Abstract
We have determined a 1.18 Å resolution X-ray crystal structure of a novel ruthenocenyle-6-aminopenicillinic acid in complex with CTX-M β-lactamase, showing unprecedented details of interactions between ruthenocene and protein. As the first product complex with an intact catalytic serine, the structure also offers insights into β-lactamase catalysis and inhibitor design.
Penicillin Binding Proteins (PBPs) are responsible for catalyzing the last steps of bacterial cell wall formation and remodeling.1, 2 PBPs are broadly categorized as high molecular mass (HMM) and low molecular mass (LMM), with HMM PBPs being bifunctional enzymes containing both a transglycosylase and transpeptidase domain, while LMM PBPs have a singular domain.1, 2 Clinically, β-lactam antibiotics have successfully inhibited PBPs since penicillin was first introduced in the 1940’s. β-Lactam antibiotics react with PBPs, leading to the formation of an acyl-enzyme adduct that remains bound to the active site and prevents PBPs from completing synthesis of the bacterial cell wall.3–5 However, the past several decades have seen a marked rise in resistance to β-lactam antibiotics, facilitated particularly by bacterial production of β-lactamases.6–11 Unlike PBPs, β-lactamases can catalyze the removal of hydrolyzed β-lactams from their active site by de-acylating the acyl-enzyme intermediate, thereby preventing inhibition of cell wall formation.12–15 In an effort to combat this increasing resistance we synthesized a novel β-lactam compound, 1, composed of 6-aminopenicillinic acid (6-APA) conjugated with a ruthenocenyl moiety (Figure 1). Details on the synthesis of compound 1 are provided in the SI. Ruthenocene is an organometallic compound belonging to the so-called metallocene complexes.16 This class of molecules consists of a metal center sandwiched between two cyclopentadienyl rings, and has chemical properties similar to aromatic compounds.16 Ferrocene is an archetypical metallocene and its medicinal applications have been intensively studied mainly in relation to anti-cancer activity.17–23 Conversely, the biological properties of ruthenocene derivatives have been investigated to a much lesser extent.21 With the increasing problem of bacterial resistance and the ensuing race to find new inhibitors, the antibacterial properties of these organometallic groups are starting to be investigated more intensely.20
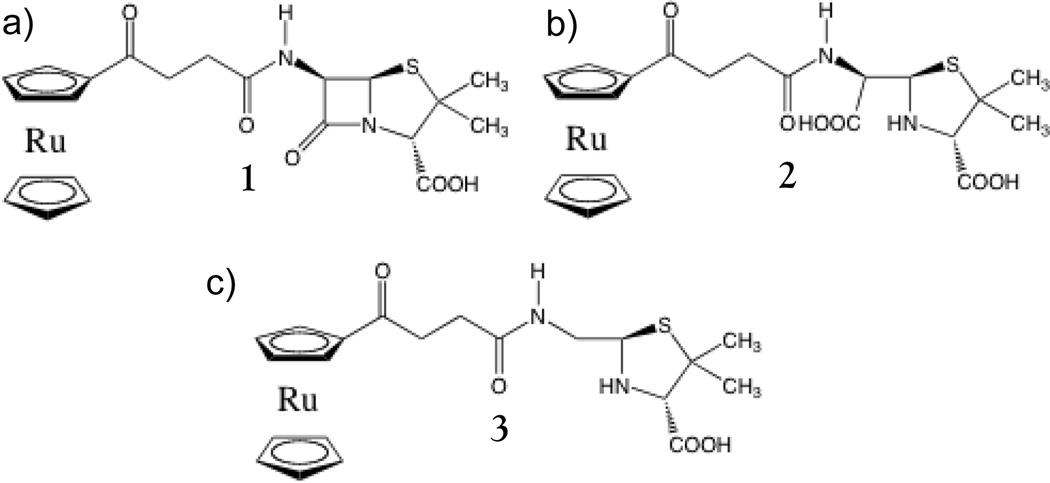
Figure 1.
Structures of 1 and hydrolyzed products. a) Intact 1. b) Hydrolyzed 1, penicilloic acid form. c) Decarboxylated and hydrolyzed 1, penilloic acid form. 2 and 3 are termed as carboxylated and decarboxylated products respectively in the text.
