Abstract
Type 1 diabetes (T1D) is a polygenic disease with multiple insulin dependent diabetes loci (Idd) predisposing humans and non-obese diabetic (NOD) mice to disease. NOD.B10 Idd9 congenic mice, in which the NOD Idd9 chromosomal region is replaced by the Idd9 from T1D-resistant C57BL/10 mice, are significantly protected from T1D development. However, the genes and pathways conferring T1D development or protection by Idd9 remain to be fully elucidated. We have developed novel NOD.B10-Idd9 (line 905) congenic mice that predominantly harbor islet-reactive CD4+ T cells expressing the BDC2.5 T cell receptor (BDC-Idd9.905 mice). To establish functional links between the Idd9 genotype and its phenotype, we used microarray analyses to investigate the gene expression profiles of ex vivo and antigen-activated CD4+ T cells from these mice and BDC2.5 (BDC) NOD controls. Among the differentially expressed genes, those located within the Idd9 region were greatly enriched in islet-specific CD4+ T cells. Bioinformatics analyses of differentially expressed genes between BDC-Idd9.905 and BDC CD4+ T cells identified Eno1, Rbbp4 and Mtor, all of which are encoded by Idd9 and part of gene networks involved in cellular growth and development. As predicted, proliferation and Th1/Th17 responses of islet-specific CD4+ T cells from BDC-Idd9.905 mice following antigen stimulation in vitro were reduced compared to BDC mice. Furthermore, proliferative responses to endogenous autoantigen and diabetogenic function were impaired in BDC-Idd9.905 CD4+ T cells. These findings suggest that differential expression of the identified Idd9 genes contributed to Idd9-dependent T1D susceptibility by controlling the diabetogenic function of islet-specific CD4+ T cells.
1. Introduction
Type 1 diabetes (T1D) is a T cell-mediated organ-specific autoimmune disease, resulting from the destruction of the insulin-producing pancreatic beta cells (1). In humans and non-obese diabetic mice (NOD), a well-established mouse model for T1D (2), multiple genetic regions, insulin dependent diabetes (Idd) loci, confer risk of T1D development (3). Various laboratories have used mice congenic for a number of such insulin dependent diabetes (Idd) loci derived either from T1D-susceptible or T1D-resistant strains to study the mechanisms by which these loci regulate T1D development and to identify causative genes (4-16).
Congenic fine-mapping approaches can be combined with DNA microarray expression analysis in models of complex trait diseases to simultaneously investigate gene expression within, as well as outside of a congenic region of interest. This approach successfully identified Cd36 as an insulin resistance gene (17). Two T1D studies profiled longitudinal gene expression in naive spleen cells from NOD mice and NOD.Idd congenic mice (18, 19). The findings of these studies were less informative than expected, suggesting that activated specific lymphocyte populations are better subjects for investigation. Accordingly, CD3-stimulated CD4+ T cells were profiled in NOD.Idd3/5 congenic mice, which identified two new T1D candidate genes (11).
Fine mapping of the Idd9 region identified four subregions that independently confer partial protection from T1D: Idd9.1, Idd9.2, Idd9.3 and Idd9.4 (20). The Idd9.1 subregion partially overlaps Idd11, a T1D-protective interval defined in NOD.B6 congenic mouse strains (13). Consistent with the notion that Idd9 encodes a number of immunologically relevant genes, NOD mice congenic for Idd9 from the T1D-resistant B10 or NOR strains display various immune-related phenotypical differences (4, 6, 7, 10, 12, 14, 21, 22).
NOD.B10 Idd9 congenic mice have the NOD-derived Idd9 region of chromosome 4 replaced with the Idd9 from T1D-resistant C57BL/10 mice, resulting in significant T1D protection (4). Differentially expressed genes within the Idd9 region may contribute to these differences. Alternatively, but not exclusively, altered expression of Idd9 genes could lead to perturbations in the expression of genes shared by both strains, but located outside of this congenic region. To identify genes and molecular pathways that potentially control the diabetogenic potential of islet-specific CD4+ T cells, we conducted microarray expression analysis of ex vivo and antigen-stimulated CD4+ T cells from newly generated BDC2.5 TCR transgenic NOD mice that contain the C57BL/10SnJ derived Idd9 region (line 905) (hereafter referred to as BDC-Idd9.905 mice) (23) and BDC2.5 TCR transgenic NOD mice (24) (hereafter referred to as BDC mice).
In this study, we report that genes involved in cellular growth and development showed significantly reduced expression in islet-specific CD4+ T cells from BDC-Idd9.905 mice compared to BDC control mice. Among these genes Eno1, Rbbp4, and Mtor were identified as novel Idd9 candidate genes. Consistent with these results, functional analyses of CD4+ T cells from BDC-Idd9.905 mice showed diminished proliferative and Th1 and Th17 cytokine responses following antigen-specific stimulation in vitro compared to BDC control mice. In addition, BDC-Idd9.905 CD4+ T cells were impaired in their proliferation to endogenous autoantigen and in their diabetogenic function. Our findings confirm the validity of using microarray gene expression analysis in CD4+ T cells from BDC-Idd9.905 congenic mice to identify Idd9 candidate genes and molecular mechanisms that control islet-specific CD4+ T cell functions.
2. Material and Methods
Mice
NOD.B10 Idd9 (NOD.B10-Idd9R905) mice were originally obtained from Taconic and the congenic interval present in the strain has been described previously (15). BDC2.5 TCR transgenic NOD mice (24) were originally obtained D. Mathis and C. Benoist (Joslin Diabetes Center, Boston, MA). Both strains were maintained in our facility. NOD mice were purchased from Taconic. Mating male BDC2.5 TCR transgenic NOD mice with female NOD.B10 Idd9 mice generated BDC2.5 TCR transgenic NOD mice containing the B10 Idd9R905 interval. BDC2.5 TCR transgenic F1 litters were identified by flow cytometric analysis of PBLs antibody-stained for CD4 and TCRVβ4 as described previously (7) and then crossed with NOD.B10 Idd9 mice. Transgenic F2 litters were screened for the homozygous presence of the B10 Idd9 interval by PCR using microsatellite markers to differentiate between the NOD and B10 genomic segments between markers D4Mit258 and D4Mit42 as described previously (7). Mice that were 6-9 weeks old and free of diabetes as determined by urine glucose measurement were used for experiments. All mice were housed at the Pennsylvania State College of Medicine specific pathogen-free (SPF) facility in accordance with Pennsylvania State Institutional Animal Care and Use Committee guidelines.
Microarray and quantitative PCR analysis
Three independent samples of single cell suspensions from two spleens pooled from BDC or BDC-Idd9.905 mice were separated into two batches each. The first batch of spleen cells (ex vivo) was stained with anti-CD4 (RM4-5) and anti-TCRVβ4 (KT4) antibodies (BD Biosciences, San Jose, CA) for 20 min at 4°C, washed and subsequently sorted for CD4+TCRVβ4+ (transgenic) T cells on a BD FACSAria III. The second batch of spleen cells (1×106 cells/ml) was stimulated with BDC2.5 mimotope p79 (1 μg/ml) in culture medium for 48 hours at 37°C. Stimulated spleen cells were then stained and sorted for CD4+TCRVβ4+ T cells as described for the first batch. Total RNA was extracted from sorted ex vivo or p79-stimulated BDC and BDC-Idd9.905 CD4+TCRVβ4+ cells using the RNeasy kit (Qiagen). RNA quality and concentration was assessed using an Agilent 2100 Bioanalyzer with RNA Nano LabChip (Agilent, Santa Clara, CA).
