Abstract
Introduction
Tumor lymphocyte infiltration is associated with clinical response to chemotherapy in estrogen receptor (ER) negative breast cancer. To identify variants in immunosuppressive pathway genes associated with prognosis after adjuvant chemotherapy for ER-negative patients, we studied stage I-III invasive breast cancer patients of European ancestry, including 9,334 ER-positive (3,151 treated with chemotherapy) and 2,334 ER-negative patients (1,499 treated with chemotherapy).
Methods
We pooled data from sixteen studies from the Breast Cancer Association Consortium (BCAC), and employed two independent studies for replications. Overall 3,610 single nucleotide polymorphisms (SNPs) in 133 genes were genotyped as part of the Collaborative Oncological Gene-environment Study, in which phenotype and clinical data were collected and harmonized. Multivariable Cox proportional hazard regression was used to assess genetic associations with overall survival (OS) and breast cancer-specific survival (BCSS). Heterogeneity according to chemotherapy or ER status was evaluated with the log-likelihood ratio test.
Results
Three independent SNPs in TGFBR2 and IL12B were associated with OS (P <10−3) solely in ER-negative patients after chemotherapy (267 events). Poorer OS associated with TGFBR2 rs1367610 (G > C) (per allele hazard ratio (HR) 1.54 (95% confidence interval (CI) 1.22 to 1.95), P = 3.08 × 10−4) was not found in ER-negative patients without chemotherapy or ER-positive patients with chemotherapy (P for interaction <10−3). Two SNPs in IL12B (r2 = 0.20) showed different associations with ER-negative disease after chemotherapy: rs2546892 (G > A) with poorer OS (HR 1.50 (95% CI 1.21 to 1.86), P = 1.81 × 10−4), and rs2853694 (A > C) with improved OS (HR 0.73 (95% CI 0.61 to 0.87), P = 3.67 × 10−4). Similar associations were observed with BCSS. Association with TGFBR2 rs1367610 but not IL12B variants replicated using BCAC Asian samples and the independent Prospective Study of Outcomes in Sporadic versus Hereditary Breast Cancer Study and yielded a combined HR of 1.57 ((95% CI 1.28 to 1.94), P = 2.05 × 10−5) without study heterogeneity.
Conclusions
TGFBR2 variants may have prognostic and predictive value in ER-negative breast cancer patients treated with adjuvant chemotherapy. Our findings provide further insights into the development of immunotherapeutic targets for ER-negative breast cancer.
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-015-0522-2) contains supplementary material, which is available to authorized users.
Introduction
Breast cancer is still the leading cause of cancer-related death in women despite improving survival rates of cancer patients due to earlier detection and expanded treatment options [1], representing nearly 15% of cancer deaths in women [2]. Although at least half of newly diagnosed patients present with early-stage breast cancer, about 20% of these women will experience recurrence at a distant site within 10 years of diagnosis despite chemotherapy and hormonal therapy options [3]. Therefore, limitations of current therapeutic modalities, in particular for estrogen receptor negative (ER-negative) tumors and ER-negative/progesterone receptor (PR-negative) with low expression of human epidermal growth factor receptor 2 (HER2) (triple-negative (TN)) tumors, have led to search for new prognostic tools and therapy targets.
Tumor immunoevasion is recognized as an emerging hallmark of cancer, in addition to the tumor-promoting inflammation [4]. Inhibition of immune response may result from an immunosuppressive state in the tumor microenvironment [5]. Two main types of immune cells involved in the immunosuppression of cancer are the regulatory T cells (Treg cells) and the myeloid derived suppressor cells (MDSCs). Treg cells refer to a subset of T lymphocytes normally expressing CD4 + CD25 + FOXP3+, which play an important role in maintenance of self-tolerance and regulation of immune response [6,7]. MDSCs are a heterogeneous population of immature myeloid cells with expression of CD11b + GR1+ including precursors of macrophages, granulocytes and dendritic cells, which are also involved in tumor immunosuppression [8,9]. Tumor infiltration by immune cells, including Treg cells and MDSCs, has been implicated in cancer patient prognosis after chemotherapy [10-13]. ER-negative tumors typically show higher levels of tumor-infiltrating lymphocytes than ER-positive tumors [14,15]. Indeed, tumor lymphocyte infiltration, including Treg cells has been associated with clinical response to chemotherapy and with prognosis in ER-negative breast cancer [12,13,15], possibly due to the sensitivity of infiltrating lymphocytes to chemotherapeutic agents [16-18].
Therefore, we hypothesized that inherited common variation in genes of the immunosuppressive pathway, including Treg cells and MDSCs, could modulate response to adjuvant chemotherapy, particularly among ER-negative breast cancer patients. We evaluated genetic associations of single nucleotide polymorphisms (SNPs) located in or near (within 50 kb upstream and downstream) 133 candidate genes of the immunosuppressive pathway with overall survival (OS) and breast cancer-specific survival (BCSS) in breast cancer patients of European ancestry from 16 Breast Cancer Association Consortium (BCAC) studies [19] and performed replications for the variants with the strongest associations using two independent patient samples.
Methods
Study sample
We selected women of European ancestry diagnosed with histologically verified primary invasive but not metastatic breast cancer (stage I to III disease) and restricted to women with available age information, because age is an important risk factor for breast cancer (flow chart of patient selection in Additional file 1: Figure S1). The cause of death for an individual patient was recorded by hospital, cancer registry or health offices in the respective studies. The majority of the studies were all carried out in developed countries where deaths were accurately and mandatorily reported and causes of death had to be reported by the physicians, thus, the vast majority of deaths were reliably captured. Follow up was censored at 10 years from study entry. Studies with fewer than 10 events for all-cause mortality within this period were excluded as well as women with missing information on ER status, adjuvant chemotherapy, vital status and cause of death. Excluded patients had a similar mean age as compared to patients included in the study (55.7 versus 56.8 years), had more family history of breast cancer (30.53% versus 23.01%), had a lower frequency in receiving adjuvant chemotherapy (30.56% versus 39.85%), and had similar distribution in tumor stage, grade, size, and ER/PR/HER2 status. A total of 11,668 patients (9,334 with ER-positive disease, 2,334 with ER-negative disease) from 16 studies in BCAC were included (Additional file 2: Table S1a). Of these patients, 4,650 patients (3,151 with ER-positive disease and 1,499 with ER-negative disease) had received adjuvant chemotherapy, 7,018 patients (6,183 with ER-positive disease and 835 with ER-negative disease) did not receive chemotherapy.
For the replication analyses in ER-negative patients who had received adjuvant chemotherapy, we used four Asian studies in BCAC as one sample set and the Prospective Study of Outcomes in Sporadic versus Hereditary breast cancer (POSH) study (consisting of early-onset patients of European ancestry) as a second sample set [20,21]. As for the discovery, we included only ER-negative patients treated with adjuvant chemotherapy and restricted follow-up to 10 years after diagnosis. Thus, 372 breast cancer patients (42 events) from the BCAC Asian studies and 127 early-onset breast cancer patients (62 events) in the POSH study were included (Additional file 2: Table S1b). All studies were approved by the relevant ethics committees and all participants had signed an informed consent (Additional file 2: Table S1a and S1b).
SNP selection and genotyping
Genes related to Treg cell and MDSC pathways were identified through an extensive and comprehensive literature review in PubMed [22-34], using the search terms immunosuppression/immunosuppressive, regulatory T cells/Treg cells/FOXP3+ T cells, myeloid derived suppressor cells/MDSCs, immunosurveillance, and tumor escape, as only the broader immune pathways were accessible in the KEGG [35] and GO [36] databases. The final candidate gene list included 133 immunosuppression-related genes (Additional file 2: Table S2). SNPs with minor allele frequency (MAF) >0.05 within 50 kb upstream and downstream of each gene were identified using HapMap CEU genotype data and dbSNP 126 as references [37].
