Abstract
SecM, a bacterial secretion monitor protein, contains a specific amino acid sequence at its C-terminus, called arrest sequence, which interacts with the ribosomal tunnel and arrests its own translation. The arrest sequence is sufficient and necessary for stable translation arrest. However, some previous studies have suggested that the nascent chain outside the ribosome affects the stability of translation arrest. To clarify this issue, we performed in vitro translation assays with HaloTag proteins fused to the C-terminal fragment of E. coli SecM containing the arrest sequence or the full-length SecM. We showed that the translation of HaloTag proteins, which are fused to the fragment, is not effectively arrested, whereas the translation of HaloTag protein fused to full-length SecM is arrested efficiently. In addition, we observed that the nascent SecM chain outside the ribosome markedly stabilizes the translation arrest. These results indicate that changes in the nascent polypeptide chain outside the ribosome can affect the stability of translation arrest; the nascent SecM chain outside the ribosome stabilizes the translation arrest.
Introduction
Recent studies have revealed that ribosomes do not always translate mRNAs at a constant rate. The biased codon usage and specific mRNA sequences can change the rate of polypeptide elongation [1, 2]. Sometimes the nascent chains halt the translation elongation or termination to regulate gene expression. This phenomenon, known as translation arrest, is observed in several species of both prokaryotic and eukaryotic organisms [3]. Translation arrest mediated by SecM in Escherichia coli has been studied most often and is best-characterized.
The E. coli SecM is a 170-amino acid (aa) secretion monitor protein. This protein regulates the translation of the downstream gene, secA, in response to protein secretion activity in the cell [4]. This gene encodes an ATPase that drives protein translocation. When a cell is secretion-competent, SecM evokes a transient translation arrest that is executed by physical pulling of Sec translocation apparatus [5]. Under such circumstances, the SecA ribosome-binding site is masked by the secondary structure of the mRNA, and the translation is suppressed [6, 7]. However, under secretion-limiting conditions, the SecM translation is subjected to prolonged arrest, allowing secA translation by altering the structure of secM-secA mRNA [7].
This arrest is evoked by the translation of the C-terminal specific sequence (150FSTPVWISQAQGIRAGP166) of SecM, called arrest sequence [8]. The ribosome stalls when the Pro166 codon is positioned at the A site. In other words, the stalled ribosome has the polypeptidyl-tRNA at the P site and unreacted Pro-tRNAPro at the A site of the ribosome [9, 10]. Mutations of the key residues in the arrest sequence reduce (Phe150, Trp155, Ile156, Gly161, Ile162 and Ala164) or impair (Arg163, Gly165 and Pro166) the translation arrest [8, 11, 12]. This sequence also causes translation arrest of unrelated proteins, which can be utilised to generate nascent chain-ribosome complexes [13–24]. Consequently, it is widely accepted that the SecM arrest sequence is sufficient and necessary for a sustained translation arrest.
However, some of the existing data suggest that the arrest sequence alone is not enough to provide a stable translation arrest [21, 22]. Evans et al. have shown that the efficiency of translation arrest changes depending on the protein displayed on the ribosome [13]. We hypothesized that the nascent chain outside the ribosome affects the efficiency of translation arrest. To test this hypothesis, we performed in vitro translation assays using HaloTag proteins fused to either the E. coli SecM C-terminal sequence containing the arrest sequence or full-length SecM (Fig. 1). As a result, we found that changes in the nascent polypeptide chain outside the ribosome can affect the stability of translation arrest and the nascent SecM chain outside the ribosome helps to stabilize the arrest.
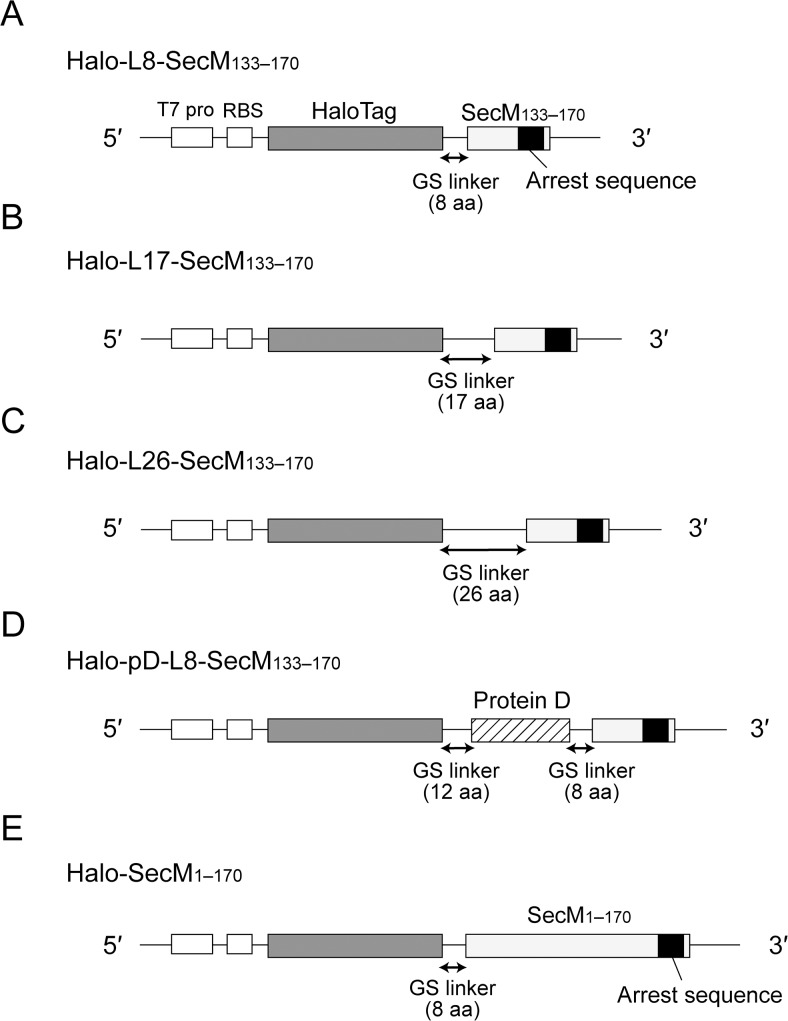
Fig 1. DNA constructs used for in vitro translation of HaloTag proteins harbouring the E. coli SecM arrest sequence.
