Abstract
Here, we present for the first time that Mycobacterium tuberculosis ParB is phosphorylated by several mycobacterial Ser/Thr protein kinases in vitro. ParB and ParA are the key components of bacterial chromosome segregation apparatus. ParB is a cytosolic conserved protein that binds specifically to centromere-like DNA parS sequences and interacts with ParA, a weak ATPase required for its proper localization. Mass spectrometry identified the presence of ten phosphate groups, thus indicating that ParB is phosphorylated on eight threonines, Thr32, Thr41, Thr53, Thr110, Thr195, and Thr254, Thr300, Thr303 as well as on two serines, Ser5 and Ser239. The phosphorylation sites were further substituted either by alanine to prevent phosphorylation or aspartate to mimic constitutive phosphorylation. Electrophoretic mobility shift assays revealed a drastic inhibition of DNA-binding by ParB phosphomimetic mutant compared to wild type. In addition, bacterial two-hybrid experiments showed a loss of ParA-ParB interaction with the phosphomimetic mutant, indicating that phosphorylation is regulating the recruitment of the partitioning complex. Moreover, fluorescence microscopy experiments performed in the surrogate Mycobacterium smegmatis ΔparB strain revealed that in contrast to wild type Mtb ParB, which formed subpolar foci similar to M. smegmatis ParB, phoshomimetic Mtb ParB was delocalized. Thus, our findings highlight a novel regulatory role of the different isoforms of ParB representing a molecular switch in localization and functioning of partitioning protein in Mycobacterium tuberculosis.
Introduction
Mycobacterium tuberculosis (M. tb), the etiologic agent of Tuberculosis, infected 8.6 million people and was responsible for the death of 1.3 million people in 2012 thus remaining a major threat to global health [1]. The emergence of multi-drug-resistant strains makes M. tb an increasing global health threat since the number of newly infected people, emphasized by AIDS infections, keeps increasing in both developing and industrialized countries (WHO). M. tb has a complex lifestyle that relies on its remarkable capacity to survive within infected host macrophages by modulating its own cell functions in order to adapt to a new environment. One of the remarkable features of mycobacteria, crucial for the persistent infection, is their ability to enter dormant, non-replicating state. However, little is known about the control of mycobacterial cell cycle.
In bacteria, the key processes of the cell cycle, chromosome replication and segregation, are tightly coupled. Unlike in eukaryotes, bacterial chromosome segregation occurs during ongoing replication. The process of replication begins in a defined locus, called origin of replication (oriC). Newly duplicated oriC regions are precisely organized, moved and anchored by the segregation machinery. Subsequently, the rest of chromosome(s) is actively transported into specific positions in future daughter cells. In many bacteria, both organization and segregation of oriC regions are governed by the ParABS partitioning system. ParBs are DNA binding proteins with specific affinity for centromere-like parS sequences; ParA homologues are cytoskeletal weak ATPases directly interacting with ParB and proposed to be the motor element of the segregation machinery via polymerization/depolymerization events [2]; and centromere-like parS sequences are palindromic sequences of 14–16 mers located in the vicinity of the oriC region.
By interacting with parS and ParA, ParB promotes the formation of a core structure called the segrosome where other proteins participating in chromosome segregation can dock. This in turn promotes oligomerization to form a larger nucleoprotein complex whose movement is governed by a cytoskeletal ParA protein [3]. The parB homologues are present in most bacterial genomes (with exception of E. coli); in Caulobacter crescentus [4,5], Streptomyces coelicolor [6,7], Corynebacterium glutamicum [8], Pseudomonas spp. [9–11], Myxococcus xanthus [12,13] and Bacillus subtilis [14–17] are non-essential for almost all bacteria except Caulobacter crescentus [4] and Myxococcus xanthus [13] while in the other species elimination of ParB leads to severe chromosome segregation defects and associated with an increased number of anucleate cells [8,9,13,18]. The segrosome is thought to organize and position the oriC proximal part of the chromosome at the cell pole of the bacterium before cell division.
While replication [19–22] and cell division [23–26] are relatively well-studied processes in mycobacteria, chromosome segregation remains to be fully understood. Previously it was shown that mycobacterial chromosomes encode proteins participating in chromosome segregation [6,27,28]. Interestingly, transposon mutagenesis experiments revealed that parB could also be essential in Mycobacterium tuberculosis H37Rv [29], but it was proved to be nonessential in M. smegmatis [30]. M. smegmatis parB mutant strain produced increased number of anucleate cells. In addition, microscopy analysis revealed that proper positioning of ParB in the cytosol of M. smegmatis depends on the presence of ParA thus highlighting the fact that the functional ParABS system is crucial to accurate chromosome segregation in mycobacteria [31]. Moreover, in vitro experiments showed that M. tuberculosis ParB interacts with parS sequences and ParA, which would enhance ParB affinity for parS sequences.
Understanding M. tuberculosis (M. tb) cell cycle is crucial to decipher the mechanism leading to its remarkable persistence within host cells. This persistence is a unique feature enabling M. tb to modulate its own cell functions in order to adapt to new environmental conditions. A well-known mechanism of signal transduction in prokaryotes and eukaryotes is phosphorylation. The Ser/Thr Protein Kinases (STPKs) have been shown to regulate major cell processes so that Ser/Thr phosphorylation has emerged as a key regulatory process in mycobacteria [32,33]. Cell division is a key cell process that undergoes a tight regulation either in time and space. Interestingly, recent studies identified several mycobacterial cell cycle proteins as mycobacterial STPKs substrates including the polar determinant DivIVA homologue named Wag31 [34], FtsZ [35], or FipA [36], a FtsZ partner. In this study, we show for the first time that M. tb partitioning protein ParB activity is regulated by Ser/Thr phosphorylation and that phosphorylation not only negatively affects ParB affinity for its specific centromere-like parS sequences, but also inhibits ParB interaction with its cytoskeletal partner ParA. Furthermore, our microscopy observations suggest that phosphorylation also triggers ParB delocalization. Consequently, phosphorylation of ParB via Ser/Thr kinase phosphorylation would inhibit the ParABS system and thus the formation of the mitotic-like apparatus, which is essential to proper chromosome segregation.
Materials and Methods
Bacterial strains and growth conditions
Strains used for cloning and expression of recombinant proteins were E. coli 10G (Lucigen) and E. coli BL21(DE3)Star (Novagen) as detailed in S1 Table. They were grown in LB medium at 37°C. Media were supplemented with ampicillin (100 μg/ml), hygromycin (200 μg/ml), kanamycin (25 μg/ml) or spectinomycin (100 μg/ml) when required. Mycobacteria strains were grown aerobically on Middlebrook 7H10 agar plates with OADC enrichment (Difco) or in Middlebrook 7H9 medium supplemented with 10% (v/v) OADC enrichment, 0.5% (v/v) glycerol and 0.05% (v/v) Tyloxapol. Hygromycin (50 μg/ml) and kanamycin (25 μg/ml) were added for the selection of appropriate strains.
