Abstract
Lactation is a dynamic process, which evolved to meet dietary demands of growing offspring. At the same time, the mother's metabolism changes to meet the high requirements of nutrient supply to the offspring. Through strong artificial selection, the strain of milk production on dairy cows is often associated with impaired health and fertility. This led to the incorporation of functional traits into breeding aims to counteract this negative association. Potentially, distributing the total quantity of milk per lactation cycle more equally over time could reduce the peak of physiological strain and improve health and fertility. During lactation many factors affect the production of milk: food intake; digestion, absorption, and transportation of nutrients; blood glucose levels; activity of cells in the mammary gland, liver, and adipose tissue; synthesis of proteins and fat in the secretory cells; and the metabolic and regulatory pathways that provide fatty acids, amino acids, and carbohydrates. Whilst the endocrine regulation and physiology of the dynamic process of milk production seems to be understood, the genetics that underlie these dynamics are still to be uncovered. Modeling of longitudinal traits and estimating the change in additive genetic variation over time has shown that the genetic contribution to the expression of a trait depends on the considered time-point. Such time-dependent studies could contribute to the discovery of missing heritability. Only very few studies have estimated exact gene and marker effects at different time-points during lactation. The most prominent gene affecting milk yield and milk fat, DGAT1, exhibits its main effects after peak production, whilst the casein genes have larger effects in early lactation. Understanding the physiological dynamics and elucidating the time-dependent genetic effects behind dynamically expressed traits will contribute to selection decisions to further improve productive and healthy breeding populations.
Keywords: time-dependent, longitudinal, lactation curve, breeding value, genome-wide association, genomic selection, genomic prediction
Introduction
Lactation is an orchestrated process aimed at providing nutrition and immune protection to the offspring; however, the mother must also retain sufficient resources to ensure her own survival. Thus, the quantity and composition of milk produced is strongly dependent on the developmental stage of the offspring and the maintenance requirements of the mother. As such, milk production is a classic exemplar of a time-dependent dynamic process.
The domestication of animals inevitably led to selective breeding for increased productivity. The uninterrupted increasing global demand for dairy products necessitated a concurrent increase in milk production. Thus, in order to meet market requirements, the dairy sector implemented selective breeding programs which have led to a doubling in the amount of milk produced per cow over the last 50 years, such that total milk production is increasing despite a decline in dairy cattle populations (Food and Agriculture Organization of the United Nations, FAO, 20121). Recently, this has included the implementation advanced breeding programs and the development of tools to utilize genetic and genomic information (Goddard and Hayes, 2007; Seefried et al., 2010). However, increasing the milk production per cow has detrimental effects on animal health and fertility (Ingvartsen et al., 2003; Oltenacu and Broom, 2010). Consequently, breeding goals were adjusted to incorporate health and fertility traits into breeding indices (Osteras et al., 2007; Boichard and Brochard, 2012).
These breeding indices have enabled dairy farmers to breed for milk production and functional traits without requiring knowledge on how these practices impact upon the dynamics of milk production or change the expression of underlying genes. However, the continuous development of genetic and genomic tools, as well as computational capacities, will allow breeders of the future to base their decisions not only on phenotypically observable traits or indirect genetic marker information but also on the direct causative genetic variants.
As with many other complex traits important in livestock production, milk production is influenced by many genetic loci that act directly, interact with each other and/or interact with the environment (Lemay et al., 2009). This makes the study of quantitative traits challenging, especially when time-dependent components are considered. This review details the most important regulators of milk production and their underlying genes in the context of the dynamics of a lactation cycle, and summarizes the efforts made to identify genetic loci affecting the dynamics of milk production during lactation.
Conflict between production and functional traits
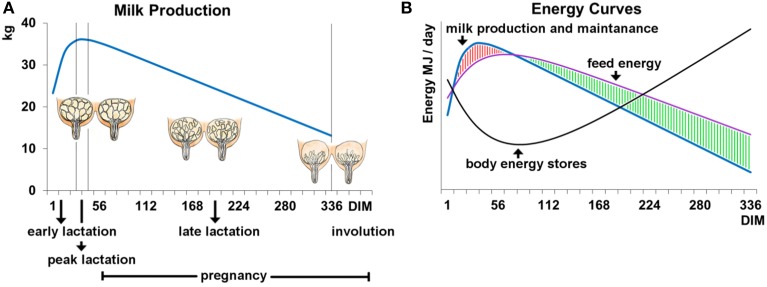
The milk production of a cow follows a dynamic curve (Figure 1A; Stanton et al., 1992). After an initial rapid increase in milk yield during early lactation, milk yield (as well as protein and fat content) peak around 6 weeks into lactation, after which production slowly decreases until the end of lactation. Dairy cows experience an energy deficiency during early and peak lactation (Figure 1B; Collard et al., 2000) due to the high energy requirements for milk production not being met because of physiological limitations which constrain food intake (i.e., bulk capacity; Allen, 1996) and mobilization of bodily energy resources. This energy deficit has been proposed to have detrimental effects on health and fertility which have been reviewed and discussed by Oltenacu and Broom (2010), and negative genetic correlations have been reported between milk production and a variety of functional traits (Zimmermann and Sommer, 1973; Dekkers et al., 1998; Ingvartsen et al., 2003; Muir et al., 2004). However, it has to be noted that total milk yield and the energy balance during early lactation seem to be independent, as correlations have been reported to be very low (Spurlock et al., 2012). Further, the negative impact of lactation on fertility may serve a functional purpose to provide optimal birth spacing for the survival of offspring. Therefore, there may be other endogenous factors yet to be discovered that negatively affect health and fertility traits.
Figure 1.
(A) Milk production and (B) energy supply and requirements during the lactation cycle of 340 days. (A) The curve represents the milk yield per day of lactation and reaches a peak production around lactation days 40–50. Shortly before lactation and until peak production the udder and the alveolar system are highly developed. In later lactation the alveolar system regresses continuously until the end of lactation and into involution. (B) The blue curve represents the energy that is needed for milk production and maintenance of vital body functions. The energy needed for milk production is highest when milk production reaches a peak. At the same time the energy taken in through food (purple curve) cannot cover the energy requirements for milk production which leads to a loss in body energy stores (black curve). This imbalance in energy homeostasis changes with the decline of milk production in late lactation.
From a nutritionist's point of view it might be necessary to reduce, rather than to increase, peak milk yield in order to decrease the energy deficiency experienced during early and peak lactation, and thereby improve health and fertility traits. However, this is in direct conflict with the desire to increase overall milk production. Therefore, an alternative method of increasing overall milk production might be via increasing production persistency. A better production persistency raises the overall gain per lactation due to an increased persistency affecting the longest part of the lactation (i.e., late lactation; Dekkers et al., 1998; Inchaisri et al., 2011). However, there are some reports indicating that a high persistency may also be antagonistic to the animal's health, and thus also needs to be considered in regards to finding an optimal persistency and lactation duration (Harder et al., 2006; Appuhamy et al., 2009).
Production persistency is most often defined as a lesser decrease in milk production after the peak, i.e., a flatter shape of the lactation curve compared to another animal or the heard average. Such calculations can be based on the difference of peak yield to a 305d measurement, on test-day deviations, or on parameters of lactation curve models (Gengler, 1996). As persistency is negatively correlated to yields, some studies prefer to calculate persistency as a linear regression of test-day deviations on days in milk to achieve a yield independent estimate (Cole and VanRaden, 2006; Cole and Null, 2009). By employing such an estimate, it would enable a breeder to select on milk yield and persistency independently; however, currently only very few breeding companies provide such estimates to their clients.