A T7 promoter (T7 pro) and a ribosome-binding site (RBS) are located upstream from the gene encoding HaloTag protein fused via a spacer sequence to the C-terminal sequence of E. coli SecM (residues 133–170; SecM133–170) or E. coli full-length SecM (residues 1–170; SecM1–170). (A) DNA construct for in vitro translation of Halo-L8-SecM133–170. The spacer sequence consists of an 8-aa glycine–serine (GS) linker (GSGGGSGS). (B) DNA construct for in vitro translation of Halo-L17-SecM133–170. The spacer sequence is a 17-aa GS linker (GSGGGSGGGSGGGSGGS). (C) DNA construct for in vitro translation of Halo-L26-SecM133–170. The spacer sequence consists of a 26-aa GS linker (GSGGGSGGGSGGGSGGGSGGGSGGGS). (D) DNA construct for in vitro translation of Halo-pD-L8-SecM133–170. The spacer sequence is composed of a 12-aa GS linker (GSGGGSGGGSMG), a monomeric version of protein D from the bacteriophage λ (residues 21–110) and an 8-aa GS linker (GSGGGSGS) [17, 20, 28]. (E) DNA construct for in vitro translation of Halo-SecM1–170. SecM1–170 is fused to the C-terminus of HaloTag via an 8-aa GS linker (GSGGGSGS). The molecular masses of Halo-L8-SecM133–170, Halo-L17-SecM133–170, Halo-L26-SecM133–170, Halo-pD-L8-SecM133–170 and Halo-SecM1–170, calculated from the deduced amino acid sequences, were 38, 39, 40, 49 and 53 kDa, respectively.
Materials and Methods
Reagents
PrimeSTAR HS DNA Polymerase was obtained from Takara Bio, Inc. The PURExpress ΔRibosome Kit, nuclease-free water and HaloTag TMR Ligand were purchased from New England Biolabs, QIAGEN and Promega, respectively. SUPERase-In RNase Inhibitor, NuPAGE 10% Bis-Tris Gel, NuPAGE MOPS SDS Running Buffer and RNAsecure were purchased from Life Technologies. Molecular weight markers (Precision Plus Protein Prestained Standards) and puromycin were purchased from Bio-Rad and Sigma-Aldrich, respectively. Anti-mouse IgG conjugated with HiLyte Fluor 555 [Anti-IgG (H + L), Mouse, Rabbit-Poly, HiLyte Fluor 555] was obtained from AnaSpec, Inc. Other reagents were purchased from Wako Pure Chemicals Industries, Ltd.
Preparation of DNA templates for in vitro translation
The T7-based expression plasmids for HaloTag proteins harbouring the E. coli SecM arrest sequence were constructed as described in S1 Document. Primer sequences used in this study are listed in S1 Table. The plasmids were used as templates for PCR amplification with primer 1, 5′-GAAATTAATACGACTCACTATAGGGG-3′ and primer 2, 5′-GCTAGTTATTGC TCAGCGG-3′. Primer 1 contained the T7 promoter sequence (underlined) and primer 2 overlapped the T7 terminator sequence. The PCR products were used as templates for in vitro translation of the proteins [25].
In vitro translation
HaloTag proteins harbouring the SecM arrest sequence were synthesized using the PURExpress ΔRibosome Kit. First, the reaction mixtures without ribosomes were assembled. The mixture for reaction with HaloTag TMR Ligand contained 4.0 μL of Solution A, 1.2 μL of Factor Mix, 2.0 μL of template DNA, 0.5 μL of 20 μM HaloTag TMR Ligand, 0.5 μL of 20 U/μL RNase inhibitor and 1.3 μL of nuclease-free water in a 10 μL reaction. The mixture for reaction without HaloTag TMR Ligand contained 4.0 μL of Solution A, 1.2 μL of Factor Mix, 2.0 μL of template DNA, 0.5 μL of 20 U/μL RNase inhibitor and 1.8 μL of nuclease-free water in a 10 μL reaction. Then, the mixtures were incubated at 37°C for 10 min to allow transcription. After the incubation, 0.5 μL of 13.3 μM ribosomes was added to the mixture, and the mixture was incubated at 37°C for 20 or 40 min to allow translation. Subsequently, puromycin was added to a final concentration of 1 mg/mL, and the mixture was incubated at 37°C. For the experiments described in Figs. 2 and 3, aliquots were withdrawn from the mixture before and 3 min after the addition of puromycin. For the experiments illustrated in Fig. 4, aliquots were withdrawn from the mixture at the indicated times after the addition of puromycin.