Cloning, expression and purification of M. tuberculosis ParB and mutant proteins
The parB gene was amplified by PCR using M. tb H37Rv chromosomal DNA as the template and the forward and reverse primers listed in S2 Table containing NdeI and HindIII restriction sites, respectively. The amplified product was digested with NdeI and HindIII, and ligated into the pETPhos plasmid [37], a variant of pET15b (Novagen) that includes a tobacco etch virus (TEV) protease site instead of the thrombin site, and an N-terminal His-tag free of Ser/Thr/Tyr residues, thus generating pETPhos_parB. The parB gene containing either the ten mutations T32A, T41A, T53A, T110A, T195A, T254A, T300A, T303A, S5A, S239A, or the mutations T32D, T41D, T53D, T110D, T195D, T254D, T300D, T303D, S5D, S239D, were both synthesized by GenScript with 5' NdeI and 3' HindIII restriction sites. These two parB mutants were cloned into the pETPhos vector, generating pETPhos_parB_ T32A/T41A/T53A/T110A/T195A/T254A/T300A/T303A/S5A/S239A and pETPhos_parB_ T32D/T41D/T53D/T110D/T195D/T254D/T300D/T303D/S5D/S239D named pEThos_parB_Ala and pETPhos_parB_Asp, respectively. A duet strategy was used to generate hyper-phosphorylated ParB protein as described previously [38]. The parB gene was cloned into the pCDFDuet-1 vector already carrying the sequence encoding the PknB kinase domain using primers listed in S2 Table, thus generating plasmid pDuet_parB, which was used to transform E. coli BL21(DE3)Star cells in order to overexpress His-tagged ParB. All constructs were verified by DNA sequencing. Recombinant strains harboring the different constructs were used to inoculate 400 ml of LB medium supplemented with ampicillin or spectinomycin, and the resulting cultures were incubated at 37°C with shaking until the optical density of the culture reached an A 600 of 0.6. IPTG (0.5 mM final) was added to induce the overexpression, and growth was continued for 3 h at 37°C. Purification of the His-tagged recombinant proteins was performed as described by the manufacturer (Clontech).
In vitro kinase assays
In vitro phosphorylation was performed with 4 μg of wild-type ParB or ParB derivatives in 20 μl of buffer P (25 mM Tris-HCl, pH 7.0; 1 mM DTT; 5 mM MgCl2; 1 mM EDTA; 50μM ATP) with 200 μCi ml-1 (65 nM) [γ-33P]ATP (PerkinElmer, 3000 Ci mmol-1), and 2 to 4 μg of kinase in order to obtain the optimal autophosphorylation activity for each mycobacterial kinase for 30 min at 37°C. Each reaction mixture was stopped by addition of an equal volume of 5 × Laemmli buffer and the mixture was heated at 100°C for 5 min. After electrophoresis, gels were soaked in 16% TCA for 10 min at 90°C, and dried. Radioactive proteins were visualized by autoradiography using direct exposure to films.
Mass spectrometry analysis
Purified His-tagged hyper-phosphorylated ParB (ParB-P) from the E. coli strain carrying pDuet_parB and co-expressing the PknB kinase domain, was subjected to mass spectrometry without further treatment. Subsequent mass spectrometric analyses were performed as previously reported [39–42]. Spectra were analyzed with the paragon algorithm from the ProteinPilot 2.0 database-searching software (Applied Biosystems) using the phosphorylation emphasis criterion against a homemade database that included the sequences of ParB and its derivatives.
Electrophoretic mobility shift assay
The DNA probe for electrophoretic mobility shift assays (EMSA) was generated by PCR using M. tb H37Rv chromosomal DNA as a template which encompassed the parS region with respective primers pair listed in S2 Table. The 5' end of the double-stranded PCR product was labeled using [γ-32P]-ATP and T4 polynucleotide kinase. A typical assay mixture contained in 20 μl: 10 mM Tris-HCl, pH 7.5; 50 mM NaCl; 1 mM EDTA; 1 mM dithiothreitol (DTT); 0.1 μg of nonspecific competitor (polydI-dC); 5% (v/v) glycerol; radioactive DNA probe (2000 cpm.ml-1) and various amounts (0,5 to 2,5μM) of the purified ParB proteins. After 20 min of incubation at room temperature, 20 μl of this mixture was loaded onto a native 5% (w/v) polyacrylamide TBE Ready Gel (Bio-Rad) and electrophoresed in 1% TBE (v/v) buffer for 1 h at 100 V.cm-1. Radioactive species were detected by autoradiography using direct exposure to films.
Overexpression of ParB and derivatives in M. smegmatis mc2155 and their purification
M. tb parB and parB_Ala genes were amplified from the corresponding pETPhos vector contructs and cloned into the shuttle vector pVV16 [43] using the primers listed in S2 Table. The resulting constructs pVV16_parB and pVV16_parB_Ala were electroporated into M. smegmatis mc 2 155. Transformants were grown and used for the purification of the His-tagged ParB proteins as described above. The purified recombinant proteins were used for immunoblotting using anti-phosphothreonine and anti-phosphoserine antibodies according to the manufacturer’s instructions (Invitrogen) and revealed using secondary antibodies labeled with IRDye 800CW infrared dyes (LiCOR).
Bacterial two-hybrid assays
M. tb parB, parB_Ala and parB_Asp genes were amplified from the corresponding pETPhos vector contructs and cloned into the pUT18C or pKT25 bacterial two-hybrid vectors as previously described [44] using the primers listed in S2 Table. The resulting constructs were chemically transformed into E. coli BTH101 [44] cells. Transformants were plated onto LB/X-Gal media supplemented with ampicillin and kanamycin and incubated at 30°C for 48 h.
Microscopy
M. tb parB, parB_Ala and parB_Asp genes were amplified from the corresponding pETPhos vector constructs and cloned into the pVV16_egfp vector with respective primers pair listed in S2 Table. The resulting constructs were electroporated into M. smegmatis mc2155ΔparB strain [30]. For protein localization, M. smegmatis mc2155ΔparB complemented strains were grown to OD600 0.8, fixed in 2% paraformaldehyde solution, washed with phosphate buffered saline (PBS; 10 mM sodium phosphate, pH 7.4, 150 mM NaCl, 15 mM KCl) and placed on poly-L-lysine coated coverslips before slides mounting using Vectashield H1000 (Vector). Fixed cells with fluorescent ParB were observed with an AxioImager Z2 (Zeiss) equipped with an apotome, using a 63X NA 1.4 apochromat DICIIIPhIII objective. Images were acquired and processed with the Zen software (Zeiss). For the detection of ParB-GFP derivatives, cytoplasmic protein extracts were isolated from M. smegmatis mc2155ΔparB complemented strains cell cultures grown until OD600 0.8 in 7H9 medium complemented with kanamycin (25μg/ml) at 37°C as previously described, and protein fractions (10 μg/lane) were separated using SDS-PAGE, blotted onto a nitrocellulose membrane, and subjected to Western blot analysis using the anti-GFP antibody (Santa Cruz).