One problem with persistent production is the requirement to dry-off a cow between lactations. However, if the production system does not require yearly calving, the duration of the lactations can be chosen according to daily yield. Subsequently, with increased lactation duration, the time point of insemination has to be postponed. Assuming that the peak production remains around 6 weeks into the lactation cycle, a later time point for insemination has the added benefit that a new pregnancy begins after the energy deficit caused by the high peak production. Thus, fertility issues potentially arising from an energy deficit will be reduced. Regardless of lactation duration, the general recommendation for days dry is still 45–70 days (Kuhn et al., 2006, 2007; Sawa et al., 2012). The potential implications of increased lactation duration on generation intervals and fewer replacement animals could be counteracted through the utilization of sexed semen to increase the ratio of female calves.
Current methods in animal breeding apply an index of traits weighted according to their economic importance as well as heritability in the breeding goal. Further, phenotypic and genetic correlations between traits within the index are included, on the one hand to increase accuracy on lowly heritable traits, and on the other hand to account for potential negative correlations (Dekkers, 2007).Whilst milk production is still the most important trait in most countries, conformation, udder health, and fertility have been added to balance the negative correlation between a high production and the animal's welfare and longevity (VanRaden, 2004; Miglior et al., 2005). However, the exact impact of such breeding indices on the shape of the lactation curve or the dynamic gene effects remains unknown. In the following section, we look at the physiological interplay that forms the lactation cycle as this is the basis of understanding which genetic factors are ultimately involved.
Physiology of a dynamic milk production
Mammogenesis
The development of the mammary gland is the primary factor affecting milk production. A well-developed mammary gland with many fully differentiated secretory cells, good blood supply, and strong connective tissue will be highly productive over a long time.
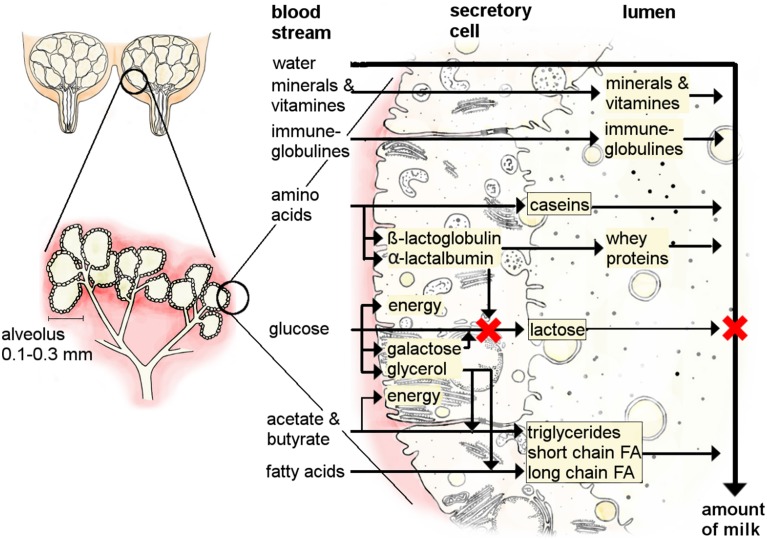
The mammary gland forms a rudimentary duct tree during fetal development in response to maternal hormones (Watson and Khaled, 2008). From birth until puberty, mammary gland growth is due to the formation of a fat-pad rather than the development of specialized mammary gland tissue (McNally and Martin, 2011). At puberty the initiation of the estrus cycle, via follicle-stimulating hormones and luteinizing hormone, stimulates the ovaries to synthesize and release estrogen and progesterone. The concurrent elevations in both estrogen and progesterone orchestrate the main growth of the mammary gland during pregnancy by ductal growth and lobular formation which leads to the formation of lobule-alveoli (Hennighausen and Robinson, 2005; Bloise et al., 2010; Koos, 2011). Alveoli are an accumulation of secretory cells grouped around a hollow center, the lumen, where the milk is stored (Figure 2). Thus, as pregnancy progresses, the adipose cells of the mammary gland are gradually replaced by specialized mammary gland tissue. Mammary gland growth continues during early lactation until peak lactation, after which the mammary gland shrinks due to the rate of secretory cell loss exceeding the rate of cell division (Figure 1A; Capuco and Akers, 1999).
Figure 2.
The process of milk secretion in the udder of a cow (partially adapted from Wattiaux, 1996). Milk is secreted in the alveoli system of the mammary gland. Several substances can pass the cell membrane from the blood stream (water, minerals, vitamins, immune-globulins), whilst others need transporters and are produced in the secretory cells (proteins, fat, lactose).
Hormones and growth factors are important in determining how many secretory cells develop, and thus how much milk can be produced in the mammary gland (Watson and Khaled, 2008; McNally and Martin, 2011). By slowing down the process of hormonal stimulation of secretory cell proliferation during late pregnancy and early lactation, and favoring an extended time during which new cells are produced, the peak milk production could be reduced and a better persistency achieved. This may also be achieved by slowing down the rate of cell death which is also regulated through a cascade of hormones and growth factors (Sureshbabu et al., 2011; Watson et al., 2011).
Milk secretion
A second crucial point for milk production concerns the quantity and quality of the secreted milk. Milk is an emulsion of fat and water containing dissolved carbohydrates, proteins, vitamins, and minerals that all have to be produced in or transported to the mammary gland. During lactation, quantitative milk yield is primarily regulated by lactose within the alveoli. Alveolar lactose influences the osmotic pressure between blood and alveoli and thereby the amount of water drawn into the alveoli (Figure 2; Zhao and Keating, 2007). Some of the substances in milk such as minerals, vitamins, or immune-globulins pass the cell membranes directly from the blood into the lumen via transporter proteins (Figure 2; Neville and Watters, 1983). The activity of these transporter proteins is increased when milk production starts to enhance the uptake of water into the secretory cells of the mammary gland (Figure 3; Zhao and Keating, 2007; Anantamongkol et al., 2010; Wickramasinghe et al., 2012). Substances such as lactose, proteins and fat have to be synthesized in the secretory cells from components such as glucose, amino acids, triglycerides, or fatty acids that stem from the dietary nutrients or body resources such as adipose tissues or skeletal muscles (Figures 2, 3; Burgoyne and Duncan, 1998; Zhao and Keating, 2007; Bionaz and Loor, 2008b). Lactose is synthesized from blood glucose and galactose (synthesized from glucose) by a lactose synthase enzyme composed of galactosyltransferase and α-lactalbumin in the golgi complex of mammary secretory cells (Figure 2). The amount of glucose in the blood is regulated by energy intake, insulin and leptin (Figure 3; Li et al., 2010).
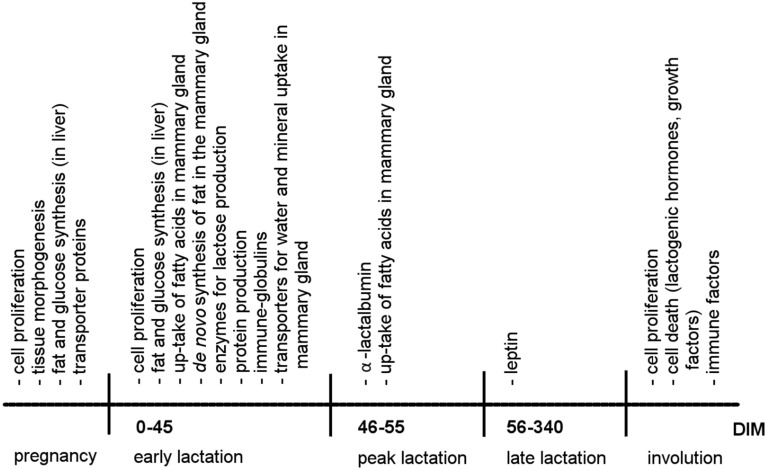
Figure 3.