Microarray analysis was performed using the Illumina MouseWG-6 v2.0 R3 Expression BeadChip Kit (Illumina, San Diego, CA) at the Pennsylvania State University College of Medicine Genome Sciences Facility. Thus, cRNA was synthesized by TotalPrep Amplification (Ambion, Austin, TX) from 500 ng of RNA according to manufacturer’s instructions. In vitro transcription (IVT) was employed to generate multiple copies of biotinylated cRNA. The labeled cRNA was purified using filtration, quantified by NanoDrop, and volume-adjusted to 750 ng/sample. Samples were fragmented, and denatured before they were hybridized to MouseWG-6 v2.0 R3 Expression BeadChips for 18 hours at 58°C. Following hybridization, the chips were washed and fluorescently labeled. Beadchips were scanned with a BeadArray Reader and resultant scan data were extracted with GenomeStudio 1.0 (Illumina, San Diego, CA) (Illumina). Analysis of expression data was performed using GeneSpring Gx11 software (Agilent Technologies, Santa Clara, CA). Expression for a transcript in a sample was considered Present/Marginal if the detection p-value was <0.15. Transcripts were then further filtered for signal level >100 in at least 50% of the values in one of the six samples. If a transcript/probe did not meet these cutoffs it was excluded from further analysis. Genelists were obtained through volcano plots between non-averaged group comparison using fold-change of 1.4 or greater and asymptotic unpaired t-test p-value computation of p<0.05 (25). The microarray data presented in this study have been submitted to the Gene Expression Omnibus at the National Center for Biotechnology Information under the accession number GSE64674 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64674).
For real-time PCR validation of microarray expression data, two to three independent cDNAs from total RNA of splenic BDC and BDC-Idd9.905 CD4+ T cells were synthesized using the QuantiTect Reverse transcription Kit according to the manufacturer’s protocol (Qiagen). Quantitative PCR was performed in three replicates with the Opticon 2 DNA Engine machine (Biorad) following manufacturer’s instructions. In brief, 12.5 μl FastStart SYBR Green master mix (Roche), 0.2 μM oligonucleotide primers, 12.4 μl of cDNA (5ng) and dH2O in a total volume of 25 μl. PCR primers were the following:
18S rRNA sense 5′-AAACGGCTACCACATCCAAG; 18S rRNA antisense 5′-CCTCCAATGGATCCTCGTTA
Eno1 sense 5′- CACCCTCTTTCCTTGCTTTG; Eno1 antisense 5′-CTTTTGCGGTGTACAGATCG
Agtrap sense 5′- GTCTACCACATGCACCGTGAA; Agtrap antisense 5′-GAGGGTCCGAAGAAATCGGG
S100pbp sense 5′- GCCTAAAAGCAATGCCTCATTTC; S100pbp antisense 5′-CAACAAGGAGTCATCCAACTCAT
Rbbp4 sense 5′- CAGCAGTAGTGGAGGACGTG; Rbbp4 antisense 5′-AGTGGCTTGGCTTGGAAGTA;
2610305D13Rik sense 5′- GGAATGTCTCAATTTTGCTCAGAG; 2610305D13Rik antisense 5′- GTCTTGTTCCAAGACCTTCTCCT
Casp9 sense 5′- TGGACCGTGACAAACTTGAG; Casp9 antisense 5′-ATCTCCATCAAAGCCGTGAC;
Rcan3 sense 5′- TTGAGGTTTCCTGTGCTGTG; Rcan3 antisense 5′-CAAAAGCAAACTGGCTCTCC
Clic4 sense 5′- TCAAGGCCGGAAGTGATGG; Clic4 antisense 5′-GGTTGTGACACTGAACACGAC
Txlna sense 5′- TTCAGCTGCAGATGGAACAG; Txlna antisense 5′-TTGTCGATATGCTCCTCACG;
Rnf19b sense 5′- GCTCAACCCACACGACATC; Rnf19b antisense 5′-GCAATAACAGCATAACCGCAGT;
Syf2 sense 5′- CGGAATGAAGCTCGTAAGCTG Syf2 antisense 5′-TCCAGACGCGCTTTCTTGG;
Mtor sense 5′- ACCGGCACACATTTGAAGAAG; Mtor antisense 5′-CTCGTTGAGGATCAGCAAGG;
Gm13212 sense 5′- TCTTGGATCAAGACAGCCAGT; Gm13212 antisense 5′-GGGATTCAAGAATCACCTTGCT
Fmr1 sense 5′- CAATGGCGCTTTCTACAAGGC; Fmr1 antisense 5′-TCTGGTTGCCAGTTGTTTTCA;
Necap2 sense 5′- ATGGAGGAGAGTGAGTACGAGT; Necap2 antisense 5′-CATTCTGAGGCCCTGTAACCA.
PCR specificity was analyzed using a melting curve analysis. The mean Ct was calculated from triplicate samples. Mean fold change in mRNA expression, normalized to 18S rRNA was calculated using the comparative cycle threshold (CT) method (2−ΔΔCT method) (26). Expression differences were analyzed for statistical significance by Student’s t test, following confirmation that they were distributed normally by Shapiro-Wilk normality test.
Bioinformatics analysis of microarray data
Lists of normalized genes that were significantly differentially expressed (FC>1.4, p<0.05) in microarray analysis were subjected to functional annotation cluster analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (27). This program groups genes according to their known biological functions (GO) to determine pathways and processes of major biological significance. In addition, lists of normalized genes were subjected to Ingenuity Pathway Analysis (IPA) 8.6 (Ingenuity Systems, Redwood City, CA) using T cell-specific filters to uncover significant gene networks.
Frequency of activated CD4+ T cells and Treg cells
The frequency of activated CD4+ T cells was determined by staining spleen cells from BDC and BDC-Idd9.905 with anti-CD4 (RM4-5) mAb and anti-CD25 (7D4), -CD62 ligand (MEL-14), -CD69 (H1.2F3) or -CD44 (IM7.8.1) mAb (BD Bioscience), followed by flow cytometric analyses as described (7). Treg cell frequency was determined by surface staining of spleen cells with anti-CD4 (RM4-5) mAb and intracellular staining with anti-Foxp3 (MF23) mAb using reagents from BD Bioscience. Stained cells were acquired on a FACS Calibur or LSR II flow cytometer (Becton Dickinson) and analyzed using FlowJo software (Tree Star).
T cell proliferation
CD4+ T cells were purified using CD4 (L3T4) microbeads to >95% purity according to the manufacturer’s instructions (Miltenyi Biotec GmbH). Purified CD4+ T cells (5 × 104 cells/well) in presence of irradiated (3,200 rad) NOD splenocytes were stimulated with different concentrations of BDC2.5 mimotope p79 (AVRPLWVRME) (28) in complete DMEM supplemented with 10% (v/v) FCS, 100 units/ml penicillin, 100 units/ml streptomycin, 50 μM 2-mercaptoethanol, 10 mM HEPES, 1mM sodium pyruvate, and 1× MEM non-essential amino acids in triplicates of 96-well plates at 37°C for 72 h. During the last 12 h of culture, [3H]-thymidine (0.5 μCi/well) was added to each well and radioactive incorporation was subsequently measured using a MicroBeta Liquid Scintillation counter (PerkinElmer, Santa Clara, CA).
Treg cell suppression assay
CD4+ T cells were purified from spleens and lymph nodes of BDC and BDC-Idd9.905 mice by depleting B220+, CD8+ and CD11b+ cells using magnetic beads (Miltenyi Biotec GmbH). The purified CD4+ T cells were subsequently stained with biotinylated anti-CD25 mAb (clone 7D4, BD Bioscience), followed by column purification using streptavidin-conjugated microbeads (Miltenyi Biotec GmbH). The CD4+CD25− fraction was used as effector T cells. The CD4+CD25+ fraction was further purified by positive selection using an MS column (Miltenyi Biotec GmbH) and utilized as Treg cells. CD4+CD25− T cells (2.5 × 104 cells/well) were cultured in triplicate with serial dilutions of CD4+CD25+ T cells in the presence of irradiated (3,200 rads) spleen cells (5 × 104 cells/well) from NOD.scid mice and BDC2.5 mimotope p79 (0.1 μg/ml) in 96-well plates. T cell proliferation was determined as described above. Suppression of T cell proliferation was calculated using the following formula: ((T cell proliferation in the absence of Tregs - T cell proliferation in the presence of Tregs) / T cell proliferation in the absence of Tregs) × 100.
ELISA
The concentration of cytokines was determined in culture supernatants of BDC and BDC-Idd9.905 spleen cells stimulated with p79. Cytokine production in 72 h culture supernatants was assayed by quantitative capture ELISA according to the manufacturer’s guidelines (BD Biosciences, San Diego, CA; R&D Systems, Minneapolis, MN (IL-17)).