Results and Discussion
ParB is phosphorylated in vitro by mycobacterial Ser/Thr protein kinases
The M. tuberculosis genome encodes eleven Ser/Thr protein kinases (STPKs) [45,46]. While these mycobacterial kinases appear to be involved in different key pathways such as cell wall metabolism, antibiotic susceptibility, and virulence [32,33,47,48], little is known about the nature of the target proteins that are phosphorylated.
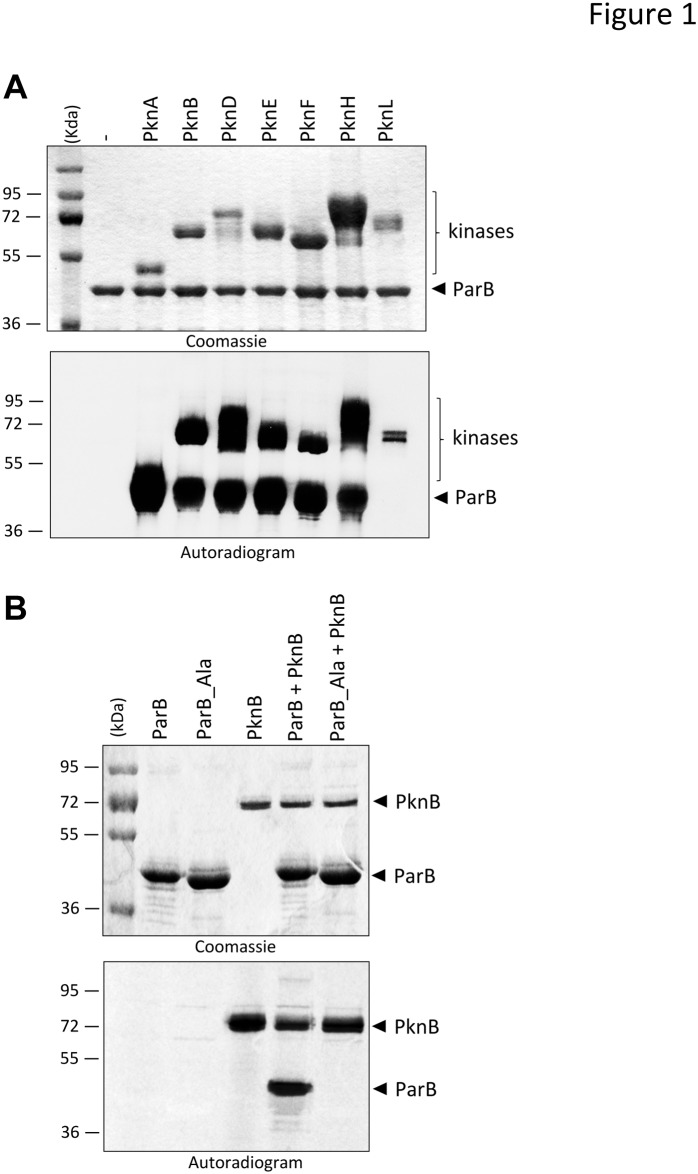
We therefore decided to investigate whether the chromosome segregation system could be regulated by phosphorylation, and decided to test the major protein ParB. This was first investigated in vitro in the presence of purified STPKs. The soluble kinase domains of several transmembrane kinases from M. tuberculosis were expressed as GST-tagged fusion proteins and purified from E. coli as reported earlier [49]. The kinase enzymes were incubated with M. tuberculosis ParB and [γ-33P]ATP, the proteins were resolved by SDS-PAGE and the protein phosphorylation status was analyzed by autoradiography. The presence of an intense radioactive signal indicated that ParB was phosphorylated by PknA, PknB, PknD, PknE, PknF and PknH (Fig. 1A). No signal was observed in the presence of PknL. As expected, no radioactive band was observed in the absence of kinase. Another finding arising from these experiments is that ParB can interact with multiple STPKs as previously shown for M. tuberculosis STPK substrates [33], suggesting that these enzymes may be regulated by multiple signals. However, it remains to be established whether this STPK cross-talk occurs in vivo, which would argue for a very complex signalling mechanism.
Fig 1. In vitro phosphorylation of ParB and mutant derivatives.
(A) in vitro phosphorylation of M. tb ParB by PknB. The soluble domains of seven recombinant M. tb STPKs (PknA to PknL) were expressed and purified as GST-tagged fusions and incubated with purified His-tagged ParB and [γ-33P]ATP. The amount of the STPKs used varied from 0,3 to 2 μg to obtain the optimal autophosphorylation activity for each kinase. Samples were separated by SDS-PAGE, stained with Coomassie Blue (upper panel), and visualized by autoradiography after overnight exposure to a film (lower panel). Upper bands reflect the phosphorylation signal of ParB, and the lower bands correspond to the autophosphorylation activity of each kinase. M, molecular mass markers. (B) in vitro phosphorylation of the ParB_Ala mutant. Purified ParB and phosphoablative ParB (ParB_Ala) were incubated with PknB and [γ33-P]ATP. Samples were separated by SDS-PAGE, stained with Coomassie Blue (upper panel), and visualized by autoradiography (lower panel) after overnight exposure to a film.
ParB is phosphorylated on serine and threonine residues
Mass spectrometry was used to identify and localize the phosphorylation site(s) on M. tuberculosis ParB. This method has been successfully used to elucidate the phosphorylation sites in a sequence-specific fashion for several M. tuberculosis STPK substrates [39,40,50–53]. Phosphorylated ParB-P was purified from E. coli co-expressing PknB and ParB (pDuet_parB) and subjected to mass spectrometric analysis after tryptic digestion. Noteworthy, we decided to use the PknB kinase previously described as involved in cell division processes among the kinases able to phosphorylate ParB in our in vitro assay [34,54,55]. A sequence coverage of 98% that included all Ser and Thr residues was obtained. The MS/MS spectra unambiguously identified the presence of ten phosphate groups (Table 1), thus indicating that ParB is phosphorylated on eight threonines, Thr32, Thr41, Thr53, Thr110, Thr195, and Thr254, Thr300, Thr303 and on two serines, Ser5 and Ser239. Moreover, in order to confirm that the phosphosites identified were not dependent on the kinase co-expressed with ParB, we performed mass spectrometry analysis when ParB was phosphorylated by PknF or PknH which resulted in the same phosphorylation sites identified. Most of the phosporylated residues are located in non-conserved regions of the ParB/Spo0J protein family, either in N- or C- terminus of the common conserved central domain (residues 60 to 251 in M. tuberculosis ParB) according to sequence alignment with the crystallised member ParB/Sp0J protein family, (i.e. Spo0J from Thermus thermophiles that shares 48% sequence identity with ParB for its core domain). The only phosphorylated residue positioned in the core domain is the threonine Thr195. This residue is located in the middle of one of the helix from the helix-turn-helix DNA-binding motif.
