Abstract
Macroalgal surfaces support abundant and diverse microorganisms within biofilms, which are often involved in fundamental functions relating to the health and defense of their seaweed hosts, including algal development, facilitation of spore release, and chemical antifouling. Given these intimate and important interactions, environmental changes have the potential to negatively impact macroalgae by disrupting seaweed–microbe interactions. We used the disappearance of the dominant canopy-forming fucoid Phyllospora comosa from the metropolitan coast of Sydney, NSW, Australia as a model system to study these interactions. We transplanted Phyllospora individuals from nearby, extant populations back onto reefs in Sydney to test whether bacterial assemblages associated with seaweed surfaces would be influenced by (i) the host itself, independently of where it occurs, (ii) the type of habitat where the host occurs, or (iii) site-specific differences. Analyses of bacterial DNA fingerprints (terminal fragment length polymorphisms) indicated that assemblages of bacteria on Phyllospora were not habitat-specific. Rather, they were primarily influenced by local, site-specific conditions with some evidence for host-specificity in some cases. This could suggest a lottery model of host-surface colonization, by which hosts are colonized by ‘suitable’ bacteria available in the local species pool, resulting in high variability in assemblage structure across sites, but where some species in the community are specific to the host and possibly influenced by differences in host traits.
Keywords: seaweed-microbe interaction, biofilm, colonization, succession
Introduction
Marine macroorganisms live in persistent contact with diverse microorganisms that are abundant and ubiquitous in the surrounding seawater (Reinheimer, 1992) and within biofilms on their surfaces (Wahl, 1989). Although our understanding of the functional importance of biofilm-associated microorganisms in the lives of the higher organisms they live upon is still evolving (Egan et al., 2008), emerging evidence points to their fundamental involvement in the development, functioning, and defense of diverse macroorganisms (e.g., Armstrong et al., 2001; Lindquist et al., 2005; Wahl et al., 2012).
The phylogenetic structure of biofilm assemblages often has a high degree of organismal- (e.g., Longford et al., 2007), species- (e.g., Taylor et al., 2004), and tissue-specificity (e.g., Thiel et al., 2007; Campbell et al., 2011; Fernandes et al., 2012). More recent work has also provided evidence for functional redundancy within microbial consortia in biofilms (Burke et al., 2011a), where various combinations of phylotypes are capable of providing a core set of functions as required by the host (Burke et al., 2011b).
Functional redundancy within microbial biofilms could be beneficial to host organisms that rely on them for development, function, or defense, if a disturbance disrupts the composition of a biofilm, because lost functions could be restored by functionally (but not necessarily taxonomically) similar microbes available in the local species pool. Marine ecosystems are undergoing rapid change on a global scale, with rates of ocean warming higher than on land (Burrows et al., 2011), and worldwide increases in coastal development (Small and Nicholls, 2003; Bullieri and Chapman, 2010) leading to widespread degradation in coastal marine ecosystems (Jackson, 2001; Lotze et al., 2006; Airoldi and Beck, 2007). Environmentally mediated impacts have already affected diverse marine macroorganisms at multiple spatial scales, including species distributional range shifts (Parmesan, 2006), habitat fragmentation (Goodsell et al., 2007), higher incidences of disease (Harvell et al., 1999), and species extinctions (Sala and Knowlton, 2006). Evidence suggests that environmental change is also having profound and alarming effects on the abundance (Sarmento et al., 2010), distribution (Cook et al., 2011), and function (Wohlers et al., 2009) of planktonic marine microorganisms. However, we know almost nothing about how environmental change can affect the composition or function of epibiotic, biofilm-associated microorganisms (Wahl et al., 2012).
Much of the available information on environmental impacts on host–biofilm interactions comes from studies into coral holobionts (Mouchka et al., 2010). The composition (Ben-Haim et al., 2003; Bourne et al., 2007) and functional gene profile (Thurber et al., 2009) of coral-associated microbial assemblages can change with changes in the environment. Higher temperatures are often correlated with the detection of pathogenic strains within coral-associated microbial communities (Ben-Haim et al., 2003; Bourne et al., 2007), the production of lytic compounds (Ben-Haim et al., 2003), or the up-regulation of genes involved in pathogenicity (Thurber et al., 2009). The involvement of bacteria (and other microorganisms) in coral bleaching and subsequent coral reef decline is now a major focus of environmental microbiology, given the ecological and socio-economic importance of these ecosystems.
On temperate coasts, macroalgae are the dominant habitat-forming primary producers, playing analogous ecological roles to corals on tropical reefs (Steneck et al., 2002). Similar to corals, large, canopy-forming macroalgae are declining from many temperate rocky reefs (e.g., Steneck et al., 2002; Airoldi and Beck, 2007; Connell et al., 2008; Wernberg et al., 2011), but these systems receive far less attention than their tropical counterparts. Consequently, our understanding of the importance of bacteria to macroalgae and the effects of environmental change on macroalgal–bacterial interactions is less well developed (see Egan et al., 2012; Minich and Dinsdale, 2014). However, recent work highlights the importance of bacteria to the development (e.g., Marshall et al., 2006), function (e.g., de Oliveira et al., 2012), and defense (Egan et al., 2008) of some seaweeds. Like corals, there is evidence that seaweed-associated bacterial communities can be species-specific (Lachnit et al., 2009), fluctuate seasonally (Lachnit et al., 2011), and as a function of host condition (Campbell et al., 2011; Fernandes et al., 2012).
When investigating microbes associated with a green alga (Ulva australis) Burke et al. (2011a) found a high degree of within-species variability with respect to biofilm composition, but conservation of the functional gene profiles of microbial assemblages among samples. Their findings suggest macroalgae like U. australis rely on functions provided by their surface biofilms. Thus, any environmentally mediated disruption to the biofilm could have negative impacts for the macroalga, if lost functions cannot be restored rapidly. Although seasonally correlated changes in macroalgal-associated biofilms have been recorded (Lachnit et al., 2011) and some laboratory experiments have demonstrated that the composition of biofilms associated with macroalgae are affected by environmental factors [e.g., temperature, Stratil et al. (2013); and salinity, Stratil et al. (2014)], these ideas have not been tested experimentally in the field.