The antibacterial properties of 1 were tested against Gram-positive methicillin-sensitive Staphylococcus aureus (MSSA), methicillin-resistant S. aureus (MRSA), S. epidermidis, Enterococcus faecalis, and twelve clinically isolated Staphylococcus strains (Table S1). This screening revealed a noticeable antibacterial activity of compound 1 against the majority of bacterial strains tested. The highest activity was observed against S. aureus ATCC® 29213™ with an MIC value of 2.0 µg ml−1, and S. epidermidis ATCC® 12228™ with an MIC value of 4.0 µg ml−1. For comparison, the MIC values of ampicillin against these two bacterial strains are 0.5 and 5.0 µg ml−1 respectively. While 1 was unable to reach the level of inhibition seen with ampicillin against the MRSA strain, the addition of the ruthenocenyl moiety to 6-APA showed much higher levels of inhibition than 6-APA alone (Table S1). The addition of the ruthenocene moiety also showed greater inhibition than the 6-APA ferrocene conjugate that we previously studied,24 indicating that the larger ruthenium ion may confer more potent antibacterial properties than the smaller iron ion. Compound 1 was tested in vitro for hemolytic activity, and no activity was observed on human erythrocytes, which is one of the conditions for safe antibiotic use. To date, 1 represents one of the most active ruthenocene conjugated antibiotics.25
To understand the molecular interactions between organometallic groups and protein residues, we next set out to obtain a crystal structure of 1 in complex with CTX-M-14 E166A β-lactamase. We chose CTX-M as its active site shares many key catalytic features with that of the PBPs,6, 26, 27 and it yields high-quality crystals, capable of diffracting to atomic resolution (<1.2 Å).28 The E166A mutation further increases the similarity of the CTX-M active site to that of PBPs by hampering deacylation in the active site, as Glu166 serves as the general base for the deacylation reaction. This can possibly trap the hydrolyzed product in its acyl-enzyme form, as was demonstrated by previous studies.29 Several crystal structures were determined, with the highest resolution data set measuring at 1.18 Å (Table S2). Upon examination, the density for a compound was clearly visible in the active site (Figure 2a) for both monomers of CTX-M in each crystallographic asymmetric unit. We expected the E166A mutation would trap 1 in the acyl-enzyme state with a covalent bond between the compound and the catalytic Ser70. However, we did not observe this form in the active site, possibly because the slightly basic pH (7.9) in our buffer sped up the deacylation process, whereas an acidic pH (4.5) was used in previous studies.29 Instead, we observed an interesting hydrolyzed product (3, Figure 1), where the carboxylate group formed from the opening of the β-lactam ring was not present. While this decarboxylated β-lactam product has been characterized in studies focusing on the degradation of penicillin,30–33 to the best of our knowledge, there is no information about its interaction with β-lactamase. Previous crystallographic analysis using inactive CTX-M S70G or AmpC S64G mutants have only shown that the original product (similar to 2, Figure 1) with the newly generated carboxylate group would be expelled from the active site due to steric clashes and electrostatic repulsion.34, 35