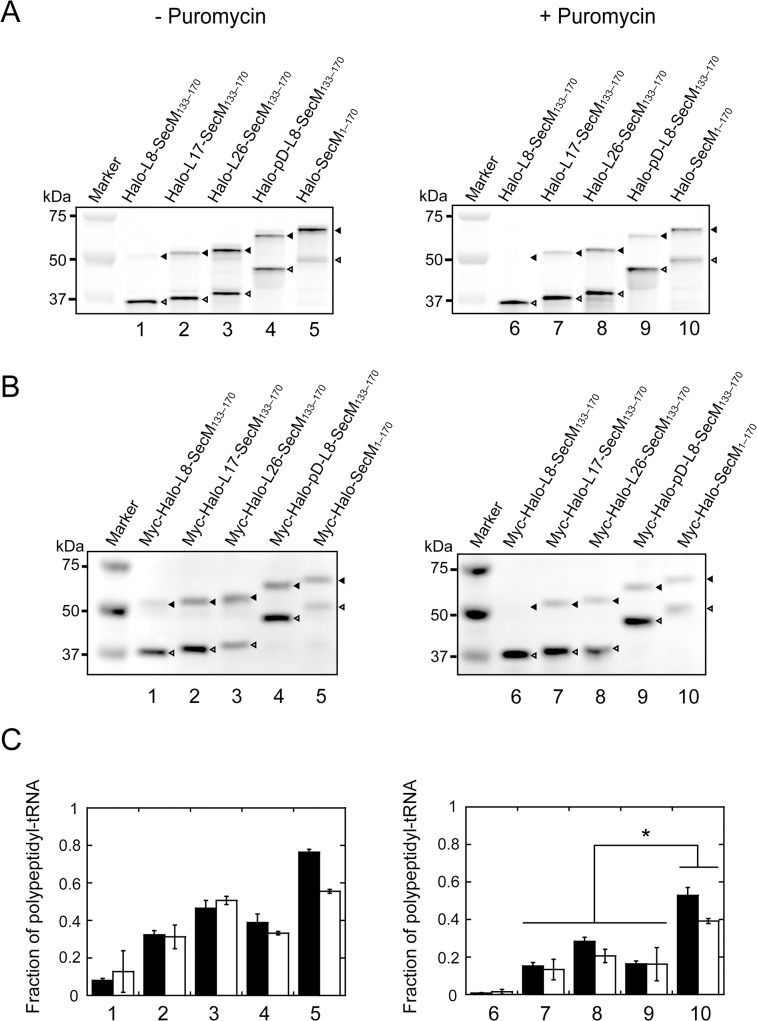
Fig 2. In vitro translation of HaloTag proteins harbouring the arrest sequence.
(A) Halo-L8-SecM133–170 (lane 1), Halo-L17-SecM133–170 (lane 2), Halo-L26-SecM133–170 (lane 3), Halo-pD-L8-SecM133–170 (lane 4) and Halo-SecM1–170 (lane 5) were translated in the presence of HaloTag TMR Ligand using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min, and the reaction mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after 3-min incubation and subjected to NuPAGE. Polypeptides labelled with HaloTag TMR Ligand were detected using Molecular Imager FX. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. The results shown are representative of three independent experiments with similar results. (B) Myc-Halo-L8-SecM133–170 (lane 1), myc-Halo-L17-SecM133–170 (lane 2), myc-Halo-L26-SecM133–170 (lane 3), myc-Halo-pD-L8-SecM133–170 (lane 4) and myc-Halo-SecM1–170 (lane 5) were translated in the absence of HaloTag TMR Ligand using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added at 0 min, and the reaction mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after a 3-min incubation and subjected to NuPAGE. Myc-tagged polypeptides were detected by western blotting with anti-c-myc-tag. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. The results shown are representative of three independent experiments with similar results. (C) Fractions of translation arrest products in the absence (left) and the presence of puromycin (right). Filled bars, fluorescence detection using HaloTag TMR Ligand; open bars, detection by western blotting. Error bars represent the standard deviation (SD) of three independent experiments. The asterisk indicates statistical significance as determined by the Student's t-test (p < 0.05).