We experimentally investigated how changing an alga’s environment affected the composition of its surface biofilms, using a large, brown fucoid alga, Phyllospora comosa (hereafter ‘Phyllospora’). Like many other large canopy-forming macroalgae, Phyllospora is showing signs of decline and has disappeared from ca. Seventy kilometers of coastline adjacent to the metropolitan area of Sydney, NSW, Australia’s largest city, but remains dominant on shallow subtidal reefs north and south of the city (Coleman et al., 2008). The local disappearance of this species was linked to poor water quality in the region due to sewage pollution. Since the 1980s, sewage treatment has improved and deep ocean outfalls have been constructed, which has vastly enhanced water quality in the region (Scanes and Philip, 1995). Despite this improvement, Phyllospora has failed to recover and remains absent in this area. Genetic analyses of populations of Phyllospora north and south of this range fragmentation suggest a high degree of connectivity and genetic exchange across its gap in distribution. We hypothesized that environmental conditions adjacent to Sydney might affect Phyllospora-associated biofilms and that this may be one factor that has contributed to its failure to re-establish naturally in the region. To investigate this, we transplanted Phyllospora individuals from extant populations back onto reefs within the Sydney region and compared the bacterial communities that developed.
Materials and Methods
Field Experiments
To determine whether bacterial communities on P. comosa individuals were (i) host-specific (i.e., that all Phyllospora individuals will have similar communities, regardless of where they occur or are moved to); (ii) influenced by a particular environment (i.e., will change when they are moved from a ‘Phyllospora’ to a ‘non-Phyllospora’ habitat); or (iii) site-specific (i.e., they will change when they are moved to a particular place, regardless of whether it is a ‘Phyllospora’ habitat or not), we transplanted adults from two extant populations on the periphery of Sydney (donor habitats) into two physically similar reef habitats within metropolitan Sydney where Phyllospora has been absent for several decades (recipient habitats; Coleman et al., 2008; Campbell et al., 2014).
Full details of the experimental design and procedure can be found in Campbell et al. (2014) but briefly, the donor populations on the periphery of Sydney were in Cronulla (Cr; 34∘03′23′′ S 151∘09′23′′ E) and Palm Beach (PB; 33∘35′58′′ S 151∘19′43′′ E). Shallow rocky reefs at these places are characterized by a mosaic of patches of Phyllospora forests (size-range: 7–40 m2), barrens, turfing corallines, and ‘fringe’ habitats, with few individuals of the kelp Ecklonia radiata. The recipient habitats in Sydney were in Long Bay (LB; 33∘57′58′′ S 151∘15′27′′ E) and Cape Banks (CB; 33∘59′57′′ S 151∘14′52′′ E). Reefs at these recipient sites are very similar to those in donor places, except that patches of Ecklonia forests are more abundant and Phyllospora forests are absent. Collections and transplantations were carried out under a Scientific Collection Permit (# P00/0054-6.0) issued to the authors by the New South Wales Department of Primary Industries (Fishing and Aquaculture).
Experiments were done twice. In the first experiment (February 28 to May 9, 2011), 40 adults were collected haphazardly (collected individuals were typically 1–3 m apart) at the same depth (1–2 m) from each donor habitat by carefully detaching the holdfast from the substratum. Individuals were kept in 50 L containers with seawater for ∼2–3 hs during transportation until reattachment.
Phyllospora individuals from the two donor habitats were randomly allocated to one of three treatments (as per Campbell et al., 2014): (i) Transplanted individuals (‘TP’; n = 20), which were moved to one of the recipient habitats in Sydney (individuals from Cronulla were moved to Long Bay, while those from Palm Beach were moved to Cape Banks); (ii) Disturbed individuals (‘D’; n = 10), which were disturbed in the same manner as required for transplantation, but were returned to their original donor habitat; or (iii) Translocated individuals (‘TL’; n = 10), which were similarly disturbed, but were taken to the other donor site (i.e., an environment in which extant, natural populations of Phyllospora persist – from Cr to PB and vice versa). Undisturbed individuals (‘U’; n = 20) were haphazardly selected and marked in situ at each donor site but otherwise not handled. Disturbance and translocation treatments allowed for us to distinguish between the effects of transplantation to a different environment from the possible effects of the transplantation procedure or the effects of simply moving the algae from one place to another, regardless of environment (Marzinelli et al., 2009).
Algae that were removed from the substratum (TP, D, and TL individuals) were re-attached using cable-ties to 0.25 m2 plastic meshes, which were 0.5–2 m apart and had been previously attached to bare rock in barren patches approximately at 1–2 m depth. Five individuals were attached to each mesh to approximate natural densities (mean density 6.7 ± SE 1.1 per 0.25 m2), creating a patch of ∼4–5 m2 area at each place. After 2 months, one blade from each of 2–5 P. comosa individuals from each treatment at each location was sampled. Sampling size varied among treatments because several individuals were lost during the experiment. Blades were selected at random from the algae and cut approximately 30 cm from the tip of the blade. Each blade was sealed underwater in an individual plastic, press-seal bag. Blades were taken to the surface, rinsed with filter-sterilized seawater to remove any unattached epibionts (Millipore 0.2 μm filter) and using a sterile cotton swab, microbial assemblages from the algal surfaces were sampled (approximately 10 cm2 of thallus surface was gently swabbed for 30 s). The cotton tip of each swab was aseptically transferred into individual sterile 2.0 ml cryogenic storage tubes. Tubes were closed the flash frozen onsite in liquid nitrogen then stored at -80∘C until processing.
The experiment was repeated in late winter/spring (started August 9, 2011). In this second experiment, algae from both donor populations were transplanted to each recipient site to test for differences between algae from different sources at the same destination. Sixty algae were collected from each donor place (Palm Beach and Cronulla) and randomly assigned to three treatments: (i) individuals TP to Long Bay (n = 20 from each donor site), (ii) individuals TP to Cape Banks (n = 20 from each donor site), (iii) TL individuals to the other donor place (n = 20). U individuals (n = 20) were haphazardly selected and marked in situ (four sub-patches of five individuals each to resemble replication in the other treatments). Algae were attached to meshes as described above. Total patch-sizes ranged between 4 and 8 m2 at each place. Bacteria on blades from each of five individuals were sampled after 5 months (January 17, 2012) as described above.