For the BCAC studies, study samples were genotyped for 211,155 SNPs using a custom Illumina iSelect array (iCOGS) designed for the Collaborative Oncological Gene-Environment Study (COGS) [19]. Of the 211,155 SNPs, 4,246 SNPs were located in the candidate genes within a window of ±50 kb. A series of centralized quality controls after genotyping led to exclusion of 243 SNPs. The exclusion criteria included a called rate <95% in all samples genotyped with iCOGS; being monomorphic; deviation from Hardy-Weinberg equilibrium (HWE) with a P-value <10−7, and concordance in duplicate samples <98%. After restricting the study sample to the subjects eligible (n = 11,668), we additionally excluded 393 SNPs with MAF <0.05 and deviation from HWE (P-value <10−7). A total of 3,610 SNPs passed all quality controls and were analyzed.
We used imputed genotype data of the POSH study. Imputation of POSH genome-wide association study (GWAS) data (genotyped using the Illumina 660-Quad SNP array, San Diego, CA, USA) was performed utilizing MACH 1.0 [38] based on the CEU population from HapMap phase 2 [37] and a posterior probability of 0.9. Imputation data were excluded based on MAF <0.01 and HWE with P-value <10−4. More details of POSH data are described elsewhere [39].
Statistical methods
Cox proportional hazard regression analysis with right truncation at 10 years after diagnosis was applied to model patient survival. Each single SNP was assessed as an ordinal variable (coded as 0, 1 and 2 respectively, according to number of minor allele). Analyses were adjusted for age at diagnosis and nine principal components to account for population substructure and stratified by study. To account for possible confounding due to differences in patient characteristics, we included tumor size, tumor grade and node status as further covariates. Delayed entry (left truncation) was used to reduce potential survival bias due to eligible patients who died before recruitment into the study or before the blood draw. Follow-up time was thus calculated from the date of interview or blood draw until event or censoring (date of last follow up). To determine the number of independent SNPs for adjustment of multiple testing, we applied the option, --indep-pairwise, in PLINK [40]. SNPs were pruned by linkage disequilibrium (LD) of r2 < 0.2 for a window size of 50 SNPs and step size of 10, yielding 699 independent SNPs. The significance threshold using Bonferroni correction corresponding to an alpha of 5% had a P-value <7.15 × 10−5.
In the primary analysis, we modeled OS in a multivariate Cox proportional hazard regression framework for ER-negative breast cancer patients separately for those who received adjuvant chemotherapy and those who did not receive adjuvant chemotherapy. To investigate whether SNP associations were restricted to ER-negative breast cancer, we assessed heterogeneity of associations between these two subgroups by using interaction terms between chemotherapy and SNPs, which were evaluated using likelihood ratio tests, comparing models with and without the interaction term. We also assessed whether selected SNPs associated with OS in ER-negative breast cancer patients who received adjuvant chemotherapy were associated with OS in ER-positive breast cancer patients treated with chemotherapy. Possible heterogeneity in the associations of SNPs with OS for patients who received chemotherapy according to ER status was assessed statistically by using interaction terms between ER status and SNPs and evaluated using likelihood ratio tests. In secondary analysis, we evaluated SNP associations with OS separately for ER-negative/PR-negative and TN breast cancer patients who received or did not receive adjuvant chemotherapy, respectively. Additionally, we assessed the associations of the SNPs with breast cancer-specific survival (BCSS) in ER-negative patients who received adjuvant chemotherapy. All statistical tests mentioned above were two-sided and conducted using SAS 9.2 (Cary, NC, USA).
For genes with multiple associated SNPs, HaploView was used to examine LD between SNPs. To identify potentially independently associated SNPs, we ran models including multiple associated SNPs within a gene. The proportional hazard assumption for the associated SNPs was assessed according to Grambsch and Therneau [41] and no significant deviation was noted. Cluster plots for the most significant SNPs were examined among BCAC samples and all showed good discrimination of three genotypes.
Meta-analyses were performed to summarize the results from the discovery and replication studies and to determine study heterogeneity using the I2 index and Q-statistics [42,43] and forest plots were generated using R (version 2.15.2).
Results
A descriptive summary of characteristics of the study population with available follow-up information is given in Table 1. There were 9,334 ER-positive breast cancer patients and 2,334 ER-negative breast cancer patients, of whom 1,904 had ER-negative/PR-negative disease and 1,007 TN disease. Of patients who had received adjuvant chemotherapy, 3,151 had ER-positive disease (376 events), 1,499 ER-negative disease (267 events), 1,271 ER-negative/PR-negative disease (221 events) and 692 TN disease (111 events).
Table 1.
Characteristics of the BCAC European study participants
| Characteristics | ER-negative patients who received chemotherapy | Percent | ER-negative patients who did not receive chemotherapy | Percent | ER-positive patients who received chemotherapy | Percent |
|---|---|---|---|---|---|---|
| Number of patients | 1499 | 100.00 | 835 | 100.00 | 3151 | 100.00 |
| Age at diagnosis (mean ± SD, years) | 51.69 ± 10.85 | 59.46 ± 12.18 | 51.74 ± 9.88 | |||
| Family history | ||||||
| No | 974 | 64.98 | 457 | 54.73 | 2233 | 70.87 |
| Yes | 275 | 18.35 | 148 | 17.72 | 596 | 18.91 |
| Missing | 250 | 16.68 | 230 | 27.54 | 322 | 10.22 |
| Tumor stage | ||||||
| 1 | 356 | 23.75 | 401 | 48.02 | 606 | 19.23 |
| 2 | 804 | 53.64 | 259 | 31.02 | 1751 | 55.57 |
| 3 | 182 | 12.14 | 58 | 6.95 | 526 | 16.69 |
| Missing | 157 | 10.47 | 117 | 14.01 | 268 | 8.51 |
| Histological grade | ||||||
| Well-differentiated | 23 | 1.53 | 97 | 11.62 | 390 | 12.38 |
| Moderately differentiated | 293 | 19.55 | 310 | 37.13 | 1624 | 51.54 |
| Poorly/undifferentiated | 1183 | 78.92 | 428 | 51.26 | 1137 | 36.08 |
| Tumor size | ||||||
| ≤2 cm | 664 | 44.30 | 528 | 63.23 | 1387 | 44.02 |
| ≥2 cm to ≤5 cm | 744 | 49.63 | 271 | 32.46 | 1490 | 47.29 |
| ≥5 cm | 91 | 6.07 | 36 | 4.31 | 274 | 8.70 |
| Lymph node status | ||||||
| Negative | 735 | 49.03 | 651 | 77.96 | 976 | 30.97 |
| Positive | 764 | 50.97 | 184 | 22.04 | 2175 | 69.03 |
| PR status | ||||||
| PR-negative | 1271 | 84.79 | 633 | 75.81 | 546 | 17.33 |
| PR-negative HER2-negative | 692 | 46.16 | 315 | 37.72 | 304 | 9.65 |
Results are presented as number of patients unless stated otherwise. BCAC, Breast Cancer Association Consortium; ER, estrogen receptor; PR, progesterone receptor; SD, standard deviation; HER2, human epidermal growth factor receptor 2.