Chronology of gene expression and physiological processes during a lactation cycle. DIM, days in milk.
Proteins and fat are important for qualitative milk yield in terms of organoleptic properties of the milk and downstream industries such as cheese and butter production (Bailey et al., 2005; Bauman et al., 2006). Caseins, α-lactalbumin and β-lactoglobulin represent the main fraction of milk proteins. They are synthesized mainly from amino acids broken down from digested food and transported through the blood stream to the secretory cells (Burgoyne and Duncan, 1998). Milk fat is composed of triglycerides, long- and short-chain fatty acids which are partly synthesized in the liver or in secretory cells of the mammary gland from short-chain dietary lipids that are obtained from the rumen, and partly from mobilized fats from bodily fat depots (Figure 2; Bionaz and Loor, 2008b).
Lactation is coupled with changes in the activity of genes in the mammary gland but also in other organs. In the liver, fat and glucose synthesis is highly increased from pregnancy to early lactation to provide fatty acids and blood glucose for milk production (Figure 3; Bell and Bauman, 1997; Casey et al., 2009), whereas fat synthesis is decreased in adipose tissue and the expression of transporter genes for the uptake of blood glucose into somatic cells is reduced to ensure that nutrients are available for milk production (Bell and Bauman, 1997; Casey et al., 2009).
In conclusion, to understand the genetics behind a lactation cycle, a number of gene pathways need to be considered. These include genes regulating food intake and blood glucose levels; the digestion, absorption, and transportation of nutrients; the activity of the secretory cells in the mammary gland, liver, and adipose tissue; the synthesis of proteins and fat in the secretory cells; and the pathways which provide triglycerides, fatty and amino acids, transporter proteins, and transcription factors.
Genetics of milk production
The establishment of the Bos taurus genome assembly (Bovine HapMap et al., 2009), along with proteome and gene expression studies, have made it possible to estimate the number of genes involved in milk production, from mammogenesis to milk secretion. Between 6000 and 19,000 genes distributed across all 29 bovine autosomes and the X-chromosome have been reported to be differentially expressed during the lactation cycle, though not exclusively in the mammary gland (Lemay et al., 2009; Wickramasinghe et al., 2012). Thus, the genes predicted to be involved (directly or indirectly) in the regulation of milk production, account for between 25 and 75% of all predicted cattle genes (Bos taurus UMD 3.1-Primary Assembly, Zimin et al., 2009). Most genes contribute to pathways that directly affect economically important traits such as milk yield and composition. A multitude of genome-wide association studies (GWAS) using high density SNP chip data have previously been conducted to narrow down regions and identify causative genes that affect milk production traits (Cole et al., 2011; Strucken et al., 2012a; Buitenhuis et al., 2014; Raven et al., 2014). Whilst regions and potential genes with effects on milk production traits have been reported for almost all bovine chromosomes, repeatedly occurring genes are located on chromosomes 27, 6, 20, and 14 (Lemay et al., 2009).
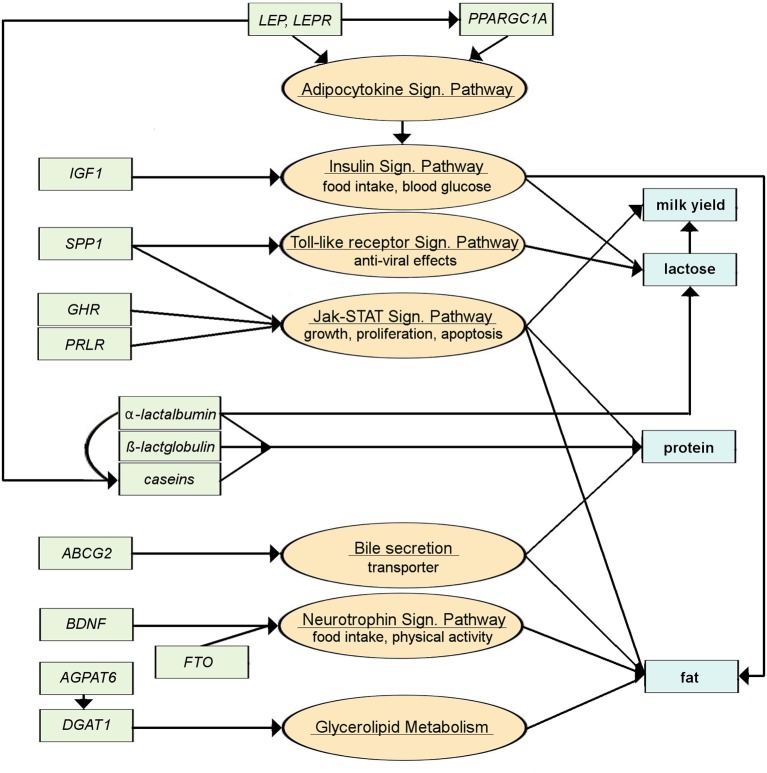
Only around a dozen candidate genes have been consistently identified between studies and described more extensively with regards to their association with the main milk production traits (Table 1). The pathways through which these genes affect milk production traits depict the variety of processes that have to be considered (Figure 4). Genes like the BDNF, FTO, or IGF1 impact upon food intake and thus nutrient and energy availability (Mullen et al., 2011; Zielke et al., 2011, 2013; Waters et al., 2012). Other genes such as GHR, PRLR, or SPP1 affect growth, proliferation, and apoptosis of cells (Viitala et al., 2006; Khatib et al., 2007; Banos et al., 2008; Lu et al., 2011a; Rahmatalla et al., 2011), whilst DGAT1 and AGPAT6 are involved directly in triglyceride synthesis (Winter et al., 2002; Bionaz and Loor, 2008a; Strucken et al., 2010a; He et al., 2011). Of further note are the casein genes which encode the major fraction of milk proteins (Velmala et al., 1995). Figure 4 provides an overview of those candidate genes and the pathways through which they affect milk production traits. To our knowledge, no genes affecting mammogenesis have been directly linked to milk production. Recently, Raven et al. (2014) included traits of the mammary system in a GWAS study which identified five regions on four different chromosomes with significant effects; however, a clear description of the phenotype (the mammary system) was lacking.
Table 1.