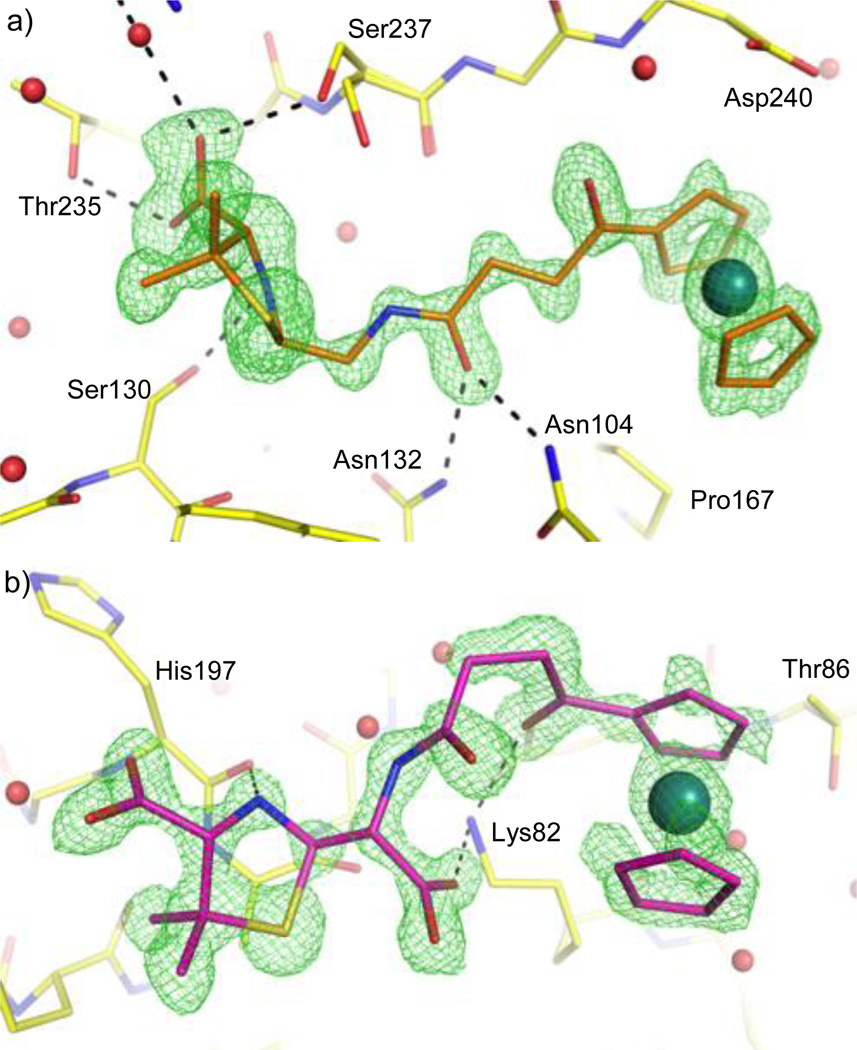
Figure 2.
Hydrolyzed products captured in and outside CTX-M-14 E166A active site. Unbiased Fo-Fc maps shown in green at 3 σ. a) 3 bound in the active site. b) 2 bound outside the active site.
Upon examining the binding of 3 to CTX-M we can see that it forms many favorable interactions with active site residues and binds deeply in the pocket. The 6-APA portion of 3 forms hydrogen bonds with Asn104, Ser130, Asn132, Thr235, and Ser237, as well as a water mediated interaction with Arg276. The bulky ruthenocenyl moiety further stabilizes the compound in the active site by stacking against the protein backbone, including the peptide bonds of Gly238-Asp240, and Asn170-Thr171. In addition, the ruthenocenyl moiety sits atop Pro167, forming favorable non-polar interactions. The fact that we only observe the decarboxylated product in the active site, as well as the seemingly favorable interactions it has with the active site, raises an interesting question as to whether the decarboxylated product can serve as a β-lactamase inhibitor. Further analysis will need to be conducted, but the decarboxylated product may provide a starting scaffold for inhibitor discovery against β-lactamase.
Close examination of the CTX-M crystal structure also revealed four other product molecules bound to the protein outside the active site and at the crystal-packing interface. Two of these product molecules still had the carboxylate group that was missing from the compounds in the active site (Figure 2b). It should also be noted that one of these two molecules (not shown) has a different chirality at the C5 atom on the five-membered ring compared with 2 and 3, suggesting the original compound stock used in our studies may contain some enantiomeric impurity. Nevertheless, our structure demonstrates that both the carboxylated (2) and decarboxylated (3) products are present in solution, and that the decarboxylated product binds preferentially to the active site. This is consistent with the hypothesis that steric clashes and electrostatic repulsion, caused by the newly generated carboxylate group, expel the original hydrolyzed product (2) from the active site during β-lactamase hydrolysis.34, 35 In the other two compounds outside the active site only the ruthenocenyl group was observed in the electron density and the rest of the compound was disordered. The first of the “free” ruthenocenyl groups is in close proximity to one of the two carboxylated products (Figure 3) and is well defined in the electron density. The second “free” ruthenocenyl group is only weakly visible in the electron density and was omitted from further discussion.