A quantile-quantile (QQ) plot for tests of associations with OS for the 3,610 evaluated SNPs in ER-negative breast cancer patients who received adjuvant chemotherapy is shown in Figure 1. Three independent genetic variants in the two genes, TGFBR2 and IL12B, showed associations with OS (P <10−3) only in ER-negative breast cancer patients who received adjuvant chemotherapy. None of the associations was significant after Bonferroni correction (P <7.15 × 10−5) (Table 2). In ER-negative breast cancer patients who did not receive chemotherapy, none of the SNPs were associated (P <10−3). The results for all assessed 3,610 SNPs in ER-negative breast cancer patients treated with adjuvant chemotherapy are summarized in Additional file 2: Table S3.
Figure 1.
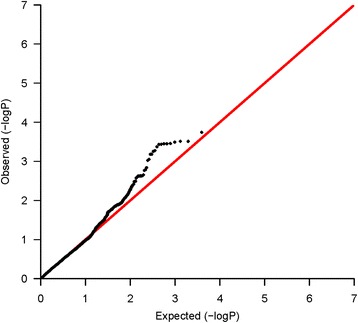
Quantile-quantile (QQ) plot of the observed P -values for associations with overall survival in estrogen receptor (ER)-negative patients who received chemotherapy. QQ plot shows the observed -log10 P-values (y axis) versus the expected -log10 P-values (x axis) for association of 3,610 SNPs in the immunosuppression pathway, with overall survival in ER-negative breast cancer patients who received adjuvant chemotherapy. The black dots indicate that there is inflation for observed associations.
Table 2.
TGFBR2 and IL12B SNPs associated with overall survival ( P -value <0.001) in ER-negative patients with chemotherapy
| Chr | Gene | SNP | Minor allele | MAF | ER-negative patients who received adjuvant chemotherapy | ER-negative patients who did not receive adjuvant chemotherapy | ER-positive patients who received adjuvant chemotherapy | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases, number | Events, number | HR a (95% CI) | P -value | Cases, number | Events, number | HR a (95% CI) | P -value | P heterogeneity b | Cases, number | Events, number | HR a (95% CI) | P -value | P heterogeneity c | |||||
| 3 | TGFBR2 | rs1367610 | C | 0.14 | 1499 | 267 | 1.54 (1.22, 1.95) | 3.08 × 10−4 | 834 | 155 | 0.78 (0.55, 1.13) | 0.191 | 8.82 × 10−4 | 3151 | 376 | 0.88 (0.70, 1.10) | 0.251 | 2.62 × 10−4 |
| 5 | IL12B | rs2546892 | A | 0.17 | 1499 | 267 | 1.50 (1.21, 1.86) | 1.81 × 10−4 | 835 | 155 | 0.99 (0.74, 1.33) | 0.968 | 0.025 | 3151 | 376 | 0.99 (0.82, 1.20) | 0.940 | 4.63 × 10−3 |
| 5 | IL12B | rs2853694 | C | 0.51 | 1499 | 267 | 0.73 (0.61, 0.87) | 3.67 × 10−4 | 835 | 155 | 1.06 (0.85, 1.33) | 0.596 | 0.020 | 3151 | 376 | 0.95 (0.82, 1.10) | 0.529 | 0.023 |
aHR adjusted for age of diagnosis, tumor size, tumor grade, node status and nine principal components to account for population substructure and stratified by study. b P-value for test of heterogeneity between ER-negative breast cancer patients who received adjuvant chemotherapy and ER-negative breast cancer patients who did not receive adjuvant chemotherapy. c P-value for test of heterogeneity between ER-negative breast cancer patients who received adjuvant chemotherapy and ER-positive breast cancer patients who received adjuvant chemotherapy. TGFBR2, transforming growth factor, beta receptor II; SNP, single nucleotide polymorphism; ER, estrogen receptor; Chr, chromosome; MAF, minor allele frequency; HR, hazard ratio; IL12B, interleukin 12B; and CI, confidence interval.
In TGFBR2, the strongest association in ER-negative patients who received chemotherapy was seen for SNP rs1367610 (G > C) (per allele hazard ratio (HR) 1.54 (95% confidence interval (CI) 1.22, 1.95), P = 3.08 × 10−4). A regional association plot for all SNPs in TGFBR2 is shown in Figure 2. The Kaplan-Meier survival curve stratified by genotype of SNP rs1367610 is shown in Figure 3. For the univariate survival curves, the P-value of the log-rank test was 2.0 × 10−4. There was no evidence of heterogeneity for the association across eight studies with at least ten events in ER-negative patients with chemotherapy (Additional file 1: Figure S2). This SNP was not associated with OS in ER-negative patients who did not receive chemotherapy (P-value for interaction = 8.82 × 10−4) or with ER-positive patients who received chemotherapy (P-value for interaction = 2.62 × 10−4). Variant alleles of nine further SNPs in TGFBR2 in strong LD with rs1367610 (r2 ≥ 0.97) were similarly associated with poorer OS in ER-negative breast cancer patients treated with chemotherapy (Additional file 2: Table S3). After accounting for rs1367610, none of other nine TGFBR2 variants showed association with OS.
Figure 2.
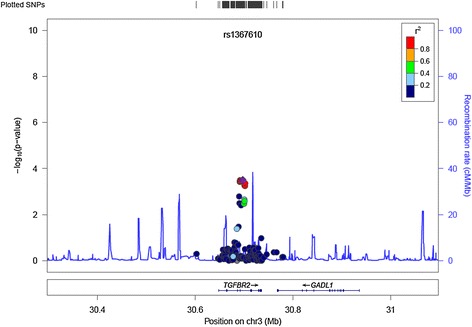
Regional association plot for single nucleotide polymorphisms (SNPs) in TGFBR2. The regional plot shows associations with overall survival in ER-negative breast cancer patients who received adjuvant chemotherapy, for all SNPs in TGFBR2. The y-axis shows the -log10 P-value. The purple diamond indicates SNP rs1367610, with the most significant association in TGFBR2. Chr, chromosome.
Figure 3.
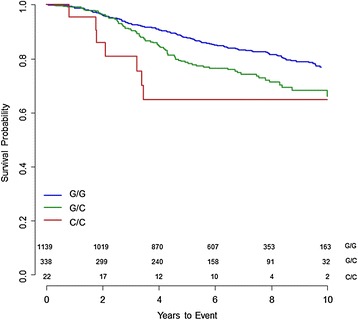
Kaplan-Meier survival curves of overall survival in estrogen receptor (ER)-negative patients who had chemotherapy for TGFBR2 rs1367610. The survival curves for TGFBR2 rs1367610 (G > C) stratified by genotype are shown for the Breast Cancer Association Consortium European sample. The P-value of the log-rank test was 2.0 × 10−4. The number of events and cases in parenthesis for each genotype are GG (180/1139, blue line), GC (80/338, green line) and CC (7/22, red line) respectively.
Two independent SNPs in IL12B (r2 = 0.20) showed associations with ER-negative disease after chemotherapy: rs2546892 (G > A) with poorer OS (HR 1.50 (95% CI 1.21, 1.86), P = 1.81 × 10−4), and rs2853694 (A > C) with improved OS (HR 0.73 (95% CI 0.61, 0.87), P = 3.67 × 10−4). These SNPs were not associated with OS in ER-negative patients who did not receive chemotherapy (P-value for interaction: 2.53 × 10−2 for rs2546892, 1.98 × 10−2 for rs2853694), or in ER-positive patients who received chemotherapy (P-value for interaction: 4.63 × 10−3 for rs2546892, 2.27 × 10−2 for rs2853694) (Table 2). Three other SNPs in IL12B (rs2853697, rs2569254 and rs3181225) in high LD with rs2546892 (r2 ≥ 0.81, Additional file 1: Figure S3a) were also associated with OS (P <10−3) (Additional file 2: Table S3). After adjusting for rs2853694, rs2546892 (but not the other three SNPs) was still associated with OS, suggesting that there are two potential independently associated variants in IL12B.