DNA Fingerprinting of Phyllospora-Associated Bacterial Assemblages
To compare the composition of bacterial assemblages from Phyllospora in different treatments, we used a polymerase chain reaction (PCR) based DNA fingerprinting technique (terminal restriction fragment length polyorphisms [TRFLP; Liu et al., 1997]). TRFLP is popularly used by molecular ecologists to characterize and compare the composition and diversity of microbial communities (Walker et al., 2004), theoretically with a species resolution, but typically to the widely accepted ‘operational taxonomic unit’ (OTU; Nocker et al., 2007). To carry-out a TRFLP analysis, phylogenetic marker genes within the sample DNA were amplified using PCR with a fluorescent dye attached to the 5′ end of the forward primer. PCR products were then digested using restriction enzymes, which resulted in DNA fragments of variable length. These fragments were then physically separated in sequencing capillaries and the labeled terminal fragments were detected using a laser, producing an electropherogram. A size-standard labeled with a different fluorophore was also analyzed, allowing the fragment lengths to be estimated with a resolution of one lase pair (Liu et al., 1997; Nocker et al., 2007). Each OTU is represented by a different fragment length and thus the composition and diversity of the community can be estimated based on the polymorphism of the terminal restriction fragment lengths from a sample of community DNA. Swab samples were thawed on ice. DNA from each sample was extracted and isolated using the Powersoil DNA Isolation Kit (Mo Bio Laboratories #12888-100) then stored in a -20∘C freezer.
PCR and Fragment Analysis
A fragment (∼500 bp) of the bacterial 16S rRNA gene was amplified in each sample using the community DNA as a template. Primers 27F and 519R (sequences 5′-AGAGTTTGATCMTGGCTCAG-3′ and 5′-GWATTACCGCGGCKGCTG-3′, respectively), which encompass the 16S Variable regions V1–V3, were used. Primers were fluorescently labeled on the 5′ end with phosphoramidite dyes (27F labeled with 6-FAM and 519R labeled with VIC; Applied Biosystems).
The PCR reaction mixtures (25 μl) contained 5 pmol of the labeled 27F and 519R primers, 12.5 μl Econotaq 2x MasterMix (Lucigen), 3 μl of community template DNA, and molecular grade H2O. DNA amplification was performed with a PCR Express thermal cycler (Thermo Hybaid) using the following program: a 3 min start at 94∘C, 30 cycles consisting of 94∘C denaturation for 30 s, 56∘C annealing for 30 s, and 72∘C extension for 5 min. The program continued with a final extension at 72∘C for 5 min. Successful amplification was verified by gel-electrophoresis of 2 μl PCR products on 1% agarose gels with gel red, visualized under UV-light. PCR products were purified and concentrated using the DNA Clean and Concentration -5 Kit (Zymo Research, D4014). Purified product was eluted in 12 μl molecular grade H2O and quantified using a nano-drop ND1000 (Thermo Scientific).
Terminal fragment length polymorphisms was conducted using the restriction enzyme HAEIII (NE Biolabs) and standard methodology. Fragments were visualized by fluorophore color of 6-FAM (blue) and VIC (green). TRFLP data was first analyzed using Peak Scanner (Applied Biosystems). Fragment size was determined by comparison to internal size standard Liz-600, and fragments of <30 or >600 bp were excluded. Using the program T-REX (BMC Bioinformatics), background noise was removed to distinguish true fragments from background fluorescence.
Statistical Analyses
Bacterial TRFs data were compared among Phyllospora from different treatments using permutational multivariate analyses of variance (Anderson, 2001) with the PERMANOVA add-on in PRIMER v6 (Anderson et al., 2007). Similarity matrices based on Bray–Curtis distances of square-root transformed relative abundances or on Jaccard distances (presence/absence) were generated for the analyses, which used 9,999 permutations of residuals under a reduced model. Multivariate dispersion, which is an estimate of variance used to test for homogeneity among groups, was also compared among treatments (for both relative abundances and presence/absence of TRFs) using the PERMDISP analysis within the PERMANOVA add-on in PRIMER v6 (Anderson et al., 2007). P-values were calculated using 9,999 permutations. To visualize multivariate patterns in bacterial TRFs assemblages, non-metric multi-dimensional scaling (nMDS) was used as an ordination method using PRIMER v6 (Clarke and Gorley, 2006).
To test hypotheses about influences of the host, the environment, or the sites on surface-associated bacterial communities, we analyzed the data from the point of view of the origin of the algae and also their destination. Analyses from the point of view of the origin of the algae had two factors: Treatment, which was fixed with four levels (first experiment: U, D, TL, TP; second experiment: U, TL, TP to LB, TP to CB), and Place of origin, random, with two levels (Cr, PB). In addition, comparisons of surface-associated bacteria between algae that ended up in the same place of destination were also conducted. Analyses from the point of view of the destination of the algae in the first experiment had two factors: Treatment, fixed with three levels (U, D, TL), and Place of destination, random with two levels (Cr, Pb). Two analyses from the point of view of the destination were done for the second experiment. The first analysis was similar to that of the first experiment, with two factors: Treatment, fixed with two levels (U, TL), and Place of destination, random with two levels (Cr, Pb). The second analysis compared bacteria on algae originally from different donor populations that ended up in the same Sydney metro destination: treatment was a fixed factor with two levels (TP from PB, TP from Cr), and Place of destination was random with two levels (CB, LB). All analyses are detailed in each Table.
The model that surface-associated bacterial communities are influenced by the host leads to the prediction that the structure and composition of bacterial communities on translocated algae to sites within the Phyllospora habitat and on those transplanted to the Sydney metro habitat will not differ from undisturbed algae at the site of origin. Under the model of differences in the environment, we predict that bacterial communities on transplanted algae to the Sydney metro habitat will differ from those translocated to sites within the Phyllospora habitat, which will remain similar to undisturbed algae at the site of origin. Finally, the model that communities are site-specific leads to the prediction that the communities on translocated algae to sites within the Phyllospora habitat and on those transplanted to the Sydney metro habitat will differ from each other and from undisturbed algae at the site of origin, and will become similar to algae in the same place of destination.
