Abstract
HIV genetic diversity is a major obstacle for vaccine development. To define whether potential T-cell epitope (PTE) peptide usage improves the detection of T cell responses in a highly diverse HIV-1 epidemic, we compared the magnitude, breadth and depth of group M PTE peptide responses to consensus M peptides in Gag and Nef proteins. Gag PTE responses were detected at a higher magnitude, more Nef PTE responses were detected at a cohort (but not individual) level, and depth was detected in both Gag and Nef responses.
The genetic diversity of HIV represents a major challenge for the development of a universal HIV vaccine. One approach that has been developed to deal with this variability is ‘mosaic’ vaccine immunogens, representing sets of bioinformatically-designed sequences that attempt to maximize coverage of HIV diversity [1-3]. By including multiple variants for a particular epitopic region, PTE peptides are designed to increase immunological recognition of HIV. Studies in non-human primates have shown that mosaic immunogens improved the breadth and depth of cellular immune responses to globally circulating HIV-1 strains compared to consensus and natural antigens [4,5], as well as eliciting antibody responses that protected from heterologous SHIV challenge [6].
Testing the recognition of variant peptide sets containing ‘potential T cell epitopes’ (PTE peptides) based on mosaic vaccines in HIV-infected individuals represents a proxy (albeit with significant limitations) for the potential cross-reactivity of mosaic immunogens to viruses circulating in a given population [7]. To date, group M PTE peptides have not been evaluated for recognition in a highly diverse HIV-1 epidemic, such as west central Africa, which is home to virtually all HIV-1 subtypes and many circulating and unique recombinant forms (CRF and URF) [8,9]. We previously characterized the high diversity of circulating strains in HIV-1-infected blood donors from Cameroon and evaluated cellular HIV responses [8,10]. Here, we assessed the recognition of PTE peptides based on mosaic group M sequences compared to consensus (CON) group M sequences in this group. Both sets of peptides were obtained from the NIH AIDS Reagent Program (https://www.aidsreagent.org), and CON M peptides were based on the 2001 HIV-1 group M consensus sequences from the Los Alamos National HIV sequence database, which included pure clades and recombinant forms.
We measured IFN-γ responses by ELISPOT to two 15-mer peptide panels: HIV-1 group M CON peptides spanning Gag (129 peptides) and Nef proteins (53); and group M PTE peptides for Gag (320) or Nef (127), including 0-6 variants (median 1) per epitopic region. To identify positive responses, PTE peptides were screened in pools arranged using the software “Deconvolute This” (Mario Roederer, Vaccine Research Center, NIH), in which each individual Gag and Nef peptide appeared 3-4 times; the CON set was arranged in a matrix of 29 pools, with each peptide appearing twice, as described in [10]. Subsequently, all potentially reactive peptides were tested individually in a confirmatory ELISpot assay. PTE peptides with one or more amino acid changes but with an overlap of at least 10 amino acids in that region were considered ‘variants’ of the same epitopic region. Twenty-four HIV-infected blood donors from Cameroon with known reactivity to CON M Gag or Nef were studied, 10 of whom had both Gag and Nef responses, 7 Gag-only and 7 were Nef-only responders. Participants had a median plasma viral load of 5.1 log RNA copies/ml (range 4-6.3) and median CD4 count of 474 cells/mm3 (range 42-1972). All individuals were antiretroviral therapy naïve at the time of analysis, and 75% were male. We have previously characterized viral diversity and HIV-specific T cell responses in this group [8,10]. Fourteen of the 17 individuals (82%) tested for Gag responses were CRF02_AG in Gag, while 9/17 (53%) of the individuals were CRF02_AG in Nef, with the remainder being infected with a wide range of pure clades, CRFs or URFs, namely D, F, CRF01_AE, CRF01_AE/F, CRF11_cpx, CRF36_cpx, CRF37_cpx, and one unclassified sequence that was an outlier of both the CRF02_AG and G clades.