In the secondary analysis of ER-negative/PR-negative patients who received chemotherapy, rs1488369 (A > G) in a further gene, CCR9, was observed to be associated with an improved OS (HR 0.72 (95% CI 0.59, 0.87), P = 8.63 × 10−4), besides SNPs in TGFBR2 and IL12B (Additional file 2: Table S4). An association was not found for ER-negative PR-negative patients without chemotherapy (P-value for interaction = 1.78 × 10−2). This SNP was associated (HR 0.75 (95% CI 0.63, 0.90, P = 1.70 × 10−3) in patients with ER-negative disease (Additional file 2: Table S3).
In TN breast cancer patients treated with chemotherapy, rs2285440 (A > C), rs1726599 (C > A) and rs6956139 (C > A) in moderate LD (r2 ≥ 0.38) located in HDAC9 showed associations with two-fold increased HRs (HR 1.92 to 2.41, P <10−3) (Additional file 1: Figure S3b, Additional file 2: Table S4). None of the three SNPs was associated with OS in TN patients who did not receive chemotherapy. SNP rs2285440 remained strongly associated (HR 2.09 (95% CI 1.06, 4.15) after adjusting for the other two SNPs. This SNP showed a weaker association in patients with ER-negative disease (HR 1.47 (95% CI 1.09, 1.98), P = 1.26 × 10−2) (Additional file 2: Table S3).
Additionally, rs9863120 (A > G) located in EIF2A was associated with a significantly improved OS in TN patients who received chemotherapy (per allele HR 0.53 (95% CI 0.38, 0.74), P = 1.87 × 10−4) but not those without chemotherapy (P-value for interaction = 8.02 × 10−4) (Additional file 2: Table S4). In patients with ER-negative disease, this SNP showed a weaker association (HR 0.78 (95% CI 0.65, 0.95), P = 1.43 × 10−2) (Additional file 2: Table S3).
We also assessed the associations of the immunosuppressive pathway SNPs with BCSS among ER-negative breast cancer patients who received chemotherapy. The results were in line with the findings of the OS analysis. The most strongly associated SNP rs1872987 in TGFBR2 with BCSS is in high LD with rs1367610 (r2 = 0.99, HR 1.69 (95% CI 1.31, 2.19), P = 6.26 × 10−5) and the same SNPs in IL12B were found to be associated. Additionally, an SNP, rs658230 (G > A) in PRKCQ, was associated with an improved BCSS (per allele HR 0.70 (95% CI 0.57, 0.86), P = 6.95 × 10−4) and an SNP rs9579165 (A > G) in FLT3 showed a poorer BCSS (per allele HR 1.87 (95% CI 1.30, 2.70), P = 7.73 × 10−4) (Additional file 2: Table S5).
We performed two independent replications for the SNPs in TGFBR2 and IL12B specifically associated with OS only in the ER-negative breast cancer patients treated with adjuvant chemotherapy using the BCAC Asian samples and the (European) POSH study. The association of TGFBR2 rs1367610 with OS in ER-negative patients after adjuvant chemotherapy was replicated in both BCAC Asian samples (HR 2.18 (95% CI 0.85, 5.60), P = 1.05 × 10−1) as well as in the POSH study (HR 1.59 (95% CI 0.94, 2.69), P = 8.39 × 10−2), and was significant (HR 1.71 (95% CI 1.08, 2.72) in the replication samples combined. IL12B rs2853694 and rs2546892 did not replicate in the two studies. Meta-analysis of the discovery and replication studies yielded for TGFBR2 rs1367610 an HR of 1.57 (95% CI 1.28, 1.94, P = 2.05 × 10−5) without evidence of heterogeneity (I2 = 0%; P heterogeneity = 0.78) (Table 3).
Table 3.
Associations of TGFBR2 and IL12B SNPs with overall survival in discovery and replication samples
| Breast cancer patients | TGFBR2 rs1367610 (G > C) | IL12B rs2546892 (G > A ) | IL12B rs2853694 (A > C) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases, number | Events, number | HR (95% CI) | P -value | Cases, number | Events, number | HR (95% CI) | P -value | Cases, number | Events, number | HR (95% CI) | P -value | |
| Discovery | ||||||||||||
| ER-negative and received chemotherapya | 1499 | 267 | 1.54 (1.22, 1.95) | 3.08 × 10−4 | 1499 | 267 | 1.50 (1.21, 1.86) | 1.81 × 10−4 | 1499 | 267 | 0.73 (0.61, 0.87) | 3.67 × 10−4 |
| I 2c = 86.7%; P heterogeneityd = 5.00 × 10−4 | I 2c = 78.5%; P heterogeneityd = 9.50 × 10−3 | I 2c = 75.9%; P heterogeneityd = 0.016 | ||||||||||
| Replication | ||||||||||||
| ER-negative and received chemotherapy | ||||||||||||
| BCAC Asian studiesa | 372 | 42 | 2.18 (0.85, 5.60) | 0.105 | 372 | 42 | 0.62 (0.30, 1.26) | 0.187 | 372 | 42 | 1.03 (0.63, 1.67) | 0.919 |
| POSH studyb | 127 | 62 | 1.59 (0.94, 2.69) | 0.084 | 127 | 62 | 1.09 (0.67, 1.78) | 0.715 | 127 | 62 | 0.87 (0.62, 1.22) | 0.408 |
| Combined replication | ||||||||||||
| ER-negative and received chemotherapy | 499 | 104 | 1.71 (1.08, 2.72) | 0.022 | 499 | 104 | 0.91 (0.61, 1.36) | 0.659 | 499 | 104 | 0.92 (0.69, 1.21) | 0.535 |
| I 2c = 0%; p heterogeneityd = 0.567 | I 2c = 40.4%; P heterogeneityd = 0.20 | I 2c = 0%; P heterogeneityd = 0.577 | ||||||||||
| Combined overall | ||||||||||||
| ER-negative received adjuvant chemotherapy | 1998 | 371 | 1.57 (1.28, 1.94) | 2.05 × 10−5 | 1998 | 371 | 1.11 (0.70, 1.76) | 0.653 | 1998 | 371 | 0.78 (0.67, 0.90) | 8.00 × 10−4 |
| I 2c = 0%; P heterogeneityd = 0.781 | I 2c = 68.3%; P heterogeneityd = 0.04 | I 2c = 6.4%; P heterogeneityd = 0.344 | ||||||||||
aHR adjusted for age of diagnosis, tumor size, tumor grade, node status and principal components to account for population substructure and stratified by study. bHR adjusted for age of diagnosis, tumor size, tumor grade, node status and metastasis status. c I 2 index derived on the basis of effect estimate and variance in each study. d P-value for test of heterogeneity between studies using the DerSimonian-Laird test. TGFBR2, transforming growth factor, beta receptor II; SNP, single nucleotide polymorphism; HR, hazard ratio; ER, estrogen receptor; BCAC, Breast Cancer Association Consortium; IL12B, interleukin 12B; CI, confidence interval; and POSH, Prospective Study of Outcomes in Sporadic versus Hereditary Breast Cancer.
Discussion
In this study, we found that common variants in TGFBR2 have prognostic value for ER-negative breast cancer patients who received adjuvant chemotherapy. Our hypothesis was confirmed that this was specific for ER-negative disease, as the TGFBR2 variants were clearly not associated with OS in ER-positive breast cancer patients who received chemotherapy. The TGFBR2 variants also have predictive value, as the association with OS in ER-negative breast cancer patients was significantly differential according to treatment with chemotherapy.
