Abstract
A striking finding in the past decade is the production of numerous non-coding RNAs (ncRNAs) from mammalian genomes. While it is entirely possible that many of those ncRNAs are transcription noises or by-products of RNA processing, increasing evidence suggests that a large fraction of them are functional and provide various regulatory activities in the cell. Thus, functional genomics and proteomics are incomplete without understanding functional ribonomics. As has been long suggested by the ‘RNA world’ hypothesis, many ncRNAs have the capacity to act like proteins in diverse biochemical processes. The enormous amount of information residing in the primary sequences and secondary structures of ncRNAs makes them particularly suited to function as scaffolds for molecular interactions. In addition, their functions appear to be stringently controlled by default via abundant nucleases when not engaged in specific interactions. This review focuses on the functional properties of regulatory ncRNAs in comparison with proteins and emphasizes both the opportunities and challenges in future ncRNA research.
Keywords: the RNA world, non-coding RNA, biological functions, regulatory mechanisms, experimental approaches, functional genomics
INTRODUCTION
A major surprise since the completion of the human genome and subsequent sequencing of all biological model organisms is the limited number of protein-coding genes, which neither correlates with the complexity of organisms nor accounts for the selection pressure during the evolution of modern organisms [1]. In humans, the protein-coding sequences occupy only ~1.5% of the genome, and when considering intervening sequences (introns) within protein-coding genes and 5′ and 3′ untranslated regions, this number goes up to only ~28%. Much of the remaining portion of the human genome used to be considered ‘junk’ DNA because ~59% are repeat sequences; however, recent analysis by the Encyclopedia of DNA Elements (ENCODE) project suggests that ~80% of the genome appears to participate in some sort of biochemical activities that might be functionally important [2]. This suggests a general paradigm for functional DNA elements embedded in the non-coding part of mammalian genomes.
While initial microarray-based results met with skepticism, the ENCODE data generated by the latest deep sequencing technologies demonstrated that at least 70% of the human genome has the capacity to produce transcripts of various sizes, many of which are conserved in animal kingdom [2]. Besides mRNAs already annotated, most other transcripts do not seem to encode for proteins and are generally referred to as non-coding RNAs (ncRNAs) [3]. Although debate continues with respect to the possibility that some of these ncRNAs may still direct synthesis of short peptides, the consensus is that they are largely non-coding, which is supported by the evidence from ribosome profiling [4] and by the large-scale proteomics analysis performed on two ENCODE cell lines [5]. While most of these ncRNAs have yet to be biochemically characterized, we are witnessing functional assignment to an increasing number of ncRNAs, leading to birth of a new discipline in biological research.
Like many emerging disciplines, the ncRNA field has received great attention in recent years from the general research community, and the progress made has been extensively reviewed from the perspective of mechanistic insights [6-8] and/or biological functions [9-11]. Instead of enumerating numerous great points that have been made in those reviews, here I highlight the biochemical property of ncRNAs in comparison with proteins to formulate ideas for future research, the uniqueness of ncRNA research, which calls for the great need to develop new experimental approaches, and the potential to exploit ncRNA as a new class of biomarkers or therapeutic targets in biomedical and biotechnological applications.
ncRNA: OLD AND NEW
ncRNAs may be new to the research community at large, but actually ancient among RNA researchers. Classic ncRNAs that have been intensively studied in the past five decades since the birth of molecular biology include small ncRNAs, such as transfer RNAs (tRNAs) for carrying amino acids, small nucleolus RNAs (snoRNAs) for RNA modifications, and small nuclear RNAs (snRNAs) for RNA splicing, and large ones, such as ribosomal RNAs (rRNAs) for protein synthesis (Box 1 and Fig. 1). These ncRNAs may be considered ‘constitutive’, because they are abundantly and ubiquitously expressed in all cell types and provide essential functions to the organism. This class may also include the telomere complex-associated guide RNA, which is essential for the end formation and maintenance of chromosomes in normal proliferating cells even though the telomere complex and the ncRNA in it are often compromised in cancer cells [12].
Box 1.
| Housekeeping ncRNAs | ||
|
| ||
| Abbreviation | Full name | Function |
| rRNA | Ribosomal RNA | Translational machinery |
| tRNA | Transfer RNA | Amino acid carriers |
| snRNA | Small nuclear RNA | RNA processing |
| snoRNA | Small nucleolar RNA | RNA modifications |
| TR | Telomere RNA | Chromosome end synthesis |
|
| ||
| Regulatory ncRNAs | ||
|
| ||
| Abbreviation | Full name | Function |
| miRNA | MicroRNAs | RNA stability and translation control |
| endo-siRNA | Endogenous siRNA | RNA degradation |
| rasiRNA | Repeat-derived RNA | Transcriptional control |
| piRNA | Piwi-associated RNA | Silencing transposon and mRNA decay |
| eRNA | Enhancer-derived RNA | Regulation of gene expression |
| PATs | Promoter-associated RNA | Transcription initiation and pause release |
| IncRNA | Long non-coding RNA | Imprinting, epigenetics, nuclear structure |
Figure 1.
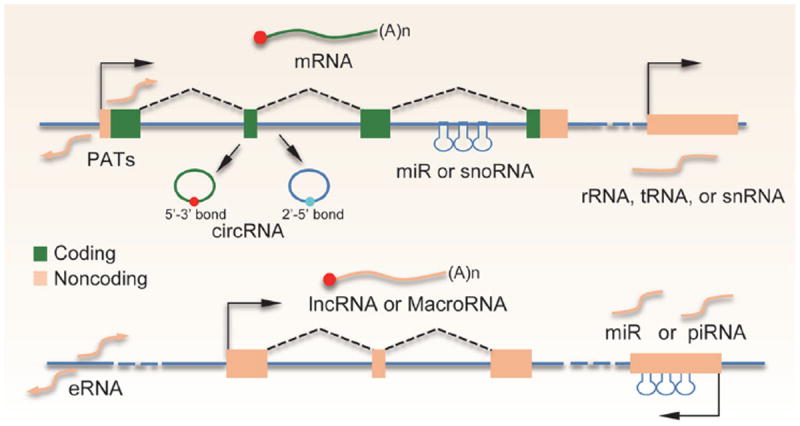
Production of distinct classes of ncRNAs from mammalian genomes. Top: protein-coding (green lines) genes produce divergent PATs at the transcription start site. Certain exonic and intronic sequences have the capacity to generate circRNAs containing either 3′–5′ or 2′–5′ phosphodiester bonds. Many intronic sequences can also encode for miRNAs or snoRNAs. Genes for rRNAs, tR-NAs, or a subfraction of snRNAs are transcribed from separate genes. Bottom: similar to protein-coding genes, transcription enhancers also produce divergent transcripts, known as eRNAs. Most of the lncRNA genes contain at least one intron and are transcribed and processed in the same way as protein-coding genes except that they do not have coding potential (yellow line). miRNAs and piRNAs can also be derived from various intergenic regions.