Major genes involved in milk production.
| Gene | Chr. | Position (bp)* | Trait | References |
|---|---|---|---|---|
| LEPR (leptin receptor) | 3 | 80,071,689–80,147,000 | Milk yield Milk fat | Banos et al., 2008 |
| LEP (leptin) | 4 | 93,249,874–93,266,624 | Milk yield Milk fat | Banos et al., 2008; Clempson et al., 2011 |
| IGF1 (insulin like growth factor 1) | 5 | 66,532,879–66,604,699 | Milk yield Milk fat | Mullen et al., 2011; Waters et al., 2012 |
| ABCG2 (ATP-binding cassette, sub-family G, member 2) | 6 | 37,959,536–38,030,585 | Milk protein Milk fat | Cohen-Zinder et al., 2005; Ron et al., 2006 |
| OPN (osteopontin) | 6 | 38,120,578–38,127,541 | Milk yield Milk protein Milk fat | Leonard et al., 2005; Khatib et al., 2007 |
| PPARGC1A (peroxisome proliferator-activated receptor gamma, coactivator 1 alpha) | 6 | 44,854,113–44,960,533 | Milk yield Milk fat | Khatib et al., 2007 |
| Casein-Cluster (CSN1S1, CSN2, CSN1S2, CSN3) | 6 | 87,141,556–87,392,750 | Milk protein | Velmala et al., 1995; Kress et al., 2011 |
| DGAT1(diacylglycerol O-acyltransferase 1) | 14 | 1,795,425–1,804,838 | Milk yield Milk fat | Winter et al., 2002; Strucken et al., 2010a |
| BDNF (brain-derived neurotrophic factor) | 15 | 59,164,519–59,200,908 | Milk fat | Zielke et al., 2011 |
| FTO (fat mass and obesity associated) | 18 | 22,118,201–22,541,540 | Milk fat Milk protein | Zielke et al., 2013 |
| GHR (growth hormone receptor) | 20 | 31,890,736–32,064,200 | Milk yield Milk protein Milk fat | Viitala et al., 2006; Banos et al., 2008; Rahmatalla et al., 2011 |
| PRLR (prolactin receptor) | 20 | 39,073,246–39,137,480 | Milk yield Milk protein Milk fat | Bole-Feysot et al., 1998; Viitala et al., 2006; Lu et al., 2011b |
| PRL (prolactin) | 23 | 35,105,135–35,113,750 | Milk yield Milk protein Milk fat | Bole-Feysot et al., 1998 |
| AGPAT6 (1-acylglycerol-3-phosphate O-acyltransferase 6) | 27 | 36,212,352–36,228,987 | Milk yield Milk fat | Bionaz and Loor, 2008a; He et al., 2011 |
Btau_4.6.1.-Primary Assembly.
Figure 4.
Simplified pathways for major genes involved in milk production. Green boxes are genes, orange circles are the pathways the genes are involved in, blue boxes are the milk production traits that are affected (information is assembled from KEGG Pathway Database, 17.11.2014; http://www.genome.jp/kegg/pathway.html and literature review; for gene names, see Table 1).
Only little is known in regards to time-dependent genetic effects causing a dynamic curve in dairy cattle but the next section summarizes the efforts and results made in this field.
Dynamic genes in animal breeding systems
Dynamic association studies
Whether a single marker for a candidate gene is used or thousands of indirect markers for a GWAS, finding associations between markers and a trait that displays dynamic expression over time can be difficult. The simplest solution may be to estimate associations over various time-points, i.e., treat each measurement as a separate phenotype. Automated milking systems could provide an accurate measurement of milk production for each day of lactation. However, this approach would mean that several hundred phenotypes would have to be analyzed. Further, whilst such measurements would provide daily milk yield, persistency cannot be estimated from a single time point. Ergo, daily measurements should not be treated as separate phenotypes. Therefore, appropriate phenotypic and genetic correlations have to be incorporated or repeated measurement analyses performed. Whilst daily measurements provide a highly accurate description of lactation performance, it might be computationally too time-consuming to be practically applied. Further, milking systems have still not penetrated the entire dairy sector and analyses solely relying on daily measurements would require additional methods to include animals with missing records. Most countries with national evaluation networks record milk production once a month. Assuming the lactation period of a cow lasts for 340 days, one record a month sums up to approximately 11 test-days. Because crucial changes such as peak yield occur roughly 6 weeks into the lactation cycle, one analysis every month could still give a fairly thorough picture of the lactation performance. However, analyzing 50k or 800k markers (the marker number of the most commonly used SNP-chip in dairy cattle at present) for thousands of animals would still take time.
Instead of using the measurements of the actual test-days, fewer parameters can be sufficient to describe an entire lactation. The profile of milk production, and its components, over the course of a single lactation has been described by various mathematical and biological functions (Pollott, 2004; Silvestre et al., 2009). Thus, using these mathematical lactation curve models provides a means of reducing the amount of time-points to a minimum of three curve parameters. These parameters describe the production curve through its properties such as slopes, apex (maximum), and level of production.
Such approaches are known as functional modeling in human genetics where it is mostly applied to map dynamic loci affecting disease traits using growth curves such as cubic splines (Hou et al., 2008; Li et al., 2009; Yang et al., 2009). In livestock research, a similar approach is known as the modeling of longitudinal or dynamic traits (Rodriguez-Zas et al., 2002; Suchocki and Szyda, 2011). In most livestock studies, the change in additive genetic variation over time was analyzed mainly for body weight and milk yield in dairy cattle, sheep, and goats (Lund et al., 2008; Roldan et al., 2008; Forni et al., 2009; Hadjipavlou and Bishop, 2009; Strucken et al., 2011). However, most of these studies used either no marker information or only a few markers on selected chromosomes to conduct their analyses.
The few results of time-dependent association studies in livestock reflect reported dynamic expressions of genes involved in milk production (Bionaz and Loor, 2008a; Verbyla and Verbyla, 2009) or add a time component to known but static effects of candidate genes such as the DGAT1 gene or the region around the casein genes. The described effects of the DGAT1 gene, with antagonistic impacts on milk yield and fat content, were shown to be detectable only after lactation day 40 (Strucken et al., 2011). This late effect points to a possible utilization of DGAT1 in changing the persistency of milk production. Markers around the casein genes had strongest effects in early lactation (Strucken et al., 2012b), which is confirmed by the higher protein content in colostrum milk. Furthermore, investigations of the genes surrounding trait-associated markers showed that a substantial number of genes with stronger effects in early lactation are involved in immune response and not directly in milk production (Strucken et al., 2012b). Even though those genes have no direct effect on milk production, immune-related genes could influence the productivity of the animal by supporting udder health in a time of high activity (Wheeler et al., 2012; Chaneton et al., 2013) and through effects on food intake (Greer et al., 2008; Laurenson et al., 2011). This adds another group of genes that have to be considered when genetic influences on milk production traits are analyzed.
In general, the highest variation in associated loci were reported for early and late lactation, suggesting that those time periods provide the best opportunity for alteration through breeding schemes. This would also serve the idea of decreasing peak production through a slower increase in early lactation and increase the persistency of production in late lactation. Furthermore, by analyzing marker associations over time, we are more likely to find genetic markers with small effects over the whole lactation but strong effects at a specific time-point as they are not masked by major candidate genes such as DGAT1. Thus, time-dependent analyses could aid in detecting the missing genetic variance that explains the observed phenotypic variation.
Finally, differences in genetic effects were not only found for different lactation stages but also between lactations, especially between the first and later lactations (Strucken et al., 2012a). These differences between the first and later lactations are also observed in phenotypic production curves (Schmidt et al., 1988). Even though most cows are in puberty and have reached a sufficient weight and size to support a pregnancy at the age of first mating, first parity cows are still growing and the mammary gland undergoes the required changes to produce milk for the first time (Taylor et al., 2003, 2004). Therefore, this ongoing development during the first parity is most likely the reason for the lower performance compared to later lactations.
Applications
In animal breeding, the ability of an animal to improve a trait in the next generation can be summarized using an estimated breeding value (EBV). The current standard for breeding value estimation is to include the animal's own performance records as well as the records of relatives, assuming that related individuals share a certain amount of genes with each other. In milk production, obviously a bull does not produce milk, and therefore its EBV is entirely dependent on milk production records of female relatives.