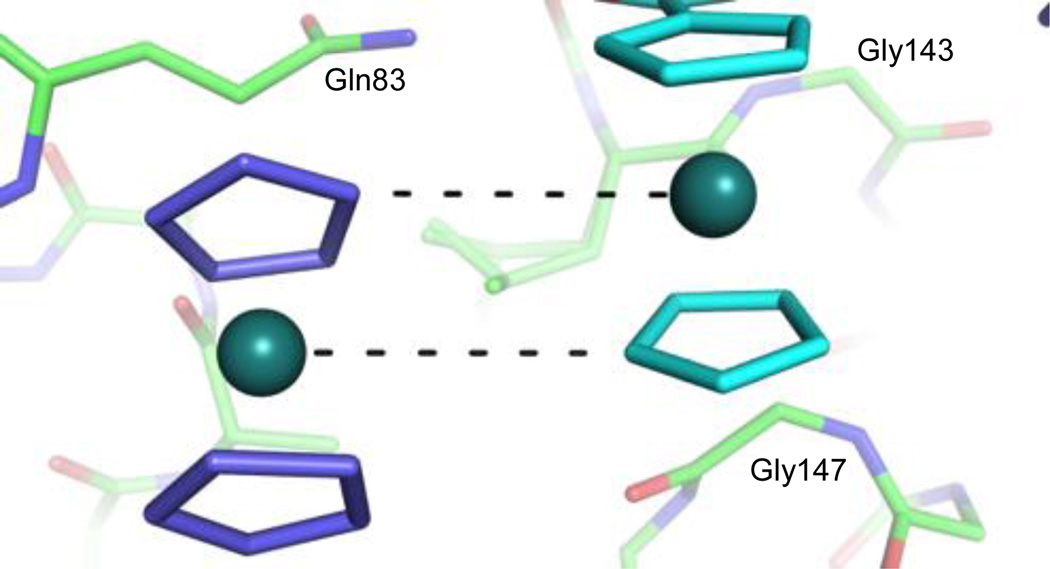
Figure 3.
Ru---H interactions between ruthenocenyl groups outside the active site. Ru---C contacts (at a distance of 4.3–4.4 Å) are shown as dashed black lines, along which the hydrogens from the cyclopentadienyl carbon atoms are projected. For the “free” ruthenocene (left), the rest of the compound is disordered.
The spatial orientation of the ruthenocenyl groups outside the active site offer valuable insights into the molecular interactions involving these organometallic groups. It has been noted in the literature that when ruthenocenyl groups are crystalized Ru---H contacts occur between the Ru of one ruthenocenyl group and the H of an adjacent ruthenocenyl group, with the ruthenocenyl groups orienting parallel and with a half molecule shift to one another.36 This parallel with a half molecule shift orientation is evident between two adjacent ruthenocenyl groups, one from a carboxylated product molecule and the other from the first “free” ruthenocenyl group (Figure 3). However, the cyclopentadienyl rings are not parallel with one another between the two ruthenocenyl groups as observed in the small molecule crystal. Based upon their orientation, it appears that the “free” ruthenocenyl group is forming two Ru---H contacts with the adjacent carboxylated inhibitor molecule. The corresponding Ru---C distances are 4.3 and 4.4 Å, and when accounting for the C---H distance of 1.083 Å, we find the Ru---H distances to be 3.2–3.3 Å, which are in close agreement with the 3.2 Å distance found in the literature.36
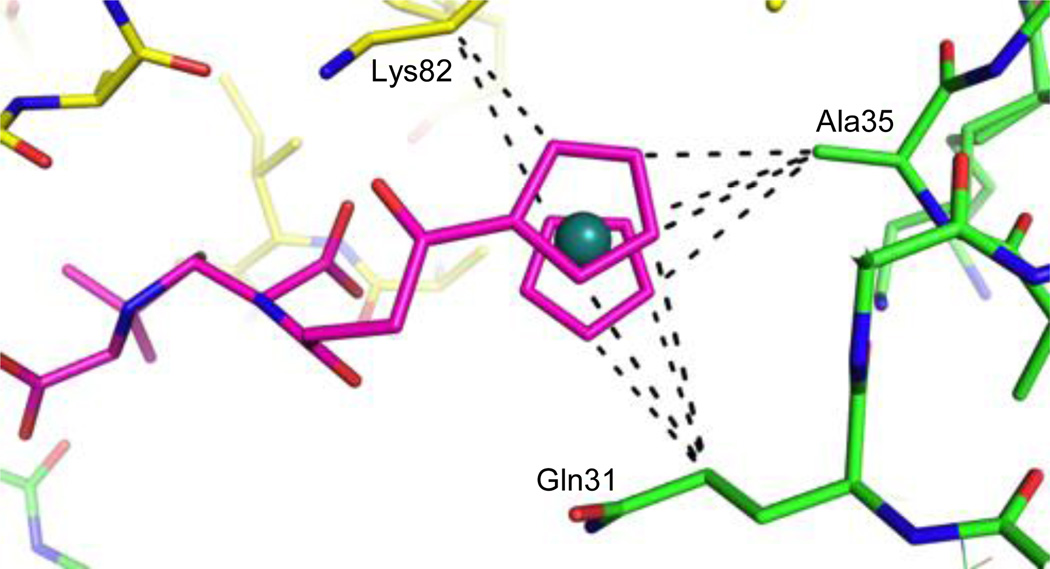
When examining why these molecules bind away from the active site, it appears that the ruthenocenyl group is engaged in many different types of interactions with the protein, including stacking interactions between the cyclopentadienyl ring and Arg191. Most interestingly, the carbon atoms of the cyclopentadienyl rings of one of the inhibitors are involved in several non-polar interactions with the protein molecules. Specifically, the product molecule is sandwiched between two protein monomers in the crystal lattice, and its carbon atoms are within optimal VDW distance of Gln31 and Ala35 of one monomer, and Lys82 of another (Figure 4). The distances between Ru and the hydrogen atoms on these protein side chain carbons are longer than 3.6–3.7 Å, which is farther than those observed between ruthenocenyl groups (Figure 3). These observations, together with the non-polar features of the side chain hydrocarbon groups, suggest that Ru---H hydrogen bonds are unlikely in this case (Figure 4).