We now have extensive knowledge about ‘tiny’ ncRNAs, such as microRNAs (miRNAs), endogenous siRNAs (endo-siRNAs), and PIWI-associated small RNAs (piRNAs) that are expressed in animals and plants (Box 1). The biogenesis, targeting, and function of these classes of ncRNAs have been extensively studied and reviewed [13-17] (see also Chen et al., this issue), and are thus not focused in this review. These small RNAs are normally processed from larger RNA precursors, either from their own transcripts or from sequences within specific protein-coding genes (Fig. 1). In contrast to these small RNAs, deep sequencing has identified an increasing number of long intergenic non-coding RNAs (lincRNAs) or simply long non-coding RNAs (lncRNAs), now listed in various databases [18,19], which has received great attention from the research community.
In general, ncRNAs have been classified based on an arbitrary size cut-off of 200 nt to separate small ncRNAs from lncRNAs. However, many ncR-NAs may fall into both sides of this cut-off, such as enhancer-associated RNAs (eRNAs), promoter-associated transcripts (PATs), and the more recently emerged circular RNAs (circRNAs) (Box 1; Fig. 1). In fact, these ncRNAs have their own structural features at each end, as eRNAs and PATs have cap, but no poly(A) tail [20], while circRNAs obviously have no ends, which add to structural characteristics of other ncRNAs after processing (e.g. snRNAs with the 5′ tri-methylated cap, miRNAs with the 5′-phosphate, etc.). These features distinguish them from the class of lncRNAs (Box 1), which are transcribed and processed in an identical way to that of protein-coding genes (e.g. capping, splicing, and polyadenylation, see Fig. 1), and as such, their genes are also associated with characteristic chromatin marks (e.g. H3K4me3 at promoters and H3K36me3 in the gene body), which have been exploited for their prediction, identification, and characterization in mammalian genomes [21].
A common feature of newly identified ncRNAs is their highly regulated expression in different cell types or during development. Our current understanding of their functions, although still quite limited, suggests that these ncRNAs may have diverse regulatory activities (Box 1). Because ncRNAs are either transcribed from specific genomic loci or derived from segments of protein-coding genes, the question is whether all expressed ncRNAs that are detectable by sensitive technologies are functional or some of them may simply reflect transcriptional noises or by-products of RNA processing [22]. A deeper question is whether the process of producing some of those ncRNAs, rather than the final products, is of biological importance because transcription of these ncRNAs is often associated with chromatin remodeling activities. Despite continuous debate on these valid questions, the field has experienced tremendous progress in elucidating the function and mechanism of various ncRNAs, particularly lncRNAs. Thus, for practical reasons, one may first focus on studying ncRNAs that have already some functional evidence, while ignoring many potential ‘junk’ RNAs, at least for the time being.
FUNCTION OF ncRNA IN COMPARISON WITH PROTEIN
The hypothesis of ‘the RNA world’ proposes that the development of life, which has to fulfill the requirement of having the ability to carry and replicate its genetic material, may begin with RNA [23,24]. ncR-NAs appear to have preserved most, if not all, of their original features and functions in modern organisms that have evolved to adopt more efficient strategies to replicate and express their genetic information along the central dogma from DNA to RNA to protein. As a result of exploring selective advantages of proteins and RNA during evolution, many functions of RNA are passed onto proteins while others are retained. In this regard, it might be informative to compare the function of ncRNAs with proteins to conceptualize ncRNA function and mechanism.
RNA as enzyme
One of the key functions of proteins is to catalyze chemical reactions. Some ncRNAs have long been known to preserve this critical function, known as catalytic RNA, such as the RNAs associated with RNase P required for tRNA processing [25] and auto-catalytic introns [26]. In fact, through in vitro selection from random sequences, one may select RNA capable of catalyzing RNA ligation [27] or polymerization [28]. Other ncRNAs preserve their catalytic function only when folded correctly with help of proteins. The best known example is rRNAs in which all key catalytic reactions in reading the coding information in mRNA are provided by the so-called RNA centers [29]. This may also be the case in the spliceosome, which is responsible for intron removal during pre-mRNA splicing and where the catalytic center may form with both RNA and proteins [30]. Therefore, although most catalytic activities of RNA have been passed onto proteins in modern organisms, at least some ncRNAs appear to have kept such function during evolution. Even so, some key functional properties of RNA are maintained in many ribonucleoprotein (RNP) machines. The best known examples are in fact miRNAs and piRNAs in argonaute-containing complexes where these tiny ncRNAs provide targeting information whereas the associated proteins execute the biochemical reactions [31,32]. We thus should not be surprised if many additional ncRNAs are found to make direct contribution to catalysis in the form of RNPs.
RNA as scaffold of molecular interactions
A major function of proteins in the cell is to engage in protein–protein, protein–DNA, and protein–RNA interactions in diverse biochemical reactions. These functions are mediated by specific domains, ~600 of which have been characterized to date among ~3000 potential ones [33-35]. In comparison, RNA seems to have similar, if not larger, capacity to perform such molecular interactions through their unique sequence motifs and secondary structures, the latter of which may adapt into different combinations when exposed to different environments or interacting with different proteins. In principle, a specific RNA moiety may interact with DNA or RNA through base-pairing whereas both primary sequences and secondary structures may serve as modules for interactions with specific proteins or protein complexes. For example, specific stem-loop domains in the 7SK RNA are known to interact with distinct protein components [36], and the lncRNA HO-TAIR uses its 5′ domain to interact with Polycomb Complex 2 (PRC2) and its 3′ domain to recruit the histone lysine 4 demethylase LSD1, thus coordinating two separate transcription repressor complexes to act on target genes [37]. The ability of a ncRNA to simultaneously engage in interactions with DNA and proteins has been exemplified with the rRNA gene-associated transcripts, which, together with the transcription factor TTF-1, recruit the DNA methyl-transferase DNMT3b to CpG islands [38]. These examples illustrate unique advantages of ncRNAs in the regulation of gene expression.
The ncRNA steroid receptor RNA activator is one of the first examples documented to function as a transcription co-activator in gene activation [39], and we now know that many other ncRNAs appear to have such enhancer function [40]. Numerous studies have exposed the mechanisms of regulatory ncRNAs in transcriptional control, including (1) transcription interference by antisense RNA [41,42] (Fig. 2a), (2) direct inhibition of Pol II activity by Alu repeat-derived transcripts [43,44] (Fig. 2b), (3) sequestration of transcriptional regulators [45] (Fig. 2c), (4) guiding transcription regulators to specific regulatory loci through RNA–DNA base-pairing interactions [38] (Fig. 2d), (5) recruitment of additional transcription regulators [37] (Fig. 2e), and (6) mediating long-distance interactions between promoter and enhancer [40,46] (Fig. 2f). Each of these action mechanisms by specific lncRNAs on their target genes has been detailed in multiple recent reviews [6-8,11]. Interestingly, a recent study showed that two lncRNAs (PRNCR1 and PCGEM1) overexpressed in prostate cancer cells interact in a consecutive fashion with the androgen receptor to promote gene expression and cell proliferation in castration-resistant prostate cancer [47]. These and other findings emphasize the involvement of extensive RNA-dependent interactions in transcriptional control.
