 h prior to treatment with 10 μM C6-ceramide, after which they were labeled for 60 min with 3H-serine as described above. Results for all data shown are the means ± SD of replicates (n = 4–6). Shown are representative results from at least three independent experiments. *Significant at P < 0.05, **significant at P < 0.005 by Student’s t-test. B: Permeabilized HBEC cultures were assayed for SPT activity. 3H-serine incorporation into total sphingolipids was measured as described in the Materials and Methods using HBEC cell lines expressing either a vector control [ORMDL3 (−)] or moderately or highly elevated levels of ORMDL3 [ORMDL3 (+)]. Shown are results from four experiments in which counts for each sample have been normalized to the vector-control samples. Each experiment included four replicates of each condition. Results for all data shown are the means ± SD of the normalized data. Replicate wells were also incubated in the presence of 1 μM myriocin as a control (not shown), which eliminated labeling by over 90%. *Significant at P < 0.05 by Student’s t-test. ORMDL3-High did not differ significantly from control. C: Cell lysates were generated from both stably transfected HBECs and transiently transfected HeLa cells that were transfected with either empty vector (V) or mouse ORMDL3 (not epitope tagged) as described in the Materials and Methods. Western blotting and densitometry were performed as described in the Materials and Methods. Numerical values are relative expression levels as compared with vector-only HBECs. D: One well of each 12-well plate used for ceramide labeling studies [see (A)] was lifted and gene expression was determined by quantitative real-time PCR as described in the Materials and Methods. Results are normalized to levels of 18S internal control and represented as fold change in mRNA expression levels as calculated by the ΔΔCT formula.
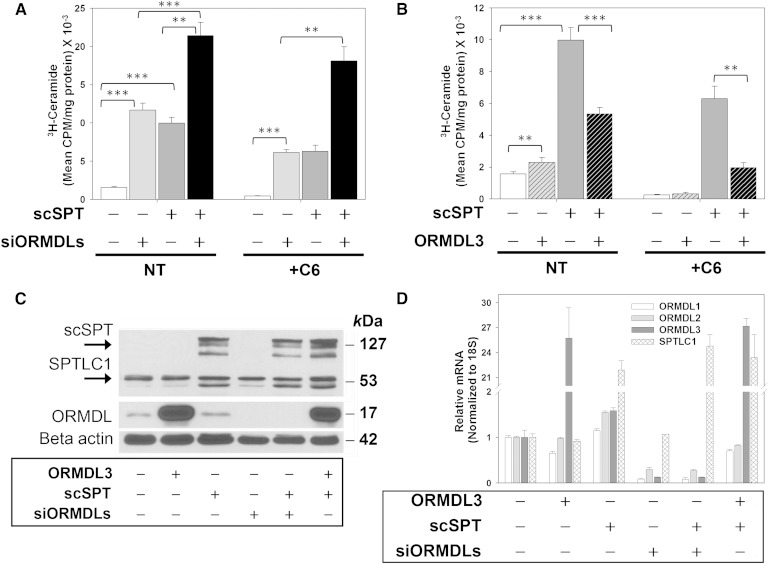
Fig. 3.
ORMDL3 overexpression inhibits SPT expressed at elevated levels in HeLa cells. A: Forty-eight hours prior to labeling total lipids with 3H-serine, HeLa cells were transfected with either scrambled or ORMDL-specific siRNA oligonucleotides targeting all three isoforms of ORMDL. Twenty-four hours after ORMDL knockdown, cells were transiently transfected with either empty vector or a construct expressing three subunits of SPT as a single polypeptide (scSPT) as described in the Materials and Methods. Labeling and lipid extraction/quantitation were performed exactly as outlined for Fig. 2. NT, not treated with C6-ceramide; +C6, treated with 10micromolar C-6 ceramide for 60 minutes as described in Methods. Open bars, vector/control siRNA transfected. Light grey bars, transfected with siRNA directed against all three ORMDLs as described in Methods. Dark grey bars, transfected with scSPT. Black bars, transfected with both scSPT and with ORMDL siRNA. B: Twenty-four hours prior to labeling total lipids with 3H-serine, HeLa cells were transiently transfected with either empty vector, mouse ORMDL3 (not epitope tagged), scSPT, or a combination of ORMDL3 and scSPT as described in the Materials and Methods. Labeling and lipid extraction/quantitation were performed exactly as outlined for (A). C: Cell lysates were generated from one well of each 12-well plate used in the ceramide labeling studies outlined above. SDS-PAGE electrophoresis followed by Western blot analysis, as described in the Materials and Methods, was performed to determine levels of protein expression for all cell lines used for ceramide labeling studies. D: Real-time PCR was performed as described for Fig. 2D. A, B: Results are representative of at least three experiments, at least four replicates/sample. Mean ± SD is shown. **Significant to P < 0.005, ***significant to P < 0.0005 by Student’s t-test.
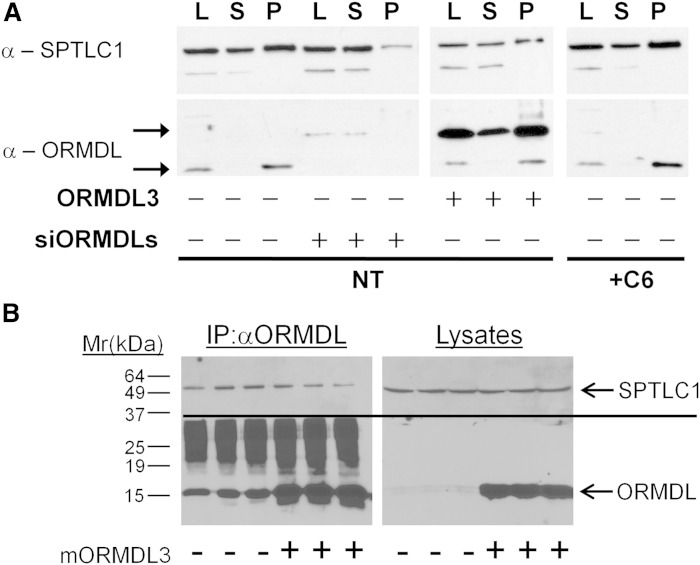
Fig. 5.