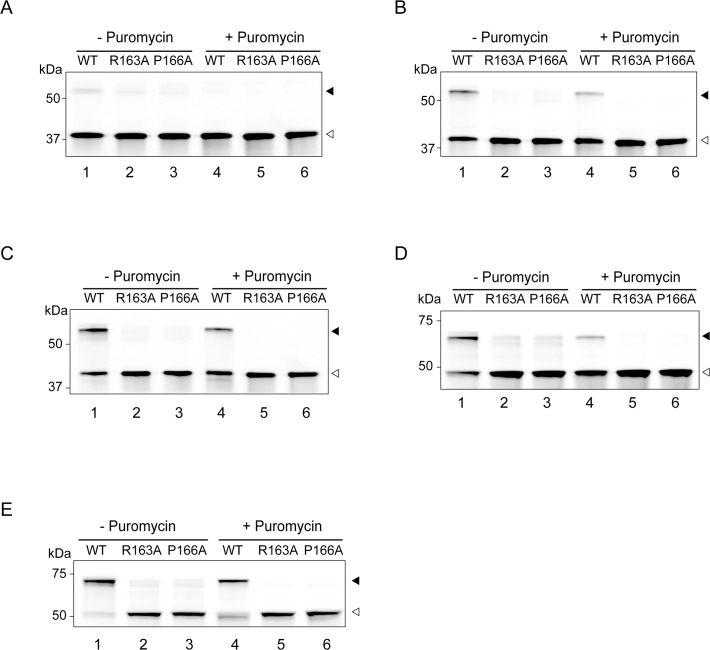
Fig 3. In vitro translation of HaloTag proteins with mutated arrest sequence.
Each protein construct, with or without a mutation (R163A or P166A) in the arrest sequence, was translated in the presence of HaloTag TMR Ligand using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added at 0 min, and the reaction mixture incubated at 37°C for 3 min. Aliquots were withdrawn before and 3 min after the addition of puromycin and subjected to NuPAGE. Polypeptides labelled with HaloTag TMR Ligand were detected using Molecular Imager FX. A, Halo-L8-SecM133–170; B, Halo-L17-SecM133–170; C, Halo-L26-SecM133–170; D, Halo-pD-L8-SecM133–170; E, Halo-SecM1–170. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. The results shown are representative of three independent experiments with similar results.
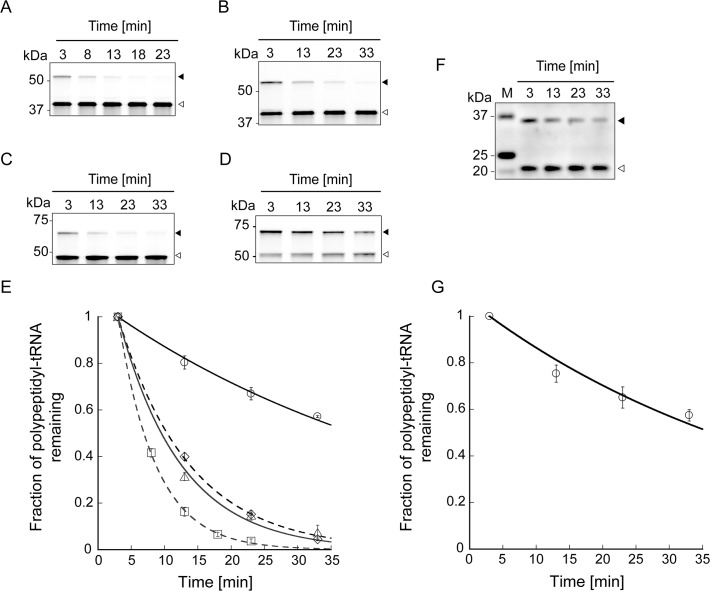
Fig 4. Lifetimes of the translation arrest of HaloTag proteins harbouring the arrest sequence.
(A-D) Time-course analyses of polypeptidyl-tRNA remaining after the addition of puromycin. Halo-L17-SecM133–170 (A), Halo-L26-SecM133–170 (B), Halo-pD-L8-SecM133–170 (C) and Halo-SecM1–170 (D) were translated in the presence of HaloTag TMR Ligand using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min, and the mixture was incubated at 37°C. Aliquots removed at the indicated time points were subjected to NuPAGE. Polypeptides labelled with HaloTag TMR Ligand were detected using Molecular Imager FX. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. (E) Plots of the fraction of polypeptidyl-tRNA remaining in the presence of puromycin as a function of time. Squares, Halo-L17-SecM133–170; diamonds, Halo-L26-SecM133–170; triangles, Halo-pD-L8-SecM133–170; circles, Halo-SecM1–170. Data points represent means ± SD of three independent experiments. The solid and dotted lines show the fit to the data obtained using a single exponential function. The lifetimes of the translation arrest of Halo-L17-SecM133–170, Halo-L26-SecM133–170, Halo-pD-L8-SecM133–170 and Halo-SecM1–170 were 5.6 ± 0.066, 11 ± 0.22, 9.4 ± 0.63 and 51 ± 1.6 min, respectively (the errors represent fitting errors). (F) Time-course analysis of myc-SecM1–170 polypeptidyl-tRNA remaining after the addition of puromycin. Myc-SecM1–170 was translated using the PURExpress ΔRibosome Kit at 37°C for 40 min. Puromycin (1 mg/mL) was added at 0 min, and the mixture was incubated at 37°C. Aliquots were withdrawn at indicated time points and subjected to NuPAGE. Myc-SecM1–170 was detected by western blotting with anti-c-myc-tag. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. (G) The fraction of myc-SecM1–170 polypeptidyl-tRNA remaining in the presence of puromycin as a function of time. Data points with error bars represent means ± SD for three independent experiments. The solid line shows the fit to the data obtained using a single exponential function. The lifetime of the translation arrest of myc-SecM1–170 was 48 min ± 4.3 min (the error corresponds to fitting error).