Table 1. Phosphoacceptors identified after purification of M. tuberculosis ParB from the E. coli BL21(DE3)star strain co-expressing M. tuberculosis PknB.
| Phosphorylated tryptic peptide sequence of ParB purified from pCDFDuet co-expressing PknB | Number of detected phosphate groups (LC/MS/MS) | Phosphorylated residue(s) |
|---|---|---|
| [15–28] ENLYFQGHMTQPpSR | 1 | S5 |
| [36–58] GLAALIPTGPADGESGPPpTLGPR | 1 | T32 |
| [42–58] PTGPADGESGPPpTLGPR | 1 | T32 |
| [59–83] MoxGSApTADVVIGGPVPDTSVM GAIYR | 1 | T41 |
| [59–83] MGSApTADVVIGGPVPDpTSVM GAIYR | 1 | T41+T53 |
| [59–83] MGSATADVVIGGPVPDpTSVMGAIYR | 1 | T53 |
| [126–143] SLAGSQpTGVRYQIVMGER | 1 | T110 |
| [211–220] SRPLIpTNMIR | 1 | T195 |
| [244–261] ALLSLEAGPEAQEELApSR | 1 | S239 |
| [244–270] ALLSLEAGPEAQEELApSRIVAEGLSVR | 1 | S239 |
| [271–285] ATEEpTVTLANHEANR | 1 | T254 |
| [271–299] ATEEpTVTLANHEANRQAHHSDATTPAPPR | 1 | T254 |
| [315–329] LSTTFDpTRVTVSLGK | 1 | T300 |
| [315–330] LSTTFDpTRVTVSLGKR | 1 | T300 |
| [315–330] LSTTFDpTRVpTVSLGKR | 1 | T300+T303 |
| [315–329] LSTTFDTRVpTVSLGK | 1 | T303 |
| [315–330] LSTTFDTRVpTVSLGK | 1 | T303 |
Sequences of the phosphorylated peptides identified in ParB as determined by mass spectrometry following tryptic digestion are indicated, and phosphorylated residues (pT or pS) are shown in bold.
Then, to confirm that only the identified residues are phosphorylated in vitro, these residues were mutated to alanine. The corresponding phosphoablative ParB_T32A/T41A/T53A/T110A/T195A/T254A/T300A/T303A/S5A/S239A mutant (ParB_Ala) was expressed as a His-tagged protein in E. coli BL21(DE3)Star harboring pETPhos_parB_Ala. The resulting ParB_Ala mutant protein was purified and incubated with PknB in the presence of [γ-33P]ATP. Following separation by SDS-PAGE and analysis by autoradiography, total abrogation of the phosphorylation signal was observed (Fig. 1B).
ParB is phosphorylated in mycobacteria
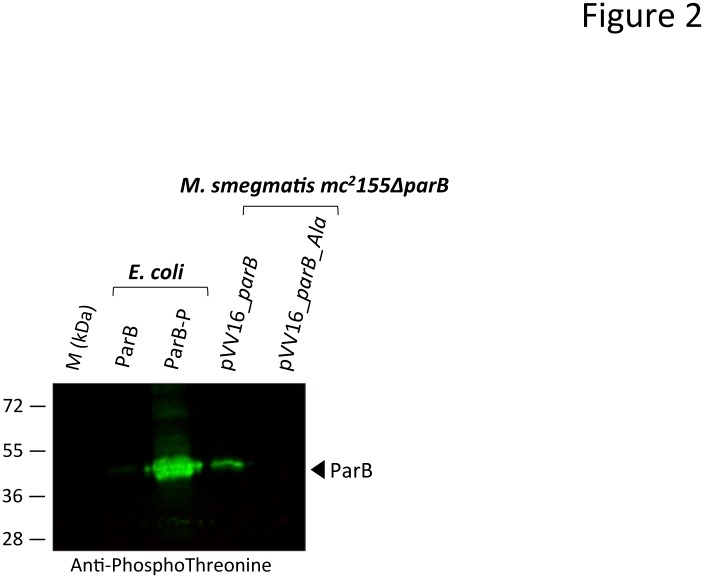
To address the relevance of our in vitro findings, we investigated the phosphorylation status of M. tb ParB in vivo. To do so, we used the convenient M. smegmatis mc 2 155 strain as a surrogate for M. tb as reviewed by Shiloh et al. [56]. First, due to the homology of ParB from M. tb and M. smegmatis (72%), we confirmed in vitro phosphorylation of M. tb ParB by PknB from M. smegmatis (data not shown). Next, we investigated the in vivo phosphorylation status by western blotting using either anti-phosphothreonine, or anti-phosphoserine antibodies [39,41]. First, the specificity of the antibodies was determined using the ParB isoforms purified from either E. coli or E. coli co-expressing PknB, based on the strategy described by Molle et al. (2010) [38]. Only the phosphorylated ParB isoform derived from pDuet_parB (ParB-P) reacted with the anti-phosphothreonine and antiphosphoserine antibodies as expected, while the unphosphorylated ParB isoform derived from the pETPhos_parB construct (ParB) failed to generate a signal with these antibodies (Fig. 2). Moreover, in order to address the role of ParB phosphorylation in mycobacteria, the genes encoding ParB and ParB_Ala (harboring Ser/Thr to Ala substitutions at ten positions as the phosphoablative control) were cloned as C-terminal His-tag fusions into the shuttle vector pVV16, and the resulting constructs, designated pVV16_parB and pVV16_parB_Ala, respectively, were introduced into M. smegmatis mc 2 155. The His-tag-purified proteins derived from these M. smegmatis mc 2 155 recombinant strains were subjected to western blotting using anti-phosphothreonine, and anti-phosphoserine antibodies. As shown in Fig. 2, a clear signal for the wild-type ParB was detected with anti-phosphothreonine and anti-phosphoserine antibodies, while no signal was observed with the ParB_Ala (Fig. 2). No signal could be detected when probing the membrane with anti-phosphotyrosine antibodies (data not shown). Overall, these results confirm the presence of phosphorylated Ser and/or Thr residues and demonstrate that ParB from M. tuberculosis is indeed being phosphorylated in M. smegmatis thus confirming the relevance to use this M. smegmatis mutant in this study.