Results
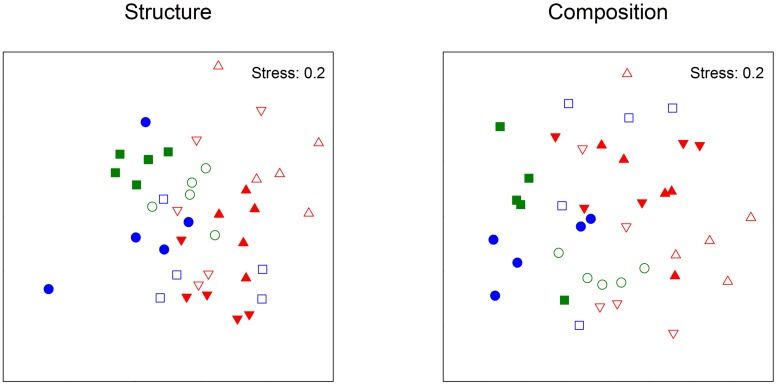
We detected a total of 795 microbial TRFs in this study. TRF sizes were searched against the RDP and SILVA databases using MICA (Shyu et al., 2007). No fragment was found to correspond with the predicted size of chloroplast or plastid 16S rRNA genes contained within the two databases. This indicates that the TRFLP profiles are not contaminated by host DNA or epiphytic algae and thus contain only bacterial or archaeal 16s rRNA gene information. In the first experiment, the structure and composition of bacterial fingerprints differed among treatments, but these differed according to the transplants place of origin (i.e., there was Treatment × Place interaction, Table 1A). Bacteria on algae transplanted from Palm Beach to Sydney metro differed from those on individuals that remained in Palm Beach undisturbed, on disturbed individuals returned to Palm Beach and on those individuals translocated to Cronulla. Although bacteria on translocated algae differed from those on undisturbed algae, there were no differences between the translocated and disturbed treatments (Table 1A). Algae originally from Cronulla had different bacterial TRFLP across all treatments (Table 1A; Figure 1).
Table 1.
PERMANOVAs based on Bray–Curtis (BC) similarity measure for square-root transformed relative abundances or Jaccard similarity measure (composition) of bacterial TRF profiles associated with Phyllospora in the first experiment from the point of view of (A) the origin of the algae or (B) the destination.∗
| Square-root transformed BC |
Jaccard |
||||||
| Source | df | MS | pseudo-F | p(perm.) | MS | pseudo-F | p(perm.) |
| (A) Origin | |||||||
| Treatment (Tr) | 3 | 2456 | 1.06 | 0.426 | 3149 | 0.95 | 0.569 |
| Place (Pl) | 1 | 1608 | 2.13 | 0.002 | 2462 | 1.68 | 0.004 |
| Tr × Pl | 3 | 2326 | 3.08 | <0.001 | 3324 | 2.27 | <0.001 |
| Residual | 26 | 756 | 1463 | ||||
| Pairwise tests | PB: TP ≠ TL = D ≠ U Cr: TP ≠ TL ≠ D ≠ U |
PB: TP ≠ TL = D ≠ U Cr: TP ≠ TL ≠ D ≠ U |
|||||
| (B) Destination | |||||||
| Tr | 2 | 2041 | 1.35 | 0.291 | 2728 | 1.23 | 0.330 |
| Pl | 1 | 2272 | 3.13 | < 0.001 | 3942 | 2.74 | < 0.001 |
| Tr × Pl | 2 | 1517 | 2.09 | 0.001 | 2226 | 1.55 | 0.002 |
| Residual | 18 | 725 | 1440 | ||||
| Pairwise tests | PB: TL = D ≠ U Cr: TL ≠ D ≠ U |
PB: TL = D ≠ U Cr: TL = U ≠ D |
|||||
∗(A) Treatment was fixed with four levels (U, Undisturbed; D, Disturbed; TL, Translocated; TP, Transplanted), Place of origin was random with two levels (CR, Cronulla; PB, Palm Beach). (B) Treatment was fixed with three levels (U, D, TL), Place of destination was random with two levels (Cr, PB). The replicates were the Phyllospora individuals (n = 2–5). P-values were calculated using 9,999 permutations under a reduced model. Bold indicates statistical significance (at alpha = 0.005).
FIGURE 1.
nMDS based on Bray-Curtis measure of square-root transformed relative abundances (structure) or Jaccard measure (composition) of bacterial TRFLP on Phyllospora comosa originally from Cronulla (empty symbols) or Palm Beach (filled symbols) in the first experiment. Treatments: Undisturbed (green symbols), Disturbed (black symbols), Translocated (blue symbols), Transplanted (red symbols). Destination places: Cronulla (circles), Palm Beach (squares), Long Bay (downward triangle), Cape Banks (upward triangle).
When we compared bacterial fingerprints from the point of view of the site of destination of transplants, algae that ended up in PB differed from those that ended up in Cr (Table 1B). At each destination place, bacteria on translocated algae were, however, different from those on undisturbed algae, except at PB where translocated algae originally from Cr did not differ from disturbed algae (which originated in Palm Beach; Table 1B). There were no differences in dispersion of the structure or composition of bacterial TRFs among treatments (PERMDISP: F3,30 = 1.92, p = 0.24; F3,30 = 1.18, p = 0.49, respectively; Figure 1).
In the second experiment, bacterial TRFs on algae originally from Palm Beach differed across all treatments. The same trend was observed for bacteria on algae originally from Cronulla, although pairwise comparisons could not resolve where these differences occurred and hence there was a significant Treatment × Place interaction (Table 2; Figure 2).
Table 2.