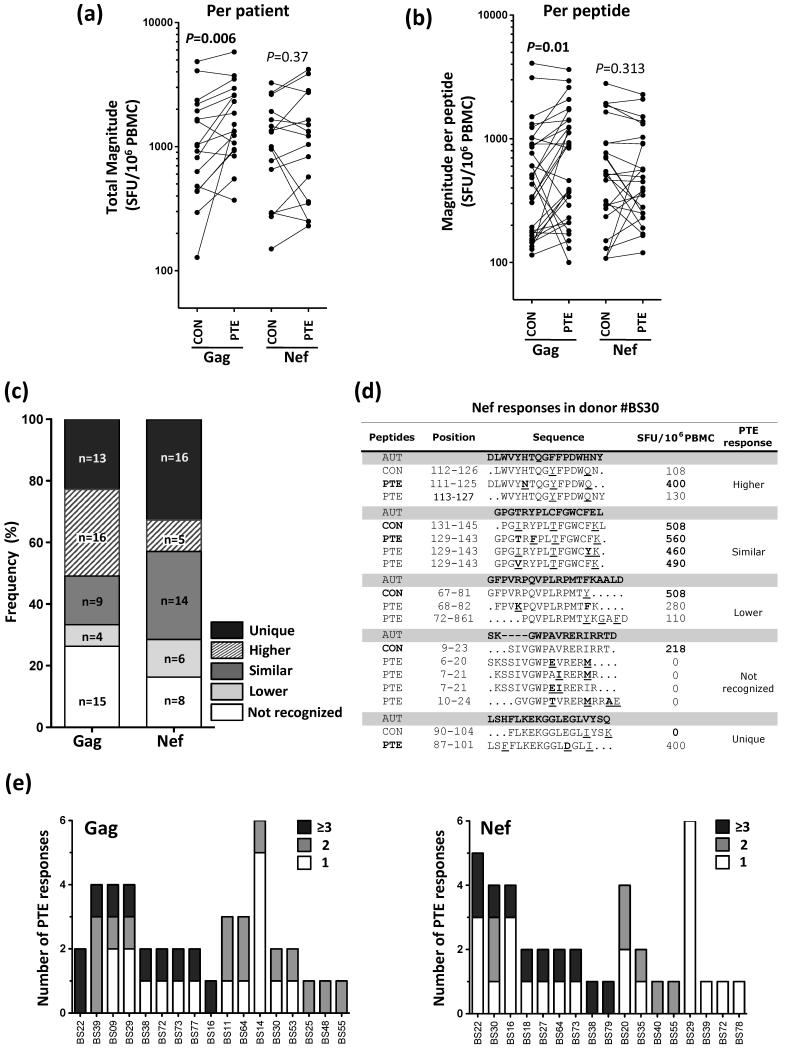
We first compared the overall magnitude of Gag or Nef-specific responses using CON and PTE peptides, within each study individual. Where a response was detected to multiple corresponding PTE variants, the highest magnitude was selected for analysis. For Gag, the magnitude of response using PTE peptides was significantly greater than that detected with CON peptides (median 1510 and 1013 SFU/106 PBMC, respectively; p=0.006) (Fig 1a). No differences were observed for Nef responses between PTE and CON panels. Since an improved response could be attributed to an increased breadth within a participant and/or an increased magnitude of response at the peptide level, we compared the magnitude for each PTE and its corresponding CON peptide. Fig 1b shows that Gag PTE peptides detect a response of greater magnitude compared to the corresponding CON peptides (median 875 and 485 SFU/106 PBMC, respectively; p=0.01). For Nef, no differences were observed (Fig 1b). Moreover, for both Gag and Nef, the breadth of responses to PTE peptides was similar to that observed for CON peptides, ranging from 1-6 peptides (median of 2; data not shown).
Fig. 1. Comparison of IFN-γ ELISPOT responses to Consensus (CON) and Potential T cell Epitope (PTE) peptides in HIV-infected individuals.
(a) Total magnitude of Gag- and Nef-specific T cell responses in 17 HIV-infected individuals using CON M or PTE M peptides. The total magnitude of response was determined by the sum of the responses to all epitopic regions. PTE peptides with an overlap of at least 10 amino acids were considered the same epitopic region. When multiple variants of a PTE peptide were recognized, the highest magnitude response was enumerated in the total magnitude. (b) Magnitude of individual Gag- and Nef-specific T cell responses detected using CON M or PTE M peptides. Only peptides that had a corresponding reactive peptide in both peptide sets are shown, which was 30 peptides for Gag and 25 for Nef. (c) Profile of Gag and Nef PTE responses compared to CON peptide responses. PTE responses were divided in five groups, namely ‘Unique’, where a PTE peptide elicited a detectable response while the corresponding CON peptide did not; ‘Higher’, where the PTE response magnitude was >30% higher than the corresponding CON peptide; ‘Similar’, where the magnitude of a PTE response was comparable to the magnitude of the corresponding CON; ‘Lower’, where the magnitude of a PTE response was >30% lower than the corresponding CON peptide; and ‘Not recognized’, where the PTE peptide did not elicit a detectable response while the corresponding CON peptide did. (d) Nef-specific responses in one donor (BS30), indicating the full spectrum of response profiles indicated in (c). Autologous viral sequences (generated by population sequencing) are indicated. Mismatches between the viral sequence and the peptide sequences are underlined, and differences in PTE peptides compared to CON are shown in bold. Amino acid position is based on HXB2. (e) Depth of Gag (left panel) and Nef (right panel) PTE responses in 17 HIV-infected individuals. The number of responses detected to PTE peptides with one variant only are shown in white, two variants in gray and three or more variants in black. Statistical comparisons were performed using a Wilcoxon matched pairs test and P values are indicated. CON, Consensus M peptide; PTE, Potential T cell Epitope peptide; AUT, autologous viral sequence.