Fig 2. Phosphorylation of ParB in mycobacteria.
E. coli harboring pETPhos_parB was used as a source of non phosphorylated ParB (ParB), and the strain harboring pDuet_parB coexpressing PknB and ParB provided the phosphorylated ParB isoform (ParB-P). ParB and ParB_Ala were produced in M. smegmatis mc 2 155ΔparB strains harboring pVV16_parB or pVV16_parB_Ala, respectively. Three μg of purified His-tagged ParB derivatives were migrated and detected on independent SDS-PAGE gels by immunoblotting using anti-phosphothreonine (middle panel) or anti-phosphoserine (lower panel) antibodies according to the manufacturer’s instructions (Invitrogen), and revealed with secondary antibodies labeled with IRDye infrared dyes (Odyssey, LiCOR). M, molecular mass markers.
Phosphorylation negatively regulates ParB DNA-binding to chromosome parS sequence
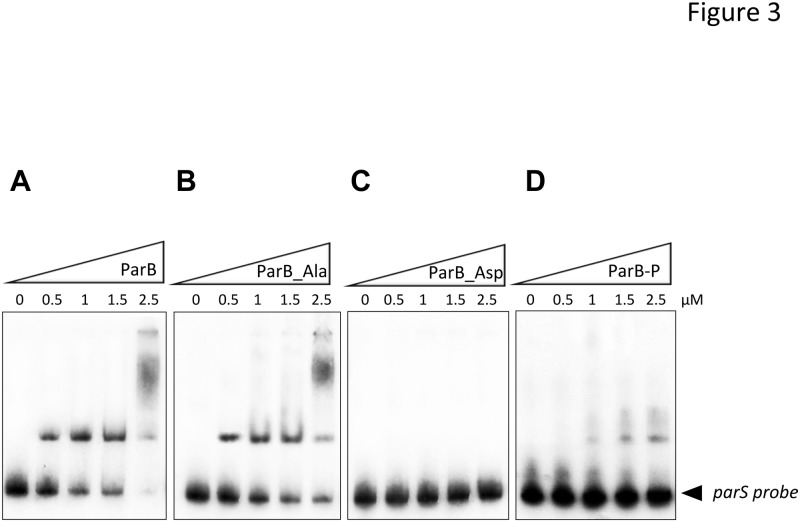
Since ParB is described as a DNA-binding protein, we decided to investigate whether phosphorylation could affect ParB binding affinity to its chromosomal target sequence, the centromere-like parS sequences. To test this hypothesis, we analyzed and compared the ability of the non-phosphorylated (ParB) and the hyper-phosphorylated form (ParB-P) to bind to the parS region, known to be the specific target of ParB as previously described (Fig. 3) [30]. Electrophoretic mobility shift assays (EMSAs) were performed and revealed that the ParB unphosphorylated isoform (ParB) binds to the parS probe in a dose-dependent manner (Fig. 3A) as reported previously [30]. In contrast, the phosphorylated isoforms (ParB_Asp and ParB-P) showed a dramatic reduction of mobility shift (Fig. 3C and 3D), supporting the view that phosphorylation negatively affects the ability of ParB to bind to its chromosomal region. As anticipated, the ParB_Ala phosphoablative mutant retained a DNA-binding activity similarly to the non-phosphorylated protein (Fig. 3B). Our results clearly suggest that phosphorylation dramatically decreases ParB affinity for parS sequences at least in vitro. Therefore phosphorylation would negatively regulate ParB DNA-binding activity in mycobacteria, potentially by phosphorylation of the residue Thr195 located within the DNA binding domain. Therefore, a single T195D mutant was generated and while ParB_T195D binding was reduced it was not abolished, suggesting that the other phosphorylation sites are required for full regulation of ParB (data not shown).
Fig 3. DNA-binding activity of ParB derivatives.
Gel electrophoretic mobility shift analysis (EMSA) of ParB binding to the parS sequence. The parS region was amplified by PCR, radioactively labeled, and incubated with 0.5, 1, 1.5, and 2.5μM of purified ParB, resolved by non-denaturing PAGE and visualized by autoradiography after overnight exposure to a film. (A) Binding of the unphosphorylated ParB (ParB), (B) ParB phosphoablative mutant (ParB_Ala), (C) ParB phosphomimetic mutant (ParB_Asp), and (D) phosphorylated ParB (ParB-P), to the parS region.
Phosphorylation inhibits ParB interaction with its cytoskeletal partner ParA
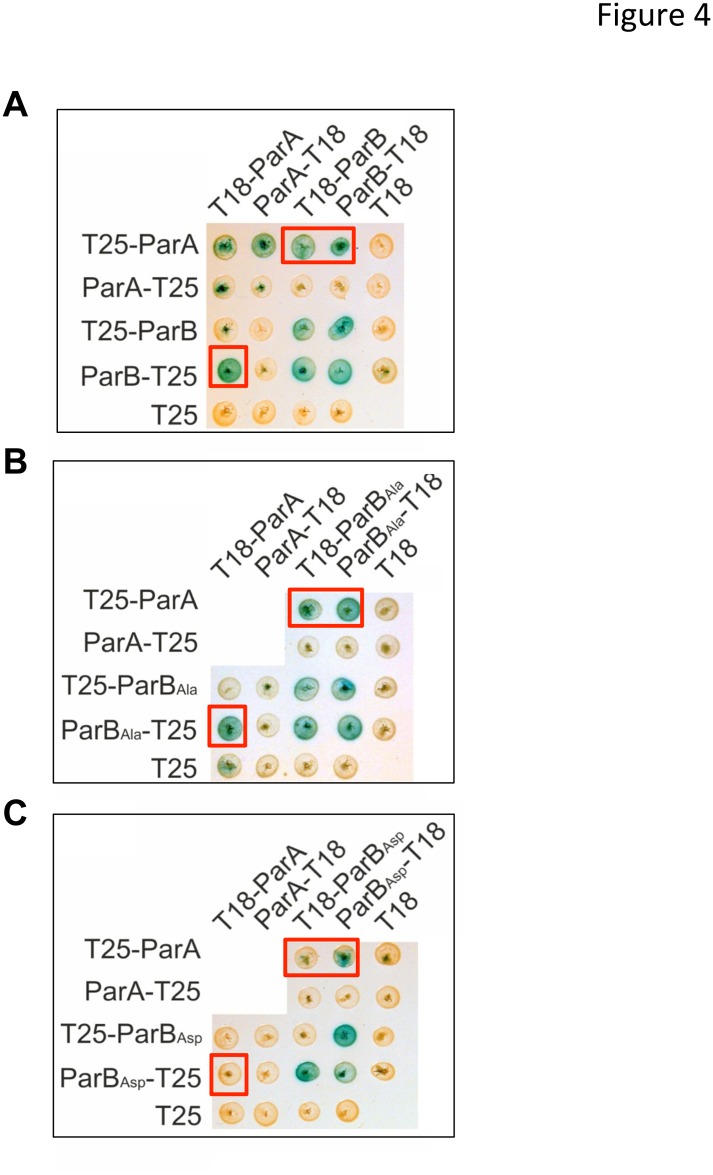
A recent study highlighted the essential role of ParA in the proper positioning and number of ParB complexes in M. smegmatis [31]. ParA localizes as pole-associated complexes connected with a patch of fluorescence accompanying two ParB complexes, positioned at 20–25% and 75–80% of the cell length. In the absence of ParA, ParB complexes are more numerous and seem to locate randomly into the cytosol highlighting the fact that ParB localization is tightly linked to its interaction with ParA. Based on these data and the localization of the phosphorylation sites along the