ORMDL3 and SPT are constitutively associated and levels of the complex are not increased by ORMDL3 overexpression or affected by elevation of cellular ceramide. A: Twenty-four hours prior to harvesting and IP, HeLa cells were transiently transfected with either scrambled siRNA oligonucleotides or siRNA olignucleotides directed against all three ORMDL isoforms (siORMDLs) as a control for the specificity of IP. A separate set of cells was transfected either with empty vector or human ORMDL3 (epitope-tagged) as described in the Materials and Methods. A third set of untransfected cells was incubated either with vehicle or 10 μM C-6 ceramide for 60 min prior to harvest. ORMDLs were immunoprecipitated and Western blotting performed as described in the Materials and Methods. Note that the overexpressed ORMDL3 is epitope tagged and so migrates at an increased molecular mass as compared with endogenous ORMDLs (P, post-IP beads; S, post-IP supernatant; L, pre-IP lysate). B: Overexpression of non-epitope-tagged mouse ORMDL3 does not increase co-IP of SPTLC1 with ORMDL. Hela cells, in triplicate, were transfected either with a vector control (−) or with mouse ORMDL3 lacking any epitope tags (+). Lysates were prepared and immunoprecipitated with anti-ORMDL antibody as described in the Materials and Methods. Immunoprecipitates and lysates were immunoblotted for ORMDL (below line) and SPTLC1 (above line) as described in the Materials and Methods.
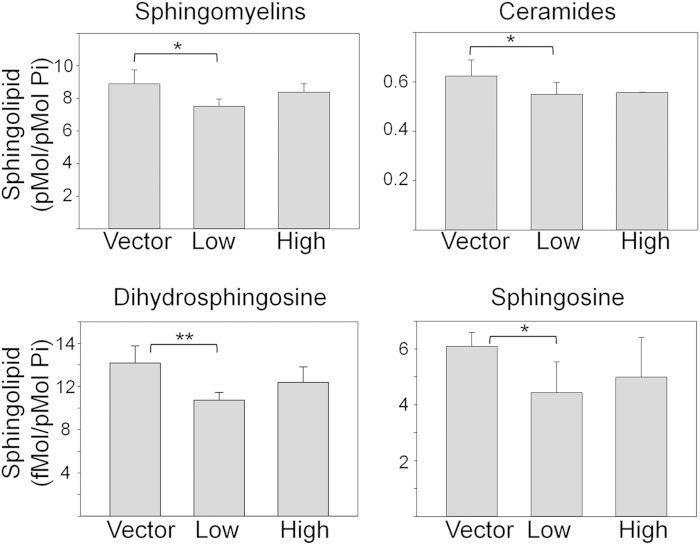
Fig. 6.
Steady state levels of sphingolipids in HBECs stably expressing ORMDL3. Stable HBEC cell lines overexpressing either empty vector or human ORMDL3 (not epitope tagged) at moderately (Low) or highly (High) elevated levels were extracted and subjected to analysis by mass spectroscopy as described in the Materials and Methods. Data represent the means ± SD of replicates (n = 6) and are representative of two independent experiments. *Significant to P < 0.05, **significant to P < 0.005 by Student’s t-test.
IP of ORMDLs in HeLa cells.
For siRNA of ORMDL1–3, HeLa cells were plated at 1 × 106 cells in collagen-coated p100 dishes overnight in complete medium. The next day, cells were transfected using 5 nM siRNA oligonucleotides for each ORMDL isoform being targeted and Lipofectamine RNAiMax transfection solution following the manufacturer’s instructions. Forty-eight hours after siRNA treatment, cells were lifted with trypsin, pelleted, washed with 1× PBS then solubilized on ice for 60 minutes with 1% digitonin IP buffer [25 mM Tris (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 15% glycerol, and 1% digitonin (w/v)]. For overexpression of recombinant ORMDL3, HeLa cells were plated at 3 × 106 cells in collagen-coated p100 dishes overnight in complete medium. The next day, cells were transfected with 25 μg plasmid DNA and 50 μl Lipofectamine® 2000 per p100 dish using the manufacturer’s instructions. Twenty-four hours after transfection, cells were lifted with trypsin, pelleted, washed with 1× PBS then solubilized for 60 minutes on ice, as described above. Solubilized cells were centrifuged at 10,000 rpm for 10 minutes at 4°C and lysates were moved to clean microfuge tubes. Lysates were then pre-cleared with Protein A beads for 1 hour at 4°C, after which beads were pelleted and discarded and cleared lysates were transferred to clean microfuge tubes. Cleared lysates were incubated on rotator at 4°C overnight with anti-ORMDL polyclonal antibody (Millipore). Following IP, samples were centrifuged at 10,000 rpm for 10 minutes at 4°C and supernatants were moved to clean microfuge tubes. Supernatants were incubated with Protein A beads (blocked with 5% BSA in IP buffer) on a rotator at 4°C for 2 hours. Beads were then pelleted, post-IP supernatants saved for Western blot analysis, and beads were washed four times with IP buffer containing 0.1% digitonin. The 2× Laemmli sample buffer was added to the washed beads and they were heated at 60°C for 30 minutes prior to SDS-PAGE (see Western blot protocol details below).