Because milk production is routinely recorded once a month in most countries, EBVs are based on these monthly test-day data. To account for the fluctuation of milk yield throughout a lactation, test-day models have been developed through the incorporation of appropriate lactation curve models (Misztal et al., 2000; Schaeffer et al., 2000; Swalve, 2000). Whilst some countries provide separate EBVs for persistency, most production EBVs are averaged over 305 days of lactation or even an average over several lactations. Therefore, the final selection decision is still based on a static value that makes it impossible to tell whether the animal had a high peak production or a good persistency. Though it should be easy for the national breeding evaluation centers to provide breeding values for certain time-periods (estimation equations implement lactation curve model), this would also increase the information output that needs to be handled and might even complicate the decision process about which animal should be used for mating.
One possible solution could be to include the shape of the production curve into the selection index and set a standard curve based on how much milk a cow can produce without inducing an energy deficiency under natural feeding conditions, and how much milk a cow should produce to be still profitable for the farmer. Based on such a standard curve, breeding values could be weighted according to their deviation from the standard curve resulting in a single EBV per animal.
A similar approach could be applied for genomically estimated breeding values (gEBVs) where the information of the genome-wide markers along with the production records of all relatives are included. gEBVs seem to be the way forward as they use the genetic constitution of the animal itself, and thus, what is actually inherited from generation to generation (Goddard and Hayes, 2007; Hayes et al., 2009; Hayes and Goddard, 2010). Nevertheless, gEBVs would require knowledge of either an optimal standard curve or the exact time-dependent genetic effects. Knowing the genetic effect of a marker enables us to simply genotype a selection candidate and sum up its genetic effects to calculate a gEBV, provided that the animals that were used to estimate the genetic effects are closely related to the selection candidate. If the actual causal mutation is known then family relations can be neglected.
Most of the reviewed studies on time-dependent genetic effects, applied a GWAS approach where the effects of each marker were estimated independently from all other markers. However, it is assumed that a quantitative trait such as milk production is shaped through the activity of many genes that might affect and even depend on each other. Thus, marker effects should not be estimated independently of all other markers in a study. A method which includes all markers at the same time has been termed Snp-BLUP, which is an extension of the original BLUP (best linear unbiased prediction) equation used to estimate the EBV of an animal (Goddard, 2009; Koivula et al., 2012).
Information obtained through dynamic GWAS or Snp-BLUP would make it possible to weight each marker according to its effects on the dynamic expression of the trait at different time-points, and thus provide a gEBV that includes the shape of the production curve. With the reducing cost of sequencing, causal genomic variants may be discovered and ultimately used in animal breeding to perform the most accurate selection possible.
Concluding remarks
Milk production is a dynamic process and factors influencing this process occur as early as the fetal development. Whilst many physiological aspects of a dynamic milk production have been discovered, research on time-dependent genetic effects is still a wide open field. The animal breeding industry considers dynamic milk production by incorporating appropriate lactation curve models into their breeding value estimates to improve accuracy. Further, through an index of traits, breeders attempt to tackle the detrimental effects of a high milk production on other functional traits. However, if we assume that some of the negative issues arising from a high milk production can be overcome by altering the shape of the production curve, the impact of such an index on the actual dynamics of the lactation cycle are poorly understood. Since genetic and genomic tools are constantly developing with whole genome sequencing already being applied, our understanding of genes, their interactions and pathways will improve and direct causative mutations might be the target of future animal breeding programs. Understanding the time-dependent effects of genes and their variants is therefore an important field to study. Finally, whilst the dynamic of milk production is an obvious example, other time-dependent traits such as growth and weight gain, marbling, or onset of puberty could also benefit from a deeper understanding of the underlying dynamic of gene effects.
Author contributions
EMS conceived of the topic and wrote the manuscript. YCSML and GAB advised and critically revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Adam Wilkins for his constructive comments on the manuscript, his teaching and time.
Footnotes
References
- Allen M. S. (1996). Physical constraints on voluntary intake of forages by ruminants. J. Anim. Sci. 74, 3063–3075. [DOI] [PubMed] [Google Scholar]
- Anantamongkol U., Charoenphandhu N., Wongdee K., Teerapornpuntakit J., Suthiphongchai T., Prapong S., et al. (2010). Transcriptome analysis of mammary tissues reveals complex patterns of transporter gene expression during pregnancy and lactation. Cell Biol. Int. 34, 67–74. 10.1042/CBI20090023 [DOI] [PubMed] [Google Scholar]
- Appuhamy J. A., Cassell B. G., Cole J. B. (2009). Phenotypic and genetic relationships of common health disorders with milk and fat yield persistencies from producer-recorded health data and test-day yields. J. Dairy Sci. 92, 1785–1795. 10.3168/jds.2008-1591 [DOI] [PubMed] [Google Scholar]
- Bailey K. W., Jones C. M., Heinrichs A. J. (2005). Economic returns to Holstein and Jersey herds under multiple component pricing. J. Dairy Sci. 88, 2269–2280. 10.3168/jds.S0022-0302(05)72903-9 [DOI] [PubMed] [Google Scholar]
- Banos G., Woolliams J. A., Woodward B. W., Forbes A. B., Coffey M. P. (2008). Impact of single nucleotide polymorphisms in leptin, leptin receptor, growth hormone receptor, and diacylglycerol acyltransferase (DGAT1) gene loci on milk production, feed, and body energy traits of UK dairy cows. J. Dairy Sci. 91, 3190–3200. 10.3168/jds.2007-0930 [DOI] [PubMed] [Google Scholar]
- Bauman D. E., Mather I. H., Wall R. J., Lock A. L. (2006). Major advances associated with the biosynthesis of milk. J. Dairy Sci. 89, 1235–1243. 10.3168/jds.S0022-0302(06)72192-0 [DOI] [PubMed] [Google Scholar]
- Bell A. W., Bauman D. E. (1997). Adaptations of glucose metabolism during pregnancy and lactation. J. Mammary Gland Biol. Neoplasia 2, 265–278. 10.1023/A:1026336505343 [DOI] [PubMed] [Google Scholar]