primary sequence, we assessed the question, if phosphorylation could affect ParB interaction with ParA. This notion was verified by bacterial two hybrid (BTH) assays (Fig. 4). ParB derivatives and ParA were fused to either T25 or T18 fragment of Bordetella pertussis adenylate cyclase. Positive signals were observed with ParB_WT and ParB_Ala versus ParA, independently of the adenylate cyclase fragment and its position in the fusion protein suggesting that ParB_WT and ParB_Ala interact with ParA in vivo. Under the same conditions, no signal was observed when the interaction between ParB_Asp and ParA was tested indicating that ParB_Asp no longer interacts with ParA. Therefore, phosphorylation seems to also regulate ParB interaction with the cytoskeletal partner protein ParA. Noteworthy, BTH experiments indicated, that all ParB derivatives tested retain the ability to dimerise. This suggests, that replacement of the phosphorylation sites by alanine or aspartate does not prevent ParB dimerization, thus these results were not due to a tertiary or quaternary structure change due to the introduction of the mutations.
Fig 4. ParA interaction with ParB derivatives.
Interactions of M. tuberculosis ParA with ParBWT (A) ParBAla (B) and ParBAsp (C) in bacterial two hybrid system. The red rectangles indicate differences in ParA-ParB interaction between WT (A), phosphoablative (B) and phosphophomimetic (C) ParB mutant proteins.
Phosphorylation induces delocalization of ParB
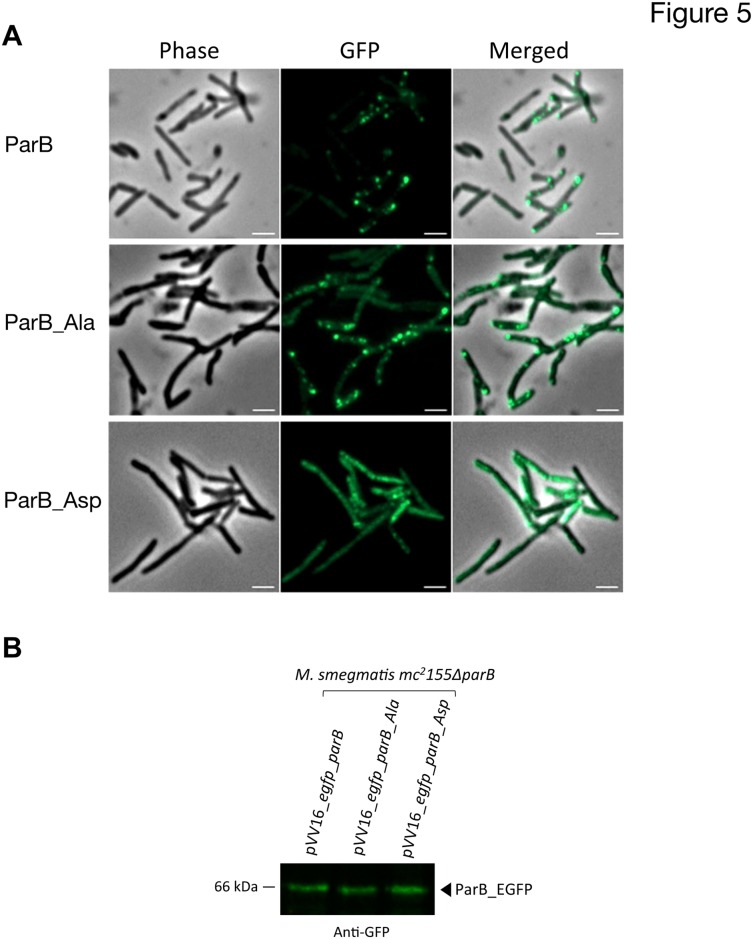
Phosphorylation appears to prevent the interaction between ParB and ParA. Since ParB location is tightly dependent on the interaction with ParA we decided to investigate if phosphorylation could affect ParB complexes occurrence, number and/or localization in vivo. Wild-type ParB, phosphoablative and phosphomimetic ParB mutants were fused to EGFP using pVV16_egfp vector under hsp60 promoter and the resulting constructs were introduced into M. smegmatis mc 2 155ΔparB strain [56]. M. tuberculosis ParB was fully functional in M. smegmatis cells, as demonstrated by the complementation of the parB deletion phenotype (2.52% anucleate cells, S3 Table). The localization of M. tuberculosis ParB resembled the native protein localization with two complexes in proximity of the cell poles (about 20 and 80% of cell length) (Fig. 5A). Such a positioning was observed in most analysed cells, however a fraction of cells with a diffuse fluorescence could also be observed. Both phosphomimetic and phosphoablative ParB mutant did not form clear foci in most of the cells but were mostly dispersed throughout the cell, either as a diffuse fluorescence (ParB_Asp) or irregularly spaced, faint foci (ParB_Ala) (Fig. 5A). This observation is in agreement with the lack of ParB_Ala and ParB_Asp complementation of segregation phenotype of M. smegmatis parB deletion mutant (S3 Table). While mislocalisation was expected in case of phosphomimetic mutant which does not bind DNA, it seems surprising for ParB_Ala. This suggests that although ParB_Ala is able to interact with DNA and ParA, the lack of phosphoregulation severely affects its localization and function. That could result from influence of phosphorylation on the dynamics of ParA-ParB-parS interactions. For example altered phosphorylation state of ParB could affect either stability of the nucleoprotein complex or ParB stimulated ParA ATPase activity changing its dynamics and localisation. Moreover, western blot analysis from ParB-GFP derivatives in M.smegmatis mc 2 155ΔparB complemented strains revealed proper expression of the GFP-recombinant ParB proteins in each strain (Fig. 5B).
Fig 5. Localization of ParB and derivatives.