Western blot analysis of protein expression levels.
Proteins were separated on 12% SDS polyacrylamide gels. Following SDS-PAGE, proteins were transferred to polyvinylidene difluoride membranes and the membranes were blocked overnight with 5% milk in TBS with 0.1% Tween20 (TBST). Blots were cut horizontally using molecular weight markers as a guide depending on the size of proteins to be probed. This allowed blots to be probed with multiple antibodies without the necessity of stripping and reprobing the blots multiple times. Anti-ORMDL (1:2,000) and anti-SPTLC1 (1:1,000) primary antibodies were incubated overnight at 4°C in TBST containing 3% milk; anti-beta actin (1:10,000) primary antibody was incubated for 1 hour at room temperature. After incubation with primary antibodies, blots were washed three times with TBST for 10 minutes per wash. Secondary antibodies (1:20,000) were incubated for 1 hour at room temperature in TBST containing 1% milk. Blots were washed three times with TBST for 10 minutes per wash. Protein bands were visualized using ECL Prime solution (GE Life Sciences, Piscataway, NJ) and exposed to film (Denville Scientific, Metuchen, NJ). Densitometry values were measured using Quantity One software and ORMDL3 levels were normalized to beta-actin for each sample.
Subcloning of the lentiviral ORMDL3 construct and generation of HBECs that stably overexpress ORMDL3.
pENTR1A Gateway® entry vector (#17398) and pLenti-CMV-puro lentiviral Gateway® destination vector (#17452) were purchased from Addgene (Cambridge, MA), as described (11). Human ORMDL3 plasmid #RC202279 (Origene, representing NM_139280) was linearized by cutting with EcoRI after which the open reading frame for ORMDL3 was amplified using CloneAmp™ HiFi PCR Premix and primers designed for use with the In-Fusion® HD Cloning Plus kit (forward, ATC CGG TAC CGA ATT GCC ATG AAT GTG GGC ACA; reverse, GTG CGG CCG CGA ATT TTA AAC GTA CTT ATT GAT TCC A). Insert and vector were gel purified prior to subcloning human ORMDL3 into pENTR1A using the In-Fusion® HD Cloning Plus kit following the manufacturer’s instructions. The human ORMDL3 open reading frame was transferred from pENTR1A entry vector to pLenti-CMV-puro destination vector using the Gateway® LR Clonase™ II enzyme mix following the manufacturer’s instructions. Sequences of all subsequent clones were verified by the University of Louisville core DNA laboratory. Human 293T cells were used to package pLenti-CMV-ORMDL3-puro into viral particles. Additional packaging plasmids expressing the viral structural proteins Gag, Pol, and Env were generous gifts of Dr. Chi Li, University of Louisville, Louisville, KY. Viral supernatants were collected every 12 hours after transfection of 293T cells, filtered to remove cell debris, then stored at −20°C until use. HBECs immortalized with telomerase (hTERT) and cyclin-dependent kinase 4 (Cdk) (9) were plated in p100 dishes at 3 × 106 cells in complete medium overnight. The next day, cells were transduced with 5 ml of viral supernatant containing either empty vector or human ORMDL3 plasmid. This was delivered in 5 ml of medium containing 8 μg/ml Polybrene® and cells were incubated at 37°C with 5% CO2 for 12 hours, after which this transduction mix was removed and fresh viral particles were added to the cells for an additional 12 hours. Viral supernatants were removed and complete medium was added to cells for overnight incubation. The next day, medium was replaced with complete medium containing 10 μg/ml puromycin to begin the selection process. Cells were maintained under puromycin selection until used for experiments as outlined. Cells were routinely collected and ORMDL3 levels were analyzed by Western blot and real-time PCR.
Isolation of total RNA and quantitative real-time PCR analysis.
For each well of cells to be analyzed, 1 ml of TRIzol® was placed directly on the cells. The TRIzol®/cell mixture was transferred to a microfuge tube and stored at −80°C until analysis. To isolate RNA, the TRIzol®/cell mixture was added to prepared phase lock gels followed by 200 μl chloroform:isoamyl alcohol then vigorously shaken for 15 seconds. Tubes were incubated at room temperature for 5 minutes then centrifuged at 16,000 g for 15 minutes (4°C) to separate phases. The upper (aqueous) phase was transferred to sterile RNase-free microfuge tubes and 500 μl isopropanol was added to each tube and incubated at room temperature for 10 minutes to precipitate RNA. Tubes were spun at 16,000 g for 20 minutes (4°C) to pellet RNA. RNA was washed with 70% ethanol then air dried and resuspended in RNase-free water (30–100 μl) and quantitated using a NanoDrop 8000. cDNA was prepared from 1 μg of total RNA using a high-capacity reverse transcriptase kit from Applied Biosystems (Life Technologies, Thermo-Fisher, Grand Island, NY) following all manufacturer’s instructions. Quantitative real-time PCR was performed using gene-specific TaqMan® probes for all three ORMDL isoforms and the large subunit #1 of SPT. TaqMan® Gene Expression Master Mix and MicroAmp® Fast Optical 96-well plates were used along with an Applied Biosystems® FAST 7500 instrument to perform real-time PCR amplification. Analysis was done using the ΔCT (Fig. 1) or ΔΔCT (Figs. 2 and 3) formula for calculating the fold change in expression levels.