TGFBR2 (3p22) encodes the transforming growth factor beta (TGF-β) receptor II, which is a transmembrane serine/threonine protein kinase receptor in the TGF-β signaling pathway [44]. As an important cytokine in tumor microenvironment, TGF-β has been considered to have a dual role in tumor suppression at early stages but then later promoting tumor invasion and metastasis [44,45]. Specifically, TGF-β functions as a stimulator in the tumor microenvironment to promote Treg cell proliferation and immune evasion [46]. An ER-negative tumor is normally associated with a higher level of infiltrating lymphocytes [14,15]. TGF-β receptor II plays a key role in the TGF-β signaling pathway, as all three TGF-β isoforms bind to this receptor [45]. Early genetic loss of TGFBR2 may lead to rapid tumor growth [45]. TGFBR2 has been identified as a susceptibility locus for breast cancer risk [19] and its expression in cancer-associated fibroblasts was found to be a prognostic marker for pre-menopausal breast cancer [47]. Since the immune-modulatory activities of TGF-ß have implications for many diseases, many drugs targeting the TGF-ß signaling have been developed. Based on our findings, it is conceivable that TGFBR2 variants may have prognostic and predictive value also for the outcome of TGF-ß signal inhibition.
TGFBR2 rs1367610 was recently reported to be possibly associated with BCSS in ER-negative patients treated with adjuvant chemotherapy using the COGS samples, however, replication in independent studies was not carried out [48]. The prior COGS study examined associations with breast cancer survival for 7,020 SNPs in 557 genes related to immune response and inflammation [48]. There were about 70 genes (1,694 SNPs) that overlapped between the two studies. The discovery sample of our study is somewhat smaller due to restriction to early breast cancer (stage I to III disease) and truncation of follow up to 10 years to minimize the influence of comorbidity on survival. However, we confirmed the prognostic value of TGFBR2 in the independent POSH study as well as in the Asian samples without study heterogeneity and also showed TGFBR2 variants to be related to both OS and BCSS.
All the top SNPs (P-value <10−3) in TGFBR2 were in the intron of this gene. According to the UCSC genome browser, the best-hit rs1367610 is located in the transcription factor binding site, and rs1019856, rs1841528 and rs6550007 are in both the DNase I hypersensitivity clusters and transcription factor binding sites. In addition, from the HaploReg online tool, we found that rs6550007 (r2 = 0.98 with rs1367610) may change the binding site of forkhead box P3 (Foxp3), which is an important transcription factor and a typical surface marker of Treg cells (Additional file 1: Figure S4). The top TGFBR2 SNPs associated with breast cancer OS are not included in the GeneVar gene expression variation database [49]. They lay in a different LD block from that of the reported breast cancer risk-associated SNPs that led to the identification of TGFBR2 as a breast cancer susceptibility locus [19]. Neither rs1367610 nor SNPs in high LD was associated with breast cancer risk in the BCAC studies. It would be worth looking for potential regulatory SNPs further than 50 kb away, and further functional analyses are necessary to identify the causal variant.
Although IL12B was found to be associated with OS and with BCSS, also reported as possibly associated in the previous publication [48], we were not able to replicate this finding using the two studies, which were smaller than the discovery sample. If a real association was overestimated in the discovery sample, a much larger study sample would be required for replication. IL12B (5q31.1-q33.1) encodes IL12 p40, which acts as a subunit of the heterodimeric structure of cytokine IL12 and IL23, two important immune cytokines in cell-mediated immunity [50,51]. IL12 and IL23 can separately promote naïve T cells into T helper (Th)1 cells and Th17 cells in vivo [51], and the balance between Th17 cells and Treg cells is a key factor in maintaining a normal immune response [52].
Further genes, LZTFL1/CCR9, HDAC9 and EIF2A, as well as PRKCQ and FLT3, were implicated to play a role in OS and/or BCSS for ER-negative patients after chemotherapy. These findings warrant follow up in large patient samples, because more variants in immunosuppressive pathway genes are potentially associated with prognosis of breast cancer.
Three GWAS studies to date have been carried out to investigate inherited genetic variants associated with overall or breast cancer-specific mortality of breast cancer [39,53,54]. In part due to the moderate study size involved, few associations have been identified and confirmed. On the other hand, a GWAS of clinical outcome in breast cancer patients who received adjuvant tamoxifen therapy identified a new locus associated with recurrence-free survival [55]. Therefore, germ-line genetic variation associated with breast cancer prognosis may be more easily detected when considering specific treatment subgroups and/or cancer subtypes.
The main strengths of this study include the uniform genotyping procedures, stringent centralized quality controls and large sample size, which provides us with sufficient statistical power to detect associations between genetic variants with moderate effects and breast cancer prognosis. The availability of centrally collated and harmonized information on molecular subtype, clinical treatment, and follow up in BCAC allowed us to assess potential differential SNP associations according to chemotherapy and also according to ER status. We used the iCOGS array with 3,610 SNPs to comprehensively assess these pathways. However, tagging SNP coverage varied across different candidate genes and could not capture variation entirely across all of the immunosuppressive pathway-related loci. Only genotyped data but not imputed data were used. As we focused on single SNP assessment and did not perform multi-marker analyses, we might have not captured all truly associated loci. Two independent study samples were employed to replicate the most promising findings. The replication in the Asian population, a different ethnic group, also suggests that the observed association with TGFBR2 variants is likely to be real. However, further genetic and functional studies are still required to identify the causal variants and the mechanisms underlying the associations observed in this study.
Conclusions
Our findings indicate that elucidating genetic variants, which influence inhibition of tumor immunity, may provide prognostic and predictive markers of chemotherapy for ER-negative breast cancer and could lead to further therapy targets.