(A) Subcellular localization of ParB-GFP in M. smegmatis mc 2 155ΔparB strain. Are shown Differential Intereference Contrast image (DIC), GFP fluorescence (GFP) and a merged image of DIC and GFP fluorescence (Merge). Localization of ParB isoforms are shown in three different panels: the upper panel shows wild-type ParB (ParB), the middle panel shows phosphoablative ParB (ParB_Ala) and the lower panel shows phosphomimetic ParB (ParB_Asp). Scalebar 2μm (B) Immunoblotting of ParB-GFP derivatives in M.smegmatis mc 2 155ΔparB complemented strains. Crude extracts of M. smegmatis mc 2 155ΔparB complemented with pVV16_egfp_parB, pVV16_egfp_parB_Ala or pVV16_egfp_parB_Asp were electrophoresed on SDS-PAGE gel, ParB-GFP derivatives were then detected by immoblotting using anti-GFP antibody according to the manufacturer’s instructions (Santa Cruz) and revealed with secondary antibodies labeled with IRDye infrared dyes (Odyssey, LiCOR).
We propose that both phosphorylated and unphosphorylated states of ParB are required for proper chromosome segregation and that a subtle balance between the phosphorylated and unphosphorylated states of ParB would tune the segregation event during the cell cycle.
Conclusion
We have used a combination of genetic and biochemical approaches to provide the first reported evidence that the partitioning protein ParB, is negatively regulated by phosphorylation. Our work suggests that a shift in the unphosphorylated/phosphorylated ParB balance in favor of the phosphorylated form rapidly leads to the delocalization of ParB into the cytosol. An hypothesis arising from this work is that a regulatory role of the different isoforms of ParB represents a molecular switch in partitioning protein localization and function. The unphosphorylated form of ParB binds to its target DNA sequences, parS, and this binding is modulated via the interaction with the cytoskeletal partner protein ParA, which is thought to enhance the binding of ParB to its DNA targets if ParB is complexed with ParA. In addition, ParB interaction with ParA is critical for a proper positioning of ParB in proximity to the cell pole. The PknB-mediated phosphorylation of ParB inactivates the DNA binding activity of this partitioning protein, as well as the interaction with ParA, thereby preventing the localization of ParB, thus the formation of the mitotic-like apparatus. Our results provide a foundation for further investigation of an important functional linkage between STPKs and the ParABS chromosome segregation system, which may contribute to proper mycobacterial cell division. Interestingly, it was shown before that ParA interacts with another phosphorylated protein Wag31 (DivIVA homologue) possibly providing another link between segregation apparatus and STPKs regulation. Although challenging, future studies will help identify extracellular cues sensed by kinases leading to the phosphorylation of ParB. This will allow us to understand how M. tuberculosis senses its environment and mediates its response in a coordinated manner to regulate chromosome partition.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors wish to thank I. Zanella-Cléon from the Mass Spectrometry Unit (CCMP), UMS3444/US8 Biosciences Gerland Lyon Sud, for her excellent expertise and technical assistance. The authors also thank the Montpellier RIO imaging facility at the University of Montpellier 2.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the National Research Agency (ANR-09-MIEN-004) and the ATIP/AVENIR Program for V.M., and the Region Languedoc-Roussilon for G.B. and from the Wroclaw Research Center EIT+ under the project Biotechnologies and advanced medical technologies—BioMed (POIG 1.1. Project 3.1.) financed from the European Regional Development Fund (Operational Program Innovative Economy, 1.1.2) for KG, JZ-C and DJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010; 328: 856–861. 10.1126/science.1185449 [DOI] [PubMed] [Google Scholar]
- 2. Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008; 135: 74–84. 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- 3. Chaudhuri BN, Dean R. The evidence of large-scale DNA-induced compaction in the mycobacterial chromosomal ParB. J Mol Biol. 2011; 413: 901–907. 10.1016/j.jmb.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 4. Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus . Cell. 1997; 88: 675–684. [DOI] [PubMed] [Google Scholar]
- 5. Mohl DA, Easter J Jr, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus . Mol Microbiol. 2001; 42: 741–755. [DOI] [PubMed] [Google Scholar]
- 6. Jakimowicz D, Chater K, Zakrzewska-Czerwinska J. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol Microbiol. 2002; 45: 1365–1377. [DOI] [PubMed] [Google Scholar]
- 7. Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol. 2000; 182: 1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donovan C, Schwaiger A, Kramer R, Bramkamp M. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum . J Bacteriol. 2010; 192: 3441–3451. 10.1128/JB.00214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J Bacteriol. 2004; 186: 6983–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Godfrin-Estevenon AM, Pasta F, Lane D. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli . Mol Microbiol. 2002; 43: 39–49. [DOI] [PubMed] [Google Scholar]
- 11. Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology. 2002; 148: 537–548. [DOI] [PubMed] [Google Scholar]
- 12. Harms A, Treuner-Lange A, Schumacher D, Sogaard-Andersen L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 2013; 9: e1003802 10.1371/journal.pgen.1003802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iniesta AA. ParABS System in Chromosome Partitioning in the Bacterium Myxococcus xanthus . PLoS One. 2014; 9: e86897 10.1371/journal.pone.0086897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewis PJ, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol Microbiol. 1997; 25: 945–954. [DOI] [PubMed] [Google Scholar]
- 15. Lin DC, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis . Proc Natl Acad Sci U S A. 1997; 94: 4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis . Cell. 1997; 88: 667–674. [DOI] [PubMed] [Google Scholar]
- 17. Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol. 2007; 64: 703–718. [DOI] [PubMed] [Google Scholar]
- 18. Ireton K, Cossart P. Interaction of invasive bacteria with host signaling pathways. Curr Opin Cell Biol. 1998; 10: 276–283. [DOI] [PubMed] [Google Scholar]
- 19. Madiraju MV, Moomey M, Neuenschwander PF, Muniruzzaman S, Yamamoto K, Grimwade JE, et al. The intrinsic ATPase activity of Mycobacterium tuberculosis DnaA promotes rapid oligomerization of DnaA on oriC . Mol Microbiol. 2006; 59: 1876–1890. [DOI] [PubMed] [Google Scholar]
- 20. Qin MH, Madiraju MV, Rajagopalan M. Characterization of the functional replication origin of Mycobacterium tuberculosis . Gene. 1999; 233: 121–130. [DOI] [PubMed] [Google Scholar]