PERMANOVAs based on BC similarity measure for square-root transformed relative abundances or Jaccard similarity measure (composition) of bacterial TRFs on Phyllospora in the second experiment from the point of view of the origin.∗
| Square-root transformed BC |
Jaccard |
||||||
|---|---|---|---|---|---|---|---|
| Source | df | MS | pseudo-F | p(perm.) | MS | pseudo-F | p(perm.) |
| Tr | 3 | 4048 | 1.85 | 0.076 | 4539 | 1.55 | 0.093 |
| Pl | 1 | 2302 | 2.41 | 0.001 | 3512 | 2.01 | < 0.001 |
| Tr × Pl | 3 | 2192 | 2.29 | <0.001 | 2933 | 1.68 | <0.001 |
| Residual | 32 | 956 | 1746 | ||||
| Pairwise tests | PB: TP–CB ≠ TP–LB ≠ TL ≠ U Cr: TP–CB ≠ TL ≠ U TP–LB ≠ TP–CB ≠ TL U = TP–LB |
PB: TP–CB ≠ TP–LB ≠ TL ≠ U Cr: TP–CB ≠ TL ≠ U TP–LB ≠ TP–CB ≠ TL U = TP–LB |
|||||
∗Treatment was fixed with four levels (U, Undisturbed; TL, Translocated; TP–CB, Transplanted to Cape Banks; TP–LB, Transplanted to Long Bay), Place of origin was random with two levels (Cr, Cronulla; PB, Palm Beach). The replicates were the Phyllospora individuals (n = 5). P-values were calculated using 9,999 permutations under a reduced model. Bold indicates statistical significance (at alpha = 0.005).
FIGURE 2.
nMDS based on Bray-Curtis measure of square-root transformed relative abundances (structure) or Jaccard measure (composition) of bacterial TRFLP on Phyllospora comosa originally from Cronulla (empty symbols) or Palm Beach (filled symbols) in the second experiment. Treatments: Undisturbed (green symbols), Translocated (blue symbols), Transplanted (red symbols). Destination places: Cronulla (circles), Palm Beach (squares), Long Bay (downward triangle), Cape Banks (upward triangle).
Bacterial TRFs on algae that ended up in PB differed from those that ended up in Cr (Table 3A). At each destination place, however, bacteria on translocated algae remained different from those on undisturbed algae (Table 3A). Bacteria on transplanted algae also differed between destination places in Sydney metro; however, bacteria on algae transplanted from Palm Beach differed from those on co-occurring algae transplanted from Cronulla at both destination places (Table 3B; Figure 2). There were no differences in dispersion of the structure or composition of bacterial TRFs among treatments (PERMDISP: F3,36 = 1.63, p = 0.28; F3,36 = 1.29, p = 0.34, respectively).
Table 3.
PERMANOVAs based on BC similarity measure for square-root transformed relative abundances or Jaccard similarity measure (composition) of bacterial TRF profiles associated with Phyllospora at their destination place in (A) donor populations or (B) Sydney metro in the second experiment.∗
| Square-root transformed BC |
Jaccard |
||||||
|---|---|---|---|---|---|---|---|
| Source | df | MS | pseudo-F | p(perm.) | MS | pseudo-F | p(perm.) |
| (A) | |||||||
| Tr | 2744 | 1.40 | 0.254 | 3578 | 1.32 | 0.259 | |
| Pl | 2929 | 3.20 | < 0.001 | 3732 | 2.19 | < 0.001 | |
| Tr × Pl | 1966 | 2.15 | 0.004 | 2702 | 1.58 | 0.004 | |
| Residual | 914 | 1706 | |||||
| Pairwise tests | PB: TL ≠ U Cr: TL ≠ U |
PB: TL ≠ U Cr: TL ≠ U |
|||||
| (B) | |||||||
| Tr | 2327 | 2.35 | 0.004 | 3126 | 1.74 | 0.006 | |
| Pl | 4288 | 4.33 | < 0.001 | 4987 | 2.79 | < 0.001 | |
| Tr × Pl | 879 | Pooled | 1875 | Pooled | |||
| Residual | 997 | 1786 | |||||
∗(A) Treatment was fixed with two levels (U, TL), Place of destination was random with two levels (Cr, PB). (B) Treatment was fixed with two levels (TP from Palm Beach or Cronulla), Place of destination was random with two levels (CB, LB). The replicates were the Phyllospora individuals (n = 5). P-values were calculated using 9,999 permutations under a reduced model. Non-significant interaction terms with p > 0.25 were pooled. Bold indicates statistical significance (at alpha = 0.005).
Discussion
To our knowledge, this is the first field manipulation of hosts which assesses impacts of environmental change on microbial communities associated with a large, habitat-forming macroalga. We found that in most cases, Phyllospora-associated microbial communities were more strongly and consistently affected by local conditions (i.e., were site-specific) than the type of environment or habitat they occurred in (i.e., ‘Phyllospora’ vs. ‘non-Phyllospora’ habitat). In the first experiment, algae that were moved from Palm Beach seemed to respond to a change of habitat (into the Sydney region) rather than to a specific site, but this pattern was not observed in individuals moved from Cronulla in the same experiment, or those from either site in the second experiment, despite some handling or ‘disturbance’ effects. This high degree of site-specificity (and lack of consistent ‘environment/habitat’ effect) suggests that simply moving algae to a different place independent of the habitat, will result in changes of their bacterial communities. We also found some evidence for host-specificity in Phyllospora-associated biofilms: algae translocated from one site to another did not adopt communities similar to undisturbed individuals at the destination site. Furthermore, communities on transplanted individuals with different origins that ended-up at the sample place, still supported different communities even after 5 months.
The lack of consistency in microbial communities associated with Phyllospora across all treatments and sites in our study does not agree with some previous studies, which compared different algal hosts and found consistent, species-specific microbial communities among places, and seasons (Lachnit et al., 2009). However, Lachnit et al. (2009, 2011) used a method with lower resolution than TRFLP fingerprinting (denaturing gradient gel electrophoresis; DGGE). Although in many studies, TRFLP and DGGE fingerprinting techniques yield similar results (Smalla et al., 2007; Campbell et al., 2011; Fernandes et al., 2012), TRFLP is a more sensitive technique (Moeseneder et al., 1999; Enwall and Hallin, 2009), and so smaller differences will be better detected in fingerprints generated by this method than DGGE, particularly in ecological studies with multiple factors.
Furthermore, Lachnit et al. (2009) compared microbial communities from several algal species. Differences among species may outweigh site- or treatment-specific differences within a single species. We did not include an ‘out-group’ (i.e., comparison with a different species or water-borne microorganisms), in part because such comparisons in our experience – particularly when using surrounding seawater as the comparator – reveal such large differences as to be uninformative with respect to the macroalgal communities (Burke et al., 2011b and below). However, comparisons with other algae could have provided a larger conceptual scale against which to compare Phyllospora individuals from different sites and/or treatments, and such comparisons are currently underway.