Figure 4.
Interactions of the ruthenocenyl group with amino acid side chains. Away from the active site, the ruthenocenyl group is sandwiched between two protein monomers and is held in place by several non-polar interactions. The inhibitor is colored magenta, while the different protein monomers are colored yellow and green. Potential C---C interactions (at a distance of 3.8–4.2 Å) are shown as dashed black lines.
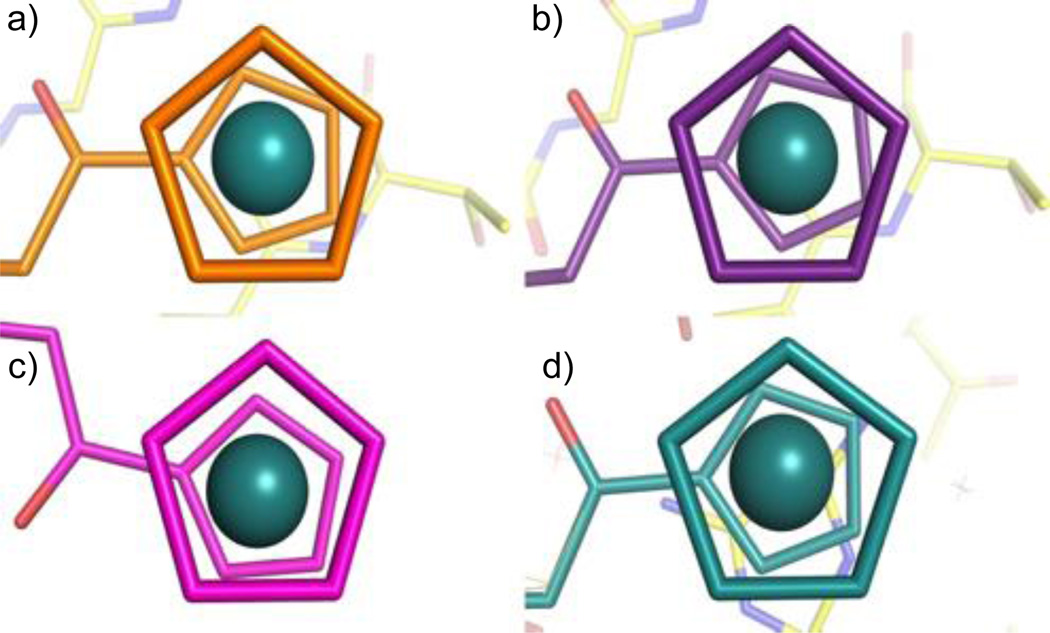
The high resolution of our crystal structure not only gives us insight as to what products are present in the active site, but also allows us to view the cyclopentadienyl rings of the ruthenocenyl group with unprecedented detail. We were able to observe a significant twisting of the two cyclopentadienyl rings in four of the compounds in the crystal structure (Figure 5), including the “free” ruthenocenyl group in the partially disordered compound (Figure 3). The rings in both of the active site product molecules adopt a slightly staggered conformation; with the bottom “floating” ring being rotated either 17.1 degrees, or 22.6 degrees in relation to the top ring. This slightly staggered conformation is also observed in one of the carboxylated product molecules that bind away from the active site. Interestingly, all of the compounds that adopt the staggered conformation show a roughly identical degree of rotation. The other carboxylated product observed in the structure has its rings in an almost eclipsed conformation, with only a very slight 3.8 degree twist of the rings evident (Figure 5c). As shown in Figures 2b and 4, the ruthenocenyl group of this inhibitor and its adjacent linker form extensive interactions with the protein, including hydrogen bonds with Lys82 and non-polar interactions with multiple residues. The two cyclopentadienyl rings are likely forced to adopt the eclipsed conformation by these interactions, suggesting that the ruthenocenyl group can adapt to the protein environment and offer more flexibility in protein-inhibitor interactions in comparison to the ferrocenyl group.