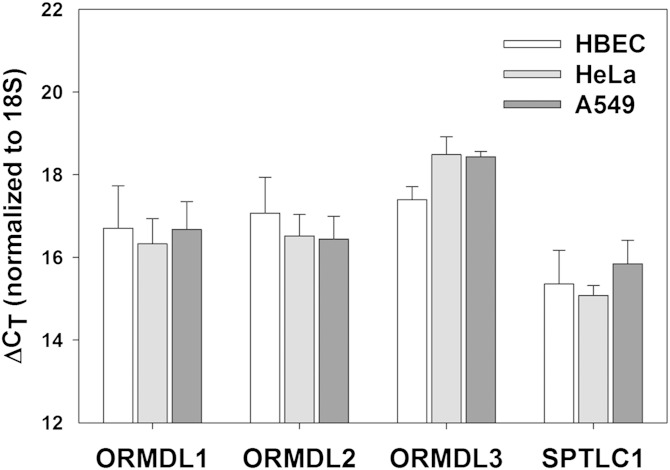
Fig. 1.
All three ORMDL isoforms, as well as subunit 1 of SPT, are expressed at similar levels in multiple cell types. Real-time PCR was performed on RNA isolated from immortalized HBECs, HeLa cells, and A549 adenocarcinoma cells as described in the Materials and Methods. Results are normalized to levels of 18S internal control and represented as ΔCT values.
Mass spectrometry analysis of steady-state sphingolipids in HBECs.
Sphingolipids were extracted from lysates as previously described (12). Prior to extraction, a mixture of C17 sphingolipids was added to each sample (125 pmol/sample, Avanti sphingolipid mix #1) as internal standards. Sphingolipids were quantitated by HPLC electrospray ionization tandem mass spectrometry using selected ion monitoring on an ABSciex 4000 Q-Trap instrument as described previously (13–15). Total phospholipids for each sample were measured using a modified Ames and Dubin assay (16), as previously described (12).
RESULTS
HeLa cells and immortalized, but untransformed, HBECs express similar levels of the three ORMDL isoforms and SPT subunit 1
Airway epithelial cells are thought to be a major cell type involved in the increased asthma risk resulting from the elevated expression of ORMDL3 induced by a risk allele in ORMDL3-adjacent sequences (17). Here we utilize a well-characterized cell line derived by immortalization of primary human airway cells by expression of telomerase (hTERT) and cyclin-dependent kinase 4 (Cdk) (9). We refer to these as HBECs. To enable comparison of ORMDL function between HBECs and the more easily manipulated HeLa cell line, we measured mRNA levels of the three ORMDL isoforms and one of the two major catalytic subunits of SPT, SPTLC1 (Fig. 1). We found that levels of these transcripts were at comparable levels when each transcript was compared between these two cell lines. Additionally, we measured levels of these transcripts in the lung adenocarcinoma cell line, A549, and found comparable levels of all transcripts in that cell line as well. Levels of these transcripts may differ among other cell lines and may change during cell differentiation (4). In the experiments outlined below, we study the role of stoichiometry in ORMDL/SPT regulation. The expression data illustrated in Fig. 1 suggests that the relative stoichiometry of these proteins is similar between these cell lines.
Overexpression of ORMDL3 does not suppress de novo sphingolipid biosynthesis at endogenous levels of SPT
The ORMDL proteins are negative regulators of SPT. It might be expected that overexpression of ORMDL3 would lead to suppression of SPT. Indeed a suppression of sphingolipid synthesis has been proposed to be the underlying biochemical effect which elevates the risk of asthma in individuals carrying the risk allele of ORMDL3 (8, 18). To test this directly, we transiently transfected HeLa cells with ORMD3 (from mouse) and established, by lentiviral infection, two HBEC cell lines stably expressing elevated levels of human ORMDL3 (Fig. 2). It should be noted that mouse and human ORMDL3 are 96% homologous and, as illustrated in Fig. 3, mouse ORMDL3 is fully functional in the human system. HBEC-ORMDL-Low expresses total ORMDL, relative to the vector-transformed cells, approximately 1.2-fold at the protein level and ORMDL3 is elevated 6-fold at the RNA level. HBEC-ORMDL-High overexpresses total ORMDL approximately 2.8-fold at the protein level and ORMDL3 is elevated 30-fold at the RNA level (Fig. 2C, D). Note that the HeLa cell transfection produces somewhat higher levels of total ORMDL (5.1 over control). Surprisingly, in neither HeLa cells nor HBECs does ORMDL3 overexpression suppress de novo sphingolipid biosynthesis (Fig. 2A). We observed that overexpression of ORMDL3 in HBECs results in a variable elevated sphingolipid biosynthesis. One of three separate experiments is shown in Fig. 2. The increase in sphingolipid synthesis ranged from 1- to 2.7-fold in HBEC-ORMDL3-Low and from 1- to 3.4-fold in HBEC-ORMDL3-High. The response in HeLa cells was similarly variable. In several experiments ORMDL3 overexpression stimulated an increase in de novo sphingolipid synthesis, as depicted in Figs. 2 and 3, but in others synthesis was unaffected. The source of this variability is not clear. We also tested whether the ORMDL-dependent inhibition of SPT in response to elevated sphingolipid levels was affected by ORMDL3 overexpression by treating the cells with a short chain (C6) ceramide in a manner that we have used previously (3) (Fig. 2A, +C6). ORMDL3 overexpression had no appreciable effect on C6-induced inhibition of de novo sphingolipid biosynthesis. To test whether effects in HBECs were directly on SPT activity, we utilized a permeabilized cell assay for SPT activity (Fig. 2B) (3). In this assay, a low level of ORMDL3 overexpression (“Low”) slightly inhibited SPT activity, however a higher level of ORMDL3 overexpression (“High”) had no significant effect on SPT activity. These data indicate that the lack of suppression of de novo ceramide synthesis by ORMDL3 overexpression as measured in intact cells is a reflection of SPT activity and not a consequence 3H-serine pool size or some other aspect of the assay used in intact cells.