Adoptive T cell transfer and T1D analysis
CD4+ T cells were purified from spleens of nondiabetic 6-9 week old BDC or BDC-Idd9.905 female mice to >95% purity by using CD4 magnetic beads (Miltenyi Biotec GmbH), following red blood cell lysis. For in vivo analysis of CD4+ T cell proliferation, CFSE-labeled CD4+ T cells (5 × 106 cells/mouse) were injected into non-diabetic NOD mice and analyzed 90 h after transfer by flow cytometry as described previously (15). For T1D analysis, 2.5 × 106 purified CD4+ T cells from each donor strain were injected (i.v.) into 6-8 week old NOD.scid female mice. Urine glucose concentration of mice was determined using Glucostix (Bayer) at least twice a week. Animals were considered diabetic when glucose concentration was >250 mg/dl at two consecutive readings. Diabetic mice also exhibited polyuria and weight loss.
Statistical Analysis
Data were analyzed as indicated by two-tailed, unpaired Student’s t-test or Mann-Whitney test. Enrichment of Idd9 genes was calculated using the hypergeometric distribution test (phyper function) in R (version 2.13.2). The Kaplan-Meier analysis with the log-rank test was used to calculate statistical difference of T1D incidence. Values of p<0.05 were considered statistically significant.
3. Results
Microarray analysis of islet-specific CD4+ T cell from BDC and BDC-Idd9.905 mice
To identify differentially expressed genes within the Idd9 T1D susceptibility locus of islet-reactive CD4+ T cells, we utilized NOD mice congenic for the C57BL/10SnJ derived Idd9 locus and transgenic for the islet-specific BDC2.5 T cell receptor. We generated these novel mice by mating BDC2.5 TCR transgenic NOD mice (24) with NOD.B10 Idd9 (line 905) mice (23) and subsequent intercrossings to obtain BDC2.5 TCR transgenic litters that were homozygous for the B10 Idd9 region, hereafter referred to as BDC-Idd9.905 mice (see Material and Methods). In contrast to previous studies by others (11, 14, 18, 19), CD4+ T cells from BDC and BDC-Idd9.905 mice allowed us to investigate differential expression of Idd9-encoded genes in CD4+ T cells that were predominantly islet-reactive.
Cytofluorometric analyses showed that spleens of both lines contained comparable frequencies (BDC: 21.2% ± 1.5% vs. BDC-Idd9.905: 21.4% ± 1.5%; n = 10, each) and absolute numbers of transgenic (CD4+TCRVβ4+) T cells (BDC: 1.1 × 107 ± 1.1 × 106 cells vs. BDC-Idd9.905: 1.1 × 107 ± 9.2 × 105 cells; n = 10, each). In addition, the vast majority of transgenic CD4+ T cells displayed a naive phenotype and the frequency of activated transgenic CD4+ T cells between the two strains were not significantly different as determined by surface expression of established T cell activation markers such as CD25, CD44, CD69 and CD62L (Supplementary Fig. 1). Taken together, these data indicated that the Idd9 locus did not mediate detectable effects on the selection and activation of islet-specific CD4+ T cells in the transgenic mice.
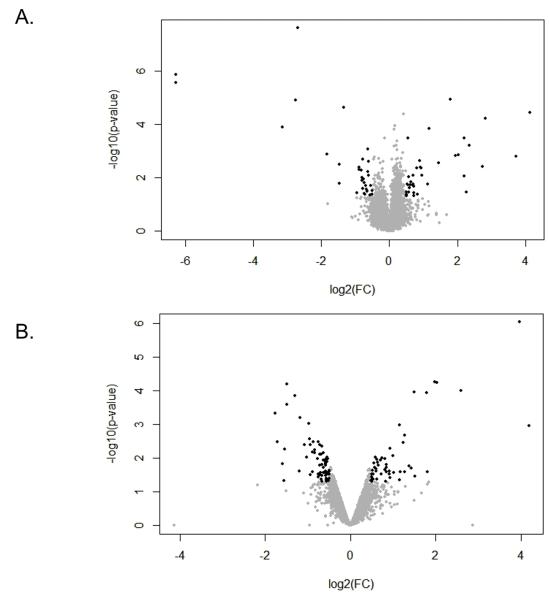
Spleen cells from BDC and BDC-Idd9.905 mice were stimulated with BDC2.5 mimotope p79 (28) for 48h or processed immediately without stimulation in three independent experiments. To reduce the chances of within strain variation, we used splenocytes from a pool of two spleens for each experiment. Ex vivo or p79-stimulated spleen cells were sorted for BDC2.5 transgenic (CD4+TCRVβ4+) cells by flow cytometry. Total RNA from these samples was then extracted and subjected to genome-wide gene expression analysis using Illumina MouseWG-6 v2.0 Expression BeadChips with three replicate microarray hybridizations performed per strain and per condition (12 hybridizations total). We scored differentially expressed genes in the microarray sets as those genes with a fold change (FC) of >1.4 and a value of p <0.05. Based on these criteria, we found 55 genes and 80 genes in ex vivo and p79-stimulated CD4+ T cells, respectively, that were expressed differentially between the two strains (Fig. 1).
Figure 1. Volcano plot of genes differentially expressed in CD4+ T cells between BDC-Idd9.905 and BDC mice.
Volcano plot showing the relative abundance of transcripts in ex vivo (A) and p79-stimulated CD4+ T cells from BDC-Idd9.905 mice compared to BDC mice. Log2 of fold change (FC) is presented on the x-axis and −log10 of p values is represented on the y-axis. Transcripts that passed the cutoff of p<0.05 and FC>1.4 (log2=0.5) were considered to be differentially expressed and are shown in black. Genes that are significantly up-regulated in BDC-Idd9.905 CD4+ T cells compared to BDC CD4+ T cells are on the right, while down-regulated genes are on the left of FC=0. Grey dots indicate genes, which did not show differential expression between the two strains.
Comparable numbers of genes were expressed at higher or lower levels (27 genes and 28 genes, respectively) in ex vivo CD4+ T cells from BDC-Idd9.905 mice (Fig. 1A). In contrast, in p79-stimulated CD4+ T cells there were more genes expressed at lower levels in BDC-Idd9.905 mice (50 genes vs. 30 genes at higher levels, Fig.1B). Of the 55 genes differentially expressed in ex vivo CD4+ T cells, 29 genes (53%) are located on chromosome 4, and 6 genes (11% ea.) are located in Idd9 or Idd11, which partially overlaps with the Idd9.1 subregion (20). The enrichment of Idd9 genes in ex vivo samples was highly significant (p = 5.3 × 10−8), as determined by hypergeometric distribution analysis. The remaining genes are distributed among all but four of the other chromosomes at an average density of 0.7 genes per chromosome (Fig. 2A). Of the 80 genes that were differentially expressed in p79-stimulated CD4+ T cells, 35 genes (44%) are located on chromosome 4, including 9 genes (11%) in Idd9 and 7 genes (9%) in Idd11 (Fig. 2B). The enrichment of Idd9 genes in p79-stimulated samples was statistically significant (p = 1.7 × 10−11). The remaining genes are distributed among all but three of the other chromosomes at an average density of 2.4 genes per chromosome, slightly more than 3-fold higher than the density of differentially expressed genes in ex vivo CD4+ T cells. Taken together, these findings demonstrate that in both ex vivo and p79-stimulated CD4+ T cells from the two strains, the Idd9/11 region and adjacent regions on chromosome 4 were highly enriched for genes that were expressed differentially.
Figure 2. Chromosomal location of differentially expressed genes in CD4+ T cells between BDC-Idd9.905 and BDC mice.
Location of the genes that were differentially expressed in ex vivo (A) and p79-stimulated (B) CD4+ T cells between BDC-Idd9.905 and BDC mice were visualized on chromosomes using chrView tool of the biological database network (bioDBnet) software. Each gene is shown as a horizontal bar on the specific chromosome.
Thirteen genes that were differentially expressed in ex vivo CD4+ T cells were also expressed differentially in p79-stimulated CD4+ T cells. Two of those genes (Agtrap, S100pbp) are located within Idd9 and 10 genes are located on chromosome 4, but outside of Idd9. Thus, the majority of differentially expressed genes in ex vivo CD4+ T cells were not identical to those in p79-stimulated CD4+ T cells, indicating that differential expression of these genes was unique to either condition. These data suggest that differential expression of the shared genes was independent of p79 stimulation.