Figure 5.
Twists of cyclopentadienyl rings in each ordered inhibitor molecule. a) Active site A, 17.1 degree twist, b) Active site B, 22.6 degree twist, c) Non-active site A, 3.8 degree twist, d) Non-active site B, 21.7 degree twist.
Conclusions
In summary, we have solved the 1.18 Å structure of a novel ruthenocene conjugated β-lactam in complex with CTX-M-14 E166A β-lactamase. To the best of our knowledge, this structure represents the first atomic resolution structure of a synthetic organometallic compound in complex with a protein. The observation of the original product (2) outside the active site supports the previous hypothesis that, during β-lactamase catalysis, the hydrolyzed product (2) is expelled from the active site due to steric clash.34, 35 Binding of the decarboxylated product (3) suggests possible product inhibition of β-lactamase, and the potential use of this product as a novel scaffold for new drug discovery against β-lactamase. The high resolution of our structure also allowed us to view the twisting of the cyclopentadienyl rings in unprecedented detail, while suggesting the utility of the ruthenocenyl group in targeting different protein hot spots and enhancing the antibacterial potency of existing drugs.
Supplementary Material
Acknowledgments
This work was supported by the NIH (AI 103158, to Y.C.) and by the National Science Centre in Cracow, Poland (Grant No. DEC-2013/11/B/ST5/00997, to K.K.). We thank Dr. Jed Fisher for insightful discussions, Dr. Derek Nichols and Kathryn Lethbridge for reading the manuscript. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, supported by the U.S. Department of Energy (Contract No. W-31-109-Eng-38).
Footnotes
Electronic Supplementary Information (ESI) available: [Compound synthesis, MIC data, and crystallographic information]. See DOI: 10.1039/c000000x/
The coordinates have been deposited in the Protein Data Bank with access code 4XXR.
Contributor Information
Konrad Kowalski, Email: kondor15@wp.pl.
Yu Chen, Email: ychen1@health.usf.edu.
Notes and References
- 1.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. FEMS microbiology reviews. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 2.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. FEMS microbiology reviews. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 3.Llarrull LI, Testero SA, Fisher JF, Mobashery S. Current opinion in microbiology. 2010;13:551–557. doi: 10.1016/j.mib.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Z, Kahne D, Kishony R. Molecular cell. 2012;48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Testero SA, Fisher JF, Mobashery S, Abraham DJ. Burger's Medicinal Chemistry and Drug Discovery. John Wiley & Sons, Inc.; 2003. [Google Scholar]
- 6.Smith JD, Kumarasiri M, Zhang W, Hesek D, Lee M, Toth M, Vakulenko S, Fisher JF, Mobashery S, Chen Y. Antimicrobial agents and chemotherapy. 2013;57:3137–3146. doi: 10.1128/AAC.00505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Therrien C, Levesque RC. FEMS microbiology reviews. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 8.Drawz SM, Bonomo RA. Clinical microbiology reviews. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaidis I, Favini-Stabile S, Dessen A. Protein science. 2014;23:243–259. doi: 10.1002/pro.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taubes G. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 11.Bush K, Fisher JF. Annual review of microbiology. 2011;65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 12.Fisher JF, Mobashery S. Current protein & peptide science. 2009;10:401–407. doi: 10.2174/138920309789351967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frere JM. Molecular microbiology. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 14.Bush K, Jacoby GA, Medeiros AA. Antimicrobial agents and chemotherapy. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adediran SA, Deraniyagala SA, Xu Y, Pratt RF. Biochemistry. 1996;35:3604–3613. doi: 10.1021/bi952107i. [DOI] [PubMed] [Google Scholar]