- Bionaz M., Loor J. J. (2008a). ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 138, 1019–1024. [DOI] [PubMed] [Google Scholar]
- Bionaz M., Loor J. J. (2008b). Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 9:366. 10.1186/1471-2164-9-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloise E., Cassali G. D., Ferreira M. C., Ciarmela P., Petraglia F., Reis F. M. (2010). Activin-related proteins in bovine mammary gland: localization and differential expression during gestational development and differentiation. J. Dairy Sci. 93, 4592–4601. 10.3168/jds.jds.2009-2981 [DOI] [PubMed] [Google Scholar]
- Boichard D., Brochard M. (2012). New phenotypes for new breeding goals in dairy cattle. Animal 6, 544–550. 10.1017/S1751731112000018 [DOI] [PubMed] [Google Scholar]
- Bole-Feysot C., Goffin V., Edery M., Binart N., Kelly P. A. (1998). Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr. Rev. 19, 225–268. 10.1210/edrv.19.3.0334 [DOI] [PubMed] [Google Scholar]
- Bovine HapMap C., Gibbs R. A., Taylor J. F., Van Tassell C. P., Barendse W., Eversole K. A., et al. (2009). Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532. 10.1126/science.1167936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitenhuis B., Janss L. L., Poulsen N. A., Larsen L. B., Larsen M. K., Sorensen P. (2014). Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genomics 15:1112. 10.1186/1471-2164-15-1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D., Duncan J. S. (1998). Secretion of milk proteins. J. Mammary Gland Biol. Neoplasia 3, 275–286. 10.1023/A:1018763427108 [DOI] [PubMed] [Google Scholar]
- Capuco A. V., Akers R. M. (1999). Mammary involution in dairy animals. J. Mammary Gland Biol. Neoplasia 4, 137–144. 10.1023/A:1018769022990 [DOI] [PubMed] [Google Scholar]
- Casey T., Patel O., Dykema K., Dover H., Furge K., Plaut K. (2009). Molecular signatures reveal circadian clocks may orchestrate the homeorhetic response to lactation. PLoS ONE 4:e7395. 10.1371/journal.pone.0007395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton L., Bonta M., Pol M., Tirante L., Bussmann L. E. (2013). Milk lactoferrin in heifers: influence of health status and stage of lactation. J. Dairy Sci. 96, 4977–4982. 10.3168/jds.2012-6028 [DOI] [PubMed] [Google Scholar]
- Clempson A. M., Pollott G. E., Brickell J. S., Bourne N. E., Munce N., Wathes D. C. (2011). Evidence that leptin genotype is associated with fertility, growth, and milk production in Holstein cows. J. Dairy Sci. 94, 3618–3628. 10.3168/jds.2010-3626 [DOI] [PubMed] [Google Scholar]
- Cohen-Zinder M., Seroussi E., Larkin D. M., Loor J. J., Everts-van der Wind A., Lee J. H., et al. (2005). Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 15, 936–944. 10.1101/gr.3806705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. B., Null D. J. (2009). Genetic evaluation of lactation persistency for five breeds of dairy cattle. J. Dairy Sci. 92, 2248–2258. 10.3168/jds.2008-1825 [DOI] [PubMed] [Google Scholar]
- Cole J. B., VanRaden P. M. (2006). Genetic evaluation and best prediction of lactation persistency. J. Dairy Sci. 89, 2722–2728. 10.3168/jds.S0022-0302(06)72348-7 [DOI] [PubMed] [Google Scholar]
- Cole J. B., Wiggans G. R., Ma L., Sonstegard T. S., Lawlor T. J., Jr., Crooker B. A., et al. (2011). Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics 12:408. 10.1186/1471-2164-12-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard B. L., Boettcher P. J., Dekkers J. C., Petitclerc D., Schaeffer L. R. (2000). Relationships between energy balance and health traits of dairy cattle in early lactation. J. Dairy Sci. 83, 2683–2690. 10.3168/jds.S0022-0302(00)75162-9 [DOI] [PubMed] [Google Scholar]
- Dekkers J. C. (2007). Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 124, 331–341. 10.1111/j.1439-0388.2007.00701.x [DOI] [PubMed] [Google Scholar]
- Dekkers J. C. M., Ten Hag J. H., Weersink A. (1998). Economic aspects of persistency of lactation in dairy cattle. Livest. Prod. Sci. 53, 237–252 10.1016/S0301-6226(97)00124-3 [DOI] [Google Scholar]
- Forni S., Gianola D., Rosa G. J., de Los Campos G. (2009). A dynamic linear model for genetic analysis of longitudinal traits. J. Anim. Sci. 87, 3845–3853. 10.2527/jas.2008-1514 [DOI] [PubMed] [Google Scholar]
- Gengler N. (1996). Persistency of lactation yields: a review. Interbull Bull. 12, 87–96. [Google Scholar]
- Goddard M. (2009). Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136, 245–257. 10.1007/s10709-008-9308-0 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J. (2007). Genomic selection. J. Anim. Breed. Genet. 124, 323–330. 10.1111/j.1439-0388.2007.00702.x [DOI] [PubMed] [Google Scholar]
- Greer A. W., Huntley J. F., Mackellar A., McAnulty R. W., Jay N. P., Green R. S., et al. (2008). The effect of corticosteroid treatment on local immune responses, intake and performance in lambs infected with Teladorsagia circumcincta. Int. J. Parasitol. 38, 1717–1728. 10.1016/j.ijpara.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Hadjipavlou G., Bishop S. C. (2009). Age-dependent quantitative trait loci affecting growth traits in Scottish Blackface sheep. Anim. Genet. 40, 165–175. 10.1111/j.1365-2052.2008.01814.x [DOI] [PubMed] [Google Scholar]
- Harder B., Bennewitz J., Hinrichs D., Kalm E. (2006). Genetic parameters for health traits and their relationship to different persistency traits in German Holstein dairy cattle. J. Dairy Sci. 89, 3202–3212. 10.3168/jds.S0022-0302(06)72595-4 [DOI] [PubMed] [Google Scholar]
- Hayes B., Goddard M. (2010). Genome-wide association and genomic selection in animal breeding. Genome 53, 876–883. 10.1139/G10-076 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E. (2009). Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92, 433–443. 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- He C., Wang C., Chang Z. H., Guo B. L., Li R., Yue X. P., et al. (2011). AGPAT6 polymorphism and its association with milk traits of dairy goats. Genet. Mol. Res. 10, 2747–2756. 10.4238/2011.November.4.8 [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G. W. (2005). Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 6, 715–725. 10.1038/nrm1714 [DOI] [PubMed] [Google Scholar]
- Hou W., Li H., Zhang B., Huang M., Wu R. (2008). A nonlinear mixed-effect mixture model for functional mapping of dynamic traits. Heredity (Edinb.) 101, 321–328. 10.1038/hdy.2008.53 [DOI] [PubMed] [Google Scholar]
- Inchaisri C., Jorritsma R., Vos P. L., van der Weijden G. C., Hogeveen H. (2011). Analysis of the economically optimal voluntary waiting period for first insemination. J. Dairy Sci. 94, 3811–3823. 10.3168/jds.2010-3790 [DOI] [PubMed] [Google Scholar]
- Ingvartsen K. L., Dewhurst R. J., Friggens N. C. (2003). On the relationship between lactational performance and health traits: is it yield or metabilic imbalance that causes diseases in dairy cattle? A position paper. Livest. Prod. Sci. 83, 277–308 10.1016/S0301-6226(03)00110-6 [DOI] [Google Scholar]