Cluster and network analysis of differentially expressed genes in islet-specific CD4+ T cells from BDC and BDC-Idd9.905 mice
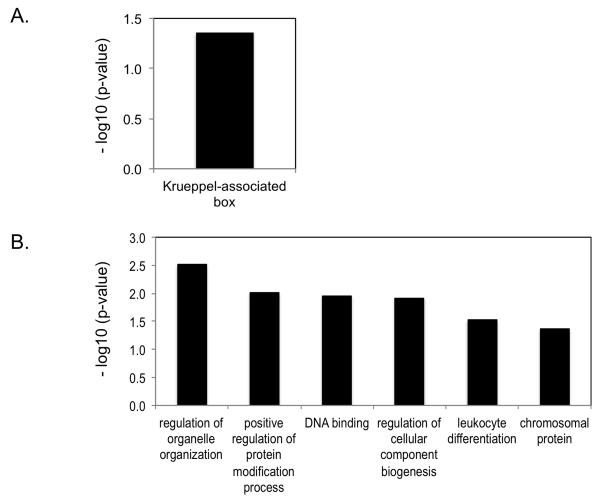
To identify biological processes that might be affected by genes expressed differentially in ex vivo or p79-stimulated BDC and BDC-Idd9.905 CD4+ T cells, we analyzed gene-ontology-based gene clustering using the Functional Annotation Clustering (FAC) tool of the Database for Annotation, Visualization and Integrated Discovery (DAVID) (27). FAC analysis of genes whose expression was reduced in ex vivo CD4+ T cells from BDC-Idd9.905 mice revealed a significant (p<0.05) enrichment in Krueppel-associated box (KRAB) genes; genes whose expression was higher in these mice did not cluster into any enriched annotation group (Fig.3A and data not shown). KRAB is a transcriptional repressor domain of many eukaryotic Krueppel-type C2H2 zinc finger proteins, which are implicated in cellular development and differentiation (29). Thus, these data suggest that islet-specific CD4+ T cells between the two strains differ in their development or differentiation. Genes that were expressed at reduced levels in p79-stimulated BDC-Idd9.905 CD4+ T cells clustered into 6 significantly (EASE score >1.3) enriched groups, consisting of regulation of organelle organization, positive regulation of protein modification process, DNA binding, regulation of cellular component biogenesis, leukocyte differentiation and chromosomal proteins (Fig. 3B). As all of these processes are involved in cell proliferation and differentiation, these results suggest that proliferation and/or differentiation of BDC-Idd9.905 CD4+ T cells in response to p79 stimulation may be reduced as compared to BDC CD4+ T cells. DAVID FAC analysis of genes that were expressed at higher levels in p79-stimulated CD4+ T cells from BDC-Idd9.905 mice did not identify any gene ontology clusters whose expression differed significantly from that in BDC CD4+ T cell (data not shown).
Figure 3. DAVID functional annotation cluster analysis of differentially expressed genes between BDC-Idd9.905 and BDC CD4+ T cells.
Analysis was performed on the 55 genes and 80 genes that showed differential expression in ex vivo (A) and p79-stimulated (B) CD4+ T cells between BDC-Idd9.905 and BDC mice. Bars describe the gene ontology terms and represent the most significantly enriched functional annotation clusters for down-regulated genes (p<0.05, FC>1.4). Significance of enrichment of gene ontology terms is shown (p<0.05) as EASE score, which is defined as the minus log10 transformation on the geometric mean of p-values (modified Fisher’s exact test) in a corresponding annotation term that associates with the gene group’s members.
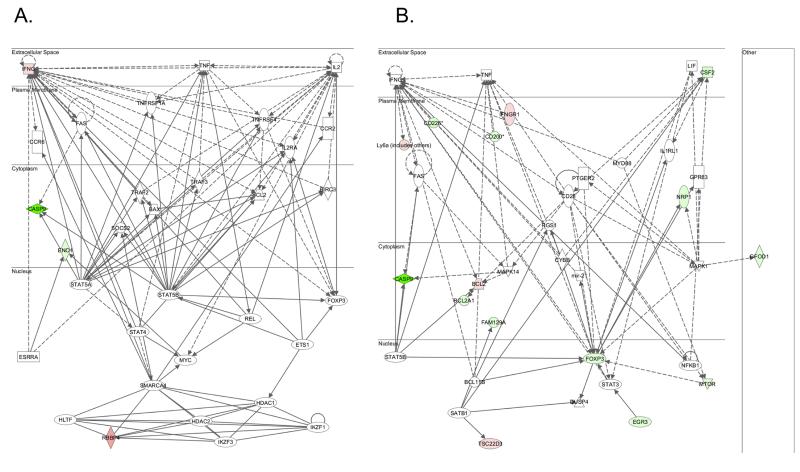
To analyze the microarray data in a tissue-relevant context, we used Ingenuity Pathway Analysis (IPA) software; a bioinformatics annotation tool that identifies molecular networks among differentially expressed genes in selected tissues. With a T cell-specific filter, the most significant gene network that IP analysis revealed in ex vivo CD4+ T cells from the two mouse strains was associated with cellular development, cellular growth and proliferation, hematological system development and function (Fig. 4A). Four focus genes of this network were among the genes we identified, and two of them, Eno1 and Rbbp4, are located within the Idd9 region.
Figure 4. Gene network of differentially expressed genes in CD4+ T cells between BDC-Idd9.905 and BDC mice.
The 55 genes and 80 genes that were differentially expressed in ex vivo (A) and p79-stimulated (B) CD4+ T cells between BDC-Idd9.905 and BDC mice, respectively were subjected to Ingenuity Pathway analysis. The most significant molecular network for each gene set is shown. Nodes represent gene products and biological relationships between two nodes are represented as a line (direct interaction: solid line; indirect interaction: broken line). Green and red nodes indicate elevated and decreased expression levels, respectively.
In p79-stimulated CD4+ T cells, the top IPA network of differentially expressed genes between the two strains was associated with cellular development and hematological system development and function (Fig. 4B). Fifteen focus genes of this network were differentially expressed in the microarray data set, four of which were expressed at higher levels in BDC-Idd9.905 CD4+ T cells and eleven of which were expressed at lower levels. One of the down-regulated genes and the only one located within the Idd9 region is Mtor, a serine/threonine protein kinase that regulates antigen responsiveness of CD4+ T cells, which in turn directs T helper (Th) effector cell differentiation (30). Other focus genes of this network that are immunologically relevant are located outside the Idd9 region (see discussion). In particular, we note that Cd226 expression in p79-stimulated BDC-Idd9.905 CD4+ T cells was reduced. CD226 triggers naive CD4+ T cell differentiation and proliferation via its association with lymphocyte function–associated antigen 1 (LFA-1) (31, 32) and is a candidate gene for T1D susceptibility in humans.(33)
Taken together, the T cell-specific IPA and DAVID analyses support the hypothesis that differential expression of Idd9 genes contribute to differential proliferation and differentiation of islet-specific CD4+ T cells between BDC-Idd9.905 and BDC mice.
Evaluation of microarray gene expression by quantitative PCR
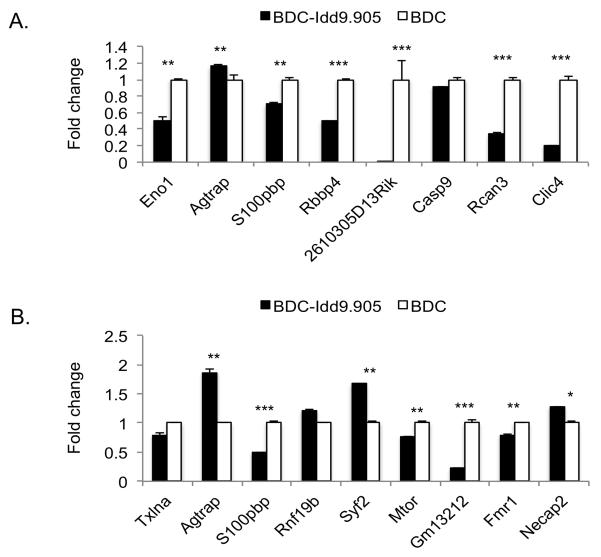
To evaluate our microarray results, we next measured gene expression in BDC and BDC-Idd9.905 CD4+ T cells by reverse transcription quantitative PCR (RT-qPCR). For the ex vivo CD4+ T cell samples, we selected five genes located within Idd9 and three genes located outside of Idd9 whose expression in the two strains differed the most. For the p79-stimulated CD4+ T cell samples, we assayed seven genes located within Idd9 and two located outside of it with the highest expression differences. Consistent with the microarray results, transcripts from the Idd9 genes Eno1 (Idd9.3) and 2610305D13Rik (Idd9.2) were more abundant in ex vivo BDC-Idd9.905 CD4+ T cells than in BDC CD4+ T cells, and more Agtrap transcripts (Idd9.2) were observed in the BDC-Idd9.905 samples (Fig. 5A, Table 1). For genes located outside of the Idd9 region, Rcan3 and Clic4 transcripts were less abundant in ex vivo CD4+ T cells from BDC-Idd9.905 than from BDC mice, as predicted by the microarray results.