In contrast, Burke et al. (2011b) compared microbial communities from replicate samples of the green alga U. australis occurring in separate rock pools at a single site and also compared algal-associated communities to those within the surrounding water column, by creating and sequencing sophisticated 16S rRNA gene clone libraries. As well as finding almost no similarity between algal-associated and water-borne microbial communities, they detected a very high degree of variability among communities associated with algal samples from different rock pools (despite the ‘out-group’ comparison with water samples). They proposed that microorganisms could colonize algal surfaces using a ‘competitive lottery model,’ in which multiple species could colonize algal surfaces, so long as a core set of functions (rather than a core phylogeny) was represented within the microbial consortia. Indeed, there was a high degree of conservation of functional gene profiles expressed by the microbial communities from replicate algal samples (Burke et al., 2011a). Interestingly, previous work on the same system using the lower-resolution technique DGGE, suggested that a temporally and spatially consistent, species-specific community was present on U. australis (Tujula et al., 2010). In the latter study, a core set of OTUs was always present, despite some spatial and temporal variation. Our results suggest that Phyllospora may require a core set of functions from its biofilm, rather than a core set of ‘species’ (or OTUs) and that local, site-specific conditions rather than habitat-type will influence the phylogenetic composition of Phyllospora’s biofilm.
However, because translocated individuals did not come to resemble undisturbed individuals and furthermore, because transplanted individuals from different origins did not conform to a ‘destination site-specific’ community, this suggests some degree of host-specificity – that is, microbial communities are influenced by more than simply the site in which they occur (or to which they are moved). This may simply be an issue of timing: longer than 5 months may be required for convergence of microbial assemblages to occur in some cases. This seems unlikely, however, given the relatively rapid rates of colonization and succession in microbial communities compared to macroorganisms. Alternatively, the starting conditions of the biofilm may influence microbial succession such that multiple, alternative stable states (rather than just one) are possible, given a set of environmental conditions and host traits. Such concepts have been widely discussed and tested in the field of classical ecology (reviewed by McCook, 1994; Young et al., 2001) and are beginning to feature in the study of microbial ecology as well (Burke et al., 2011a). The fact that there was some degree of host-specificity suggests that a component of the community may be influenced by differences in host traits. Host genetic variation can strongly influence the structure of microbiomes associated with plants (e.g., Bulgarelli et al., 2013) and animals, including mice (Benson et al., 2010) and humans (Arumugam et al., 2011). Our results suggest that Phyllospora may also have a core microbiome that can be influenced by local, site-specific conditions, and host traits.
Because crayweed remains absent from the Sydney region, we hypothesized that the environment in Sydney may impact seaweed-associated microbial communities, which may, in turn, have contributed to its failure to recover in the region. We found no consistent evidence that the environment within Sydney had any significant impact on Phyllospora-associated microbes. Previously, we reported that individuals transplanted from extant populations to these recipient sites within the Sydney region had survivorship rates comparable to those in natural populations (Campbell et al., 2014). Together with these observations, this suggests that environmental conditions within Sydney are now suitable to support Phyllospora again. Furthermore, they suggest that individuals transplanted to or recruiting naturally onto reefs in Sydney will not develop a ‘Sydney-specific’ biofilm, rather they will adopt a biofilm specific to the local conditions (whether it is within or outside of the Sydney region).
It is possible, however, that at the time of its disappearance, the habitat within Sydney had a negative impact on Phyllospora’s interactions with its biofilm. Then, a high volume of poorly treated sewage was released adjacent to the shorelines where this species was dominant (Coleman et al., 2008). Ecotoxicological experiments with Phyllospora suggest that it is physiologically sensitive to high levels of nutrients (Burridge et al., 1995). The production of metabolites and other natural products by macroalgae strongly influences the composition and maintenance of biofilms on their surfaces (Egan et al., 2012). Thus an environmentally mediated change in physiology (e.g., due to high levels of pollution) might have indirectly altered Phyllospora’s surface-associated microbial community, which may have contributed to is decline. Understanding the role of microorganisms in the health of important, habitat-forming organisms is essential to understand the processes that affect their persistence, and to inform efforts to restore populations of these organisms if they decline. The emergence of molecular techniques to rapidly (and with increasing affordability) assess the composition of microbial assemblages associated with these organisms and even characterize their functional gene profiles, facilitates the inclusion of microbes into more ecological studies.
Most studies on holobionts to date have shown either strong host-specificity of surface-associated microbial communities, suggesting that hosts require a core set of specific taxa, or high variability of surface-associated microbial communities, suggesting that hosts may be colonized by taxonomically distinct bacteria available in the local species pool and where a core set of specific functions is more important for the host than a core set of specific taxa. Our results suggest that a combination of both processes influence bacteria on the Phyllospora holobiont. Thus, although some component of the community may vary across sites depending on the available taxa, other components of the community may be driven by specific traits of the host.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Torsten Thomas for assistance help with TRF analyses and the volunteers who helped with the fieldwork component of this project, which was funded by the Centre for Marine Bio-Innovation and the Evolution and Ecology Research Centre at the University of New South Wales. This is contribution # 149 from the Sydney Institute of Marine Science.