To define in more detail the differences between reactivity to PTE and CON peptides, the PTE responses were divided into five profiles: i) ‘Unique’, where a PTE peptide elicited a detectable response while the corresponding CON peptide did not, ii) ‘Higher’, where the PTE response magnitude was at >30% higher than the corresponding CON peptide, iii) ‘Similar’, where the magnitude of a PTE response was comparable to the magnitude of the corresponding CON, iv) ‘Lower’, where the magnitude of a PTE response was >30% lower than the corresponding CON peptide, and v) ‘Not recognized’, where the PTE peptide did not elicit a detectable response while the corresponding CON peptide did (Fig 1c). For Gag, the proportion of PTE peptides eliciting ‘Unique’ responses (13/57, 22.8%) was similar to the proportion of PTE peptides that were not recognized (15/57, 26.3%), confirming that PTE peptide usage did not increase the breadth of Gag responses detected. Among the Gag responses detected with both PTE and CON peptide sets (29/57, 50.8%), 55% were of higher magnitude using PTE peptides (at a median of 2.3-fold greater, ranging from 1.4 to 9.6), and 13.8% were of lower magnitude using PTE peptides (at a median of 0.3-fold lower, ranging from 0.2 to 0.4). For Nef, two-thirds of the PTE responses detected were ‘Unique’ (16/49, 32.6%) or ‘Similar’ (14/49, 28.6%), with twice the number of ‘Unique’ PTE responses as those not recognized compared to their CON counterparts (16 vs 8; Fig 1c). Of note, these different profiles of PTE responses could all be observed within a single individual simultaneously, as shown in Fig 1d. Overall, these data demonstrate that the use of Gag M PTE peptides led to detection of a greater magnitude of response compared to CON M peptides, but not a greater breadth of response. For Nef, no benefit was observed in either the magnitude or breadth of the responses when PTE peptides were used to detect responses in this cohort.
Lastly, we evaluated the depth of the response elicited by PTE peptides, defined as the number of PTE variants recognized per epitopic region. For Gag, all individuals exhibited depth for at least one PTE peptide response, and in 53% (9/17) of the participants, three or more variants were recognized (Fig 1e). Similarly for Nef, 76% of the individuals (13/17) exhibited depth, with 53% recognizing three or more PTE variants.
Overall, our data testing the detection of PTE peptides in a highly diverse HIV-1 epidemic demonstrate that these peptides offer a modest advantage over consensus peptides, including greater response magnitudes detected to Gag, the detection of more unique Nef responses at the cohort level, and the ability to cross-detect multiple variants in both Gag and Nef. The enhanced response magnitudes detected to Gag PTE peptides compared to CON peptides, which was not observed for Nef, could be explained by the greater overall sequence conservation of Gag compared to Nef. In contrast, the greater number of unique Nef epitopes detected using PTE peptides compared with CON peptides, which was not observed for Gag, may be attributed to the more divergent viral sequences infecting the donors tested for Nef responses, compared to the lower diversity within the Gag donors, the majority of whom were infected with CRF02_AG. In contrast to our results, two similar studies assessing Nef reactivity in HIV-1 clade B-infected individuals and clade B PTE or variant B peptides found a greater advantage for detecting Nef responses [7,11]. This may be attributed to the greater differences between matching a single clade to a population infected with that clade, compared to our cohort that were infected with a diverse range of group M viruses, including some that may be under-represented in sequence databases. A limitation of our study worth noting was that the majority of individuals tested for Gag responses were infected with CRF02_AG, which is not representative of the extensive diversity in Cameroon, where we have recently described CRF02_AG accounting for only 50% of the circulating viruses [8]. We can speculate that if we had used consensus peptides for CRF02_AG in our present study group, we would have detected a higher magnitude of responses than CON M, based on extrapolation of our previous data comparing clade C consensus and group M CON peptides in a clade C-infected population from South Africa [10,12]. Clade-matching the study population and the peptides may thus balance out the advantage offered by the PTE M set with respect to detecting higher magnitude responses. On the other hand, a PTE set based on CRF02_AG Gag, containing the major variants of CRF02_AG, would be expected to increase both the magnitude and breadth of responses detected in this study group, compared to CRF02_AG CON Gag or CON M Gag, based on the findings of Malhotra and colleagues [7]. Ultimately, the utility of vaccine designs encompassing the diversity of HIV, such as the mosaic approach, compared to those focusing the immune response to conserved regions of HIV [13], will only be elucidated in clinical efficacy trials of vaccine candidates.