- Khatib H., Zaitoun I., Wiebelhaus-Finger J., Chang Y. M., Rosa G. J. (2007). The association of bovine PPARGC1A and OPN genes with milk composition in two independent Holstein cattle populations. J. Dairy Sci. 90, 2966–2970. 10.3168/jds.2006-812 [DOI] [PubMed] [Google Scholar]
- Koivula M., Stranden I., Su G., Mantysaari E. A. (2012). Different methods to calculate genomic predictions–comparisons of BLUP at the single nucleotide polymorphism level (SNP-BLUP), BLUP at the individual level (G-BLUP), and the one-step approach (H-BLUP). J. Dairy Sci. 95, 4065–4073. 10.3168/jds.2011-4874 [DOI] [PubMed] [Google Scholar]
- Koos R. D. (2011). Minireview: putting physiology back into estrogens' mechanism of action. Endocrinology 152, 4481–4488. 10.1210/en.2011-1449 [DOI] [PubMed] [Google Scholar]
- Kress C., Kieu K., Droineau S., Galio L., Devinoy E. (2011). Specific positioning of the casein gene cluster in active nuclear domains in luminal mammary epithelial cells. Chromosome Res. 19, 979–997. 10.1007/s10577-011-9249-9 [DOI] [PubMed] [Google Scholar]
- Kuhn M. T., Hutchison J. L., Norman H. D. (2007). Dry period length in US Jerseys: characterization and effects on performance. J. Dairy Sci. 90, 2069–2081. 10.3168/jds.2006-702 [DOI] [PubMed] [Google Scholar]
- Kuhn M. T., Hutchison J. L., Norman H. D. (2006). Effects of length of dry period on yields of milk fat and protein, fertility and milk somatic cell score in the subsequent lactation of dairy cows. J. Dairy Res. 73, 154–162. 10.1017/S0022029905001597 [DOI] [PubMed] [Google Scholar]
- Laurenson Y. C., Bishop S. C., Kyriazakis I. (2011). In silico exploration of the mechanisms that underlie parasite-induced anorexia in sheep. Br. J. Nutr. 106, 1023–1039. 10.1017/S0007114511001371 [DOI] [PubMed] [Google Scholar]
- Lemay D. G., Lynn D. J., Martin W. F., Neville M. C., Casey T. M., Rincon G., et al. (2009). The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 10:R43. 10.1186/gb-2009-10-4-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S., Khatib H., Schutzkus V., Chang Y. M., Maltecca C. (2005). Effects of the osteopontin gene variants on milk production traits in dairy cattle. J. Dairy Sci. 88, 4083–4086. 10.3168/jds.S0022-0302(05)73092-7 [DOI] [PubMed] [Google Scholar]
- Li M., Li Q., Gao X. (2010). Expression and function of leptin and its receptor in dairy goat mammary gland. J. Dairy Res. 77, 213–219. 10.1017/S0022029910000063 [DOI] [PubMed] [Google Scholar]
- Li N., Das K., Wu R. (2009). Functional mapping of human growth trajectories. J. Theor. Biol. 261, 33–42. 10.1016/j.jtbi.2009.07.020 [DOI] [PubMed] [Google Scholar]
- Lu A., Hu X., Chen H., Dong Y., Pang Y. (2011a). Single nucleotide polymorphisms of the prolactin receptor (PRLR) gene and its association with growth traits in Chinese cattle. Mol. Biol. Rep. 38, 261–266. 10.1007/s11033-010-0103-5 [DOI] [PubMed] [Google Scholar]
- Lu A., Hu X., Chen H., Dong Y., Zhang Y., Wang X. (2011b). Novel SNPs of the bovine PRLR gene associated with milk production traits. Biochem. Genet. 49, 177–189. 10.1007/s10528-010-9397-1 [DOI] [PubMed] [Google Scholar]
- Lund M. S., Sorensen P., Madsen P., Jaffrezic F. (2008). Detection and modelling of time-dependent QTL in animal populations. Genet. Sel. Evol. 40, 177–194. 10.1186/1297-9686-40-2-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally S., Martin F. (2011). Molecular regulators of pubertal mammary gland development. Ann. Med. 43, 212–234. 10.3109/07853890.2011.554425 [DOI] [PubMed] [Google Scholar]
- Miglior F., Muir B. L., Van Doormaal B. J. (2005). Selection indices in Holstein cattle of various countries. J. Dairy Sci. 88, 1255–1263. 10.3168/jds.S0022-0302(05)72792-2 [DOI] [PubMed] [Google Scholar]
- Misztal I., Strabel T., Jamrozik J., Mantysaari E. A., Meuwissen T. H. (2000). Strategies for estimating the parameters needed for different test-day models. J. Dairy Sci. 83, 1125–1134. 10.3168/jds.S0022-0302(00)74978-2 [DOI] [PubMed] [Google Scholar]
- Muir B. L., Fatehi J., Schaeffer L. R. (2004). Genetic relationships between persistency and reproductive preformance in first-lactation Canadian Holsteins. J. Dairy Sci. 87, 3029–3037. 10.3168/jds.S0022-0302(04)73435-9 [DOI] [PubMed] [Google Scholar]
- Mullen M. P., Berry D. P., Howard D. J., Diskin M. G., Lynch C. O., Giblin L., et al. (2011). Single nucleotide polymorphisms in the Insulin-like Growth Factor 1 (IGF-1) gene are associated with performance in Holstein-Friesian dairy cattle. Front. Genet. 2:3. 10.3389/fgene.2011.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. C., Watters C. D. (1983). Secretion of calcium into milk: review. J. Dairy Sci. 66, 371–380. 10.3168/jds.S0022-0302(83)81802-5 [DOI] [PubMed] [Google Scholar]
- Oltenacu P. A., Broom D. M. (2010). The impact of genetic selection for increased milk yield on the welfare of dairy cows. Anim. Welfare 19, 39–49. [Google Scholar]
- Osteras O., Solbu H., Refsdal A. O., Roalkvam T., Filseth O., Minsaas A. (2007). Results and evaluation of thirty years of health recordings in the Norwegian dairy cattle population. J. Dairy Sci. 90, 4483–4497. 10.3168/jds.2007-0030 [DOI] [PubMed] [Google Scholar]
- Pollott G. E. (2004). Deconstructing milk yield and composition during lactation using biologically based lactation models. J. Dairy Sci. 87, 2375–2387. 10.3168/jds.S0022-0302(04)73359-7 [DOI] [PubMed] [Google Scholar]
- Rahmatalla S. A., Muller U., Strucken E. M., Reissmann M., Brockmann G. A. (2011). The F279Y polymorphism of the GHR gene and its relation to milk production and somatic cell score in German Holstein dairy cattle. J. Appl. Genet. 52, 459–465. 10.1007/s13353-011-0051-3 [DOI] [PubMed] [Google Scholar]
- Raven L. A., Cocks B. G., Hayes B. J. (2014). Multibreed genome wide association can improve precision of mapping causative variants underlying milk production in dairy cattle. BMC Genomics 15:62. 10.1186/1471-2164-15-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Zas S. L., Southey B. R., Heyen D. W., Lewin H. A. (2002). Detection of quantitative trait loci influencing dairy traits using a model for longitudinal data. J. Dairy Sci. 85, 2681–2691. 10.3168/jds.S0022-0302(02)74354-3 [DOI] [PubMed] [Google Scholar]
- Roldan D. L., Rabasa A. E., Saldano S., Holgado F., Poli M. A., Cantet R. J. (2008). QTL detection for milk production traits in goats using a longitudinal model. J. Anim. Breed. Genet. 125, 187–193. 10.1111/j.1439-0388.2007.00714.x [DOI] [PubMed] [Google Scholar]
- Ron M., Cohen-Zinder M., Peter C., Weller J. I., Erhardt G. (2006). Short communication: a polymorphism in ABCG2 in Bos indicus and Bos taurus cattle breeds. J. Dairy Sci. 89, 4921–4923. 10.3168/jds.S0022-0302(06)72542-5 [DOI] [PubMed] [Google Scholar]
- Sawa A., Bogucki M., Neja W. (2012). Dry period length and performance of cows in the subsequent production cycle. Archiv. Tierzucht. 55, 140–147. [Google Scholar]