Figure 2.
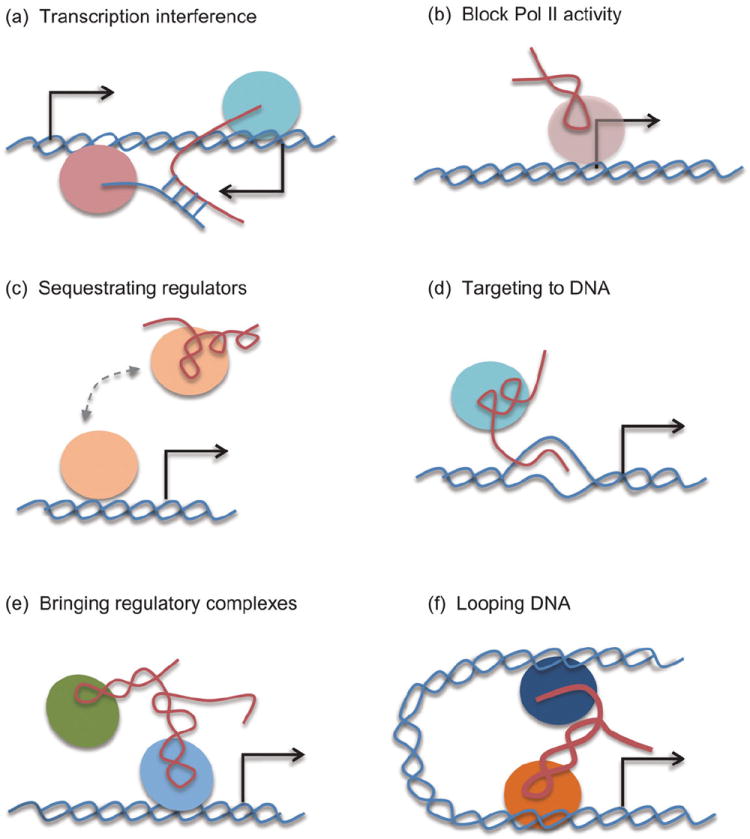
Modes of ncRNA action on genomic DNA in regulated gene expression. lncRNAs are best characterized for their interactions with transcriptional regulators on functional DNA elements. (a) Various antisense transcripts, which appear to be quite widespread in humans and mice [42], may act as ncRNAs to interfere with Pol II elongation [41]. (b) Repeat-derived ncRNAs to block transcription. The prototype ncRNAs in this class are some transcribed Alu sequences, which bind to and interfere with Pol II function at gene promoters [44]. (c) A ncRNA may function as a decoy to compete for a specific transcription factor. The prototype for this mode is PANDA in sequestering the transcription factor NF-YA [45]. (d) A ncRNA may also facilitate the recruitment of a transcription regulator to a specific target site by engaging base-pairing interactions with genomic DNA. The prototype for this mode is the rRNA gene PATs [38]. (e) A ncRNA may bridge protein–protein interactions between transcription regulators to enhance their activities on a common DNA target. The prototype for this mode is the ncRNA HOTAIR in bridging PRC2 and the lysine demethylase LSD1 to mediate gene silencing [37]. (f) A ncRNA may mediate long-distance interactions between promoter and enhancer during transcription activation. Both cis-acting eRNAs and lncRNAs have been demonstrated to play such a role [40,46,47,51].
Cis-acting RNA as regulatory signal
A common property associated with many regulatory ncRNAs is their action in cis, meaning that they function at the genomic loci where they are transcribed [40], which is likely due to their rapid turnover once released from the site of synthesis. An analogy may be made in this case with secreted proteins synthesized on endoplasmic reticulum (ER), where the signal peptide guides the protein during translation into the lumen of ER and then removed by peptidase [48]. Some promoter-proximal ncR-NAs appear to interfere in cis with transcription either through direct interaction with core components of the transcription machinery [49] or through separate RNA-binding proteins (RBPs) [50]. Certain lncRNAs, such as HOTTIP, appear to also act in cis because of the difficulty in restoring their functional requirement with exogenous transcripts [51]. However, inactivation of most lncRNAs by RNAi seems to invoke genome-wide responses, implying that those lncRNAs may function in trans to module gene expression in multiple locations in the genome [52].
One particular type of ncRNAs that function exclusively near the site of their production is enhancer-transcribed ncRNAs (or eRNAs) [53,54]. Recent studies demonstrated that eRNA production is essential for activating their targeted promoters [20,46,55,56]. As enhancer activities may reflect binding and activity of Pol II, which has been shown to induce chromatin remodeling [57] and promote DNA looping between enhancer and promoter [56], the question is whether or not the process of such transcriptional activities might be more functionally relevant than the RNA products. A BoxB-λN tethering strategy was first used to demonstrate HOTTIP in coordinating long-range chromatin interactions [51], and a recent study also took this approach to show that eRNA mediates DNA looping between enhancer and promoter [46].
Another class of potential cis-acting ncRNAs is PATs. Interestingly, most mammalian genes appear to express divergent transcripts from their promoters, a phenomenon that is not evident in yeast or Drosophila [58,59]. Currently, little is known about the function of these ncRNAs transcribed in the opposite direction of the genes. Interestingly, the antisense transcripts tend to lack U1-binding sites whereas the sense transcripts lack the polyadenylation signals [60]. These features might be responsible for the termination of antisense transcription while allowing sense transcription to proceed, as U1 is known to protect the genome by preventing premature transcriptional termination [61]. The sense PATs may also represent aborted transcription products of paused Pol II immediately downstream of mammalian promoters [62]. Interestingly, one such RNA signal has been well studied in HIV-1, where it attracts the HIV tat protein to bind and recruit additional transcription activators, particularly pTEFb, a Pol II CTD kinase, to release paused Pol II into the gene body [63]. A recent study indicates that many cellular genes may employ a similar mechanism through the splicing factor (SRSF2) to facilitate pause release of Pol II from gene promoter into gene body [64], thus suggesting a general role of PATs in providing signals for Pol II to enter productive elongation. It has also been demonstrated that nascent RNA from the gene body near the transcription start site may provide cis signals for the Polycomb Complexes to bind [65]. Another important message from these studies is that parts of pre-mRNAs from protein-coding genes may also be considered as a new class of ncRNAs in regulated transcription.