SDS-PAGE analysis
To detect polypeptidyl-tRNA, translation products were subjected to SDS-PAGE at a neutral pH (NuPAGE) [9]. The Laemmli sample buffer (62.5 mM Tris-HCl, pH6.8; 2% SDS; 10% glycerol and 5% β-mercaptoethanol) was treated with RNAsecure for 10 min at 60°C to inactivate contaminating RNase [26]. The translation products were denatured with the buffer without heat treatment [26] and separated on a NuPAGE 10% Bis-Tris gel in MOPS running buffer at 4°C. Polypeptides labelled with HaloTag TMR Ligand were visualized using Molecular Imager FX (Bio-Rad). Myc-tagged polypeptides were detected by western blotting using anti-c-myc antibody (9E10) and anti-mouse IgG conjugated with HiLyte Fluor 555. Band intensities were measured using ImageJ (http://imagej.nih.gov/ij/). The fraction of the translation arrest product (f) was calculated using the following Equation 1:
| (1) |
, where I a and I r are the band intensities of the translation arrest product and released product, respectively.
Lifetime analysis of translation arrest
The lifetimes of translation arrest of HaloTag proteins harbouring the SecM arrest sequence were determined by fitting a single exponential curve to the plot of the fraction of polypeptidyl-tRNA remaining in the presence of puromycin against incubation time. Data fitting was performed using the KaleidaGraph program (Synergy Software).
Results and Discussion
We found that translation of SecM C-terminal peptide was arrested in a similar manner to that of a native SecM (S1 Fig.), indicating that the arrest sequence is necessary and sufficient for translation arrest, which is consistent with previous studies [8, 27, 28]. However, some of the existing data suggest that the translation of proteins fused to the arrest sequence is not stably arrested [13, 21, 22]. We hypothesized that the nascent chain outside the ribosome affects the efficiency of translation arrest.
DNA constructs for in vitro translation of HaloTag proteins harbouring the E. coli SecM arrest sequence
To examine the effect of the nascent chain outside the ribosome on SecM-mediated translation arrest, we prepared DNA constructs for in vitro transcription and translation of HaloTag harbouring the E. coli SecM arrest sequence (Fig. 1). The constructs contained a T7 promoter and ribosome-binding site upstream of the gene encoding HaloTag protein. The HaloTag sequence was fused to either the C-terminal sequence of SecM (residues 133–170; SecM133–170) or full-length SecM (residues 1–170; SecM1–170), via a spacer sequence. HaloTag is a 34-kDa monomeric protein derived from a bacterial haloalkane dehalogenase, which was engineered to form efficiently a covalent bond with the HaloTag ligand [29]. SecM133–170, which contains the arrest sequence (residues 150–166), is probably within the ribosomal exit tunnel upon translation arrest [11]. The spacer sequence provides sufficient distance between the HaloTag protein and ribosome to allow the correct protein folding [19, 30]. Halo-L8-SecM133–170, Halo-L17-SecM133–170 and Halo-L26-SecM133–170 have spacers comprising 8-, 17- and 26-aa glycine–serine (GS) linker, respectively (Fig. 1A, B and C). Halo-pD-L8-SecM133–170 has a spacer comrpising a 12-aa GS linker, a monomeric version of protein D from the bacteriophage λ (residues 21–110; pD) and an 8-aa GS linker [17, 20, 30] (Fig. 1D). Halo-SecM1–170 has an 8-aa GS spacer (Fig. 1E). The molecular masses of Halo-L8-SecM133–170, Halo-L17-SecM133–170, Halo-L26-SecM133–170, Halo-pD-L8-SecM133–170 and Halo-SecM1–170, which were calculated from the deduced aa sequence, were 38, 39, 40, 49 and 53 kDa, respectively. The constructs with an N-terminal myc-tag were also prepared.
In vitro translation of HaloTag proteins harbouring the E. coli SecM arrest sequence
The DNA constructs were transcribed and translated with or without HaloTag TMR Ligand using a reconstituted in vitro transcription–translation system from E. coli [31]. HaloTag is a protein with improved folding and solubility [32], which rapidly reacts with its ligand (k on = 2 × 107 M−1 s−1) [29]. If nascent HaloTag is correctly folded outside the ribosome exit tunnel, it will be covalently labelled with its fluorescent ligand within 1 s. The translation products with or without translation inhibitor, puromycin (a structural analogue of aminoacyl-tRNA) were subjected to SDS-PAGE at a neutral pH (NuPAGE) to detect the product of translation arrest, polypeptidyl-tRNA [9, 26] (Fig. 2A and 2B). Polypeptides were visualized using HaloTag TMR Ligand or western blotting with anti-c-myc-tag antibody to detect the incorrectly folded products. The fractions of the translation arrest products obtained with or without puromycin are shown in Fig. 2C.