References
- Airoldi L., Beck M. W. (2007). “Loss, status and trends for coastal marine habitats of Europe,” in Oceanography and Marine Biology Vol. 45 eds Gibson R. N., Atkinson R. J. A., Gordon J. D. M. (Boca Raton, FL: Crc Press–Taylor & Francis Group) 345–405. [Google Scholar]
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26 32–46. [Google Scholar]
- Anderson M. J., Gorley R. N., Clarke K. R. (2007). Permanova+ for Primer: Guide to Software and Statistical Methods. Plymouth: Primer-E. [Google Scholar]
- Armstrong E., Yan L., Boyd K. G., Wright P. C., Burgess J. G. (2001). The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461 37–40 10.1023/A:1012756913566 [DOI] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473 174–180 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y., Zicherman-Keren M., Rosenberg E. (2003). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69 4236–4242 10.1128/AEM.69.7.4236-4242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Kelly S. A., Legge R., Ma F., Low S. J., Kim J., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107 18933–18938 10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D., Iida Y., Uthicke S., Smith-Keune C. (2007). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2 350–363 10.1038/ismej.2007.112 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Bullieri F., Chapman M. G. (2010). The introduction of coastal infrastructure as a driver of change in marine environments. J. Appl. Ecol. 47 26–35 10.1111/j.1365-2664.2009.01751.x [DOI] [Google Scholar]
- Burke C., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2011a). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. U.S.A. 108 14288–14293 10.1073/pnas.1101591108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke C., Thomas T., Lewis M., Steinberg P. D., Kjelleberg S. (2011b). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5 590–600 10.1038/ismej.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge T. R., Lavery T., Lam P. K. (1995). Acute toxicity tests using Phyllospora comosa (Labillardiere) C. Agardh (Phaeophyta: Fucales) and Allorchestes compressa Dana (Crustacea: Amphipoda). Bull. Environ. Contam. Toxicol. 55 621–628 10.1007/BF00196045 [DOI] [PubMed] [Google Scholar]
- Burrows M. T., Schoeman D. S., Buckley L. B., Moore P., Poloczanska E. S., Brander K. M., et al. (2011). The pace of shifting climate in marine and terrestrial ecosystems. Science 334 652–655 10.1126/science.1210288 [DOI] [PubMed] [Google Scholar]
- Campbell A. H., Harder T., Nielsen S., Kjelleberg S., Steinberg P. D. (2011). Climate change and disease: bleaching in a chemically-defended seaweed. Glob. Chang. Biol. 17 2958–2970 10.1111/j.1365-2486.2011.02456.x [DOI] [Google Scholar]
- Campbell A. H., Marzinelli E. M., Vergés A., Coleman M. A., Steinberg P. D. (2014). Towards restoration of missing underwater forests. PLoS ONE 9:e84106 10.1371/journal.pone.0084106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R., Gorley R. N. (2006). PRIMER v6: User Manual/Tutorial. Plymouth: PRIMER-E. [Google Scholar]
- Coleman M. A., Kelaher B. P., Steinberg P. D., Millar A. J. K. (2008). Absence of a large brown macroalga on urbanised reefs around Sydney, Australia and evidence for historical decline. J. Phycol. 44 897–901 10.1111/j.1529-8817.2008.00541.x [DOI] [PubMed] [Google Scholar]
- Connell S. D., Russell B. D., Turner D. J., Shepherd S. A., Kildea T., Miller D., et al. (2008). Recovering a lost baseline: missing kelp forests from a metropolitan coast. Mar. Ecol. Prog. Ser. 360 63–72 10.3354/meps07526 [DOI] [Google Scholar]
- Cook K., Vanderklift M. A., Poor A. G. B. (2011). Strong effects of herbivorous amphipods on epiphyte biomass in a temperate seagrass meadow. Mar. Ecol. Prog. Ser. 442 263–289 10.3354/meps09446 [DOI] [Google Scholar]
- de Oliveira L. S., Gregoracci G. B., Silva G. G. Z., Salgado L. T., Filho G. A., Alves-Ferreira M., et al. (2012). Transcriptomic analysis of the red seaweed Laurencia dendroidea (Florideophyceae, Rhodophyta) and its microbiome. BMC Genomics 13:487 10.1186/1471-2164-13-487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2012). The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37 462–476 10.1111/1574-6976.12011 [DOI] [PubMed] [Google Scholar]
- Egan S., Thomas T., Kjelleberg S. (2008). Unlocking the diversity and biotechnological potential of marine surface-associated microbial communities. Curr. Opin. Microbiol. 11 219–225 10.1016/j.mib.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Enwall K., Hallin S. (2009). Comparison of T-RFLP and DGGE techniques to assess denitrifier community composition in soil. Lett. Appl. Microbiol. 48 145–148 10.1111/j.1472-765X.2008.02498.x [DOI] [PubMed] [Google Scholar]
- Fernandes N., Steinberg P. D., Rusch D., Kjelleberg S., Thomas T. (2012). Community structure and functional gene profile of bacteria on healthy and diseased thalli or the red seaweed Delisea pulchra. PLoS ONE 7:e50854 10.1371/journal.pone.0050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell P. J., Chapman M. G., Underwood A. J. (2007). Differences between biota in anthropogenically fragmented habitats and in naturally patchy habitats. Mar. Ecol. Prog. Ser. 351 15–23 10.3354/meps07144 [DOI] [Google Scholar]
- Harvell C. D., Kim K., Burkholder J. M., Colwell R. R., Epstein P. R., Grimes D. J., et al. (1999). Emerging marine diseases – climate links and anthropogenic factors. Science 285 1505–1510 10.1126/science.285.5433.1505 [DOI] [PubMed] [Google Scholar]
- Jackson J. B. C. (2001). What was natural in the coastal oceans? Proc. Natl. Acad. Sci. U.S.A. 98 5411–5418 10.1073/pnas.091092898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T., Blumel M., Imhoff J. F., Wahl M. (2009). Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Auquat. Biol. 5 181–186. [Google Scholar]
- Lachnit T., Meske D., Wahl M., Harder T., Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13 655–665 10.1111/j.1462-2920.2010.02371.x [DOI] [PubMed] [Google Scholar]
- Lindquist N., Barber P. H., Weisz J. B. (2005). Endosymbiotic microbes as food and defence for marine isopods: unique symbioses in a hostile environment. Proc. R. Soc. B 272 1209–1216 10.1098/rspb.2005.3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Marsh T. L., Cheng H., Forney L. J. (1997). Characterisation of microbial diversity by determining terminal restriction fragemnt length polymorphisms of genes encoding 16S RNA. Appl. Environ. Microbiol. 63 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longford S. R., Tujula N. A., Crocetti G. R., Holmes A. J., Holmstrom C., Kjelleberg S., et al. (2007). Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 48 217–229 10.3354/ame048217 [DOI] [Google Scholar]