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians, and administrative staff who have enabled this work to be carried out. This study would not have been possible without the contributions of the following: Kyriaki Michailidou, (BCAC), Andrew Berchuck (OCAC), Rosalind A Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog and Ken Offit (CIMBA), Andrew Lee, Ed Dicks, Graig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F Nielsen, Borge G Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M Cunningham, Sharon A Windebank, Christopher A Hilker, Jeffret Meyer and the staff of Mayo Clinic Genotyping Core Facility. ABCS would like to thank Annegien Broeks, Emiel Rutgers, Laura van ’t Veer, Richard van Hien, Frans Hogervorst, Senno Verhoef. HEBCS would like to thank Kirsimari Aaltonen, Karl von Smitten, Sofia Khan, Tuomas Heikkinen and Irja Erkkilä. KBCP thanks Eija Myöhänen and Helena Kemiläinen. LMBC thanks Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts. MARIE thanks Judith Heinz, Nadia Obi, Alina Vrieling, Muhabbet Celik, Til Olchers and Stefan Nickels. NBCS would like to thank the following clinical collaborators: Professor Per Eystein Lønning, MD (Section of Oncology, Institute of Medicine, University of Bergen and Department of Oncology, Haukeland University Hospital, Bergen, Norway), Professor Emeritus Sophie D Fosså, MD (National Resource Centre for Long-term Studies after Cancer, Rikshospitalet-Radiumhospitalet Cancer Clinic Montebello, Oslo, Norway), Head physician Tone Ikdahl, MD (Department of Oncology, Oslo University Hospital, Oslo, Norway), Dr Lars Ottestad, MD (Department of Genetics and Department of Oncology, Oslo University Hospital Radiumhospitalet), Dr Marit Muri Holmen, MD (Department of Radiology, Oslo University Hospital Radiumhospitalet, Oslo, Norway), Dr Vilde Haakensen, MD (Department of Genetics and Department of Oncology, Oslo University Hospital Radiumhospitalet and Institute for Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway), Professor Bjørn Naume, MD (Division of Cancer Medicine and Radiotherapy, Department of Oncology, Oslo University Hospital Radiumhospitalet, Oslo, Norway), Professor Eiliv Lund (Department of Community Medicine, Faculty of Health Sciences, University of Tromsø - The Arctic University of Norway, Tromsø, Norway), Associate Professor Åslaug Helland, MD (Department of Genetics, Institute for Cancer Research and Department of Oncology, Oslo University Hospital Radiumhospitalet, Oslo, Norway and Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway), Professor Inger Torhild Gram, MD (Department of Community Medicine, Faculty of Health Sciences, University of Tromsø and Norwegian Centre for Integrated Care and Telemedicine, University Hospital of North Norway, Tromsø, Norway), Professor Emeritus Rolf Kåresen, MD (Department of Breast and Endocrine Surgery, Institute for Clinical Medicine, Ullevaal Hospital, Oslo University Hospital and Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway), Dr. Ellen Schlichting, MD (Department for Breast and Endocrine Surgery, Oslo University Hospital Ullevaal, Oslo, Norway), Professor Toril Sauer, MD (Department of Pathology at Akershus University hospital, Lørenskog, Norway), Dr Olav Engebråten, MD (Institute for Clinical Medicine, Faculty of Medicine, University of Oslo and Department of Oncology, Oslo University Hospital, Oslo, Norway), Dr Margit Riis, MD (Department of Surgery, Akershus University Hospital and Department of Clinical Molecular Biology (EpiGen), Institute of Clinical Medicine, Akershus University Hospital, University of Oslo, Lørenskog, Norway). OFBCR thanks Teresa Selander and Nayana Weerasooriya. PBCS thanks Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao and Michael Stagner, RBCS would like to thank Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk and the Erasmus MC Family Cancer Clinic. SASBAC would like to thank the Swedish Medical Research Counsel. SEARCH thanks Craig Luccarini, Caroline Baynes, Don Conroy, Anne Stafford, Sue Irvine, Barbara Perkins, Val Rhenius. SKKDKFZS thank all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to the study. kConFab/AOCS wish to thank Heather Thorne, Eveline Niedermayr, Charmaine Smith, Sandra Picken, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the kConFab Follow Up Study for their contributions to this resource, and the many families who contribute to kConFab. MYBRCA thanks Phuah Sze Yee, Peter Kang, Kang In Nee, Kavitta Sivanandan, Shivaani Mariapun, Yoon Sook-Yee, Daphne Lee and Teh Yew Ching for DNA extraction and patient recruitment. Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement number 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The BCAC is funded by CR-UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST program (BM0606). The ABCS study was supported by the Dutch Cancer Society (grants NKI 2007-3839; 2009 4363); BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007); and the Dutch National Genomics Initiative. The work of the BBCC study was partly funded by ELAN-Fond of the University Hospital of Erlangen. The HEBCS study was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. Financial support for KARBAC study was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institute, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation. The KBCP study was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, and by the strategic funding of the University of Eastern Finland. The LMBC study is supported by the Stichting tegen Kanker (232–2008 and 196–2010). The MARIE study was supported by the Deutsche Krebshilfe e.V. (70-2892-BR I, 106332, 108253, 108419), the Hamburg Cancer Society, the German Cancer Research Center and the Federal Ministry of Education and Research (BMBF) Germany (01KH0402). The MCBCS study was supported by the NIH grants CA128978, CA116167, CA176785 and NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and the Breast Cancer Research Foundation and a generous gift from the David F and Margaret T Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. The NBCS study was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to ALBD, FUGE-NFR 181600/V11 to VNK and a Swizz Bridge Award to ALBD. The OFBCR study was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The PBCS study was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The RBCS study was funded by the Dutch Cancer Society (DDHK 2004–3124, DDHK 2009–4318). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G Komen Breast Cancer Foundation. The SEARCH study is funded by a program grant from Cancer Research UK (C490/A10124)] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The SKKDKFZS study is supported by the German Cancer Research Center. The kConFab study is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. The kConFab follow-up study has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA). KAP is a National Breast Cancer Foundation Fellow (Australia). The HERPACC study was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, Culture and Technology of Japan, by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from Ministry Health, Labour and Welfare of Japan, by Health and Labour Sciences Research Grants for Research on Applying Health Technology from Ministry Health, Labour and Welfare of Japan and by National Cancer Center Research and Development Fund. The MYBRCA study is funded by research grants from the Malaysian Ministry of Science, Technology and Innovation (MOSTI), Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation (CARIF). Additional controls were recruited by the Singapore Eye Research Institute, which was supported by a grant from the Biomedical Research Council (BMRC08/1/35/19/550), Singapore and the National medical Research Council, Singapore (NMRC/CG/SERI/2010). The SEBCS study was supported by the BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0000347). The TWBCS study is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. The POSH study was supported by Funding Breast Cancer Campaign (NOV210PR62) and Cancer Research UK (C1275/A9896). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. Douglas F Easton is a Principal Research Fellow of Cancer Research UK. The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- BCAC
Breast Cancer Association Consortium
- BCSS
breast cancer-specific survival
- CCR9
chemokine (C-C motif) receptor 9
- CI
confidence interval
- COGS
Collaborative Oncological Gene-Environment Study
- EIF2A
eukaryotic translation initiation factor 2 alpha
- ER
estrogen receptor
- FLT3
fms-related tyrosine kinase 3
- Foxp3
forkhead box P3
- GWAS
genome-wide association studies
- HDAC9
histone deacetylase 9
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- HWE
Hardy-Weinberg equilibrium
- IL12B
interleukin 12B
- LD
linkage disequilibrium
- LZTFL1
leucine zipper transcription factor-like 1
- MAF
minor allele frequency
- MDSCs
myeloid derived suppressor cells
- OS
overall survival
- POSH
Prospective Study of Outcomes in Sporadic versus Hereditary Breast Cancer Study
- PR
progesterone receptor
- PRKCQ
protein kinase C, theta
quantile-quantile
- SD
standard deviation
- SNP
single nucleotide polymorphism
- TGFBR2
transforming growth factor, beta receptor II
- TGF-β
transforming growth factor beta
- Th
T helper
- TN
triple negative
- Treg cells
regulatory T cells
Additional files
Flow chart of patient selection. Figure S2 Forest plot of eight studies with at least ten events for TGFBR2 rs1367610. Figure S3a Linkage disequilibrium of five top single nucleotide polymorphisms (SNPs) in IL12B associated with estrogen receptor (ER)-negative patients with chemotherapy. Figure S3b linkage disequilibrium of seven top SNPs in HDAC9 associated with triple-negative patients with chemotherapy. Figure S4 UCSC browser graphic for TGFBR2 rs1367610.
Description of studies included: (S1a) Sixteen Breast Cancer Association Consortium (BCAC) European studies in the discovery analysis; (S1b) Four BCAC Asian studies and Prospective Study of Outcomes in Sporadic versus Hereditary Breast Cancer (POSH) study in the replication analysis. Table S2 List of 133 candidate genes related to the immunosuppressive pathway by chromosomal position. Table S3 Associations of 3610 single nucleotide polymorphisms (SNPs) in the immunosuppressive pathway with overall survival in estrogen receptor (ER)-negative patients with chemotherapy. Table S4 Top SNPs associated with overall survival of ER-negative/progesterone receptor (PR)-negative and triple-negative patients who received chemotherapy. Table S5 Top SNPs associated with breast cancer-specific survival of ER-negative patients who received chemotherapy.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
J Lei, AR, and SB participated in the design of the study, performed the statistical analysis, and drafted the manuscript. KBM, ELG, DFE, MKS, and JCC conceived the study, and participated in its design and coordination and helped to draft the manuscript. SR, PDPP, PS, PAF, ILA, VNK, FJC, UH, MJH, HN, UE, MKB, JD, QW, AL, AM, DL, MGC, PH, GCT, MS, RL, LH, ABE, MWB, JAK, GG, ST, GIGA, ALBD, SN, JEO, EH, CV, DT, HUU, TR, AJ, CHMD, MMATL, TAM, KA, CB, SM, VMK, JMH, VK, SH, HW, AS, JF, SJC, J Lissowska, J Li, KH, KAP, kConFab Investigators, SL, SC, SAJB, DK, JYC, SKP, KYY, CNH, PEW, MFH, CYS, SHT, NAMT, CHY, GFH, KM, H Ito, H Iwata, KT, AMD, JB, KC, LES, TM, WJT, and DE participated in the study design and helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ information
kConFab Investigators: Research Department, Peter MacCallum Cancer Center, East Melbourne, Victoria 3002, Australia. Georgia.Trench@qimrberghofer.edu.au.
Contributor Information
Jieping Lei, Email: j.lei@dkfz-heidelberg.de.
Anja Rudolph, Email: a.rudolph@dkfz-heidelberg.de.
Kirsten B Moysich, Email: Kirsten.Moysich@roswellpark.org.
Sajjad Rafiq, Email: S.Rafiq@soton.ac.uk.
Sabine Behrens, Email: S.Behrens@dkfz-heidelberg.de.
Ellen L Goode, Email: egoode@mayo.edu.
Paul PD Pharoah, Email: paul.pharoah@medschl.cam.ac.uk.
Petra Seibold, Email: p.seibold@dkfz-heidelberg.de.
Peter A Fasching, Email: peter.fasching@uk-erlangen.de.
Irene L Andrulis, Email: Andrulis@lunenfeld.ca.
Vessela N Kristensen, Email: v.n.kristensen@medisin.uio.no.
Fergus J Couch, Email: couch.fergus@mayo.edu.
Ute Hamann, Email: u.hamann@dkfz-heidelberg.de.
Maartje J Hooning, Email: m.hooning@erasmusmc.nl.
Heli Nevanlinna, Email: heli.nevanlinna@hus.fi.
Ursula Eilber, Email: u.eilber@dkfz-heidelberg.de.
Manjeet K Bolla, Email: mkh39@medschl.cam.ac.uk.
Joe Dennis, Email: jgd29@cam.ac.uk.
Qin Wang, Email: qw232@medschl.cam.ac.uk.
Annika Lindblom, Email: annika.lindblom@ki.se.
Arto Mannermaa, Email: arto.mannermaa@uef.fi.
Diether Lambrechts, Email: Diether.Lambrechts@med.kuleuven.be.
Montserrat García-Closas, Email: montse.garciaclosas@icr.ac.uk.
Per Hall, Email: Per.Hall@ki.se.
Georgia Chenevix-Trench, Email: Georgia.Trench@qimr.edu.au.
Mitul Shah, Email: ms483@medschl.cam.ac.uk.
Robert Luben, Email: robert.luben@phpc.cam.ac.uk.
Lothar Haeberle, Email: lothar.haeberle@uk-erlangen.de.
Arif B Ekici, Email: Arif.Ekici@humgenet.uni-erlangen.de.
Matthias W Beckmann, Email: Fk-direktion@uk-erlangen.de.
Julia A Knight, Email: knight@lunenfeld.ca.
Gord Glendon, Email: gglendon@uhnresearch.ca.
Sandrine Tchatchou, Email: sandrine@lunenfeld.ca.
Grethe I Grenaker Alnæs, Email: greal@rr-research.no.
Anne-Lise Borresen-Dale, Email: a.l.borresen-dale@medisin.uio.no.
Silje Nord, Email: silje.nord@rr-research.no.
Janet E Olson, Email: olsonj@mayo.edu.
Emily Hallberg, Email: Hallberg.emily@mayo.edu.
Celine Vachon, Email: vachon.celine@mayo.edu.
Diana Torres, Email: d.torreslopez@dkfz-heidelberg.de.
Hans-Ulrich Ulmer, Email: hu.ulmer@klinikum-mittelbaden.de.
Thomas Rüdiger, Email: Thomas.Ruediger@klinikum-karlsruhe.de.
Agnes Jager, Email: a.jager@erasmusmc.nl.
Carolien HM van Deurzen, Email: c.h.m.vandeurzen@erasmusmc.nl.
Madeleine MA Tilanus-Linthorst, Email: m.tilanus-linthorst@erasmusmc.nl.
Taru A Muranen, Email: taru.a.muranen@helsinki.fi.
Kristiina Aittomäki, Email: kristiina.aittomaki@hus.fi.
Carl Blomqvist, Email: carl.blomqvist@helsinki.fi.
Sara Margolin, Email: Sara.Margolin@karolinska.se.
Veli-Matti Kosma, Email: veli-matti.kosma@uef.fi.
Jaana M Hartikainen, Email: jaana.hartikainen@uef.fi.
Vesa Kataja, Email: vesa.kataja@ksshp.fi.
Sigrid Hatse, Email: Sigrid.Hatse@med.kuleuven.be.
Hans Wildiers, Email: Hans.Wildiers@uzleuven.be.
Ann Smeets, Email: Ann.Smeets@uzleuven.be.
Jonine Figueroa, Email: figueroaj@mail.nih.gov.
Stephen J Chanock, Email: chanocks@mail.nih.gov.
Jolanta Lissowska, Email: lissowsj@coi.waw.pl.
Jingmei Li, Email: jingmei@gmail.com.
Keith Humphreys, Email: Keith.Humphreys@ki.se.
Kelly-Anne Phillips, Email: Kelly.Phillips@petermac.org.
Sabine Linn, Email: s.linn@nki.nl.
Sten Cornelissen, Email: s.cornelissen@nki.nl.
Sandra Alexandra J van den Broek, Email: s.vd.broek@nki.nl.
Daehee Kang, Email: dhkang@snu.ac.kr.
Ji-Yeob Choi, Email: jiyeob.choi@gmail.com.
Sue K Park, Email: suepark@snu.ac.kr.
Keun-Young Yoo, Email: kyyoo@plaza.snu.ac.kr.
Chia-Ni Hsiung, Email: chiani@ibms.sinica.edu.tw.
Pei-Ei Wu, Email: peiei@gate.sinica.edu.tw.
Ming-Feng Hou, Email: mifeho@kmu.edu.tw.
Chen-Yang Shen, Email: bmcys@ibms.sinica.edu.tw.
Soo Hwang Teo, Email: soohwang.teo@carif.com.my.
Nur Aishah Mohd Taib, Email: nuraish@gmail.com.
Cheng Har Yip, Email: chenghar.yip@gmail.com.
Gwo Fuang Ho, Email: gwoho@um.edu.my.
Keitaro Matsuo, Email: keitarom@med.kyushu-u.ac.jp.
Hidemi Ito, Email: hidemi@aichi-cc.jp.
Hiroji Iwata, Email: hiwata@aichi-cc.jp.
Kazuo Tajima, Email: ktajima@clin.medic.mie-u.ac.jp.
Alison M Dunning, Email: amd24@medschl.cam.ac.uk.
Javier Benitez, Email: jbenitez@cnio.es.
Kamila Czene, Email: Kamila.Czene@ki.se.
Lara E Sucheston, Email: lara.sucheston@roswellpark.org.
Tom Maishman, Email: T.Maishman@soton.ac.uk.
William J Tapper, Email: w.j.tapper@soton.ac.uk.
Diana Eccles, Email: D.M.Eccles@soton.ac.uk.
Douglas F Easton, Email: dfe20@medschl.cam.ac.uk.
Marjanka K Schmidt, Email: mk.schmidt@nki.nl.
Jenny Chang-Claude, Email: j.chang-claude@dkfz-heidelberg.de.
References
- 1.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Ann Oncol. 2014;25:1650–1656. doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 2.Breast Cancer - Estimated Incidence, Mortality and Prevalence Worldwide in 2012. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Accessed 23 June 2014.
- 3.Early Breast Cancer Trialists’ Collaborative G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. [DOI] [PubMed]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 6.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 8.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 11.Lee HJ, Seo JY, Ahn JH, Ahn SH, Gong G. Tumor-associated lymphocytes predict response to neoadjuvant chemotherapy in breast cancer patients. J Breast Cancer. 2013;16:32–39. doi: 10.4048/jbc.2013.16.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 13.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 15.Calabro A, Beissbarth T, Kuner R, Stojanov M, Benner A, Asslaber M, et al. Effects of infiltrating lymphocytes and estrogen receptor on gene expression and prognosis in breast cancer. Breast Cancer Res Treat. 2009;116:69–77. doi: 10.1007/s10549-008-0105-3. [DOI] [PubMed] [Google Scholar]
- 16.Ghiringhelli F, Larmonier N, Schmitt E, Parcellier A, Cathelin D, Garrido C, et al. CD4 + CD25+ regulatory T cells suppress tumor immunity but are sensitive to cyclophosphamide which allows immunotherapy of established tumors to be curative. Eur J Immunol. 2004;34:336–344. doi: 10.1002/eji.200324181. [DOI] [PubMed] [Google Scholar]
- 17.Menard C, Martin F, Apetoh L, Bouyer F, Ghiringhelli F. Cancer chemotherapy: not only a direct cytotoxic effect, but also an adjuvant for antitumor immunity. Cancer Immunol Immunother. 2008;57:1579–1587. doi: 10.1007/s00262-008-0505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutsiak ME, Semnani RT, De Pascalis R, Kashmiri SV, Schlom J, Sabzevari H. Inhibition of CD4(+)25+ T regulatory cell function implicated in enhanced immune response by low-dose cyclophosphamide. Blood. 2005;105:2862–2868. doi: 10.1182/blood-2004-06-2410. [DOI] [PubMed] [Google Scholar]
- 19.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eccles D, Gerty S, Simmonds P, Hammond V, Ennis S, Altman DG, et al. Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH): study protocol. BMC Cancer. 2007;7:160. doi: 10.1186/1471-2407-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copson E, Eccles B, Maishman T, Gerty S, Stanton L, Cutress RI, et al. Prospective observational study of breast cancer treatment outcomes for UK women aged 18–40 years at diagnosis: the POSH study. J Natl Cancer Inst. 2013;105:978–988. doi: 10.1093/jnci/djt134. [DOI] [PubMed] [Google Scholar]
- 22.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills KH. Regulatory T, cells: friend or foe in immunity to infection? Nat Rev Immunol. 2004;4:841–855. doi: 10.1038/nri1485. [DOI] [PubMed] [Google Scholar]
- 24.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 25.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 26.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrand-Rosenberg S. Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev. 2008;18:11–8. doi: 10.1016/j.gde.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18:349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Driessens G, Kline J, Gajewski TF. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. 2009;229:126–144. doi: 10.1111/j.1600-065X.2009.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29:309–316. doi: 10.1007/s10555-010-9223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilczynski JR, Duechler M. How do tumors actively escape from host immunosurveillance? Arch Immunol Ther Exp (Warsz). 2010;58:435–448. doi: 10.1007/s00005-010-0102-1. [DOI] [PubMed] [Google Scholar]
- 32.Poschke I, Mougiakakos D, Kiessling R. Camouflage and sabotage: tumor escape from the immune system. Cancer Immunol Immunother. 2011;60:1161–1171. doi: 10.1007/s00262-011-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieg C, Boyman O. The role of chemokines in cancer immune surveillance by the adaptive immune system. Semin Cancer Biol. 2009;19:76–83. doi: 10.1016/j.semcancer.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Sakaguchi S, Vignali DA, Rudensky AY, Niec RE, Waldmann H. The plasticity and stability of regulatory T cells. Nat Rev Immunol. 2013;13:461–467. doi: 10.1038/nri3464. [DOI] [PubMed] [Google Scholar]
- 35.KEGG: Kyoto Encyclopedia of Genes and Genomes. http://www.genome.jp/kegg/. Accessed 08 May 2014.
- 36.Gene Ontology Consortium. http://www.geneontology.org/. Accessed 09 May 2014.
- 37.International HapMap Project. www.hapmap.org. Accessed 23 June 2014.
- 38.University of Michigan: Center for Statistical Genetics. MACH 1.0. http://www.sph.umich.edu/csg/abecasis/MACH/index.html. Accessed 30 April 2014.
- 39.Rafiq S, Tapper W, Collins A, Khan S, Politopoulos I, Gerty S, et al. Identification of inherited genetic variations influencing prognosis in early-onset breast cancer. Cancer Res. 2013;73:1883–1891. doi: 10.1158/0008-5472.CAN-12-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. doi: 10.1093/biomet/81.3.515. [DOI] [Google Scholar]
- 42.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 43.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 44.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 45.Akhurst RJ, Derynck R. TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 46.Ferretti G, Felici A, Cognetti F, Mandala M. Transforming growth factor-beta signaling and regulatory T cells. J Clin Oncol. 2007;25:4695–4696. doi: 10.1200/JCO.2007.13.2894. [DOI] [PubMed] [Google Scholar]
- 47.Busch S, Acar A, Magnusson Y, Gregersson P, Ryden L, Landberg G. TGF-beta receptor type-2 expression in cancer-associated fibroblasts regulates breast cancer cell growth and survival and is a prognostic marker in pre-menopausal breast cancer. Oncogene. 2013;34:27–38. doi: 10.1038/onc.2013.527. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Lindstrom LS, Foo JN, Rafiq S, Schmidt MK, Pharoah PD, et al. 2q36.3 is associated with prognosis for oestrogen receptor-negative breast cancer patients treated with chemotherapy. Nat Commun. 2014;5:4051. [DOI] [PMC free article] [PubMed]
- 49.Wellcome Trust Sanger Institute. Genevar (GENe Expression VARiation). http://www.sanger.ac.uk/resources/software/genevar/. Accessed 30 April 2014.
- 50.Kaarvatn MH, Vrbanec J, Kulic A, Knezevic J, Petricevic B, Balen S, et al. Single nucleotide polymorphism in the interleukin 12B gene is associated with risk for breast cancer development. Scand J Immunol. 2012;76:329–335. doi: 10.1111/j.1365-3083.2012.02736.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Massey D, Tremelling M, Parkes M. Genetics of inflammatory bowel disease: clues to pathogenesis. Br Med Bull. 2008;87:17–30. doi: 10.1093/bmb/ldn031. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, Wang C, Luo J, Zhao X, Wang L, Li X. Combination with methotrexate and cyclophosphamide attenuated maturation of dendritic cells: inducing Treg skewing and Th17 suppression in vivo. Clin Dev Immunol. 2013;2013:238035. doi: 10.1155/2013/238035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azzato EM, Pharoah PD, Harrington P, Easton DF, Greenberg D, Caporaso NE, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shu XO, Long J, Lu W, Li C, Chen WY, Delahanty R, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 2012;72:1182–1189. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiyotani K, Mushiroda T, Tsunoda T, Morizono T, Hosono N, Kubo M, et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum Mol Genet. 2012;21:1665–1672. doi: 10.1093/hmg/ddr597. [DOI] [PubMed] [Google Scholar]