Translation of Halo-L8-SecM133–170 produced one faint and one strong band with apparent molecular masses of approximately 53 and 38 kDa, corresponding to the arrested polypeptidyl-tRNA and the polypeptides released from the ribosome, respectively (Fig. 2A and 2C, lane 1). The arrested polypeptidyl-tRNA is not effectively released from the ribosome after the addition of puromycin because Pro-tRNAPro occupies the A site of the ribosome and blocks the access of puromycin to this site [9–11]. However, the arrested polypeptidyl-tRNA disappeared almost completely within 3 min after the addition of puromycin (Fig. 2A and 2C, lane 6). The same results were obtained by western blotting for Halo-L8-SecM133–170 with an N-terminal myc-tag (Fig. 2B and 2C, lane 1 and 6). These results indicate that HaloTag-L8-SecM133–170 has little arrest potential.
Translation of Halo-L17-SecM133–170, Halo-L26-SecM133–170 and Halo-pD-L8-SecM133–170 produced two bands: the upper band derived from the arrested polypeptidyl-tRNA and the lower, the polypeptide released from the ribosome (Fig. 2A and 2C, lane 2–4). In each construct, approximately half of the upper band survived the treatment with puromycin (Fig. 2A and 2C, lane 2–4 and 7–9). Comparable results were obtained by western blotting using the myc-tagged constructs (Fig. 2B and 2C, lane 2–4 and 7–9). These results suggest that the Halo-L17-SecM133–170, Halo-L26-SecM133–170 and Halo-pD-L8-SecM133–170 have a weak arrest potential and that their translation can be arrested in a manner similar to that of a native SecM.
Translation of Halo-SecM1–170 yielded strong and weak bands corresponding to the arrested polypeptidyl-tRNA and polypeptide released from the ribosome, respectively (Fig. 2A and 2C, lane 5). Most of the arrested polypeptidyl-tRNA was resistant to puromycin (Fig. 2A and 2C, lane 5 and 10), showing that Halo-SecM1–170 is more likely to be translated as the arrested form than other constructs. Similar results were obtained using western blotting (Fig. 2B and 2C, lane 5 and 10).
In vitro translation of HaloTag proteins with mutated arrest sequence
Similar experiments were performed using the constructs with R163A or P166A mutation in the arrest sequence (Fig. 3). Previous studies have shown that R163A and P166A mutations completely abolish SecM-mediated translation arrest [8, 11, 12]. Introduction of R163A and P166A mutations into the constructs eliminated the bands derived from the arrested polypeptidyl-tRNA (Fig. 3). These results indicate that the translation of Halo-L17-SecM133–170, Halo-L26-SecM133–170, Halo-pD-L8-SecM133–170 and Halo-SecM1–170 is arrested in an arrest sequence-dependent manner. These data, in conjunction with the data shown in Figs. 2 and 3, demonstrate that the efficiency of translation arrest can be affected by the nascent chain outside the ribosome in an arrest sequence-dependent manner.
Lifetime of the translation arrest of HaloTag proteins harbouring the arrest sequence
Next, to evaluate the stability of the translation arrest of Halo-L17-SecM133–170, Halo-L26-SecM133–170, Halo-pD-L8-SecM133–170 and Halo-SecM1–170, the lifetimes of the translation arrest were examined (Fig. 4). The translation products were treated with 1 mg/mL puromycin at 37°C to inhibit translation. Aliquots of the mixture, obtained at the indicated time points, were subjected to NuPAGE (Fig. 4A–D). When the translation arrest is released and the peptide elongation resumes, puromycin can enter the A site of the ribosome, producing polypeptidyl-puromycin. Therefore, the arrested polypeptidyl-tRNA is converted to the polypeptide and released from the ribosome as the incubation progresses. The lifetime of each translation arrest was determined by fitting a single exponential curve to the plot of the fraction of polypeptidyl-tRNA remaining in the presence of puromycin against incubation time with puromycin (Fig. 4E). The results were as follows: 5.6 ± 0.066 min for Halo-L17-SecM133–170, 11 ± 0.22 min for Halo-L26-SecM133–170, 9.4 ± 0.63 min for Halo-pD-L8-SecM133–170 and 51 ± 1.6 min for Halo-SecM1–170 (the errors represent fitting errors). The lifetimes of Halo-SecM1–170 and N-terminal myc-tagged SecM (48 ± 4.3 min) were similar (Fig. 4G), indicating that their translation is arrested in the same manner. We concluded that the changes in the nascent chain outside the ribosome affects the stability of translation arrest and the nascent SecM chain outside the ribosome stabilizes the translation arrest.