Acknowledgements
We thank Mrs Kathryn Norman for administrative assistance. HIV-1 Group M Consensus and PTE Gag and Nef peptide sets were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIH. This study was approved by the National Ethics Committee of the Cameroonian Ministry of Health and the University of Cape Town Faculty of Health Sciences Research Ethics Committee.
Source of funding: This research was supported by the International Atomic Energy Agency (Technical Co-operation Project RAF/6/029), the Poliomyelitis Research Foundation (PRF) of South Africa and the University of Cape Town, for collaborative projects with partners in the Global South. M.T. was supported by a Carnegie Corporation Fellowship and received a training fellowship from The World Academy of Sciences (TWAS). W.A.B. is a Wellcome Trust Intermediate Fellow in Public Health and Tropical Medicine.
Footnotes
Data from this study were presented previously at the Keystone Symposia, Banff, Alberta, Canada, March 8-14, 2014; Poster Number: 3051.
Conflict of interest
There are no conflicts of interest.
References
- 1.Nickle DC, Rolland M, Jensen MA, Pond SLK, Deng W, Seligman M, et al. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2006;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 3.Thurmond J, Yoon H, Kuiken C, Yusim K, Perkins S, Theiler J, et al. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics. 2008;24:1639–40. doi: 10.1093/bioinformatics/btn251. [DOI] [PubMed] [Google Scholar]
- 4.Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med. 2010;16:319–23. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santra S, Liao H-X, Zhang R, Muldoon M, Watson S, Fischer W, et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med. 2010;16:324–8. doi: 10.1038/nm.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al. Protective Efficacy of a Global HIV-1 Mosaic Vaccine against Heterologous SHIV Challenges in Rhesus Monkeys. Cell. 2013;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra U, Li F, Nolin J, Allison M, Zhao H, Mullins JI, et al. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J Virol. 2007;81:5225–37. doi: 10.1128/JVI.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tongo M, Martin DP, Zembe L, Mpoudi-Ngole E, Williamson C, Burgers WA. Characterization of HIV-1 gag and nef in Cameroon: further evidence of extreme diversity at the origin of the HIV-1 group M epidemic. Virol J. 2013;10:29. doi: 10.1186/1743-422X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr JK, Wolfe ND, Torimiro JN, Tamoufe U, Mpoudi Ngole E, Eyzaguirre L, et al. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: Evolutionary relics? Retrovirology. 2010;7:1–8. doi: 10.1186/1742-4690-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tongo M, Zembe L, Ebong E, Roux S, Bekker L-G, Williamson C, et al. Striking lack of T cell immunodominance in both a multiclade and monoclade HIV-1 epidemic: Implications for vaccine development. Vaccine. 2014;32:2328–36. doi: 10.1016/j.vaccine.2014.02.063. [DOI] [PubMed] [Google Scholar]
- 11.Rolland M, Frahm N, Nickle DC, Jojic N, Deng W, Allen TM, et al. Increased breadth and depth of cytotoxic T lymphocytes responses against HIV-1-B Nef by inclusion of epitope variant sequences. PLoS One. 2011;6:e17969. doi: 10.1371/journal.pone.0017969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zembe L, Burgers WA, Jaspan HB, Bekker LG, Bredell H, Stevens G, et al. Intra- and inter-clade cross-reactivity by HIV-1 Gag specific T-cells reveals exclusive and commonly targeted regions: implications for current vaccine trials. PLoS One. 2011;6:e26096. doi: 10.1371/journal.pone.0026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson KE, Barouch DH. A global approach to HIV-1 vaccine development. Immunol Rev. 2013;254:295–304. doi: 10.1111/imr.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]