Trans-acting RNA as molecular sink
The molecular sink mechanism is a key strategy for proteins to function in signaling networks in mammalian cells. This concept has also been well documented with many RNA motifs in mRNAs as well as in transcripts from transcribed pseudogenes in mammalian genomes [66,67], again indicating that some parts of mRNAs also function as ncRNAs in nature. These RNA elements have been shown to sequester specific miRNAs to prevent their action on other target mRNAs, but the stoichiometry between competing ncRNAs and target RNAs has to be considered in each case for the physiological relevance of any sequestration effect detected [68]. Some specific lncRNAs have also been shown to sponge miRNA [69] and titrate transcription activators to inhibit cell cycle progression under starvation conditions [70] or in response to DNA damage [45]. Therefore, the entire repertoire of expressed RNAs, whether they are mRNAs or ncRNAs, may participate in diverse RNA–RNA or RNA–protein interaction networks to regulate various cellular activities.
Interestingly, analysis of poly(A−) RNA, which has been largely ignored in the past, revealed many stable ncRNA species, which have been abundantly detected in the oocyte nucleus [71]. One of the general mechanisms for these ncRNAs to remain stable may be that their ends are somehow sealed. Three strategies have been elucidated for stabilization of such ncRNAs. One is to ligate their 5′ and 3′ ends, thus forming circRNAs (see Fig. 1) [72,73]. This likely results from the action of the spliceosome, leading to the ligation of the upstream 3′ splice site to the downstream 5′ splice site of an exon, although the precise mechanism for their production remains to be understood. Interestingly, through characterizing poly(A−) RNAs, another strategy to ‘seal’ the ends was recently revealed, which is to prevent de-branching on some released introns [74]. This type of intron-derived circRNAs is thus sealed by the 2′–5′ phosphodiester bond formed at the branch-point during pre-mRNA splicing (see Fig. 1). The third strategy to protect the RNA ends is via some stable RNA moieties, such as those found in snoRNAs [75] or the formation of a triple helical structure, such as that characterized at the ends of the stable MALAT-1 RNA [76,77] and some virus-derived ncRNAs [78]. Such RNA structures, either alone or in complex with specific RBPs, protect the RNA from degradation after release from their pre-mRNA precursors.
Functionally, one specific circRNA has been shown to contain an array of binding sites for miRNAs, thus serving as a molecular sink to prevent the miRNAs from interacting with their targets [72,73]. The snoRNA-protected intronic ncRNAs appear to trap a critical RNA binding protein RBFox2, thus titrating its active pool for regulated splicing in the cell [75]. In fact, the classic RNA that serves as a molecular sink is the very abundant 7SK RNA, which has been well characterized to bridge pTEFb to its inhibitor HEXIM1 in the inactive pool of the CTD kinase in the cell [63]. It is unlikely, however, that a molecular sink is the only function associated with various stable ncRNAs. For example, the intron-derived circRNAs sealed by the 2′–5′ phosphodiester bond appear to play a positive role in transcription of their host genes, although the mechanism has remained elusive [74]. This finding further highlights the functional importance of various sequences in the pre-mRNA of protein-coding genes, as they not only give rise to miRNAs and snoRNAs, but also produce various circRNAs that appear to have both cis and trans functions.
RNA as ligand
Both small molecules and proteins are well known for their abilities to bind and induce conformational changes of their protein partners, thereby invoking signaling. ncRNAs appear to have a similar role in modulating protein conformation. One such example is a DNA damage-induced ncRNA from the cyclin D1 promoter-proximal region. This ncRNA binds to the RNA binding protein TLS to induce its conformational changes to unmask another domain in the protein for additional protein–protein interactions to take place, eventually leading to transcriptional repression [50].
The miRNA Let-7 appears to also act like a ligand in activating the Toll-like receptor 7, which appears to be a critical event in Let-7-induced neurodegeneration [79]. Small RNAs as ligands have also been exemplified by piRNAs, which, upon incorporating into the PIWI complex, induce conformational changes of the PIWI protein (MIWI in mice) to permit its ubiquitination by a specific E3 ligase [80]. This ncRNA-induced signaling event appears to play a vital role in spermiogenesis by triggering the eventual clearance of the piRNA machinery, a pathway proven to be essential for producing mature sperms in the testis. These findings illustrate that ncRNAs can function as ligands to regulate the conformation of their target proteins to trigger the next set of molecular interactions in some important biological processes. Future structural studies of RNPs may elucidate detailed mechanisms underlying such ncRNA-induced molecular switches.
RNA as organizer of cellular structures
Many ncRNAs are quite large in size and have been referred to as macroRNAs. The best example is the nuclear enriched abundant transcript 1 (NEAT-1). NEAT-1 has two isoforms (the larger one is ~23 kb in length and the smaller one is 3.7 kb in human, 3.2 kb in mouse), both of which are localized to a specific nuclear domain known as paraspeckles [81,82]. The function of paraspeckles is largely known, although a more recent study suggests an active role of NEAT-1 in facilitating the expression of some antiviral genes [83]. A large number of RBPs have been identified to be part of this nuclear structure, although a few core factors, such as Nono, PSP1, and PSF, appear to be selectively concentrated in this nuclear domain [84]. Many repeat-containing RNAs have been shown to associate with this structure, suggesting that the domain might arise from clustering some specific classes of ncRNAs along with their RBPs [85,86]. The larger NEAT-1 isoform appears to play a critical role in organizing such clusters, as targeted degradation of this ncRNA disrupted the structure [87,88], and ectopic expression of this large, but not small, NEAT-1 isoform was sufficient to induce de novo formation of a paraspeckle-like structure around it [89].
The name of paraspeckle is due to the spatial relationship of the domain to another nuclear domain known as speckles [90]. As numerous factors implicated in the splicing reaction have been localized to this structure, it has been a cellular hallmark for the splicing machinery [91]. However, its primary function in pre-mRNA splicing has long been a subject of debate. A popular view is that this domain serves as a storage site for splicing factors; however, increasing evidence points to a more active role of the domain in gene expression via coordinating transcription and splicing reactions at its vicinity, thus suggesting that this nuclear domain may play a larger role in organizing the genome for concerted transcription and post-transcriptional processing events [92,93]. Interestingly, another large lncRNA, known as NEAT-2/MALAT-1 of ~7.5 kb in size, lies in the heart of individual nuclear speckles. The initial MALAT-1 transcript contains a tRNA-like structure at its 3′ end, which is processed to produce the mature MALAT-1 retained in the nucleus, releasing the tRNA-like small RNA to the cytoplasm [94]. Unlike NEAT-1, mature MALAT-1 does not seem to be responsible for the formation or maintenance of nuclear speckles [95]. However, depletion of this large lncRNA has been shown to affect specific events associated with nuclear speckles, such as SR protein phosphorylation [96], implying that the lncRNA is involved in various protein–protein interactions to facilitate the establishment and dynamics of this non-membrane-bound organelle in the nucleus. Interestingly, NEAT-2/MALAT-1 was originally identified as a nuclear ncRNA that was dramatically elevated in tumor cells [97], which appears to be important for metastasis of lung cancer [98], indicating that this macroRNA may have an active role in cancer initiation and/or progression through its function in regulated gene expression. It is however important to point out that knockout of either NEAT-1 or NEAT-2/MALAT-1 produced no obvious phenotypic defects, indicating that these ncRNAs are not essential for mouse development [95,99].