The effect of the nascent chain outside the ribosome on the translation arrest
We showed that the changes in the nascent chain outside the ribosome affects the stability of translation arrest and the nascent SecM chain helps to stabilize the translation arrest (Figs. 2–4). We can assume that this nascent chain stabilizes the translation arrest by interacting with the ribosome. The majority of the annotated SecM proteins are basic proteins; the E. coli SecM also shares this inherent property (pI = 9.98). Because the ribosome has a highly negatively charged surface, it is likely that the nascent SecM chain associates directly with the ribosome via electrostatic interactions, resulting in a sustained translation arrest. This translation arrest induces the expression of the secA gene, which is downstream of secM [7]. Then, the SecA proteins in association with SecYEG translocon disrupt the interaction between SecM and ribosome in the Sec-mediated export process [7, 8]. The residues 100–109 in SecM might work as ‘release mediator’ to alleviate the arrest [33]. We can conclude that the nascent SecM chain outside the ribosome plays an important role in the regulation of the translation arrest.
We also observed that the efficiency and stability of translation arrest correlate with the length of the spacer sequence between HaloTag protein and SecM C-terminal sequence (Figs. 2 and 4). In barnase-ubiquitin fusion proteins, the conformational strain can be spread over the entire polypeptide when the protein folds [34, 35]. Thus, it is likely that upon translation of Halo-L8-SecM133–170, Halo-L17-SecM133–170, Halo-L26-SecM133–170 or Halo-pD-L8-SecM133–170, the nascent SecM133–170 is pulled by the extension of the GS linker when HaloTag and pD fold cotranslationally. The pulling force might decrease with the increasing linker length [34]. Woolhead et al. [11] have reported that the C-terminal sequence of SecM adopts a compact conformation in the ribosome exit tunnel, which is essential for the translation arrest. The pulling force exerted on the nascent chain would interfere with the conformational transition of the SecM C-terminal sequence. It has been suggested that the SecM arrest sequence is susceptible to a pulling force exerted by the nascent chain outside the ribosome [22, 24]. In the light of these reports and our own findings, SecM appears to be involved in a remarkably sophisticated system regulating its translation.
Supporting Information
(DOCX)
(A) Wild-type and mutant SecM proteins were translated in the presence of [35S]-methionine (0.31 μCi/μL) using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min and the mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after 3-min incubation and separated on a NuPAGE 12% Bis-Tris gel in MES running buffer at 4°C. Polypeptides were detected by autoradiography. (B) The C-terminal peptide of SecM (residues 133–170; SecM133–170), with or without a mutation (R163A or P166A) in the arrest sequence, was translated in the presence of [35S]-methionine (0.31 μCi/μL), using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min and the reaction mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after 3-min incubation and separated on a NuPAGE 12% Bis-Tris gel in MES running buffer at 4°C. Polypeptides were detected by autoradiography. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. Single asterisks indicate bands corresponding to the methionine-charged tRNA and a double asterisk indicates bands corresponding to a translation by-product (probably peptidyl-tRNA).
(TIF)
Restriction enzyme recognition sites are underlined. The bold sequences encode myc-tag. The mutated codons are indicated as boxed nucleotides.
(DOCX)
Acknowledgments
The authors would like to thank Drs Koreaki Ito and Shinobu Chiba from Kyoto Sangyo University and Dr Takashi Kanamori from GeneFrontier Corporation for useful discussion, Dr Shigeo Murata and Kazutaka Sahara from The University of Tokyo for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Grants-in-Aid for Scientific Research on Priority Areas (22020006 to RI), Young Scientists (B) (24710247 to RI), Scientific Research (B) (24370061 to TF) and Exploratory Research (25650046 to TF) from Japan Society for the Promotion of Science (http://www.jsps.go.jp/english/index.html). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484: 538–541. 10.1038/nature10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem. 2013; 82: 171–202. 10.1146/annurev-biochem-080211-105026 [DOI] [PubMed] [Google Scholar]
- 4. Oliver D, Norman J, Sarker S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J Bacteriol. 1998; 180: 5240–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol. 2003; 185: 6719–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McNicholas P, Salavati R, Oliver D. Dual regulation of Escherichia coli secA translation by distinct upstream elements. J Mol Biol. 1997; 265: 128–141. [DOI] [PubMed] [Google Scholar]
- 7. Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001; 7: 185–192. [DOI] [PubMed] [Google Scholar]
- 8. Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002; 108: 629–636. [DOI] [PubMed] [Google Scholar]
- 9. Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006; 22: 545–552. [DOI] [PubMed] [Google Scholar]
- 10. Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem. 2006; 281: 34258–34268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006; 22: 587–598. [DOI] [PubMed] [Google Scholar]
- 12. Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009; 34: 201–211. 10.1016/j.molcel.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans MS, Ugrinov KG, Frese MA, Clark PL. Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nat Methods. 2005; 2: 757–762. [DOI] [PubMed] [Google Scholar]