- 16.Long NJ. Metallocenes-An Introduction to Sandwich Complexes. Oxford: Blackwell Science; 1998. [Google Scholar]
- 17.Gasser G, Ott I, Metzler-Nolte N. Journal of medicinal chemistry. 2011;54:3–25. doi: 10.1021/jm100020w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamels D, Dansette PM, Hillard EA, Top S, Vessières A, Herson P, Jaouen G, Mansuy D. Angewandte Chemie International Edition. 2009;48:9124–9126. doi: 10.1002/anie.200903768. [DOI] [PubMed] [Google Scholar]
- 19.Ornelas C. New Journal of Chemistry. 2011;35:1973–1985. [Google Scholar]
- 20.Patra M, Gasser G, Metzler-Nolte N. Dalton transactions. 2012;41:6350–6358. doi: 10.1039/c2dt12460b. [DOI] [PubMed] [Google Scholar]
- 21.Salmon AJ, Williams ML, Hofmann A, Poulsen SA. Chemical communications. 2012;48:2328–2330. doi: 10.1039/c2cc15625c. [DOI] [PubMed] [Google Scholar]
- 22.van Staveren DR, Metzler-Nolte N. Chemical reviews. 2004;104:5931–5985. doi: 10.1021/cr0101510. [DOI] [PubMed] [Google Scholar]
- 23.Reiter C, Capci Karagoz A, Frohlich T, Klein V, Zeino M, Viertel K, Held J, Mordmuller B, Emirdag Ozturk S, Anil H, Efferth T, Tsogoeva SB. European journal of medicinal chemistry. 2014;75:403–412. doi: 10.1016/j.ejmech.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 24.Skiba J, Rajnisz A, de Oliveira KN, Ott I, Solecka J, Kowalski K. European journal of medicinal chemistry. 2012;57:234–239. doi: 10.1016/j.ejmech.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Wenzel M, Patra M, Senges CHR, Ott I, Stepanek JJ, Pinto A, Prochnow P, Vuong C, Langklotz S, Metzler-Nolte N, Bandow JE. ACS Chemical Biology. 2013;8:1442–1450. doi: 10.1021/cb4000844. [DOI] [PubMed] [Google Scholar]
- 26.Massova I, Mobashery S. Antimicrobial agents and chemotherapy. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamski CJ, Cardenas AM, Brown NG, Horton LB, Sankaran B, Prasad BV, Gilbert HF, Palzkill T. Biochemistry. 2014 doi: 10.1021/bi501195g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Bonnet R, Shoichet BK. Journal of the American Chemical Society. 2007;129:5378–5380. doi: 10.1021/ja0712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimamura T, Ibuka A, Fushinobu S, Wakagi T, Ishiguro M, Ishii Y, Matsuzawa H. Journal of biological chemistry. 2002;277:46601–46608. doi: 10.1074/jbc.M207884200. [DOI] [PubMed] [Google Scholar]
- 30.Humber DC, Bamford MJ, Bethell RC, Cammack N, Cobley K, Evans DN, Gray NM, Hann MM, Orr DC, Saunders J, et al. Journal of medicinal chemistry. 1993;36:3120–3128. doi: 10.1021/jm00073a011. [DOI] [PubMed] [Google Scholar]
- 31.Nagele E, Moritz R. Journal of the American Society for Mass Spectrometry. 2005;16:1670–1676. doi: 10.1016/j.jasms.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande A, Baheti K, Chatterjee N. Current science. 2004;87:1684–1695. [Google Scholar]
- 33.Bentley R. Journal of Chemical Education. 2004;81:1462. [Google Scholar]
- 34.Delmas J, Leyssene D, Dubois D, Birck C, Vazeille E, Robin F, Bonnet R. Journal of molecular biology. 2010;400:108–120. doi: 10.1016/j.jmb.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 35.Beadle BM, Trehan I, Focia PJ, Shoichet BK. Structure. 2002;10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 36.Borissova AO, Antipin MY, Perekalin DS, Lyssenko KA. CrystEngComm. 2008;10:827–832. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.