Contrary to the nuclear structures associated with active gene expression, other nuclear domains are functionally linked to gene repression, such as the Polycomb body in the nucleus, which contains protein complexes responsible for depositing repressive marks, such as H3K27me3, to chromatin. This domain contains numerous ncRNAs, including Tug 1 [100]. While the precise role of this lncRNA has remained unclear, its association with the Polycomb body may compete with some common gene expression regulators that are partitioned between active and repressive domains in the nucleus, and regulated exchange between these domains appears to be a key event in switching the functional states of many genes [101]. Therefore, specific lncRNAs may provide signals or docking sites for regulatory proteins or protein complexes, thereby contributing to the organization of the human genome in the 3D space of the nucleus. More recently, repeat-derived ncRNAs were suggested to be a key part of nuclear scaffold for maintaining chromosome territories [102]. Together, various nuclear domain-associated lncRNAs may be considered as part of nuclear skeleton in analogy with the cytoskeleton in the cytoplasm.
Secreted RNA as potential hormone
ncRNAs are made in the nucleus either from their own genes or genomic loci or processed from their host genes. As cells have very active machineries to degrade most transcribed RNAs, functional ncRNAs must have evolved some strategies to survive various RNA surveillance mechanisms. As described above, some ncRNAs have specific structures to protect their ends to make them inaccessible to exonucleases while others may gain protection by forming specific RNPs. A fraction of ncRNAs are able to not only survive degradation in the cell, but also make it to the extracellular space. So far, this has been documented for miRNAs, which appear to be assembled into microvesicles for secretion [103]. We are still early in understanding how some miRNAs are imported or assembled into microvesicles for secretion, and how the specificity, if any, might be established in such a process. In any case, the detection of secreted miRNAs in the circulation system seems to provide a unique set of biomarkers for disease diagnosis [104-106]. A more important question is what these secreted miRNAs might do in the circulation system. Do they function as hormones to act in distal organs? Initial studies provide some evidence for such a possibility [107,108]. Remarkably, some exogenous miRNAs from food supply might also have such a role [109], although the finding has remained to be substantiated [110]. Overall, the idea that RNAs can function as hormones has remained as a hypothetic function for secreted miRNAs.
In concluding this section, I wish to make the point that our current knowledge has significantly expanded the function of RNAs as information carriers. They appear to be able to perform a large array of cellular functions that have been ascribed to proteins. Importantly, we are still glimpsing at the tip of iceberg, despite the impression that many working principles have been elucidated with specific ncRNA examples.
STRATEGIES FOR FUNCTIONAL AND MECHANISTIC STUDIES OF ncRNA
Small ncRNAs, particularly miRNAs, are well known for their roles in diverse biological pathways. The existing examples of characterized lncRNAs have also demonstrated their widespread participation in biological functions, ranging from dosage compensation [111,112], cell cycle control [45,113], stem cell maintenance and differentiation [52,114,115], development [116-118], and cancer etiology and progression [47,119,120]. Given their functional resemblance to proteins, essentially all experimental strategies developed to decipher protein functions may be applied to ncRNA research; however, because of their uniqueness as a linear chain of nucleic acids and the ability to fold into multiple secondary and tertiary structures, new approaches are also needed to study their functions and action mechanisms. In this section, I briefly discuss some common and unique approaches developed for ncRNA research (Box 2).
Box 2.
|
Detection of binding events
| |||
| Abbreviation | Full name | Application | Ref |
| CLIP | Crosslinking IP | Protein-RNA interactions | 142 |
| ChIRP | Chromatin isolation by RNA purification | RNA interaction with genomic DNA | 130 |
| CHART | Capture hybridization analysis of RNA targets | RNA interaction with genomic DNA and proteins | 128 |
| CLASH | Crosslinking, ligation, sequencing of hybrids | RNA-RNA interactions | 33 |
|
| |||
|
Measurement of functional consequences
| |||
| Abbreviation | Full name | Application | Ref |
| RNA-seq | RNA sequencing | Gene expression profiling | 121, 122 |
| GRO-seq | Global nuclear Run-On | Detection of nascent RNA | 123 |
| Bru-seq | BrU labeling/chasing | Time course analysis of RNA | 124 |
| RP | Ribosome profiling | Studying translational control | 4, 161 |
|
| |||
|
Probing RNA secondary structure
| |||
| Abbreviation | Full name | Application | Ref |
| SHAPE | Selective 2’-hydroxyl acylation analyzed by primer extension | Chemical probing of RNA secondary structure | 132 |
| PARS | Parallel analysis of RNA structure | Enzymatic probing of RNA secondary structure | 133 |
| DMS-seq | Dimethyl sulphate-modified RNA for sequencing | Probing unpaired adenine and cytosine in RNA | 134, 135 |
|
| |||
|
Characterization of RNA binding proteins
| |||
| Abbreviation | Full name | Application | Ref |
| RNAComplete | RNA complete | RNA binding specificity | 144 |
| SeqRS | RNA sequence specificity landscapes | RNA binding affinity | 143 |
| IC | Interactome capture | mRNA bound proteins | 139 |
| MS2-TRAP | MS2-targged RNA affinity purification | Capture RBPs on RNA | 136 |
| RAT | RNA affinity in tandem | Capture RBPs on RNA | 137 |
| Csy4 Select | Csy4 selection of RNA-protein complexes | Capture RBPs on RNA | 157 |
|
| |||
|
Genome editing
| |||
| Abbreviation | Full name | Application | Ref |
| TALEN | Transcription activator-like effector nucleases | Targeted gene mutation | 156 |
| CRISPR | Clustered, regularly interspaced short palindromic repeats | Targeted gene deletion and insertion | 138, 157 |
| CRISPRi | CRISPR interference | Interrogate genomic loci to modulate gene expression | 158, 161 |
Experimental approaches to defining ncRNA function
As with protein-coding genes, one of the most important experimental approaches to study ncRNAs nowadays is to determine their unique expression patterns associated with a specific biological question under investigation and to conduct loss-of-function studies in a particular biological setting. Using modern genomics strategies, it has become a routine to profile gene expression by RNA-seq in any given biological system [121,122], which may be combined with various affinity methods to detect RNA (both coding and non-coding) at different stages of gene expression [123,124]. The identification of the entire set of expressed lncRNAs would allow comparison under different experimental conditions or between different cell types to identify differentially expressed lncRNAs [116,125]. The challenge is to determine on which specific lncRNA(s) to further study. Currently, most studies focus on differentially expressed lncRNAs that are expressed with sufficient abundance. By using siRNA or antisense oligonucleotides (ASO), the latter of which appear to be more efficient in depleting lncRNAs via endogenous RNase H activities [126], one can efficiently deplete specific lncRNAs to evaluate their functional requirement. If resources are available or permit, this loss-of-function approach may be applied genome-wide to obtain a comprehensive set of lncRNAs involved in some defined biological processes, as exemplified on stem cells [52].