Figure 5. Validation of differential expression of selected genes by quantitative PCR.
Selected genes that showed differential expression (FC >1.4, p<0.05) in microarray analysis of ex vivo (A) and p79-stimulated (B) CD4+ T cells between BDC-Idd9.905 and BDC mice were validated by RT-qPCR. Data were normalized to 18S rRNA expression and shown as mean fold change in BDC-Idd9.905 samples relative to the mean fold change in BDC samples ± S.D. (error bars). *p<0.05, **p<0.01, ***p<0.001 (Student’s t test).
Table 1.
Expression of selected genes in ex vivo BDC-Idd9.905 vs. BDC CD4+ T cells by microarray analysis
| Gene | Entrez gene ID | Location | FC Microarray | p-value |
|---|---|---|---|---|
| 2610305D13Rik | 112422 | Idd9.2 | −78.0 | 2.8E-06 |
| Eno1 | 13806 | Idd9.3 | −1.5 | 8.4E-04 |
| S100pbp | 74648 | Idd9.1 | 1.9 | 2.3E-03 |
| Agtrap | 11610 | Idd9.2 | 2.7 | 2.8E-03 |
| Rbbp4 | 19646 | Idd9.1 | 6.7 | 3.8E-03 |
| Rcan3 | 53902 | C4 | −6.5 | 2.5E-08 |
| Casp9 | 12371 | C4 | −77.7 | 1.3E-06 |
| Clic4 | 29876 | C4 | −3.5 | 0.001 |
RNA from ex vivo BDC or BDC-Idd9.905 CD4+ T cells was processed for microarray as described in Material and Methods. Data show fold changes (FC) of indicated gene transcripts (BDC-Idd9.905/BDC) as determined by microarray hybridization using Illumina MouseWG-6 v2.0 R3 Expression BeadChips.
In p79-stimulated CD4+ T cells, the RT-qPCR results were consistent with the differences in RNA expression from the Idd9 genes Mtor (Idd9.2), Agtrap (Idd9.2), Gm13212 (Idd9.2) and Syf2 (Idd9.1) that we observed in the microarray analysis between the two strains (Fig. 5B, Table 2). Expression of Fmr1 and Necap2, both located outside the Idd9 region, also recapitulated the microarray results.
Table 2.
Expression of selected genes in p79-stimulated BDC-Idd9.905 vs. BDC CD4+ T cells by microarray analysis
| Gene | Entrez gene ID | Location | FC Microarray | p-value |
|---|---|---|---|---|
| GM13212 | 433801 | Idd9.2 | −2.5 | 4.1E-05 |
| Agtrap | 11610 | Idd9.2 | 3.9 | 2.0E-04 |
| Mtor | 56717 | Idd9.2 | −1.5 | 1.9E-03 |
| Txlna | 109658 | Idd9.1 | −1.6 | 2.2E-03 |
| Rnf19b | 75234 | Idd9.1 | −1.9 | 3.9E-03 |
| S100pbp | 74648 | Idd9.1 | 1.4 | 5.2E-03 |
| Necap2 | 66147 | C4 | 15.6 | 1.0E-05 |
| Fmr1 | 14265 | CX | −2.8 | 1.5E-05 |
RNA from p79-stimulated BDC or BDC-Idd9.905 CD4+ T cells was processed for microarray as described in Material and Methods. Data show fold changes (FC) of indicated gene transcripts (BDC-Idd9.905/BDC) as determined by microarray hybridization using Illumina MouseWG-6 v2.0 R3 Expression BeadChips.
In contrast to the microarray results, S100pbp and Rbbp4 (both Idd9.1) were expressed at lower levels in ex vivo BDC-Idd9.905 CD4+ T cells based on RT-qPCR analysis. Interestingly, S100pbp expression was reported to be down-regulated in regulatory CD4+ T cells of NOD.B10 Idd9 (strain 905) mice (20). Levels of Casp 9 and Rnf19b as well as Txlna (both Idd9.1) RNA, were indistinguishable between the two strains in ex vivo and p79-stimulated CD4+ T cells, respectively. Expression of S100pbp was reduced in p79-stimulated CD4+ T cells from BDC-Idd9.905 mice relative to BDC mice, which was different to the microarray results.
In summary, the results of our real-time RT-qPCR analysis showed that combining BDC-Idd9.905 mice and microarray gene expression analysis was successful in identifying genes that are candidates for mediating Idd9-dependent differences in basic cellular activities of islet-specific CD4+ T cells, such as proliferation and differentiation.
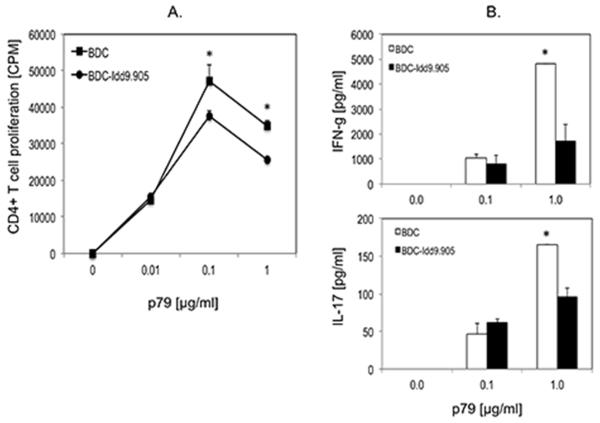
Impaired antigen-specific proliferation and pro-inflammatory cytokine responses by BDC-Idd9.905 CD4+ T cells
Bioinformatics analysis indicated that the expression of genes involved in cellular growth and development were down-regulated in p79-stimulated BDC-Idd9.905 CD4+ T cells. Thus, we hypothesized that BDC-Idd9.905 CD4+ T cells show reduced antigen-specific T cell proliferation and Th effector cell responses as compared to those from BDC mice. To test this hypothesis, we stimulated purified CD4+ T cells from BDC or BDC-Idd9.905 mice with the BDC2.5 mimotope p79 at different concentrations and determined T cell proliferation and Th cytokine responses by thymidine uptake assay and ELISA, respectively. Indeed, p79-stimulated CD4+ T cells from BDC-Idd9.905 mice proliferated significantly less than BDC CD4+ T cells (Fig. 6A). In addition, we measured significantly lower concentrations of pro-inflammatory IFN-γ and IL-17 in supernatants of stimulated BDC-Idd9.905 CD4+ T cells than BDC CD4+ T cells at the highest antigen concentrations, indicating that antigen-specific Th1 and Th17 effector responses were impaired in BDC-Idd9.905 CD4+ T cells (Fig. 6B). We did not observe detectable differences in the concentration of the Th2 signature cytokine IL-4 in the stimulated spleen cell culture supernatants of either strain (data not shown). We next investigated whether reduced pro-inflammatory responses by BDC-Idd9.905 CD4+ T cells may be the result of increased frequencies or enhanced function of Treg cells in BDC-Idd9.905 mice. Comparison of splenic CD4+Foxp3+ Treg cells in 6-9 week old BDC and BDC-Idd9.905 mice, however, did not reveal significant differences (Supplementary Fig. 2). Furthermore, CD4+CD25+ Treg cells displayed comparable suppressive function on p79-specific proliferation of CD4+CD25− effector T cells in vitro (Supplementary Fig. 3).