- Lotze H. K., Lenihan H. S., Bourque B. J., Bradbury R. H., Cooke R. G., Kay M. C., et al. (2006). Depletion, degradation, and recovery potential of estuaries and coastal seas. Science 312 1806–1809 10.1126/science.1128035 [DOI] [PubMed] [Google Scholar]
- Marshall K., Joint I., Callow M., Callow J. (2006). Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb. Ecol. 52 302–310 10.1007/s00248-006-9060-x [DOI] [PubMed] [Google Scholar]
- Marzinelli E. M., Zagal C. J., Chapman M. G., Underwood A. J. (2009). Do modified habitats have direct or indirect effects on epifauna? Ecology 90 2948–2955 10.1890/08-1893.1 [DOI] [PubMed] [Google Scholar]
- McCook L. J. (1994). Understanding ecological community succession: causal models and theories, a review. Vegetatio 110 115–147 10.1007/BF00033394 [DOI] [Google Scholar]
- Minich J., Dinsdale E. (2014). “Experimental metagenomics: influence of pulses of carbon dioxide on Kelp forest microbial ecology,” in Encyclopedia of Metagenomics ed.Nelson K. E.(New York: Springer; ), 1–8. [Google Scholar]
- Moeseneder M. M., Arrieta J. M., Muyzer G., Winter C., Herndl G. J. (1999). Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient Gel electrophoresis. Appl. Environ. Microbiol. 65 3518–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchka M. E., Hewson I., Harvell C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50 662–674 10.1093/icb/icq061 [DOI] [PubMed] [Google Scholar]
- Nocker A., Burr M., Camper A. (2007). Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54 276–289 10.1007/s00248-006-9199-5 [DOI] [PubMed] [Google Scholar]
- Parmesan C. (2006). Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37 637–669 10.1146/annurev.ecolsys.37.091305.110100 [DOI] [Google Scholar]
- Reinheimer G. (1992). Aquatic Microbiology, 4th Edn. New York: Wiley. [Google Scholar]
- Sala E., Knowlton N. (2006). Global marine biodiversity trends. Annu. Rev. Environ. Resour. 31 93–122 10.1146/annurev.energy.31.020105.100235 [DOI] [Google Scholar]
- Sarmento H., Montoya J. M., Vázquez-Domínguez E., Vaqué D., Gasol J. M. (2010). Warming effects on marine microbial food web processes: how far can we go when it comes to predictions? Philos. Trans. R. Soc. B Biol. Sci. 365 2137–2149 10.1098/rstb.2010.0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanes P. R., Philip N. (1995). Environmental impact of deepwater discharge of sewage off Sydney, NSW, Australia. Mar. Pollut. Bull. 31 343–346 10.1016/0025-326X(96)81926-6 [DOI] [Google Scholar]
- Shyu C., Soule T., Bent S. J., Foster J. A., Forney L. J. (2007). MICA: a Web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA. J. Microb. Ecol. 53 562–570 10.1007/s00248-006-9106-0 [DOI] [PubMed] [Google Scholar]
- Small C., Nicholls R. J. (2003). A global analysis of human settlement in coastal zones. J. Coastal Res. 19 584–599. [Google Scholar]
- Smalla K., Oros-Sichler M., Milling A., Heuer H., Baumgarte S., Becker R., et al. (2007). Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: do the different methods provide similar results? J. Microbiol. Methods 69 470–479 10.1016/j.mimet.2007.02.014 [DOI] [PubMed] [Google Scholar]
- Steneck R. S., Graham M. H., Bourque B. J., Corbett D., Erlandson J. M., Estes J. A., et al. (2002). Kelp forest ecosystems: biodiversity, stability, resilience and future. Environ. Conserv. 29 436–459 10.1017/S0376892902000322 [DOI] [Google Scholar]
- Stratil S. B., Neulinger S. C., Knecht H., Friedrichs A. K., Wahl M. (2013). Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2 338–349 10.1002/mbo3.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratil S. B., Neulinger S. C., Knecht H., Friedrichs A. K., Wahl M. (2014). Salinity affects compositional traits of epibacterial communities on the brown macroalga Fucus vesiculosus. FEMS Microbiol. Ecol. 88 272–279 10.1111/1574-6941.12292 [DOI] [PubMed] [Google Scholar]
- Taylor M. W., Schupp P. J., Dahllof I., Kjelleberg S., Steinberg P. D. (2004). Host specificity in marine sponge-associated bacteria and potential implications for marine microbial diversity. Environ. Microbiol. 6 121–130 10.1046/j.1462-2920.2003.00545.x [DOI] [PubMed] [Google Scholar]
- Thiel V., Neulinger S. C., Staufenberger T., Schmaljohann R., Imhoff J. F. (2007). Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol. Ecol. 59 47–63 10.1111/j.1574-6941.2006.00217.x [DOI] [PubMed] [Google Scholar]
- Thurber R. V., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R. A., Angly F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11 2148–2163 10.1111/j.1462-2920.2009.01935.x [DOI] [PubMed] [Google Scholar]
- Tujula N. A., Crocetti G. R., Burke C., Thomas T., Holmström C., Kjelleberg S. (2010). Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4 301–311 10.1038/ismej.2009.107 [DOI] [PubMed] [Google Scholar]
- Wahl M. (1989). Marine epibiosis. I. Fouling and antifouling: some basic aspects. Mar. Ecol. Prog. Ser. 58 175–189 10.3354/meps058175 [DOI] [Google Scholar]
- Wahl M., Goecke F., Labes A., Dobretsov S., Weinberger F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292 10.3389/fmicb.2012.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. M., Spencer J. F., Ragout de Spencer A. L. (2004). Environmental Microbiology: Methods and Protocols. Totowa, NJ: Humana Press. 10.1385/1592597653 [DOI] [Google Scholar]
- Wernberg T., Russell B. D., Thomsen M. S., Gurgel C. F. D., Bradshaw C. J. A., Poloczanska E. S., et al. (2011). Seaweed communities in retreat from ocean warming. Curr. Biol. 21 1828–1832 10.1016/j.cub.2011.09.028 [DOI] [PubMed] [Google Scholar]
- Wohlers J., Engel A., Zöllner E., Breithaupt P., Jürgens K., Hoppe H.-G., et al. (2009). Changes in biogenic carbon flow in response to sea surface warming. Proc. Natl. Acad. Sci. U.S.A. 106 7067–7072 10.1073/pnas.0812743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. P., Chase J. M., Huddleston R. T. (2001). Community succession and assembly: comparing, contrasting and combining paradigms in the context of ecological restoration. Ecol. Restor. 19 5–18. [Google Scholar]