The hard part of ncRNA research is to probe for the mechanism and explore new regulatory concepts. The cellular localization of specific ncRNAs may be first determined to obtain an approximation of their functional sites. As mRNAs are known to display remarkable localization patterns in the cell [127], the localization of ncRNAs, particularly lncRNAs, might be informative to their cellular functions. To understand the function of a specific lncRNA, it is often important to identify its protein partners. Furthermore, if the lncRNA under investigation acts in the nucleus to regulate gene expression, one will also need to determine its target genes. To identify protein partners, antibodies are very useful tools for protein research, but for lncRNA, one has to rely on some entirely distinct approaches. One such approach is to use affinity tagged (such as biotin) oligos to capture specific lncRNA followed by deep sequencing of linked DNA and/or by mass spectrometric analysis of associated proteins, a method known as CHART-seq [128], which has been applied to elucidate two-step spreading of Xist ncRNA complexes during X-chromosome inactivation [129]. A related method called ChIRP-seq was developed in parallel to survey lncRNA occupancy on genomic DNA [130]. This technique has been applied to probe the genomic interaction of the 7SK complex on so-called anti-pause enhancers [131].
To efficiently use this approach, it would be helpful to know the exposed RNA regions in the cell by probing RNA structure in living cells [132,133]. Two recent studies reported a more robust method based on dimethyl sulfate modification of exposed adenines and cytosines followed by deep sequencing of RNA containing the modified residues to achieve high-resolution mapping of the RNA secondary structure [134,135]. These new approaches will greatly accelerate the discovery of regulatory events on RNA targets by both ncRNAs and specific RBPs.
Another approach is to epitope tag an lncRNA with an MS2 moiety, thus permitting the capture of the lncRNA-containing RBP with an MS2 fusion protein [136]. An analogous strategy is to use an RNA tag that contains two specific hairpins, thus allowing tandem affinity purification of RNA–protein complexes [137]. This RNA-tagging strategy, however, can be problematic if the lncRNA only acts in cis or the overexpressed transcript does not effectively get assembled into its native RNP complexes. This problem can be addressed by using the latest genome editing technology to tag specific ncRNA genes [138] (see below). Given the nucleic acid nature of lncRNA, future studies may also pursue chemical engineering methods to take advantage of specific sequences or structure moieties to introduce affinity groups for lncRNA localization and affinity purification.
Studying ncRNA from the angle of RBPs
It is conceivable that lncRNA functions are mostly mediated by specific RBPs, and, thus, focusing on specific RBPs of interest may be an effective route to study lncRNA function and mechanism in general. Recent studies indicate that mammalian genomes may express at least 1000 RBPs [139], many of which may not even carry annotated RNA-binding domains [140]. As a matter of fact, we do not know the exact distinction between DNA-binding proteins and RBPs, as they have been traditionally studied based on their interactions with DNA or RNA. As a result, some DNA-binding proteins may also bind RNA and the converse may also be true. For example, two recent studies demonstrated that the PRC2, which is responsible for depositing the repressive H3K27me3 mark on histone, actually has high affinity for RNA [141], explaining its extensive interaction with nascent RNA in the cell [65].
An important point is that the cross-linking immunoprecipitation (CLIP) technology and various variants of the approach have demonstrated effectiveness in identifying protein-associated RNAs and mapping such interactions in the genome [142]. Efficient and high-throughput methods have also been developed to determine the RNA binding specificity of RBPs [143,144], and an increasing number of RBPs have been mapped to mammalian genomes using CLIP technologies. Although most published studies to date have been focused on understanding the function of RBPs in RNA metabolism, such as pre-mRNA splicing, the available mapping data indicate that many RBPs also show extensive interactions with diverse lncRNAs [145]. As the CLIP data accumulate and have been organized in the database [146], one may mine such data to identify proteins mapped to specific lncRNAs under investigation. With candidate RBPs and lncRNAs in hand, loss-of-function studies can then be performed to identify common targets for further mechanistic dissection, as exemplified by the study of p53-regulated gene expression that involves both an lncRNA (lincRNA-p21) and a specific RBP (hnRNP K) [147].
Challenges in structural analysis of RNPs
A common approach in mechanistic studies of proteins or protein complexes is to define specific protein domains engaged in a particular molecular interaction and probe a detailed interaction mechanism in crystal structure. Similar approaches are clearly needed for understanding RNA–protein interactions. The challenge in dissecting RNA domains involved in such an interaction with specific proteins has been showcased with HOTAIR, an lncRNA that interacts with two different chromatin remodeling complexes through distinct RNA segments [37]. However, there is a great uncertainty in dissecting domains with in vitro transcribed RNA, as RNA may adopt into distinct secondary structures when made in vitro versus produced inside cells where specific RBPs may be assembled onto the RNA during transcription and/or processing, which may take place in a sequential fashion. This may make it difficult to reconstitute RNPs that contain multiple protein components for biochemical studies.
In the protein world, ultimate mechanistic insights are obtained from NMR or crystallography. The structure of the largest RNA machine—the ribosomes in complex with tRNA and mRNA—has been resolved at the atomic levels [148,149], and similarly, structures of miRNAs in argonaute proteins have been determined [150-152]. The structural approach has also been applied to an H/ACA box snoRNP particle [153] and a spliceosome sub-complex [154]. In general, however, it has been quite difficult to obtain crystals of many other RNPs, such as the spliceosome, in part because of insufficient materials one can purify from the cell or the lack of ability to preserve relatively stable structures during the purification process for crystallization. The common practice in protein crystallization is to use recombinant proteins, but in light of various potential problems in assembling RNPs in vitro, it will be a major challenge to reconstitute large RBPs for structural studies.
Genome engineering to determine ncRNA function
Similar to investigating protein functions in biology, the decisive information is obtained in many cases by gene targeting, which has recently been applied to a set of lncRNAs [155]. We are at the dawn of applying this genetic approach to ncRNA research, especially in light of the recent development of the powerful TALEN and CRISPR/Cas technologies for genome engineering [138,156]. For instance, the CRISPR technology has been used to tag an lncRNA in its expression unit in the genome to allow capture of specific RNA–protein complexes assembled in vivo [157]. In this elegantly designed strategy, a small RNA hairpin is first inserted in the front of specific ncRNA under investigation in the genome by CRISPR. An inactive version of the Cys4 nuclease is next used to efficiently capture the hairpin as part of RNA hybrid along with associated proteins. The affinity-purified RNP is then released for biochemical analysis by using imidazole to activate the Csy4 nuclease. The CRISPR technology can also be used to selectively remove specific ncRNA sequences embedded in their host genes, such as those transcribed as part of introns, to study their functional requirements. Recently, a catalytic inactive form of Cas9 was exploited to develop the CRISPRi system [158,159], which permits both positive and negative modulation of endogenous genes [160] and real-time imaging dynamic movement of specific genomic loci [161]. It is anticipated that the rapidly evolving CRISPR-based genome editing technologies will find wide applications in studying genomic sequences encoding for both small and large ncRNA in the near future.
ncRNA as an integral part of genomics and proteomics
It has become increasingly evident that ncRNAs provide diverse regulatory functions in the cell, and regulatory RNA networks in general represent a crucial interphase between genomics and proteomics (Fig. 3). Both small and large ncRNAs are subjected to regulation by diverse mechanisms to control their expression, biogenesis, and degradation, all of which have been well documented with miRNAs and piRNAs [15,31]. As many lncRNAs are expressed from their own genes, a battery of transcription factors are likely involved in the regulation of these lncRNAs during development or in different cell types in a similar way to the regulation of protein-coding genes.