Overexpression of ORMDL3 suppresses the activity of overexpressed SPT
One explanation for the lack of suppression of sphingolipid biosynthesis by ORMDL3 overexpression is that ORMDLs are present in functional excess relative to SPT. To test this, we overexpressed, in HeLa cells, a construct that results in the expression of a single polypeptide that is a fusion of the two major catalytic subunits of SPT (SPTLC1 and -2) and a small subunit that enhances SPT activity (ssSPTa) (SPTLC2-ssSPTa-SPTLC1) (Fig. 3) (10). The predicted size of this construct is 129 kDa. We observe a triplet of bands close to this molecular mass when immunoblots are probed with an antibody directed against SPTLC1. The origin of the multiple bands is not clear. We termed this construct scSPT. Expression of scSPT resulted in an approximately 5-fold increase in de novo ceramide biosynthesis (Fig. 3A, B). We did not detect any change in ORMDL protein or RNA resulting from this overexpression (Fig. 3C, D, note that this antibody detects all three ORMDL isoforms). Depletion of all three ORMDL isoforms by siRNA transfection strongly enhances ceramide biosynthesis at both endogenous levels of SPT and at the elevated levels of synthesis resulting from the scSPT expression (Fig. 3A, siORMDLs). ORMDL levels are strongly reduced by siRNA transfection (Fig. 3C, D). Treatment of cells with a short chain ceramide (C6-ceramide) results in an ORMDL-dependent homeostatic inhibition of de novo ceramide biosynthesis (Fig. 3A) (3). The enhanced ceramide biosynthesis resulting from scSPT expression is also depressed by C6-ceramide treatment; however, the response is blunted compared to the almost complete inhibition of ceramide biosynthesis observed at endogenous SPT levels (Fig. 3A, compare untreated controls plus and minus C6-ceramide to scSPT plus and minus C6-ceramide). siRNA depletion of the ORMDL proteins reduces the C6-ceramide response at endogenous levels of SPT by approximately 50%, but virtually eliminates the C6-ceramide response of cells expressing scSPT. These data suggest that at elevated levels of SPT there is insufficient ORMDL to achieve sphingolipid regulation of SPT.
As noted above (Fig. 2), we observed that overexpression of ORMDL3 does not result in the expected suppression of sphingolipid synthesis in either HeLa cells or airway epithelial cells and hypothesized that this was due to a functional excess of ORMDL relative to SPT. To test this concept, we expressed scSPT in HeLa cells and then co-transfected the cells either with a control vector or a vector expressing ORMDL3 (Fig. 3B). As before, at endogenous SPT levels, ORMDL3 overexpression did not inhibit sphingolipid synthesis, in fact in the experiment depicted in Fig. 3, sphingolipid synthesis was slightly elevated. However, strikingly, ORMDL3 expression significantly inhibited de novo ceramide synthesis in scSPT-expressing cells and enhanced the C6-ceramide inhibition of ceramide synthesis in those cells. This strongly supports the notion that SPT/ORMDL stoichiometry determines whether SPT activity is regulated by levels of ORMDL expression.
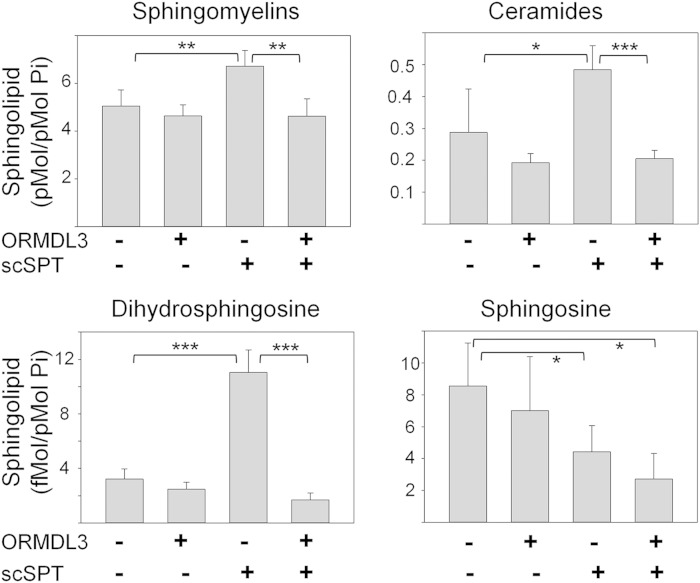
Mass spectroscopic analysis was performed on HeLa cell cultures overexpressing either the scSPT, ORMDL3, or both to examine whether the effects on de novo sphingolipid synthesis, depicted in Fig. 3B, are reflected in changes in steady-state levels of those lipids (Fig. 4). The changes in levels of dihydrosphingosine, ceramides, and sphingomyelins closely mirror the changes in the rates of sphingolipid synthesis. ORMDL3 overexpression did not significantly change levels of these lipids. A similar result has recently been reported in HEK293 cells (4). scSPT expression by itself led to an almost 3-fold increase in dihydrosphingosine levels. Dihydrosphingosine is an early intermediate in the de novo biosynthetic pathway, so it is not surprising that increased SPT activity would elevate dihydrosphingosine levels. SPT overexpression also elevated levels of ceramides and sphingomyelins, indicating that increased SPT activity is distributed throughout the sphingolipid network. In contrast to the lack of effect of ORMDL3 overexpression alone, overexpression of ORMDL3 completely reversed the increased sphingolipid levels resulting from SPT overexpression. This result confirms that while at endogenous levels of SPT, ORMDL levels are sufficient to control SPT activity; elevation of SPT upsets that ratio, which is corrected by overexpression of ORMDL3. Similar results have recently been reported in HEK293 cells (19). Changes in levels of sphingosine do not follow the pattern of the other sphingolipids, being reduced by SPT overexpression. Sphingosine is generated by degradation of ceramide by ceramidases and is consumed by the ceramide synthases and sphingosine kinases. The lack of elevation of sphingosine with SPT overexpression may be explained by changes in activity of these enzymes.