Figure 6. Reduced proliferation and Th1 and Th17 responses in BDC-Idd9.905 CD4+ T cells following stimulation with BDC2.5 mimotope.
Purified CD4+ T cells from BDC-Idd9.905 or BDC mice were stimulated with indicated concentrations of BDC2.5 mimotope p79 in presence of irradiated NOD spleen cells as APCs for 3 days. (A) CD4+ T cell proliferation was determined by [3H]thymidine incorporation assay and shown as mean counts per minute (CPM) of triplicate cultures. (B) Concentrations of indicated Th cytokines in supernatants of p79-stimulated CD4+ T cell cultures were assayed in duplicate by ELISA. One of three independent experiments each with similar data is shown. Error bars represent SD. * p < 0.03 (A) * p < 0.02 (B) (Student’s t test).
Collectively, these data demonstrate that BDC-Idd9.905 CD4+ T cells are hyporesponsive to p79-specific proliferation and Th1 and Th17 cytokine production, which did not appear to be associated with increased numbers and function of Treg cells. Thus, these findings support the results of our bioinformatics analyses and predict that islet-specific BDC-Idd9.905 CD4+ T cells may show inferior diabetogenic function as compared to those from BDC mice.
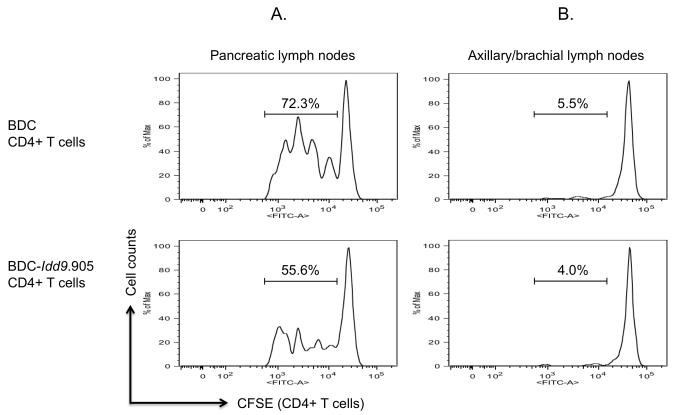
BDC-Idd9.905 CD4+ T cells show impaired proliferation to endogenous autoantigen and reduced capacity to mediate T1D
Islet-specific CD4+ T cells in NOD mice are primed in the PLN where they encounter pancreatic autoantigen (34). To investigate the effect of B10 Idd9 on proliferation of islet-specific CD4+ T cells to endogenous autoantigen, we injected CFSE-labeled CD4+ T cells from BDC or BDC-Idd9.905 mice into non-diabetic NOD recipient mice. Four days later, proliferation of transferred CD4+ T cells from PLN of recipients was examined by assessing CFSE dilution by flow cytometry. Notably, the frequency of proliferating BDC-Idd9.905 CD4+ T cells was markedly reduced compared to BDC CD4+ T cells (Fig. 7A). As a control, islet-specific CD4+ T cells from both strains failed to substantially proliferate in axillary/brachial lymph nodes, which do not drain the PLN and therefore do not contain pancreatic antigens (Fig. 7B).
Figure 7. Impaired proliferation of BDC-Idd9.905 CD4+ T cells to endogenous autoantigen.
BDC-Idd9.905 and BDC CD4+ T cells were CFSE-labeled and transferred (5 × 106, i.v.) into non-diabetic NOD mice. After 90 hours, proliferation of transferred CD4+ T cells from pancreatic lymph nodes (A) and from axillary/brachial lymph nodes (B) as control was determined by assessing CFSE dilution by flow cytometry. One of three independent experiments each with similar data is shown. Numbers in histograms represent percentages of CFSE+ CD4-gated T cells that underwent cell divisions.
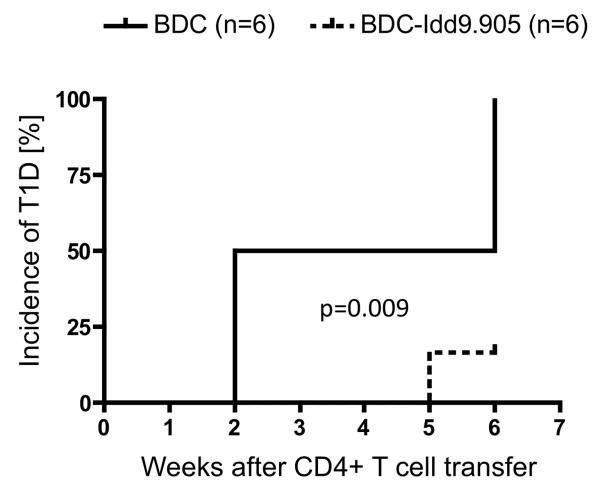
To investigate whether inefficient priming/proliferation of BDC-Idd9.905 CD4+ T cells to autoantigen results in impaired ability to mediate T1D, we next transferred CD4+ T cells from young non-diabetic BDC-Idd9.905 and BDC control mice into NOD SCID mice, which develop neither spontaneous insulitis nor diabetes (20). All (100%) of the recipients (6/6 mice) that received BDC CD4+ T cells developed T1D, whereas only 17% of the recipients (1/6 mice) that had received BDC-Idd9.905 CD4+ T cells developed T1D by the end of the experiment (50 days). These results were statistically significant (p=0.009) as determined by Kaplan-Meier analysis and log-rank test (Fig. 8). T1D severity was comparable in diabetic mice of both groups as determined by urine glucose concentration (data not shown). Taken together, reduced p79-specific responses by BDC-Idd9.905 CD4+ T cells in vitro, correlated with reduced proliferation to endogenous autoantigen and with impaired diabetogenic function of BDC-Idd9.905 CD4+ T cells.
Figure 8. Impaired diabetogenic function of BDC-Idd9.905 CD4+ T cells.
Kaplan-Meier analysis of T1D in NOD SCID mice following injection (i.v.) of purified CD4+ T cells (2.5 × 106 cells/mouse) from non-diabetic BDC or BDC-Idd9.905 mice. Recipient mice (n=6 mice/group) were examined for T1D for indicated time by measuring urine glucose concentrations at least twice a week. Mice with glucose concentration ≥250 mg/dl at two consecutive time points were diagnosed as diabetic. Mean data from one experiment are shown. p = 0.009 (log-rank test).
4. Discussion
We have used BDC and novel BDC-Idd9.905 mice to determine differences in global gene expression mediated by the Idd9 from the T1D-susceptible NOD strain or the T1D-resistant C57BL/10 strain in islet-specific CD4+ T cells, respectively. We found 55 and 80 genes to be differentially expressed in ex vivo and BDC2.5 mimotope (p79)-stimulated CD4+ T cells from BDC-Idd9.905 mice and BDC mice, respectively, as determined by microarray gene expression analysis (Fig. 1). Notably, differentially expressed genes were greatly enriched (~20%) for those genes located within the Idd9 and Idd11 region, overlapping the Idd9.1 region on chromosome 4 (Fig. 2). RT-qPCR analysis validated differential expression of 63% of selected genes, including Eno1, (ex vivo CD4+ T cells) and Mtor (p79-stimulated CD4+ T cells) (Fig. 5). The correlation coefficients for the agreement between microarray and RT-qPCR data can range markedly between values of −0.48 to 0.93 (35, 36). A number of factors could be responsible for the observation that RT-qPCR did not validate the expression of some differentially expressed microarray genes in our study. These factors may include microarray genes exhibiting small degrees of change, generally less than 2-fold, increased distance between the location of the PCR primers and microarray probes on a given gene or low intensity spots on microarrays (37, 38).