Figure 3.
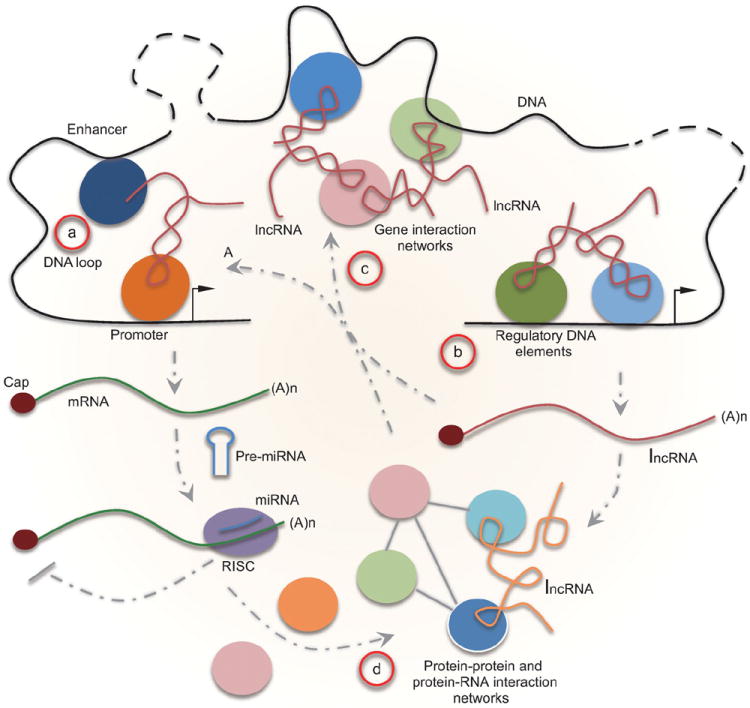
ncRNAs as integrated parts of gene networks. (a) ncRNAs mediate promoter–enhancer interactions to regulate the expression of various protein-coding genes. Protein-coding transcripts are also subjected to regulation by miRNAs to fine tune protein synthesis in the cytoplasm. (b) ncRNA genes produce various regulatory ncRNAs, which then participate in regulated expression of both protein-coding and non-coding genes. (c) ncRNAs may play a critical role in the organization of the genome in the nucleus to coordinate the expression of gene clusters. (d) Regulated gene expression at both the transcriptional and post-transcriptional levels determines the cell type-specific proteome and ncRNAs may also be extensively involved in protein interaction networks, which together contribute to gene networks in the cell.
Most lncRNAs have been characterized by their functions in the nucleus, and their interactions with various nuclear machineries may thus contribute to their nuclear retention. However, many lncRNAs are also detectable in the cytoplasm and clearly function there, as demonstrated with the BACE1-antisense transcript (BACE1-AS) and an Alu-containing lncRNA in the regulation of mRNA stability [162,163]. Because premature stop codons in mRNA trigger the nonsense-mediated RNA decay (NMD) [164], this raises the question of how various lncRNAs escape such a pathway. One possibility is that lncRNAs are not scanned by ribosome beyond immediate 5′ sequences [4,165], as the translation process is known to activate the NMD pathway [166]. However, the key NMD initiator Upf1 appears to have the capacity to bind mRNAs as well as lncRNAs in a translation-independent manner [167]. At this point, we have little knowledge about whether cytoplasmic lncRNAs are sensitive to NMD, which represents an interesting subject for future studies.
One exciting future research area is to decipher the contribution of lncRNAs to local and long-distance genomic interactions (Fig. 3a,b). Functional studies of eRNAs and certain lncRNAs have exemplified the critical role of ncRNAs in mediating enhancer–promoter interactions [46,56,168]. Recent studies suggest that the Xist complex explores some larger genomic domains to help spread the transcription repressor complex during X-chromosome inactivation [129,169]. This strategy may also be exploited for establishing both active and repressive domains that involve genomic segments separated by long linear distance on the same chromosomes or even from different chromosome, which may in turn contribute to the organization of the genome in the 3D space of the nucleus [170,171] (Fig. 3c). Research along this direction may represent a new frontier of ncRNA cell biology.
The intersection of ncRNA research with gene networks has well been established for miRNAs [172]. It is easily imaginable for numerous RNA-dependent protein–protein and protein–DNA interactions to exist in the cell, but systematic effort has yet to be undertaken to study such RNA-dependent interactions (Fig. 3d). Thus, analysis of gene networks would be incomplete without incorporating regulatory ncRNAs into various biological pathways. Towards this general goal, all classes of ncRNAs and their expression patterns have been organized in an integrated database [173]. Such a systems biology approach will greatly accelerate research on ribonomics and its integration with functional genomics and proteomics.
CONCLUSIONS
ncRNAs have undoubtedly become one of the ‘hot’ spots in modern biological and biomedical research. The existing data have abundantly demonstrated the connection of ncRNAs to diverse disciplines in biology, and have illuminated regulatory paradigms that have been largely attributed to proteins. As ncRNAs can be efficiently targeted by stable ASO, this approach may be explored as a method to target specific regulatory ncRNAs to understand their biological functions and action mechanisms in basic research and develop novel strategies for disease intervention in clinical applications. The era of ncRNA research has resulted in and benefited from the rapid advance in genomics technologies and informatics approaches that have been developed in recent years. However, we are clearly facing new challenges in dissecting the dark matter in the genome and understanding their mechanisms. Like many breakthroughs made in the history of life science, both opportunities and challenges equalize, which is up to prepared minds to seize the moment in order to make new breakthroughs.
Acknowledgments
The author wishes to thank Yang Shi for critical comments, and Xiao Li and Patrick Menzies for proofreading the manuscript. The author is also grateful to reviewers of this article who made numerous insightful suggestions.