Fig. 4.
Steady state levels of sphingolipids in HeLa cells resulting from SPT and ORMDL3 expression reflect changes in de novo synthesis. HeLa cells were transfected with control plasmids or plasmids encoding a scSPT construct and/or mouse ORMDL3 (not epitope tagged). Sphingolipids were extracted and subjected to analysis by mass spectroscopy as described in the Materials and Methods. *Significant to P < 0.05, **significant to P < 0.005, ***significant to P < 0.0005 by Student’s t-test. Unmarked bars did not differ from the vector control at P > 0.05.
SPT/ORMDL complex formation is not enhanced by ORMDL overexpression
The measurement of de novo sphingolipid biosynthesis depicted in Fig. 3 indicated that ORMDLs are in functional excess relative to SPT at endogenous levels. To establish whether this is reflected by the formation of stable SPT/ORMDL complexes, we examined the levels of these complexes by co-IP of SPT with anti-ORMDL antibodies. Overexpressed ORMDL has been shown to form a complex with SPT (1). We confirmed that this interaction takes place at endogenous levels of these proteins (Fig. 5). Specificity of this precipitation was confirmed by performing IPs using lysates prepared from cells depleted of ORMDL proteins by siRNA transfection (Fig. 5). To assess whether increased expression of ORMDL3 might elevate SPT/ORMDL complex formation, we compared the level of SPT immunoprecipitated by anti-ORMDL antibodies at endogenous levels of ORMDLs to that when ORMDL3 levels were elevated by overexpression (Fig. 5). Note that for the overexpression shown in Fig. 5A, we utilized an epitope-tagged version of human ORMDL3 that migrates somewhat more slowly in SDS-polyacrylamide gels than the native protein. We find that this epitope-tagged version is as efficient as the untagged mouse ORMDL3 in functional assays such as that depicted in Fig. 3 (data not shown). Overexpression of untagged mouse ORMDL3 (Fig. 5B) gives identical IP results to the epitope-tagged human ORMDL3 (Fig. 5A). Despite strong overexpression of ORMDL3, SPT incorporation into ORMDL immunocomplexes was not enhanced over levels found with endogenous levels of ORMDLs. This mirrors the functional test depicted in Fig. 3 and supports the notion that ORMDLs are in excess relative to SPT. We also examined whether the efficiency of ORMDL/SPT complex formation was affected under conditions which trigger ORMDL inhibition of SPT. Cells were treated with C6-ceramide, which strongly depresses de novo sphingolipid synthesis (3) (Fig. 3) and lysates were then prepared and subjected to IP with an anti-ORMDL antibody. We were unable to detect any change in the level of SPT co-precipitation with ORMDL when cells were pretreated with C6-ceramide. This result indicates that ORMDL inhibition is not a product of increased ORMDL/SPT association, but rather a change in the nature of that association within the complex.
Low-level ORMDL3 overexpression in airway epithelial cells results in slightly reduced mass levels of sphingolipids
We have established that slight overexpression of ORMDL3 does not inhibit sphingolipid biosynthesis in airway epithelial cells (Fig. 2). Considering the complexity of sphingolipid metabolism, it was important to establish how ORMDL3 overexpression impacted steady state levels of these lipids. Sphingolipids were extracted from vector control HBECs and two HBEC cell lines expressing ORMDL3 at either moderate or highly elevated levels and lipids were analyzed by mass spectroscopy (Fig. 6). ORMDL3 overexpression slightly depressed levels of all sphingolipid species measured, but only reached statistical significance in the HBECs expressing slightly elevated ORMDL3 (Low). This trend was also observed in the HBECs expressing higher ORMDL3 levels (High), but the differences were not statistically significant. Levels of sphingosine-1-phosphate and dihydrosphingosine-1-phosphate were below the levels of detection in this analysis. The distribution of individual ceramide molecular species did not differ between samples (data not shown) except for C16-dihydroceramide, which was slightly, but significantly, reduced in the ORMDL3-Low and ORMDL3-High cell lines.
DISCUSSION
These studies were designed to examine the concept that increased ORMDL3 expression elevates asthma risk by depressing sphingolipid biosynthesis, as suggested by earlier publications (8, 18). Our results present a complex picture. We clearly find that increased ORMDL3 levels do not suppress sphingolipid synthesis, as previously assumed, at least in the cell types that we have examined. The lack of suppression of sphingolipid biosynthesis by elevated ORMDL3 expression is a consequence of the fact that in the two cell types studied, there is sufficient ORMDL, at normal levels, to maximally form stable ORMDL/SPT complexes. Only when we artificially elevate SPT levels do we see that increased ORMDL expression inhibits de novo sphingolipid biosynthesis. These results mirror those of two publications that emerged while this manuscript was in revision, which reported that elevating ORMDL levels had no effect on sphingolipid biosynthesis and steady state levels in HEK293 cells (4, 19). Gupta et al. (19) also reported that overexpression of ORMDL suppressed sphingolipid synthesis resulting from SPT overexpression. Interestingly, these investigators report that elevated expression of SPT in HEK293 cells results in increased ORMDL protein expression via a post-transcriptional mechanism (19). This effect depends on the activity of SPT. One possible explanation for this observation is that at endogenous levels ORMDL is in stoichiometric excess relative to SPT and that excess ORMDL is unstable. In this model, elevated SPT, in combination with sphingolipid, forms an ORMDL-stabilizing complex and thus increases the overall levels of ORMDL protein. This is consistent with our finding that ORMDL3 transfection of HeLa cells results in only a 5-fold increase in ORMDLs while ORMDL3 message expression is elevated by 80-fold (Fig. 2C, D). However, it should be noted that we do not see an effect on endogenous ORMDL protein levels upon increased SPT expression (Fig. 3C). This may reflect a difference in endogenous levels of ORMDLs in the HeLa cells used in the experiments reported here and the HEK293 cells utilized by Gupta et al. (19). Alternatively the timing and levels of SPT expression may be important for the effect of SPT expression on ORMDL levels. Our measurements were performed on cells transiently transfected for 24 hours, whereas those of Harmon and colleagues utilized induction for 42 hours in a tetracycline-inducible system.