- 21. Rajagopalan M, Qin MH, Nash DR, Madiraju MV. Mycobacterium smegmatis dnaA region and autonomous replication activity. J Bacteriol. 1995; 177: 6527–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zawilak A, Kois A, Konopa G, Smulczyk-Krawczyszyn A, Zakrzewska-Czerwinska J. Mycobacterium tuberculosis DnaA initiator protein: purification and DNA-binding requirements. Biochem J. 2004; 382: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, et al. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006; 188: 1856–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dziadek J, Madiraju MV, Rutherford SA, Atkinson MA, Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002; 148: 961–971. [DOI] [PubMed] [Google Scholar]
- 25. Huang Q, Tonge PJ, Slayden RA, Kirikae T, Ojima I. FtsZ: a novel target for tuberculosis drug discovery. Curr Top Med Chem. 2007; 7: 527–543. [DOI] [PubMed] [Google Scholar]
- 26. Rajagopalan M, Maloney E, Dziadek J, Poplawska M, Lofton H, Chauhan A, et al. Genetic evidence that mycobacterial FtsZ and FtsW proteins interact, and colocalize to the division site in Mycobacterium smegmatis . FEMS Microbiol Lett. 2005; 250: 9–17. [DOI] [PubMed] [Google Scholar]
- 27. Casart Y, Gamero E, Rivera-Gutierrez S, Gonzalez YMJA, Salazar L. par genes in Mycobacterium bovis and Mycobacterium smegmatis are arranged in an operon transcribed from "SigGC" promoters. BMC Microbiol. 2008; 8: 51 10.1186/1471-2180-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maloney E, Madiraju M, Rajagopalan M. Overproduction and localization of Mycobacterium tuberculosis ParA and ParB proteins. Tuberculosis (Edinb). 2009; 89 Suppl 1: S65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003; 48: 77–84. [DOI] [PubMed] [Google Scholar]
- 30. Jakimowicz D, Brzostek A, Rumijowska-Galewicz A, Zydek P, Dolzblasz A, Smulczyk-Krawczyszyn A, et al. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis . Microbiology. 2007; 153: 4050–4060. [DOI] [PubMed] [Google Scholar]
- 31. Ginda K, Bezulska M, Ziolkiewicz M, Dziadek J, Zakrzewska-Czerwinska J, Jakimowicz D. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol. 2013; 87: 998–1012. 10.1111/mmi.12146 [DOI] [PubMed] [Google Scholar]
- 32. Wehenkel A, Bellinzoni M, Grana M, Duran R, Villarino A, Fernandez P, et al. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim Biophys Acta. 2008; 1784: 193–202. [DOI] [PubMed] [Google Scholar]
- 33. Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010; 75: 1064–1077. 10.1111/j.1365-2958.2009.07041.x [DOI] [PubMed] [Google Scholar]
- 34. Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005; 19: 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakur M, Chakraborti PK. GTPase Activity of Mycobacterial FtsZ Is Impaired Due to Its Transphosphorylation by the Eukaryotic-type Ser/Thr Kinase, PknA. J Biol Chem. 2006; 281: 40107–40113. [DOI] [PubMed] [Google Scholar]
- 36. Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, Kundu M, et al. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One. 2010; 5: e8590 10.1371/journal.pone.0008590 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Canova MJ, Kremer L, Molle V. pETPhos: a customized expression vector designed for further characterization of Ser/Thr/Tyr protein kinases and their substrates. Plasmid. 2008; 60: 149–153. 10.1016/j.plasmid.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 38. Molle V, Leiba J, Zanella-Cleon I, Becchi M, Kremer L. An improved method to unravel phosphoacceptors in Ser/Thr protein kinase-phosphorylated substrates. Proteomics. 2010; 10: 3910–3915. 10.1002/pmic.201000316 [DOI] [PubMed] [Google Scholar]
- 39. Corrales RM, Molle V, Leiba J, Mourey L, de Chastellier C, Kremer L. Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phagosome maturation block. J Biol Chem. 2012; 287: 26187–26199. 10.1074/jbc.M112.373209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slama N, Leiba J, Eynard N, Daffe M, Kremer L, Quemard A, et al. Negative regulation by Ser/Thr phosphorylation of HadAB and HadBC dehydratases from Mycobacterium tuberculosis type II fatty acid synthase system. Biochem Biophys Res Commun. 2011; 412: 401–406. 10.1016/j.bbrc.2011.07.051 [DOI] [PubMed] [Google Scholar]
- 41. Leiba J, Syson K, Baronian G, Zanella-Cleon I, Kalscheuer R, Kremer L, et al. Mycobacterium tuberculosis maltosyltransferase GlgE, a genetically validated antituberculosis target, is negatively regulated by Ser/Thr phosphorylation. J Biol Chem. 2013; 288: 16546–16556. 10.1074/jbc.M112.398503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leiba J, Hartmann T, Cluzel ME, Cohen-Gonsaud M, Delolme F, Bischoff M, et al. A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J Biol Chem. 2012; 287: 43607–43619. 10.1074/jbc.M112.418913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000; 275: 30092–30099. [DOI] [PubMed] [Google Scholar]
- 44. Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli . Methods Enzymol. 2000; 328: 59–73. [DOI] [PubMed] [Google Scholar]
- 45. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 46. Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis . Trends Microbiol. 2000; 8: 238–244. [DOI] [PubMed] [Google Scholar]
- 47. Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004; 304: 1800–1804. [DOI] [PubMed] [Google Scholar]
- 48. Papavinasasundaram KG, Chan B, Chung JH, Colston MJ, Davis EO, Av-Gay Y. Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J Bacteriol. 2005; 187: 5751–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Molle V, Brown AK, Besra GS, Cozzone AJ, Kremer L. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J Biol Chem. 2006; 281: 30094–30103. [DOI] [PubMed] [Google Scholar]
- 50. Veyron-Churlet R, Molle V, Taylor RC, Brown AK, Besra GS, Zanella-Cleon I, et al. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem. 2009; 284: 6414–6424. 10.1074/jbc.M806537200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barthe P, Roumestand C, Canova MJ, Kremer L, Hurard C, Molle V, et al. Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure. 2009; 17: 568–578. 10.1016/j.str.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 52. Veyron-Churlet R, Zanella-Cleon I, Cohen-Gonsaud M, Molle V, Kremer L. Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J Biol Chem. 2010; 285: 12714–12725. 10.1074/jbc.M110.105189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Canova MJ, Kremer L, Molle V. The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J Bacteriol. 2009; 191: 2876–2883. 10.1128/JB.01569-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Plocinska R, Martinez L, Gorla P, Pandeeti E, Sarva K, Blaszczyk E, et al. Mycobacterium tuberculosis MtrB Sensor Kinase Interactions with FtsI and Wag31 Proteins Reveal a Role for MtrB Distinct from That Regulating MtrA Activities. J Bacteriol. 2014; 196: 4120–4129. 10.1128/JB.01795-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006; 152: 493–504. [DOI] [PubMed] [Google Scholar]
- 56. Shiloh MU, Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010; 13: 86–92. 10.1016/j.mib.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper.