SUMMARY
Avian influenza viruses that cause infection and are transmissible in humans involve changes in the receptor binding site (RBS) of the viral hemagglutinin (HA) that alter receptor preference from α2-3-linked (avian-like) to α2-6-linked (human-like) sialosides. A human case of avian-origin H6N1 influenza virus was recently reported, but the molecular mechanisms contributing to it crossing the species barrier are unknown. We find that, although the H6 HA RBS contains D190V and G228S substitutions that potentially promote human receptor binding, recombinant H6 HA preferentially binds α2-3-linked sialosides, indicating no adaptation to human receptors. Crystal structures of H6 HA with avian and human receptor analogs reveal that H6 HA preferentially interacts with avian receptor analogs. This binding mechanism differs from other HA subtypes due to a unique combination of RBS residues, highlighting additional variation in HA-receptor interactions and the challenges in predicting which influenza strains and subtypes can infect humans and cause pandemics.
INTRODUCTION
Although a multitude of different avian influenza A viruses have been isolated, only three (H1N1, H2N2 and H3N2) have been able to adapt for transmission in the human population and these viruses have led to influenza pandemics. To circulate in the human population, an avian influenza virus has to acquire the ability to bind human receptors and lose affinity for avian receptors before being able to transmit from human to human. In the last two decades, an apparent increase has been observed in the reported cases of new avian-origin influenza subtypes infecting humans, including H9N2 (Peiris et al., 1999), H5N1 (‘bird flu’) (Beigel et al., 2005), H7N9 (Centers for Disease and Prevention, 2013), and very recently H6N1 (Shi et al., 2013a; Yuan et al., 2013) and H10N8 (Chen et al., 2014). This increase in novel zoonotic infections raises concern about the emergence of avian subtypes that could give rise to novel human pandemics.
On June 21 2013, the first case of a human infection by an avian-origin H6N1 influenza A virus (A/Taiwan/2/2013, Taiwan2) was reported by the Taiwan Centers for Disease Control (http://www.cdc.gov.tw/english/info.aspx?treeid=bc2d4e89b154059b&nowtreeid=ee0a2987cfba3222&tid=E36A5E9AB3D3A216). The patient, a 20 years old female, who had not been exposed to live poultry, was hospitalized after she developed a high fever, cough, headache, and muscle ache. She later fully recovered and no human-to-human transmission of the H6N1 virus was reported (Shi et al., 2013a; Yuan et al., 2013). Phylogenetic analysis of the viral genes from this H6N1 human isolate suggested that an avian virus A/Chicken/Taiwan/A2837/2013 was the possible origin of seven of eight genes of the Taiwan2 (H6N1) virus (for HA sequence see Figure S1A), whereas the source of the eighth gene, coding for PB2, was probably from A/Chicken/Taiwan/0101/2012 (H5N2) (Shi et al., 2013a; Yuan et al., 2013). H6N1 avian viruses are frequently isolated from birds and have been circulating worldwide, including in North America, Europe and Asia (Cheung et al., 2007; Jonassen and Handeland, 2007; Lee et al., 2006; Senne, 2003; Suss et al., 1994). In Taiwan, the H6N1 virus has been enzootic since the early 1970s (Lee et al., 2006). Although to date only this one case of human infection by an H6N1 virus has been reported, more widespread exposure to H6 viruses likely has already occurred as antibodies against H6 influenza A viruses have been detected in veterinarians in the United States and in workers in live poultry markets in China (Myers et al., 2007; Shortridge, 1992). Furthermore, virus replication studies show that some of the Taiwanese H6N1 viruses can replicate in mice without adaptation (Lee et al., 2006).
Influenza virus is an enveloped virus that contains two surface glycoproteins in its membrane envelope: hemagglutinin (HA) and neuraminidase (NA). The HA is the major glycoprotein and the target of neutralizing antibodies. The HA is a homotrimer where each monomer is synthesized as an inactive single chain precursor (HA0) that is proteolytically cleaved into two subunits: an N-terminal HA1 and a C-terminal HA2. The mature HA protein is responsible for attachment of the virus to its natural receptors, which are terminal sialic acids (N-acetylneuraminic acid (NeuAc)) on glycoprotein and glycolipids on the host cell, and for fusion of the viral envelope with the host cell in the low pH of endosomal compartments (Skehel and Wiley, 2000). So far 18 HA subtypes (16 avian and 2 bat subtypes) have been identified (Tong et al., 2013), based on serology and antigenic properties, and these can be phylogenetically divided into two groups (Air, 1981; Nobusawa et al., 1991). The H6 HA is within group 1 that also includes H1, H2, H5 and H9 subtypes that have caused human infections (Figure S1A).
The specificity of the HA for glycan receptors is believed to be a key determinant of the viral host range (Matrosovich et al., 2009), where the most important element for host specificity is the linkage between the terminal sialic acid and the penultimate galactose. HAs from avian viruses are characterized by their preference for α2-3-linked sialosides whereas HAs from human viruses are specific for α2-6-linked sialosides. This specificity difference contributes to the inability of most avian influenza viruses to transmit in the human population (Parrish and Kawaoka, 2005) and changes in the binding specificity from α2-3 to α2-6 sialosides is therefore believed to be essential to cross the species barrier and become human transmissible.
The HA receptor binding site (RBS) is located in the head domain of the HA1 subunit and consists of three conserved secondary elements, the 130- and 220- loops and the 190-helix, and a number of conserved residues in the heart of the binding site (Ha et al., 2001; Skehel and Wiley, 2000). Four key RBS residues have been implicated in the avian-human specificity switch (Matrosovich et al., 1997). In avian viruses, the 220-loop contains Gly225, Gln226 and Gly228, whereas Glu190 is present in the 190-helix (H3 numbering). For H1 subtypes, Glu190Asp and Gly225Asp substitutions are critical for attaining specificity for human receptors, as in the 1918 and 2009 pandemic H1N1 strains (Matrosovich et al., 2000; Stevens et al., 2006b; Tumpey et al., 2007). In contrast, for the 1957 H2N2 and 1968 H3N2 pandemic viruses, Gln226Leu and Gly228Ser substitutions were responsible for the switch between avian-type and human-type receptor specificity (Connor et al., 1994; Matrosovich et al., 2000). These examples have not proven predictive of the amino-acid changes required for conversion of receptor specificity in other subtypes. For example, for avian H5N1 virus, Glu190Asp and Gly225Asp mutations abolished receptor binding, while Gln226Leu and Gly228Ser mutations produced partial recognition of α2-6 linked receptors, but α2-3 binding was not lost (Stevens et al., 2006b). Further mutations at Asn158 and Asn224, or at Gln196, or combined with the loss of the N-linked glycan at Asn158, were sufficient to switch receptor specificity for H5 viruses to become transmissible in mammals (Chen et al., 2012; de Vries et al., 2014; Herfst et al., 2012; Imai et al., 2012).
RESULTS
Structural Characterization of Taiwan2 H6N1 HA
The H6N1 A/Taiwan/2/2013 virus is one of several emerging influenza viruses over the past two years, including H7N9 and H10N8, which have caused human infection, but have not yet spread in the human population (Vachieri et al., 2014). The frequency of these infections has elevated concerns that these viruses will acquire human-type receptor specificity allowing them to transmit between humans.
To analyze the receptor binding of H6N1, we first performed sequence alignment of the Taiwan2 H6N1 HA with human pandemic strains and with avian subtypes from emerging viruses that have caused sporadic outbreaks of human infections over the last few years. Taiwan2 H6 HA contains a unique combination of four key residues in the RBS not seen previously in human influenza viruses or avian viruses that sporadically infect humans (Figure S1B). The H6 190-helix contains an unusual aliphatic substitution of Val at position 190 instead of an acidic residue in all other avian and human subtypes (i.e. Glu or Asp for avian and human subtypes, respectively). In the 220-loop, H6 HA contains the G228S substitution that is associated with the receptor specificity switch of the H2 and H3 human pandemic viruses (Connor et al., 1994; de Graaf and Fouchier, 2014). In addition, two other substitutions could influence receptor binding. At position 222, H6 HA has a small hydrophobic Ala instead of a charged or polar residue (Lys or Gln) in all other subtypes, except for H3 where it is an aromatic residue (Trp). Taiwan/2/2013 H6 HA also contains Leu at position 186 instead of Pro, as in A/Chicken/Taiwan/A2837/2013 (Shi et al., 2013a) (Figure S1B).
To investigate the structural features and properties of this H6 HA, its crystal structure was determined at 2.5 Å resolution (Table S1). Overall, the structure is similar to other known influenza A HAs (Figure 1A) and the disposition of the HA head relative to the stem is most similar to H2N2 HA (PDB entry 3KU5). Six potential N-linked glycosylation sites per HA protomer are predicted (Wang et al., 2014) (5 in HA1), but interpretable electron density consistent with glycosylation is observed only at Asn21 (protomer C), Asn32 (A) and Asn 169 (A and C) from HA1 and Asn154 (C) from HA2, but not at Asn291 and Asn296, presumably due to disorder (i.e. for the trimer, for eighteen potential glycosylation sites, only five glycans could be modeled) (Figure 1A). Superposition of the H6 HA on other avian and human HAs confirms that H6 is structurally closest to H1, H2 and H5 (Group 1 HAs) [Cα root mean square deviations (RMSD) of 0.8–1.3 Å compared to 2.4–2.5 Å for Group 2 H3 and H7]. Similar results were also obtained by superposition of the HA1, HA2 and RBS subdomains (Figure 1C and Table S2). The H6 differs from other HAs due to insertion of Asp at position 144a, and a one-residue insertion of Asp at position 157a in the 150-loop compared to H1, H2, H3 and H5 HAs (Figure S1B and Figure 1C). A similar insertion of residues in the 150-loop was found for H7 HA, which contains two additional residues at positions 158a and 158b. The H6 RBS, like all other influenza A HAs, has a conserved floor of Tyr98, Trp153, His183 and Tyr195 (Skehel and Wiley, 2000) for binding the sialic acid moiety of the receptor. The surrounding 130-loop, 150-loop, 220-loop and 190-helix (Figure 1B) delineate the sides and ends of the RBS.
Figure 1. Crystal Structure of Taiwan2 H6N1 HA.
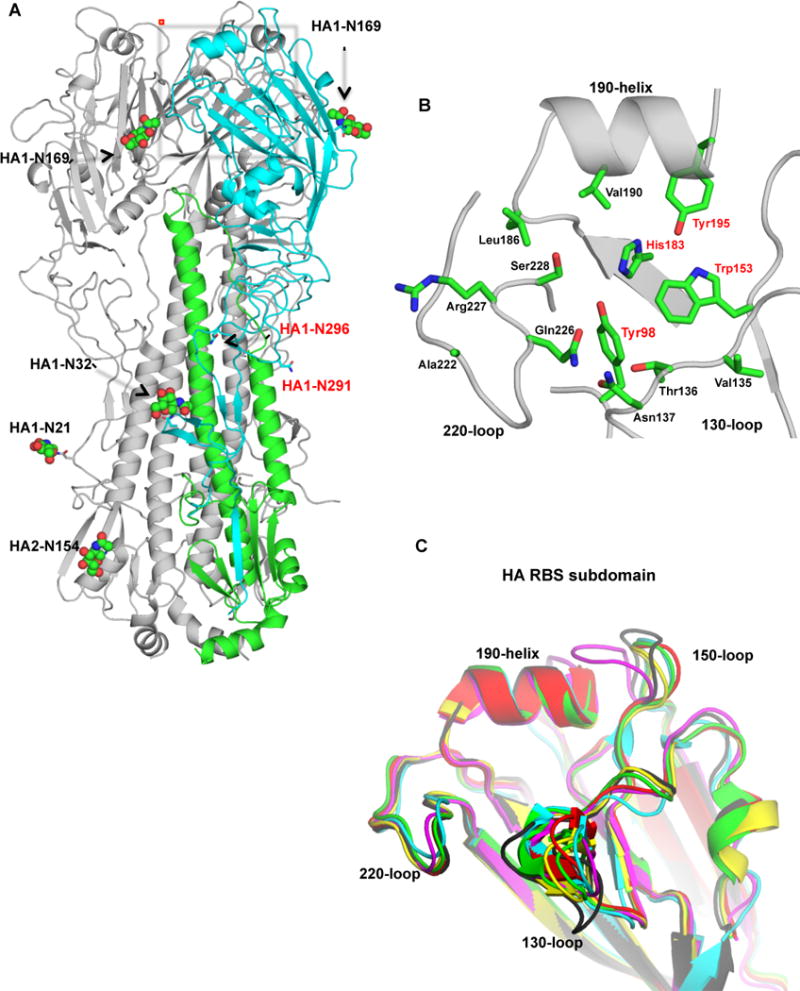
(A) Schematic representation of the H6 HA trimer. One protomer is colored in cyan and green for the HA1 and the HA2 subunits and the RBS is marked with red rectangle. N-linked glycans that could be modeled in the electron density maps are in green spheres and the corresponding asparagine is shown in sticks and labeled in black. Two other potential glycosylation sites (Asn291 and Asn 296) are labeled in red. (B) Schematic representation of the H6 HA RBS with sticks representing key residues for receptor binding (colored in green and labeled). The four highly conserved residues in the RBS are labeled in red. The H6 RBS contains Val in position 190 and Ser in position 228. (C) Superposition of the RBS subdomains of H6 HA (black) and pandemic H1 (yellow, PDB 3AL4), H2 (red, PDB 3KU5), H3 (cyan, PDB 4FNK), A/Vietnam/CL12/2004 H5 (green, PDB 2FKO) and A/Shanghai/2/2013 H7 (magenta, PDB 4N5J) HAs (see also Figure S1, Tables S1 and S2).
Receptor Specificity of Taiwan2 H6N1 HA
To determine the receptor specificity, the Taiwan2 H6 HA was expressed in insect cells and analyzed on a custom sialoside glycan array comprised of diverse α2-3 and α2-6 sialosides that correspond to N-linked and O-linked glycans, as well as linear fragments of glycans, on mammalian glycoproteins and glycolipids (Figure 2) (Stevens et al., 2006a) (Raman et al. 2014). The use of recombinant HA protein eliminates possible interference from the neuraminidase on the virus that preferentially cleaves α2-3 linked sialosides. Selective binding was observed to α2-3 sialosides with no detectable binding to α2-6 glycans (Figure 2). More specifically, the H6 HA bound preferentially to long branched O-linked and N-linked glycans terminating with α2-3 linked sialic acids (numbers 24–27) and fucosylated glycan structures (Figure 2A). Recombinant H6 HA expressed in mammalians cells bound with reduced avidity to the same glycans (Figure S2), a property observed previously for the H7N9 HA (Xu et al., 2013) that we attribute to the larger glycans expressed by mammalian cells (Stevens et al., 2006a).
Figure 2. Receptor Binding Specificity of Taiwan2 H6N1 HA.
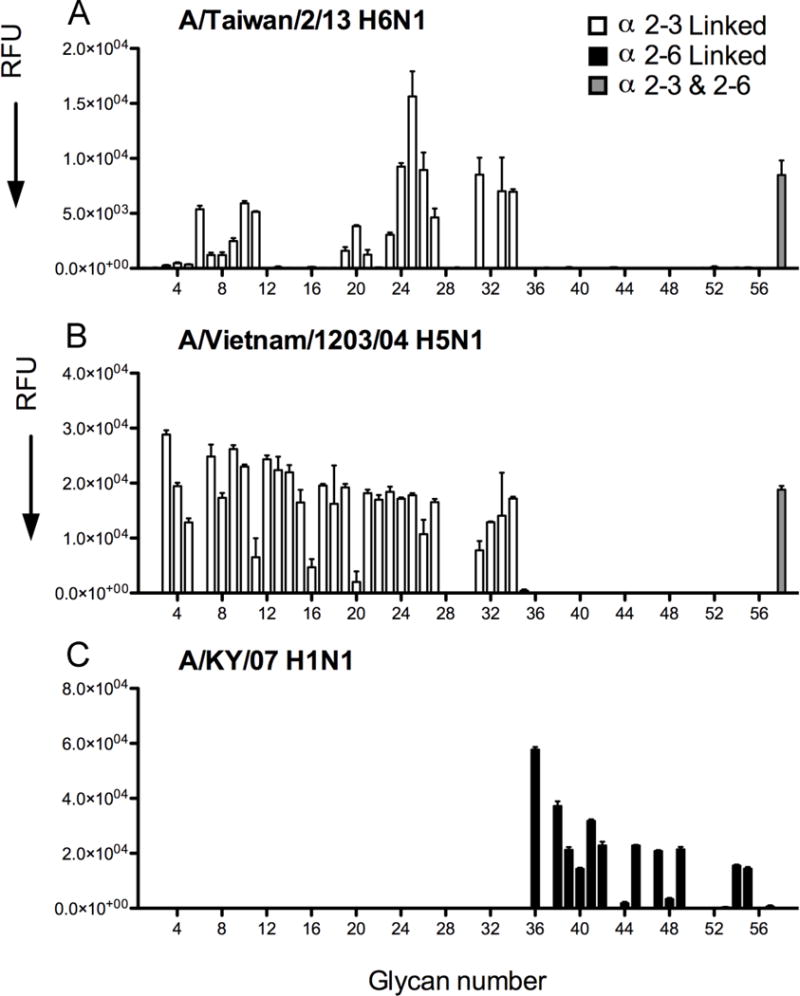
(A) Glycan microarray analysis of recombinant H6 HA protein expressed in insect cells demonstrates specific binding to a subset of α2-3 sialosides and to a mixed biantennary glycan. (B and C) The HAs of the A/Vietnam/1203/04 H5N1 (B) and H1N1 seasonal strain KY/07 (C) served as controls for α2-3 and α2-6 sialoside binding specificity. The mean signal and standard error were calculated from six independent replicates on the array. α2-3 linked sialosides in white bars (glycans 3 to 35 on the x axis), α2-6 linked sialosides in black (glycans 36 to 56), and mixed biantennary glycans containing both α2-3 and α2-6–linked sialylated glycans in gray bars (glycans 57 and 58). Glycans 1 and 2 are non-sialylated controls glycans (see also Figure S2 and Table S3).
Structural Characterization of Taiwan2 H6N1 HA in Complex with Avian Receptor Analogs
Crystal structures of H6 HA in complex with avian receptor analogs 3′-SLN (NeuAcα2-3Galβ1-4GlcNAc) and LSTa (NeuAcα2-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc) were determined at 2.3 Å and 2.4 Å, respectively (Figure 3 and Table S1). Electron density for 3′-SLN and LSTa was observed in only one RBS of the trimer as the two other binding sites are blocked by the HA1 C-terminus of a symmetry-related molecule in the crystal. As in other HA subtypes, Sia-1 of 3′-SLN and LSTa forms the usual seven conserved hydrogen bonds with the 220-loop and 130-loop (Figure 3B, C). Other conserved interactions include a hydrogen bond between the Tyr98 hydroxyl and the sialic acid (Sia-1) 8-hydroxyl and hydrophobic interactions between Trp153 and Sia-1 C8 and C9 (Figure 3B, C). In addition, the H6 RBS contains substitutions at two residues that normally contribute to Sia-1 binding in avian HA subtypes, E190V and G228S. The Gly228Ser substitution, which is also found in human H2 and H3 isolates, eliminates a conserved hydrogen bond between the Sia-1 9-hydroxyl group and the Gly228 main-chain carbonyl. Instead, the Ser228 hydroxyl hydrogen bonds with the Sia-1 9-hydroxyl that results in a small displacement of Sia-1 away from the 220-loop towards the 130-loop. Furthermore, the Glu190Val substitution removes a conserved polar interaction of avian receptor analogs with the 190-helix between Sia-1 and the Glu190 carboxyl.
Figure 3. Crystal Structures of the H6 HA in Complex with Avian Receptor Analogs.
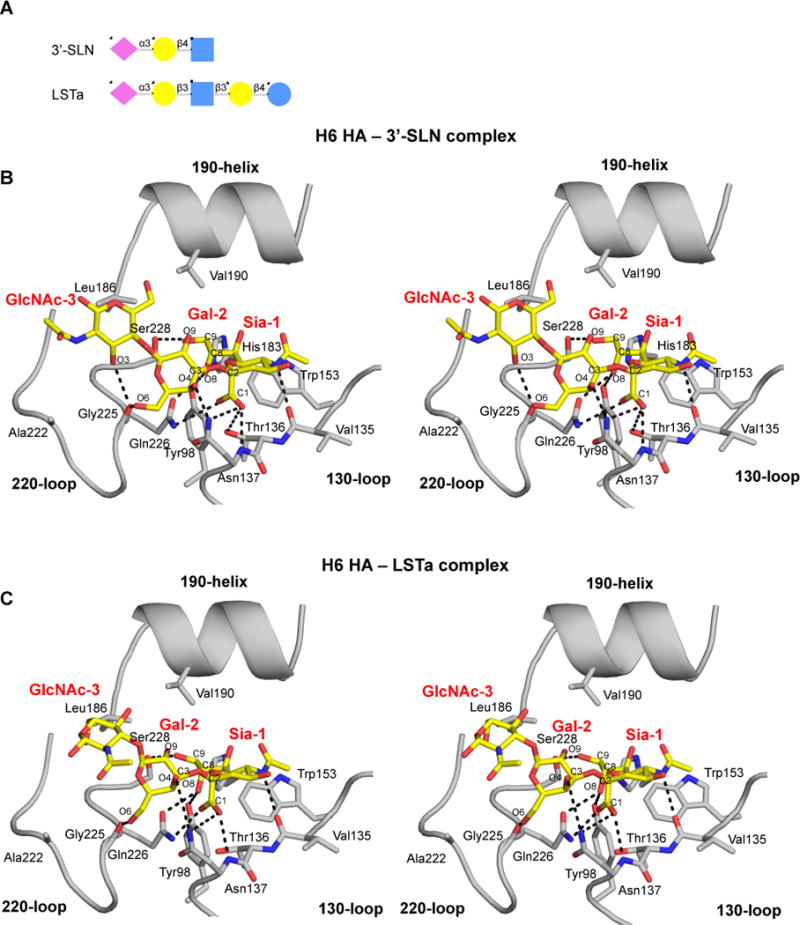
(A) Cartoon representation of the glycan structures of avian receptor analogs 3′-SLN and LSTa. Sia is the abbreviation for sialic acid, Gal for galactose, GlcNAc for N-acetylglucosamine and Glc for Glucose. (B and C) Stereo representations of the interactions between the HA H6 RBS and the avian receptor analogs. The conserved secondary elements of the HA RBS (130-loop, 190-helix and 220-loop) are labeled and shown in cartoon representation. Selected residues and receptor analogs are labeled and shown in sticks. The RBS is colored in gray and the receptor analogs in yellow. Hydrogen bond interactions of Sia-1 and Gal-2 and GlcNAc-3 of 3′-SLN (B) and LSTa (C) with the H6 HA RBS (see also Figures S3 and S4).
The second (Gal) and third (GlcNAc) sugars in 3′-SLN and LSTa exit the RBS above the 220-loop (Figure 3B, C). For avian HA subtypes, cis and trans conformations of Sia-Gal bond (C1Sia-C2Sia-O-C3Gal) of avian receptors are found (Figure S4A, B). Most analogs bind in a trans conformation that is stabilized by hydrogen bonding of Gal-2 to Gln226 and, in some structures, also to Glu190 (Lin et al., 2009; Liu et al., 2009; Xiong et al., 2013a; Xu et al., 2013). For ferret-transmissible H5 (Xiong et al., 2013a; Zhang et al., 2013) and human H7N9 isolates (Shi et al., 2013b; Xiong et al., 2013b; Xu et al., 2013), the cis conformation is stabilized by hydrogen bonding of the Gal-2 6-hydroxyl to the Gly225 main chain and by hydrophobic interaction between Gal-2 C3 and Leu226 (except for the A/Shanghai/1/2013 H7N9 HA L226Q mutant in complex with 3′-SLNLN where the hydrophobic face of Gal-2 makes hydrophobic interactions with other hydrophobic residues of the RBS (PDB entry 4LKG) (Shi et al., 2013b)). In addition, GlcNAc-3 makes a polar interaction with Lys/Gln222 (for H5 and H7 HAs respectively) (Figure S4).
In H6 HA, 3′-SLN and LSTa bind in a cis conformation, although no hydrophobic residue at position 226 or polar residue at position 222 is present (Gln226 and Ala222 in H6). The cis conformation similar to the conformation in receptor structures with HAs of ferret-transmissible H5 (Xiong et al., 2013a; Zhang et al., 2013) and human H7N9 isolates (Shi et al., 2013b; Xiong et al., 2013b; Xu et al., 2013) and stabilized by interaction of Gal-2 with Gly225 and by hydrogen bonding of Gal-2 O4 to Asn137 (Figure 3B, C). A similar interaction between Gal-2 and the 130-loop has only been reported for an avian analog complex with the transmissible H5 mutant (Zhang et al., 2013). For the 3′-SLN complex, a hydrogen bond is formed between the GlcNac-3 3-hydroxyl to the Gly225 main chain. In addition, hydrophobic interactions are made from GlcNAc-3 C8 to Arg227 CD in the 3′-SLN and from GlcNAc-3 C6 to Leu186 in the LSTa complex (Figure 3B, C).
Structural Characterization of Taiwan2 H6N1 HA in Complex with a Human Receptor Analog
Binding of human receptor analogs to the H6 HA is not detected on the glycan array. Although intrinsic differences in affinity for α2-3 and α2-6 linked sialosides are usually small, preferential binding to α2-3 linked sialosides over α2-6 linked sialosides is observed as a consequence of the avidity amplification resulting from the HA complex used in the experiment (Sauter et al., 1989), which mimics the multivalent interactions of the HAs on the viral surface. The very weak monovalent binding of α2-6 linked sialosides can be detected in the crystal soaked at high concentrations of ligand (5mM; HA:ligand stoichiometry of 1:55), helping to clarify the observed preference in receptor specificity (Xu et al., 2012). In the H6 HA structure with 6′-SLN at 2.2 Å (Figure 4 and Table S1), electron density was found for only two sugars, compared to three sugars observed with α2-3 linked analogs, consistent with the much weaker interaction of α2-6 sialosides (Figure 2). Hydrogen bonding interactions of Sia-1 with the RBS are largely conserved as in the 3′-SLN and LSTa complexes, except for the polar interaction between the Sia-1 carboxyl and Asn137 due to a side-chain rotamer change (Figure 4A) (Lin et al., 2009; Lin et al., 2012; Liu et al., 2009; Shi et al., 2013b; Xu et al., 2012). The difference in the phi angle may prevent a steric clash between Gln226 and Gal-2 C5 and enable hydrogen bond interactions between Gln226 and Gly225 and Gal-2 of 6′-SLN. Similar interactions with a human receptor analog were observed in an avian AH-H7N9 L226Q mutant with 6′-SLNLN (PDB entry 4LKK) (Xiong et al., 2013b). Furthermore, mutation of Leu226 to Gln results in changes in the phi and psi angles of the Sia-Gal linkage compared to the AH-H7N9 wild type. Consequently, Gal-2 hydrogen bonds with Gln226 and Gly225 main chain (Figure S4) (Xiong et al., 2013b). This conformation enables hydrogen bonds to be formed between the Gal-2 4-hydroxyl of 6′-SLN with Gln226 and Gly225 main-chain carbonyl (Figure 4B).
Figure 4. Crystal Structure of the H6 HA in Complex with a Human Receptor Analog.
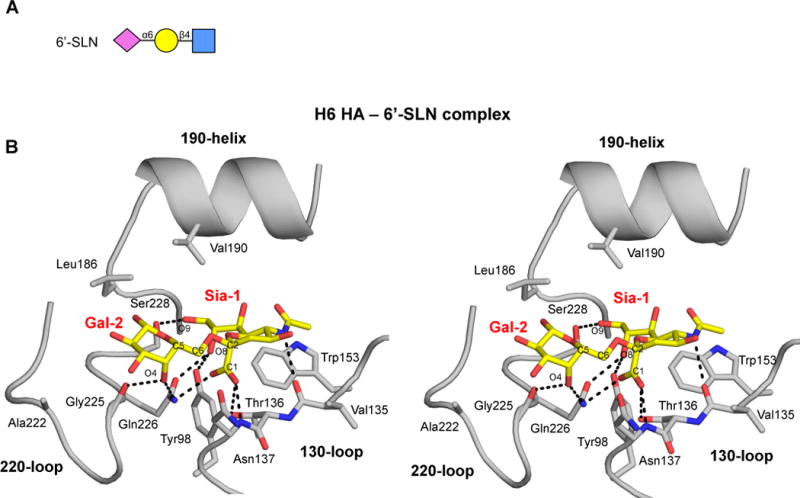
(A) Cartoon representation of the glycan structure of human receptor analog 6′-SLN. (B) Stereo representation of the hydrogen-bond interactions of Sia-1 and Gal-2 of 6′-SLN with the H6 HA RBS. The conserved secondary elements of the HA RBS (130-loop, 190-helix and 220-loop), as well as Tyr 98 and Trp153, are labeled and shown in cartoon representation (see also Figures S3 and S4).
DISCUSSION
The first H6 flu virus was isolated in 1965 from turkeys and since then from a broader range of avian species. Recent biological characterization of 256 H6 viruses from live poultry markets in China revealed that around 34% of H6N2 and H6N6 viruses could bind human-type receptors while maintaining an overall preference for avian receptors (Wang et al., 2014). In addition, most of these viruses can replicate in the respiratory system of mice and guinea pigs without preadaptation (Wang et al., 2014). Although the Gly228Ser mutation was not detected in these isolates, an Ala128Ser substitution, previously associated with human-like receptor binding, was present in a few viruses, but this mutation does not increase binding to human-type receptors (Wang et al., 2014).
Our structural and biological studies indicate that the A/Taiwan/2/2013 H6N1 virus isolated from an infected human retains the receptor binding properties of an avian virus. Although the H6N1 HA RBS maintains the overall architecture of other HAs, it contains unique substitutions or combinations of residues (Leu186, Val190, Ala222 and Ser228) that influence interaction with receptor analogs. Substitution of the highly conserved acidic residue at position 190 to Val in concert with acquisition of Leu186 increases the hydrophobicity of the 190-helix and precludes formation of hydrogen bonds between receptor analogs and the 190-helix. However, the less frequent cis conformation for avian receptors promotes interaction between Gal-2 of avian analogs and Asn137, thereby enhancing binding with the 130-loop, previously only seen for LSTa in complex with the HA of a transmissible H5 mutant (Zhang et al., 2013). The H6 crystal structure reveals weaker interaction with 6′-SLN (Figure 4 and Figure S3), as reflected by electron density for only two sugars and higher B values in the crystal structure for the ligand versus protein (ratio of the B-values for receptor homolog/ HA protein is 1.23 for 3′-SLN and 1.57 for LSTa compared to 1.72 for 6′-SLN). These structural studies of the H6 HA in complex with tri- and penta-saccharides therefore reveal the structural basis for the preferential specificity for avian versus human receptor analogs. However, other aspects required for transmission could also be impacted by additional characteristics of natural glycan receptors on human lung tissues that have still to be characterized.
In the last two years, an unprecedented number of zoonotic infections of avianorigin viruses have been detected in the human population (H7N9, H10N8 and H6N1) that may be a reflection of new zoonotic events, increased surveillance, or better detection methods and technologies, or a combination of all of these. Notwithstanding, these increasing numbers of zoonotic infections raise concern for the emergence of new pandemic viruses that contain these HA and NA subtypes (Xu et al., 2013; Vachieri et al., 2014; Zhang et al., 2015). Of primary concern is the potential of these viruses to switch their specificity from avian-type to human-type receptors and, as a result, potentially acquire the ability to transmit from human-to-human and thereby facilitate spread in the human population (de Graaf and Fouchier, 2014). Human H7N9 and H10N8 isolates have to date largely retained avian-type receptor specificity (Vachieri et al., 2014; Xu et al., 2013; Zhang et al., 2015) and here we show that the recently isolated H6N1 virus also retains avian-type receptor specificity. Receptor binding site mutations that switch specificity from avian-type to human-type receptors in the H1, H2 and H3 subtypes that transmit in humans are well documented (Connor et al., 1994; de Graaf and Fouchier, 2014; Stevens et al., 2006a). Furthermore, mutations that produce a receptor switch in the H5N1 virus have been demonstrated to increase transmission in ferrets (de Graaf and Fouchier, 2014; Imai et al., 2012). Notwithstanding, no clear rules have emerged for what it takes to switch receptor specificity for the other 14 HA subtypes (12 avian). Thus, until further insights and better methods for predicting receptor specificity are developed, constant surveillance is required to monitor any changes in avian and other zoonotic viruses that could increase their potential for transmission in the human population.
EXPERIMENTAL PROCEDURES
Expression and Purification of H6 in Insect Cells
The H6 HA cDNA of H6N1 A/Taiwan/2/13 [Global Initiative on Sharing All Influenza Data (GISAID) isolate ID: EPI_ISL_143275] was synthesized by Life Technologies (USA) and cloned into a pFastBac vector. Wild-type H6 HA was expressed in Hi5 insect cells with an N-terminal gp67 signal peptide, a C-terminal thrombin cleavage site, a foldon trimerization sequence, and a His6-tag and expressed as described previously (Stevens et al., 2006b). The expressed HA0 was purified via a His-tag affinity purification, dialyzed against 20 mM Tris-HCl pH 8.0, 100 mM NaCl, and then cleaved by trypsin (New England Biolabs, Ipswich, Massachusetts) to produce uniformly cleaved (HA1/HA2) and to remove the trimerization domain and His6-tag. The digested protein was purified further by gel filtration chromatography using a Superdex-200 column (Pharmacia). The HA protein eluted as a trimer and was concentrated to 5 mg/ml. Additional details, as well as expression of H1 and H5 HAs as controls, are given in the Supplemental Experimental Procedures.
Crystallization and Structural Determination of H6 HA
Crystals of H6 HA were obtained using the vapor diffusion sitting drop method (drop size 4 μl) at 20 °C against a reservoir solution containing 10mM NiCl2, 0.1M Tris pH 8.5, 20% (w/v) MPEG 2000 and 20% glycerol. Complexes with receptor analogs were obtained by soaking HA crystals in the reservoir solution that contained glycan ligands in a final concentration of 5 mM. Prior to data collection, the crystals were flash cooled in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source (APS) and at the Stanford Synchrotron Radiation Lightsource (SSRL). The H6 apo structure was solved by molecular replacement with an H1 HA structure (PDB entry 4M4Y) as a search model. The H6 HA apo structure then became the starting model for structure determination of the H6 HA-glycan complex structures.
Glycan Microarray Analysis of HAs Expressed in Insect and Mammalian Cells
Purified, soluble trimeric HA was pre-complexed with horseradish peroxidase (HRP)-linked anti-Strep-tag mouse antibody and with Alexa647-linked anti-mouse IgG (4:2:1 molar ratio) prior to incubation for 15 min on ice in 100 μl PBS-T, and incubated on the array surface in a humidified chamber for 90 minutes. Slides were subsequently washed and dried by centrifugation and immediately scanned for FITC signal. Fluorescent signal intensity was measured using Imagene and mean intensity minus mean background was calculated. Additional details are given in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
This work was funded in part by National Institutes of Health Grants R56 AI099275 (to I.A.W) and AI099274 (to J.C.P). RPdV is a recipient of a Rubicon grant from the Netherlands Organization for Scientific Research (NWO). We thank R. Stanfield, X. Dai and M. Elsliger for crystallographic and computational support, H. Tien of the Robotics Core at the Joint Center for Structural Genomics for automated crystal screening (supported by NIH Grant U54 GM094586), the staff at APS beamline 23ID-B (GM/CA CAT) and SSRL beamlines 11–1 and 12–2. GM/CA CAT is funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy (DOE), Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. The SSRL is a Directorate of Stanford Linear Accelerator Center National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. DOE Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, NIGMS (including P41GM103393) and the National Center for Research Resources (NCRR, P41RR001209). This is manuscript 28035 from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 4XKD for Taiwan2 H6 HA in apo form and 4XKE, 4XKF and 4XKG in complex with 3′-SLN, LSTa and 6′-SLN.
Supplemental Information includes four figures and three tables and can be found within this article online at http://dx.doi……..
AUTHOR CONTRIBUTIONS
Project design by N.T., R.P.dV., X.Z., J.C.P. and I.A.W.; X-ray structure determination and analysis by N.T., X.Z., and W.Y.; glycan array studies by R.P.dV. and R.M.; manuscript written by N.T., R.P.dV., X.Z., J.C.P. and I.A.W.
References
- Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci USA. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- Centers for Disease, C., and Prevention. Emergence of avian influenza A(H7N9) virus causing severe human illness – China, February–April 2013. MMWR Morb Mortal Wkly Rep. 2013;62:366–371. [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CL, Vijaykrishna D, Smith GJ, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Zhu X, McBride R, Rigter A, Hanson A, Zhong G, Hatta M, Xu R, Yu W, Kawaoka Y, et al. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol. 2014;88:768–773. doi: 10.1128/JVI.02690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen CM, Handeland K. Avian influenza virus screening in wild waterfowl in Norway, 2005. Avian Dis. 2007;51:425–428. doi: 10.1637/7555-033106R1.1. [DOI] [PubMed] [Google Scholar]
- Lee MS, Chang PC, Shien JH, Cheng MC, Chen CL, Shieh HK. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 2006;50:561–571. doi: 10.1637/7640-050106R.1. [DOI] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Wang G, Li A, Zhang Q, Wu C, Zhang R, Cai Q, Song W, Yuen KY. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci USA. 2012;109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci USA. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Stech J, Klenk HD. Influenza receptors, polymerase and host range. Rev Sci Tech. 2009;28:203–217. doi: 10.20506/rst.28.1.1870. [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- Myers KP, Setterquist SF, Capuano AW, Gray GC. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin Infect Dis. 2007;45:4–9. doi: 10.1086/518579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Raman R, Tharakaraman K, Shriver Z, Jayaraman A, Sasisekharan V, Sasisekharan R. Glycan receptor specificity as a useful tool for characterization and surveillance of influenza A virus. Trends Microbiol. 2014;22:632–641. doi: 10.1016/j.tim.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter NK, Bednarski MD, Wurzburg BA, Hanson JE, Whitesides GM, Skehel JJ, Wiley DC. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- Senne DA. Avian influenza in the Western Hemisphere including the Pacific Islands and Australia. Avian Dis. 2003;47:798–805. doi: 10.1637/0005-2086-47.s3.798. [DOI] [PubMed] [Google Scholar]
- Shi W, Shi Y, Wu Y, Liu D, Gao GF. Origin and molecular characterization of the human-infecting H6N1 influenza virus in Taiwan. Protein & Cell. 2013a;4:846–853. doi: 10.1007/s13238-013-3083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science. 2013b;342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- Shortridge KF. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nature Rev Microbiol. 2006a;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006b;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Suss J, Schafer J, Sinnecker H, Webster RG. Influenza virus subtypes in aquatic birds of eastern Germany. Arch Virol. 1994;135:101–114. doi: 10.1007/BF01309768. [DOI] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- Vachieri SG, Xiong X, Collins PJ, Walker PA, Martin SR, Haire LF, Zhang Y, McCauley JW, Gamblin SJ, Skehel JJ. Receptor binding by H10 influenza viruses. Nature. 2014;511:475–477. doi: 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- Wang G, Deng G, Shi J, Luo W, Zhang G, Zhang Q, Liu L, Jiang Y, Li C, Sriwilaijaroen N, et al. H6 influenza viruses pose a potential threat to human health. J Virol. 2014;88:3953–3964. doi: 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013a;497:392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013b;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86:982–990. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science. 2013;342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- Zhang H, de Vries RP, Tzarum N, Zhu X, Yu W, McBride R, Paulson JC, Wilson IA. A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.02.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Shi Y, Lu X, Shu Y, Qi J, Gao GF. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340:1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