Whereas we focus on ORMDL3 here, our previous results established that the other two ORMDL isoforms have redundant functions in the homeostatic regulation of sphingolipid biosynthesis. Only when all three isoforms are depleted by siRNA transfection in HeLa cells is ORMDL-dependent regulation of sphingolipid biosynthesis lost (3). Harmon and colleagues report that all three ORMDL isoforms equivalently inhibit overexpressed SPT (19). The independent function of the three isoforms is therefore unresolved. Kiefer et al. (4) find that overexpression of all three isoforms has no effect on sphingolipid synthesis under normal conditions, but report that when cells are supplemented with palmitate, the CoA form of which is one of the substrates for SPT, they can distinguish an effect of co-expression of all three ORMDL isoforms from expression of any single isoform. The significance of this finding will require further investigation of the ORMDL/SPT complex and its regulation.
Our results, and those inferred from results in yeast (1), indicate that the level of ORMDL regulation of SPT is not a result of the level of ORMDL expression and ORMDL/SPT complex formation, but rather that ORMDLs exist in both non-inhibitory and inhibitory states in stable ORMDL/SPT complexes. In yeast, Orm inhibitory activity is constitutive, but is relieved by phosphorylation of the Orms (1, 20). However, the mammalian ORMDLs, although otherwise highly homologous to the yeast proteins, lack the amino terminal sequences that are phosphorylated in the Orms (1). Further, we have been unable to detect ORMDL phosphorylation under any conditions (data not shown). Therefore the mechanism that triggers ORMDL inhibitory activity in response to elevated cellular sphingolipid levels remains a mystery. We have previously shown that the sphingolipid that activates the ORMDL-dependent homeostatic inhibition of SPT is ceramide or a higher order sphingolipid such as sphingomyelin or glycosphingolipid (3). These lipids may be sensed by an as yet unidentified protein that interacts with the ORMDL/SPT complex. Alternately, the ORMDLs may directly sense sphingolipid levels.
Other cell types relevant to the asthmatic phenotype, such as smooth muscle or immune cells, may have intrinsically lower levels of ORMDLs relative to SPT and therefore elevation of ORMDL3 would have a inhibitory effect, such as that we see with enforced SPT expression. Careful examination of these cell types will be required to establish whether this is the case. However, it does not appear that this simple explanation for ORMDL3-mediated effects on asthma risk holds for the airway epithelium. Instead our data indicates that ORMDL3 has additional cellular roles that should be examined. Our finding that elevation of ORMDL3 in airway epithelial cells slightly depresses steady-state levels of sphingolipids while de novo synthesis is unaffected or possibly increased suggests that elevated ORMDL3 affects sphingolipid metabolism beyond the initiating step or by indirect mechanisms, a notion that will require further study.
Additional work will be required to establish the role of ORMDL3-mediated alteration of sphingolipid metabolism in asthma. The cell lines used here were cultured under standard conditions and may not reflect the environment of the asthmatic tissue. Pharmacological and genetic inhibition of SPT in the intact animal induces airway hypersensitivity (18), a key feature of asthma. However, how this relates to increased risk of asthma is not yet clear. The cell types and specific sphingolipid species involved have yet to be determined. Although we find that elevated ORMDL3 expression does not depress de novo sphingolipid synthesis, we do find that steady-state levels of sphingolipids are slightly reduced. Future studies will be required to establish whether this is the key feature of ORMDL3 overexpression that increases risk for asthma. An understanding of the involvement of sphingolipids in the asthmatic phenotype is under development. The potent signaling lipid sphingosine-1-phosphate stimulates histamine release from mast cells (21) and an inhibitor of sphingosine kinase, which generates sphingosine-1-phosphate, reduces airway hypersensitivity in a murine asthma model (22). The sensitivity induced by elevated sphingosine-1-phosphate does not easily fit with the increased hypersensitivity induced by SPT inhibition (18) and the reduced sphingolipid levels noted in these studies presented here.
It is important to recognize that although the association of elevated asthma risk with SNPs that raise ORMDL3 levels is strong and reproducible, the increase in risk is moderate and the prevalence of the risk alleles of these SNPs is very high (6). This indicates that there are important environmental and genetic factors that interact with ORMDL3 elevation to increase asthma risk. The challenge will be to unravel the contribution that ORMDL3 makes to increase the risk for this prevalent disease.
Acknowledgments
The authors thank Dr. Eric Campeau (University of Massachusetts) for advice on use of the lentiviral vectors and the depositing of those vectors at Addgene (Cambridge, MA). We also thank Dr. Sony Sunkara (University of Kentucky) for assistance in mass spectroscopic analysis.