FUNDING
Research in the author’s research has been supported by grants from National Institutes of Health.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Birney E, Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–29. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, Russell P, Ingolia NT, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bánfai B, Jia H, Khatun J, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Song X, Glass CK, et al. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 15.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piR-NAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng JC, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol. 2013;25:190–4. doi: 10.1016/j.ceb.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 18.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burge SW, Daub J, Eberhardt R, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MT, Cho H, Lesch HP, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–5. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–5. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 23.Joyce GF. RNA evolution and the origins of life. Nature. 1989;338:217–24. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 24.Gibert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- 25.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333–41. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–8. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 27.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–7. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 28.Johnston WK, Unrau PJ, Lawrence MS, et al. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–25. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 29.Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–35. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 30.Galej WP, Oubridge C, Newman AJ, et al. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–43. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helwak A, Kudla G, Dudnakova T, et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler-Bauer A, Zheng C, Chitsaz F, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–52. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punta M, Penny CC, Ruth YE, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bateman A, Coggill P, Finn RD. DUFs: families in search of function. Acta Crystallogr F. 2010;66:1148–52. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol. 2006;26:630–42. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Gene Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 40.Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hongay CF, Grisafi PL, Galitski T, et al. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–45. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 42.Osato N, Suzuki Y, Ikeo K, et al. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bevis M, Wahr J, Khan SA, et al. Bedrock displacements in Greenland manifest ice mass variations, climate cycles and climate change. Proc Natl Acad Sci USA. 2012;109:11944–8. doi: 10.1073/pnas.1204664109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Notani D, Ma Q, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter P, Blobel G. Mechanism of protein translocation across the endoplasmic reticulum. Biochem Soc Symp. 1982;47:183–91. [PubMed] [Google Scholar]
- 49.Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, Garcia-Bassets I, Benner C, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melo CA, Drost J, Wijchers PJ, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–35. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Lai F, Orom UA, Cesaroni M, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez-Elsner T, Gou D, Kremmer E, et al. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–23. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 58.Seila AC, Calabrese JM, Levine SS, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almada AE, Wu X, Kriz AJ, et al. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–3. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaida D, Berg MG, Younis I, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji X, Zhou Y, Pandit S, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–68. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaneko S, Son J, Shen SS, et al. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest-and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gardner EJ, Nizami ZF, Talbot CC, Jr, et al. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Gene Dev. 2012;26:2550–9. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 73.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 75.Yin QF, Yang L, Zhang Y, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–30. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 76.Brown JA, Valenstein ML, Yario TA, et al. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci USA. 2012;109:19202–7. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilusz JE, JnBaptiste CK, Lu LY, et al. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Gene Dev. 2012;26:2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitton-Fry RM, DeGregorio SJ, Wang J, et al. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–7. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–35. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 80.Zhao S, Gou LT, Zhang M, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24:13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 81.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–44. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fox AH, Lam YW, Leung AK, et al. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 83.Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 84.Naganuma T, Nakagawa S, Tanigawa A, et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–34. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prasanth KV, Prasanth SG, Xuan Z, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 86.Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wrna.5. [DOI] [PubMed] [Google Scholar]
- 87.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao YS, Sunwoo H, Zhang B, et al. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–41. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 91.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong XY, Wang P, Han J, et al. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakagawa S, Naganuma T, Shioi G, et al. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–9. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall LL, Carone DM, Gomez AV, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–19. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–44. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 104.Kosaka N, Yoshioka Y, Hagiwara K, et al. Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front Genet. 2013;4:173. doi: 10.3389/fgene.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gandhi R, Healy B, Gholipour T, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729–40. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 107.Png KJ, Halberg N, Yoshida M, et al. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–4. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 108.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by mi-croRNA. Cell Res. 2012;22:107–26. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dickinson B, Zhang Y, Petrick JS, et al. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–7. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- 111.Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 112.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 113.Nadal-Ribelles M, Solé C, Xu Z, et al. Control of Cdc28 CDK1 by a stress-induced lncRNA. Mol Cell. 2014;53:549–61. doi: 10.1016/j.molcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klattenhoff CA, Scheuermann JC, Surface LE, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yildirim E, Kirby JE, Brown DE, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–42. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 122.Zhou Y, Li HR, Huang J, et al. Multiplex analysis of polyA-linked sequences (MAPS): an RNA-seq strategy to profile poly(A+) RNA. Methods Mol Biol. 2014;1125:169–78. doi: 10.1007/978-1-62703-971-0_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–2. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paulsen MT, Veloso A, Prasad J, et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc Natl Acad Sci USA. 2013;110:2240–5. doi: 10.1073/pnas.1219192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sheik Mohamed J, Gaughwin PM, Lim B, et al. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–37. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maamar H, Cabili MN, Rinn J, et al. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Gene Dev. 2013;27:1260–71. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lecuyer E, Yoshida H, Parthasarathy N, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 128.Simon MD, Wang CI, Kharchenko PV, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Simon MD, Pinter SF, Fang R, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–9. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chu C, Qu K, Zhong FL, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu W, Ma Q, Wong K, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–95. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spitale RC, Crisalli P, Flynn RA, et al. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wan Y, Qu K, Zhang QC. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–9. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ding Y, Tang Y, Kwok CK, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 135.Rouskin S, Zubradt M, Washietl S, et al. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yoon JH, Srikantan S, Gorospe M. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58:81–7. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–80. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 140.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Davidovich C, Zheng L, Goodrich KJ, et al. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Konig J, Zarnack K, Luscombe NM, et al. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 143.Campbell ZT, Bhimsaria D, Valley CT, et al. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–81. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ray D, Kazan H, Chan ET, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–70. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 145.Xiao R, Tang P, Yang B, et al. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol Cell. 2012;45:656–68. doi: 10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li JH, Liu S, Zhou H, et al. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Korostelev A, Trakhanov S, Laurberg M, et al. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 149.Selmer M, Dunham CM, Murphy FV, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 150.Wang Y, Juranek S, Li H, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–61. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y, Juranek S, Li H, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–6. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Y, Sheng G, Juranek S, et al. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–13. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–7. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 154.Lin PC, Xu RM. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J. 2012;31:1579–90. doi: 10.1038/emboj.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei C, Liu J, Yu Z, et al. TALEN or Cas9—rapid, efficient and specific choices for genome modifications. J Genet Genomics. 2013;40:281–9. doi: 10.1016/j.jgg.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 157.Lee HY, Haurwitz RE, Apffel A, et al. RNA-protein analysis using a conditional CRISPR nuclease. Proc Natl Acad Sci USA. 2013;110:5416–21. doi: 10.1073/pnas.1302807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Larson MH, Gilbert LA, Wang X, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–96. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–65. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chew GL, Pauli A, Rinn JL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5″ leaders of coding RNAs. Development. 2013;140:2828–34. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Popp MW, Maquat LE. The dharma of nonsense-mediated mRNA decay in Mammalian cells. Mol Cells. 2014;37:1–8. doi: 10.14348/molcells.2014.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zund D, Gruber AR, Zavolan M, et al. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol. 2013;20:936–43. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
- 168.Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–30. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Engreitz JM, Pandya-Jones A, McDonel P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hu Q, Kwon YS, Nunez E, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nunez E, Fu XD, Rosenfeld MG. Nuclear organization in the 3D space of the nucleus—cause or consequence? Curr Opin Genet Dev. 2009;19:424–36. doi: 10.1016/j.gde.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bu D, Yu K, Sun S, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–15. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]