- Schaeffer L. R., Jamrozik J., Kistemaker G. J., Van Doormaal B. J. (2000). Experience with a test-day model. J. Dairy Sci. 83, 1135–1144. 10.3168/jds.S0022-0302(00)74979-4 [DOI] [PubMed] [Google Scholar]
- Schmidt G. H., Van Vleck L. D., Hutjens M. F. (1988). Book Principles of Dairy Science. New Jersey, NJ: Prentice Hall Inc. [Google Scholar]
- Seefried F., Liu Z., Thaller G., Reinhardt F. (2010). Die genomische zuchtwertschätzung bei der Rasse Deutsch Holstein. Züchtungskunde 1, 14–21. [Google Scholar]
- Silvestre A. M., Martins A. M., Santos V. A., Ginja M. M., Colaço J. A. (2009). Lactation curves for milk, fat and protein in dairy cows: a full approach. Livest. Sci. 122, 308–313 10.1016/j.livsci.2008.09.017 [DOI] [Google Scholar]
- Spurlock D. M., Dekkers J. C., Fernando R., Koltes D. A., Wolc A. (2012). Genetic parameters for energy balance, feed efficiency, and related traits in Holstein cattle. J. Dairy Sci. 95, 5393–5402. 10.3168/jds.2012-5407 [DOI] [PubMed] [Google Scholar]
- Stanton T. L., Jones L. R., Everett R. W., Kachman S. D. (1992). Estimating milk, fat, and protein lactation curves with a test day model. J. Dairy Sci. 75, 1691–1700. 10.3168/jds.S0022-0302(92)77926-0 [DOI] [PubMed] [Google Scholar]
- Strucken E. M., Bortfeldt R. H., de Koning D. J., Brockmann G. A. (2012a). Genome-wide associations for investigating time-dependent genetic effects for milk production traits in dairy cattle. Anim. Genet. 43, 375–382. 10.1111/j.1365-2052.2011.02278.x [DOI] [PubMed] [Google Scholar]
- Strucken E. M., Bortfeldt R. H., Tetens J., Thaller G., Brockmann G. A. (2012b). Genetic effects and correlations between production and fertility traits and their dependency on the lactation-stage in Holstein Friesians. BMC Genet 13:108. 10.1186/1471-2156-13-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucken E. M., de Koning D. J., Rahmatalla S. A., Brockmann G. A. (2011). Lactation curve models for estimating gene effects over a timeline. J. Dairy Sci. 94, 442–449. 10.3168/jds.2009-2932 [DOI] [PubMed] [Google Scholar]
- Strucken E. M., Rahmatalla S., Koning D.-J. D., Brockmann G. A. (2010a). Haplotype analysis and linkage disequilibrium for DGAT1. Arch. Anim. Breed. 53, 247–255. [Google Scholar]
- Suchocki T., Szyda J. (2011). Statistical modelling of growth using a mixed model with orthogonal polynomials. J. Appl. Genet. 52, 95–100. 10.1007/s13353-010-0012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshbabu A., Tonner E., Flint D. J. (2011). Insulin-like growth factor binding proteins and mammary gland development. Int. J. Dev. Biol. 55, 781–789. 10.1387/ijdb.113364as [DOI] [PubMed] [Google Scholar]
- Swalve H. H. (2000). Theoretical basis and computational methods for different test-day genetic evaluation methods. J. Dairy Sci. 83, 1115–1124. 10.3168/jds.S0022-0302(00)74977-0 [DOI] [PubMed] [Google Scholar]
- Taylor V. J., Beever D. E., Bryant M. J., Wathes D. C. (2003). Metabolic profiles and progesterone cycles in first lactation dairy cows. Theriogenology 59, 1661–1677. 10.1016/S0093-691X(02)01225-6 [DOI] [PubMed] [Google Scholar]
- Taylor V. J., Beever D. E., Bryant M. J., Wathes D. C. (2004). First lactation ovarian function in dairy heifers in relation to prepubertal metabolic profiles. J. Endocrinol. 180, 63–75. 10.1677/joe.0.1800063 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M. (2004). Invited review: selection on net merit to improve lifetime profit. J. Dairy Sci. 87, 3125–3131. 10.3168/jds.S0022-0302(04)73447-5 [DOI] [PubMed] [Google Scholar]
- Velmala R., Vilkki J., Elo K., Maki-Tanila A. (1995). Casein haplotypes and their association with milk production traits in the Finnish Ayrshire cattle. Anim. Genet. 26, 419–425. 10.1111/j.1365-2052.1995.tb02694.x [DOI] [PubMed] [Google Scholar]
- Verbyla K. L., Verbyla A. P. (2009). Estimated breeding values and association mapping for persistency and total milk yield using natural cubic smoothing splines. Genet. Sel. Evol. 41:48. 10.1186/1297-9686-41-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitala S., Szyda J., Blott S., Schulman N., Lidauer M., Maki-Tanila A., et al. (2006). The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics 173, 2151–2164. 10.1534/genetics.105.046730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. M., Berry D. P., Mullen M. P. (2012). Polymorphisms in genes of the somatotrophic axis are independently associated with milk production, udder health, survival and animal size in Holstein-Friesian dairy cattle. J. Anim. Breed. Genet. 129, 70–78. 10.1111/j.1439-0388.2011.00938.x [DOI] [PubMed] [Google Scholar]
- Watson C. J., Khaled W. T. (2008). Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 135, 995–1003. 10.1242/dev.005439 [DOI] [PubMed] [Google Scholar]
- Watson C. J., Oliver C. H., Khaled W. T. (2011). Cytokine signalling in mammary gland development. J. Reprod. Immunol. 88, 124–129. 10.1016/j.jri.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Wattiaux M. A. (1996). Milk secretion in the udder of a dairy cow. Dairy Essentials 77–80 Available online at: http://babcock.wisc.edu/sites/default/files/de/en/de_20.en.pdf
- Wheeler T. T., Smolenski G. A., Harris D. P., Gupta S. K., Haigh B. J., Broadhurst M. K., et al. (2012). Host-defence-related proteins in cows' milk. Animal 6, 415–422. 10.1017/S1751731111002151 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe S., Rincon G., Islas-Trejo A., Medrano J. F. (2012). Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics 13:45. 10.1186/1471-2164-13-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A., Kramer W., Werner F. A., Kollers S., Kata S., Durstewitz G., et al. (2002). Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. U.S.A. 99, 9300–9305 10.1073/pnas.142293799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wu R., Casella G. (2009). Nonparametric functional mapping of quantitative trait loci. Biometrics 65, 30–39. 10.1111/j.1541-0420.2008.01063.x [DOI] [PubMed] [Google Scholar]
- Zhao F. Q., Keating A. F. (2007). Expression and regulation of glucose transporters in the bovine mammary gland. J. Dairy Sci. 90 (Suppl. 1), E76–E86. 10.3168/jds.2006-470 [DOI] [PubMed] [Google Scholar]
- Zielke L. G., Bortfeldt R. H., Reissmann M., Tetens J., Thaller G., Brockmann G. A. (2013). Impact of variation at the FTO locus on milk fat yield in Holstein dairy cattle. PLoS ONE 8:e63406. 10.1371/journal.pone.0063406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke L. G., Bortfeldt R. H., Tetens J., Brockmann G. A. (2011). BDNF contributes to the genetic variance of milk fat yield in German Holstein cattle. Front. Genet. 5:2–16. 10.3389/fgene.2011.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A. V., Delcher A. L., Florea L., Kelley D. R., Schatz M. C., Puiu D., et al. (2009). A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10:R42. 10.1186/gb-2009-10-4-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann E., Sommer H. (1973). Zum laktationsverlauf von kuehen in hochleistungsherden und dessen beeinflussung durch nichterbliche Faktoren. Zuechtungskunde 45, 75–88. [Google Scholar]