Footnotes
Abbreviations:
- C6
- ceramide-N-hexanoyl-D-erythro-sphingosine
- HBEC
- human bronchial epithelial cell
- IP
- immunoprecipitation
- ORMDL
- ORM1 (Saccharomyces cerevisiae)-like protein
- scSPT
- single-chain serine palmitoyltransferase
- SPT
- serine palmitoyltransferase
- SPTLC1
- serine palmitoyltransferase long chain base subunit 1
- SPTLC2
- serine palmitoyltransferase long chain base subunit 2
- ssSPTa
- serine palmitoyltransferase small subunit a
- TBST
- TBS with 1% Tween20
This work was supported by a grant from the Kentucky Lung Cancer Research Program, by a Fellowship from the James Graham Brown Cancer Center to D.S., and by funds from the James Graham Brown Cancer Center, the Center for Environmental Genomics and Integrative Biology, and the Clinical and Translational Research Pilot program of the University of Louisville School of Medicine to B.W. A.J.M. and M.S. were supported by grants from the National Institutes of Health and Veterans Administration (HL120507 and RR024598), and work utilized resources from the Lexington Veterans Affairs Medical Center. T.M.D. was supported by a grant from the National Institutes of Health (R21HD080181).
REFERENCES
- 1.Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han S., Lone M. A., Schneiter R., Chang A. 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA. 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siow D. L., Wattenberg B. W. 2012. Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287: 40198–40204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiefer K., Carreras-Sureda A., Garcia-López R., Rubio-Moscardó F., Casas J., Fabriás G., Vicente R. 2015. Coordinated regulation of the orosomucoid-like gene family expression controls de novo ceramide synthesis in mammalian cells. J. Biol. Chem. 290: 2822–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffatt M. F., Kabesch M., Liang L., Dixon A. L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. 2007. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 448: 470–473. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Moffatt M. F., Cookson W. O. 2012. Genetic and genomic approaches to asthma: new insights for the origins. Curr. Opin. Pulm. Med. 18: 6–13. [DOI] [PubMed] [Google Scholar]
- 7.Dixon A. L., Liang L., Moffatt M. F., Chen W., Heath S., Wong K. C., Taylor J., Burnett E., Gut I., Farrall M., et al. 2007. A genome-wide association study of global gene expression. Nat. Genet. 39: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 8.Levy B. D. 2013. Sphingolipids and susceptibility to asthma. N. Engl. J. Med. 369: 976–978. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez R. D., Sheridan S., Girard L., Sato M., Kim Y., Pollack J., Peyton M., Zou Y., Kurie J. M., Dimaio J. M., et al. 2004. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 64: 9027–9034. [DOI] [PubMed] [Google Scholar]
- 10.Gable K., Gupta S. D., Han G., Niranjanakumari S., Harmon J. M., Dunn T. M. 2010. A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J. Biol. Chem. 285: 22846–22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. 2009. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 4: e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siow D. L., Anderson C. D., Berdyshev E. V., Skobeleva A., Pitson S. M., Wattenberg B. W. 2010. Intracellular localization of sphingosine kinase 1 alters access to substrate pools but does not affect the degradative fate of sphingosine-1-phosphate. J. Lipid Res. 51: 2546–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deevska G. M., Sunkara M., Morris A. J., Nikolova-Karakashian M. N. 2012. Characterization of secretory sphingomyelinase activity, lipoprotein sphingolipid content and LDL aggregation in ldlr-/- mice fed on a high-fat diet. Biosci. Rep. 32: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salous A. K., Panchatcharam M., Sunkara M., Mueller P., Dong A., Wang Y., Graf G. A., Smyth S. S., Morris A. J. 2013. Mechanism of rapid elimination of lysophosphatidic acid and related lipids from the circulation of mice. J. Lipid Res. 54: 2775–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider G., Bryndza E., Abdel-Latif A., Ratajczak J., Maj M., Tarnowski M., Klyachkin Y. M., Houghton P., Morris A. J., Vater A., et al. 2013. Bioactive lipids S1P and C1P are prometastatic factors in human rhabdomyosarcoma, and their tissue levels increase in response to radio/chemotherapy. Mol. Cancer Res. 11: 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ames B. N., Dubin D. T. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235: 769–775. [PubMed] [Google Scholar]
- 17.Miller M., Tam A. B., Cho J. Y., Doherty T. A., Pham A., Khorram N., Rosenthal P., Mueller J. L., Hoffman H. M., Suzukawa M., et al. 2012. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc. Natl. Acad. Sci. USA. 109: 16648–16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worgall T. S., Veerappan A., Sung B., Kim B. I., Weiner E., Bholah R., Silver R. B., Jiang X. C., Worgall S. 2013. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci. Transl. Med. 5: 186ra67. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S. D., Gable K., Alexaki A., Chandris P., Proia R. L., Dunn T. M., Harmon J. M. 2015. Expression of the ORMDLS, modulators of serine palmitoyltransferase, is regulated by sphingolipids in mammalian cells. J. Biol. Chem. 290: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J. 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 108: 19222–19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolly P. S., Bektas M., Olivera A., Gonzalez-Espinosa C., Proia R. L., Rivera J., Milstien S., Spiegel S. 2004. Transactivation of sphingosine-1-phosphate receptors by FcepsilonRI triggering is required for normal mast cell degranulation and chemotaxis. J. Exp. Med. 199: 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price M. M., Oskeritzian C. A., Falanga Y. T., Harikumar K. B., Allegood J. C., Alvarez S. E., Conrad D., Ryan J. J., Milstien S., Spiegel S. 2013. A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in a mast cell-dependent murine model of allergic asthma. J. Allergy Clin. Immunol. 131: 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]