Bioinformatics analyses using DAVID and T cell-specific IPA revealed that genes associated with cellular growth and development, which included Idd9-encoded Eno1 and Rbbp4, were most significantly enriched among the down-regulated genes in ex vivo BDC-Idd9.905 CD4+ T cells (Fig. 4 and Fig. 5). Enolase1 (Eno1, Idd9.3), which was expressed at lower levels in ex vivo BDC-Idd9.905 CD4+ T cells, encodes a multifunctional enzyme that plays a role in growth control, glycolysis and allergic responses (39). Notably, Eno1 has been implicated in autoimmune diseases such as systematic lupus erythematous and rheumatoid arthritis because antibodies against this protein have been found in patients affected by these diseases (40). Furthermore, elevated expression of Eno1 in inflammatory cells has been reported to promote migration to inflammatory sites (41). Rbbp4 is a ubiquitously expressed and highly conserved nuclear protein that mediates chromatin assembly in DNA replication (42). The gene encoding retinoblastoma binding protein (Rbbp4, Idd9.1) was down-regulated in ex vivo BDC-Idd9.905 CD4+ T cells as determined by RT-qPCR. Interestingly, a previous gene expression analysis in CD4+ T cells from NOD and congenic NOD mice did not find significant differences in Rbbp4 gene expression between the two strains (20). It is possible that Rbbp4 expression differences in autoreactive CD4+ T cells may have been masked by those in non-autoreactive CD4+ T cells present in the study by Hamilton-Williams et al.
Mtor was the only focus gene encoded by Idd9 (Idd9.2) that was part of the most significant gene network in p79-stimulated CD4+ T cells identified by T cell-specific IPA. Mtor, which was down-regulated in p79-stimulated BDC-Idd9.905 CD4+ T cells, is a serine/threonine protein kinase that regulates cell growth and proliferation in response of growth factor signals and insulin. Importantly, Mtor is a critical regulator of T helper cell functions and differentiation (30). Mtor-deficient CD4+ T cells differentiate into Foxp3+ Treg cells, while failing to develop into Th1, Th2 or Th17 effector cells (43). Consistent with lower Mtor expression in BDC-Idd9.905 CD4+ T cells, functional analysis in vitro demonstrated that they displayed reduced Th1 and Th17 effector responses following antigen-specific stimulation relative to BDC CD4+ T cells. Differential expression of Mtor between NOD mice and C57BL/6 mice, which share Idd9 sequence identity with C57BL/10 mice could be due to several SNPs that exist in the intron region of Mtor (20). Interestingly, Hamilton-Williams et al. did not detect any differences in Mtor mRNA expression in naive or activated CD8+ T cells between NOD.B10 Idd9 and NOD mice (20). Their results combined with ours suggest that Mtor expression differs between CD4+ and CD8+ T cells subsets of these strains. Lower expression of Mtor as well as 2610305D13Rik, a KRAB-containing zinc finger protein with unknown function that this study also identified, was reported in B cells from T1D-protected NR4 mice, which are congenic for the NOR resistance locus 2 that overlaps with the Idd9.2 region (14). Thus, based on those and our data, lower expression of Mtor and 2610305D13Rik does not appear to be restricted to CD4+ T cells from T1D-protected mice.
Our bioinformatics results predicted that islet-specific BDC-Idd9.905 CD4+ T cells were impaired in their ability to proliferate and to develop effector T helper cell responses relative to BDC CD4+ T cells. Indeed, subsequent functional analyses validated these predictions by demonstrating that BDC-Idd9.905 CD4+ T cells were hyporesponsive in p79-specific proliferation as well as Th1 and Th17 cytokine responses in vitro as compared to BDC CD4+ T cells. Using a bone marrow transplantation approach, Chen et al. reported that CD4+ T cells containing the NOR-derived Idd9/11 loci selectively impaired the pathogenic potential by an undefined intrinsic factor(s) (10). Interestingly, reduced diabetogenic activity of these CD4+ T cells resulted from lower production of IFN-γ compared to NOD CD4+ T cells. Our study, using a different T1D-protected mouse strain and experimental approaches altogether, confirms and extends those findings by showing that antigen-specific IL-17 production, in addition to IFN-γ production, was reduced in BDC-Idd9.905 CD4+ T cells relative to BDC CD4+ T cells. Moreover, our analyses revealed Idd9 genes that are candidates for mediating these differential responses in CD4+ T cells. In contrast to the pathogenic role of Th17 cells in a number of autoimmune disease models, including EAE and RA, the role of Th17 cells in T1D has been unclear. A previous study showed that Th17 cells were involved in the T1D pathogenesis in NOD mice (44), while another study reported that the conversion of Th17 cells to a Th1-like phenotype in vivo underlies their diabetogenic function (45). Thus, it is conceivable that the Idd9 genes identified in our study impair the diabetogenic function BDC-Idd9.905 CD4+ T cells by negatively regulating the differentiation of Th1 and Th17 cells or the conversion of Th17 to Th1 cells in vivo.
Interestingly, the Idd9.1 locus has been implicated to control the suppressive activity of regulatory T cells (12). Notably, splenic Treg cells from BDC and BDC-Idd9.905 CD4+ mice were represented at comparable numbers in both strains and displayed similar suppressive functions. These findings are consistent with the results from a study in NOD and NOR mice, which found similar frequency and suppressive activity of Tregs between those strains (10). Taken together, our study and the study by Chen et al. suggest that genes within Idd9.2 and Idd9.3 compensate for or neutralize the Idd9.1-mediated effects on the suppressive activity of Treg cells.
A previous report showed that expression of protective Idd9 alleles by CD4+ T cells was sufficient to restore CD8+ T cell tolerance in NOD mice, albeit with contribution from non-lymphoid cells (21). Our study cannot exclude the possibility that T cell-extrinsic effects mediated by APCs contributed to differential gene expression between CD4+ T cells from BDC and BDC-Idd9.905 mice. Nevertheless, BDC-Idd9.905 CD4+ T cells proliferated less in response to antigen presented by NOD APCs in vitro as well as in vivo compared to BDC CD4+ T cells. Furthermore, BDC-Idd9.905 CD4+ T cells were less diabetogenic compared to BDC CD4+ T cells when they were exposed to APCs in NOD recipient mice. Taken together, these data indicate that Idd9-mediated effects on NOD APCs were insufficient to restore the proliferative responses and diabetogenic function of BDC CD4+ T cells to BDC-Idd9.905 CD4+ T cells.
In conclusion, by combining microarray gene expression and bioinformatics analyses we have identified Eno1, Rbbp4 and Mtor as Idd9 genes involved in T cell proliferation and differentiation that were differentially expressed between BDC-Idd9.905 and BDC CD4+ T cells. We propose that these Idd9 candidate genes contribute to Idd9-dependent differences in antigen-specific responses and diabetogenic function of islet-reactive CD4+ T cells between BDC-Idd9.905 and BDC mice.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Willard Freeman and Robert Brucklacher at the PSU College of Medicine Genome Sciences Facility for their assistance with the microarray analysis. We thank Drs. David Spector and Laura Carrel for their critical review of the manuscript.
This study was supported by funds from Pennsylvania State College of Medicine and Ono Pharmaceutical Co., Ltd. to HPW. LSW is supported by Wellcome Trust Grant 096388 and JDRF International Grant 9-2011-253. The Cambridge Institute for Medical Research is in receipt of Wellcome Trust Strategic Award 100140.
Footnotes
DISCLOSURES
The authors have no financial and commercial conflicts of interest.