- 14. Matsuura T, Yanagida H, Ushioda J, Urabe I, Yomo T. Nascent chain, mRNA, and ribosome complexes generated by a pure translation system. Biochem Biophys Res Commun. 2007; 352: 372–377. [DOI] [PubMed] [Google Scholar]
- 15. Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem Biophys Res Commun. 2007; 352: 270–276. [DOI] [PubMed] [Google Scholar]
- 16. Contreras-Martínez LM, DeLisa MP. Intracellular ribosome display via SecM translation arrest as a selection for antibodies with enhanced cytosolic stability. J Mol Biol. 2007; 372: 513–524. [DOI] [PubMed] [Google Scholar]
- 17. Uemura S, Iizuka R, Ueno T, Shimizu Y, Taguchi H, Ueda T, et al. Single-molecule imaging of full protein synthesis by immobilized ribosomes. Nucleic Acids Res. 2008; 36: e70 10.1093/nar/gkn338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osada E, Shimizu Y, Akbar BK, Kanamori T, Ueda T. Epitope mapping using ribosome display in a reconstituted cell-free protein synthesis system. J Biochem. 2009; 145: 693–700. 10.1093/jb/mvp027 [DOI] [PubMed] [Google Scholar]
- 19. Rutkowska A, Beerbaum M, Rajagopalan N, Fiaux J, Schmieder P, Kramer G, et al. Large-scale purification of ribosome-nascent chain complexes for biochemical and structural studies. FEBS Lett. 2009; 583: 2407–2413. 10.1016/j.febslet.2009.06.041 [DOI] [PubMed] [Google Scholar]
- 20. Iizuka R, Funatsu T, Uemura S. Real-time single-molecule observation of green fluorescent protein synthesis by immobilized ribosomes. Methods Mol Biol. 2011; 778: 215–228. 10.1007/978-1-61779-261-8_14 [DOI] [PubMed] [Google Scholar]
- 21. Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011; 9: e1000581 10.1371/journal.pbio.1000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ismail N, Hedman R, Schiller N, von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol. 2012; 19: 1018–1022. 10.1038/nsmb.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jha SS, Komar AA. Using SecM arrest sequence as a tool to isolate ribosome bound polypeptides. J Vis Exp. 2012; 64: e4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cymer F, von Heijne G. Cotranslational folding of membrane proteins probed by arrest-peptide-mediated force measurements. Proc Natl Acad Sci U S A. 2013; 110: 14640–14645. 10.1073/pnas.1306787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iizuka R, Yamagishi-Shirasaki M, Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem. 2011; 414: 173–178. 10.1016/j.ab.2011.03.036 [DOI] [PubMed] [Google Scholar]
- 26. Ito K, Chadani Y, Nakamori K, Chiba S, Akiyama Y, Abo T, et al. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS ONE. 2011; 6: e28413 10.1371/journal.pone.0028413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawrence MG, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol. 2008; 190: 5862–5869. 10.1128/JB.00632-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsai A, Kornberg G, Johansson M, Chen J, Puglisi JD. The dynamics of SecM-induced translational stalling. Cell Rep. 2014; 7: 1521–1533. 10.1016/j.celrep.2014.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008; 3: 373–382. 10.1021/cb800025k [DOI] [PubMed] [Google Scholar]
- 30. Matsuura T, Plückthun A. Selection based on the folding properties of proteins with ribosome display. FEBS Lett. 2003; 539: 24–28. [DOI] [PubMed] [Google Scholar]
- 31. Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001; 19: 751–755. [DOI] [PubMed] [Google Scholar]
- 32. Ohana RF, Encell LP, Zhao K, Simpson D, Slater MR, Urh M, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr Purif. 2009; 68: 110–120. 10.1016/j.pep.2009.05.010 [DOI] [PubMed] [Google Scholar]
- 33. Nakamori K, Chiba S, Ito K. Identification of a SecM segment required for export-coupled release from elongation arrest. FEBS Lett. 2014; 588: 3098–3103. 10.1016/j.febslet.2014.06.038 [DOI] [PubMed] [Google Scholar]
- 34. Cutler TA, Loh SN. Thermodynamic analysis of an antagonistic folding-unfolding equilibrium between two protein domains. J Mol Biol. 2008; 371: 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mills BM, Chong LT. Molecular simulations of mutually exclusive folding in a two-domain protein switch. Biophys J. 2011; 100: 756–764. 10.1016/j.bpj.2010.12.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(A) Wild-type and mutant SecM proteins were translated in the presence of [35S]-methionine (0.31 μCi/μL) using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min and the mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after 3-min incubation and separated on a NuPAGE 12% Bis-Tris gel in MES running buffer at 4°C. Polypeptides were detected by autoradiography. (B) The C-terminal peptide of SecM (residues 133–170; SecM133–170), with or without a mutation (R163A or P166A) in the arrest sequence, was translated in the presence of [35S]-methionine (0.31 μCi/μL), using the PURExpress ΔRibosome Kit at 37°C for 20 min. Puromycin (1 mg/mL) was added to the reaction mixture at 0 min and the reaction mixture was incubated at 37°C for 3 min. Aliquots were withdrawn before the addition of puromycin and after 3-min incubation and separated on a NuPAGE 12% Bis-Tris gel in MES running buffer at 4°C. Polypeptides were detected by autoradiography. Black and white arrowheads indicate the translation arrest products (polypeptidyl-tRNA) and released products, respectively. Single asterisks indicate bands corresponding to the methionine-charged tRNA and a double asterisk indicates bands corresponding to a translation by-product (probably peptidyl-tRNA).
(TIF)
Restriction enzyme recognition sites are underlined. The bold sequences encode myc-tag. The mutated codons are indicated as boxed nucleotides.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.