REFERENCES
- 1.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 3.Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol. 2008;100:151–175. doi: 10.1016/S0065-2776(08)00806-7. [DOI] [PubMed] [Google Scholar]
- 4.Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity. 2000;13:107–115. doi: 10.1016/s1074-7613(00)00012-1. [DOI] [PubMed] [Google Scholar]
- 5.Cannons JL, Chamberlain G, Howson J, Smink LJ, Todd JA, Peterson LB, Wicker LS, Watts TH. Genetic and functional association of the immune signaling molecule 4-1BB (CD137/TNFRSF9) with type 1 diabetes. J Autoimmun. 2005;25:13–20. doi: 10.1016/j.jaut.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Silveira PA, Chapman HD, Stolp J, Johnson E, Cox SL, Hunter K, Wicker LS, Serreze DV. Genes within the Idd5 and Idd9/11 diabetes susceptibility loci affect the pathogenic activity of B cells in nonobese diabetic mice. J Immunol. 2006;177:7033–7041. doi: 10.4049/jimmunol.177.10.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldner H, Sobel RA, Price N, Kuchroo VK. The autoimmune diabetes locus Idd9 regulates development of type 1 diabetes by affecting the homing of islet-specific T cells. J Immunol. 2006;176:5455–5462. doi: 10.4049/jimmunol.176.9.5455. [DOI] [PubMed] [Google Scholar]
- 8.Ivakine EA, Gulban OM, Mortin-Toth SM, Wankiewicz E, Scott C, Spurrell D, Canty A, Danska JS. Molecular genetic analysis of the Idd4 locus implicates the IFN response in type 1 diabetes susceptibility in nonobese diabetic mice. J Immunol. 2006;176:2976–2990. doi: 10.4049/jimmunol.176.5.2976. [DOI] [PubMed] [Google Scholar]
- 9.Vallois D, Grimm CH, Avner P, Boitard C, Rogner UC. The type 1 diabetes locus Idd6 controls TLR1 expression. J Immunol. 2007;179:3896–3903. doi: 10.4049/jimmunol.179.6.3896. [DOI] [PubMed] [Google Scholar]
- 10.Chen YG, Scheuplein F, Osborne MA, Tsaih SW, Chapman HD, Serreze DV. Idd9/11 genetic locus regulates diabetogenic activity of CD4 T-cells in nonobese diabetic (NOD) mice. Diabetes. 2008;57:3273–3280. doi: 10.2337/db08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irie J, Reck B, Wu Y, Wicker LS, Howlett S, Rainbow D, Feingold E, Ridgway WM. Genome-wide microarray expression analysis of CD4+ T Cells from nonobese diabetic congenic mice identifies Cd55 (Daf1) and Acadl as candidate genes for type 1 diabetes. J Immunol. 2008;180:1071–1079. doi: 10.4049/jimmunol.180.2.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanouchi J, Puertas MC, Verdaguer J, Lyons PA, Rainbow DB, Chamberlain G, Hunter KM, Peterson LB, Wicker LS, Santamaria P. Idd9.1 locus controls the suppressive activity of FoxP3+CD4+CD25+ regulatory T-cells. Diabetes. 2010;59:272–281. doi: 10.2337/db09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan IK, Mackin L, Wang N, Papenfuss AT, Elso CM, Ashton MP, Quirk F, Phipson B, Bahlo M, Speed TP, Smyth GK, Morahan G, Brodnicki TC. A recombination hotspot leads to sequence variability within a novel gene (AK005651) and contributes to type 1 diabetes susceptibility. Genome Res. 2010;20:1629–1638. doi: 10.1101/gr.101881.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stolp J, Chen YG, Cox SL, Henck V, Zhang W, Tsaih SW, Chapman H, Stearns T, Serreze DV, Silveira PA. Subcongenic analyses reveal complex interactions between distal chromosome 4 genes controlling diabetogenic B cells and CD4 T cells in nonobese diabetic mice. J Immunol. 2012;189:1406–1417. doi: 10.4049/jimmunol.1200120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry GJ, Budgeon LR, Cooper TK, Christensen ND, Waldner H. The type 1 diabetes resistance locus B10 Idd9.3 mediates impaired B-cell lymphopoiesis and implicates microRNA-34a in diabetes protection. Eur J Immunol. 2014 doi: 10.1002/eji.201344116. [DOI] [PubMed] [Google Scholar]
- 16.Lebailly B, He C, Rogner UC. Linking the circadian rhythm gene Arntl2 to interleukin 21 expression in type 1 diabetes. Diabetes. 2014;63:2148–2157. doi: 10.2337/db13-1702. [DOI] [PubMed] [Google Scholar]
- 17.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 18.Eaves IA, Wicker LS, Ghandour G, Lyons PA, Peterson LB, Todd JA, Glynne RJ. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res. 2002;12:232–243. [PubMed] [Google Scholar]
- 19.Eckenrode SE, Ruan Q, Yang P, Zheng W, McIndoe RA, She JX. Gene expression profiles define a key checkpoint for type 1 diabetes in NOD mice. Diabetes. 2004;53:366–375. doi: 10.2337/diabetes.53.2.366. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton-Williams EE, Rainbow DB, Cheung J, Christensen M, Lyons PA, Peterson LB, Steward CA, Sherman LA, Wicker LS. Fine mapping of type 1 diabetes regions Idd9.1 and Idd9.2 reveals genetic complexity. Mamm Genome. 2013;24:358–375. doi: 10.1007/s00335-013-9466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton-Williams EE, Wong SB, Martinez X, Rainbow DB, Hunter KM, Wicker LS, Sherman LA. Idd9.2 and Idd9.3 protective alleles function in CD4+ T-cells and nonlymphoid cells to prevent expansion of pathogenic islet-specific CD8+ T-cells. Diabetes. 2010;59:1478–1486. doi: 10.2337/db09-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kachapati K, Adams DE, Wu Y, Steward CA, Rainbow DB, Wicker LS, Mittler RS, Ridgway WM. The B10 Idd9.3 locus mediates accumulation of functionally superior CD137(+) regulatory T cells in the nonobese diabetic type 1 diabetes model. J Immunol. 2012;189:5001–5015. doi: 10.4049/jimmunol.1101013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamberlain G, Wallberg M, Rainbow D, Hunter K, Wicker LS, Green EA. A 20-Mb region of chromosome 4 controls TNF-alpha-mediated CD8+ T cell aggression toward beta cells in type 1 diabetes. J Immunol. 2006;177:5105–5114. doi: 10.4049/jimmunol.177.8.5105. [DOI] [PubMed] [Google Scholar]
- 24.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 25.Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0062. research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Judkowski V, Pinilla C, Schroder K, Tucker L, Sarvetnick N, Wilson DB. Identification of MHC class II-restricted peptide ligands, including a glutamic acid decarboxylase 65 sequence, that stimulate diabetogenic T cells from transgenic BDC2.5 nonobese diabetic mice. J Immunol. 2001;166:908–917. doi: 10.4049/jimmunol.166.2.908. [DOI] [PubMed] [Google Scholar]
- 29.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibuya K, Shirakawa J, Kameyama T, Honda S, Tahara-Hanaoka S, Miyamoto A, Onodera M, Sumida T, Nakauchi H, Miyoshi H, Shibuya A. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med. 2003;198:1829–1839. doi: 10.1084/jem.20030958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dardalhon V, Schubart AS, Reddy J, Meyers JH, Monney L, Sabatos CA, Ahuja R, Nguyen K, Freeman GJ, Greenfield EA, Sobel RA, Kuchroo VK. CD226 is specifically expressed on the surface of Th1 cells and regulates their expansion and effector functions. J Immunol. 2005;175:1558–1565. doi: 10.4049/jimmunol.175.3.1558. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoglund P, Mintern J, Waltzinger C, Heath W, Benoist C, Mathis D. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J Exp Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckman KB, Lee KY, Golden T, Melov S. Gene expression profiling in mitochondrial disease: assessment of microarray accuracy by high-throughput Q-PCR. Mitochondrion. 2004;4:453–470. doi: 10.1016/j.mito.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 36.Etienne W, Meyer MH, Peppers J, Meyer RA., Jr. Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques. 2004;36:618–620. 622. doi: 10.2144/04364ST02. 624-616. [DOI] [PubMed] [Google Scholar]
- 37.Wurmbach E, Yuen T, Sealfon SC. Focused microarray analysis. Methods. 2003;31:306–316. doi: 10.1016/s1046-2023(03)00161-0. [DOI] [PubMed] [Google Scholar]
- 38.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R. alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. 2012;2012:156795. doi: 10.1155/2012/156795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundberg K, Kinloch A, Fisher BA, Wegner N, Wait R, Charles P, Mikuls TR, Venables PJ. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58:3009–3019. doi: 10.1002/art.23936. [DOI] [PubMed] [Google Scholar]
- 41.Bae S, Kim H, Lee N, Won C, Kim HR, Hwang YI, Song YW, Kang JS, Lee WJ. alpha-Enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. J Immunol. 2012;189:365–372. doi: 10.4049/jimmunol.1102073. [DOI] [PubMed] [Google Scholar]
- 42.Qian YW, Wang YC, Hollingsworth RE, Jr., Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 43.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain R, Tartar DM, Gregg RK, Divekar RD, Bell JJ, Lee HH, Yu P, Ellis JS, Hoeman CM, Franklin CL, Zaghouani H. Innocuous IFNgamma induced by adjuvant-free antigen restores normoglycemia in NOD mice through inhibition of IL-17 production. J Exp Med. 2008;205:207–218. doi: 10.1084/jem.20071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bending D, De la Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.