Summary
In trypanosomes, mRNAs are processed by spliced leader (SL) trans splicing, in which a capped SL, derived from SL RNA, is spliced onto the 5′ end of each mRNA. This process is mediated by the spliceosome, a large and dynamic RNA-protein machinery consisting of small nuclear ribonucleoproteins (snRNPs) and non-snRNP proteins. Due to early evolutionary divergence, the amino acid sequences of trypanosome splicing factors exhibit limited similarity to those of their eukaryotic orthologs making their bioinformatic identification challenging. Most of the ~ 60 protein components that have been characterized thus far are snRNP proteins because, in contrast to individual snRNPs, purification of intact spliceosomes has not been achieved yet. Here, we characterize the non-snRNP PRP19 complex of Trypanosoma brucei. We identified a complex that contained the core subunits PRP19, CDC5, PRL1, and SPF27, as well as PRP17, SKIP and PPIL1. Three of these proteins were newly annotated. The PRP19 complex was associated primarily with the activated spliceosome and, accordingly, SPF27 silencing blocked the first splicing step. Interestingly, SPF27 silencing caused an accumulation of SL RNA with a hypomethylated cap that closely resembled the defect observed previously upon depletion of the cyclin-dependent kinase CRK9, indicating that both proteins may function in spliceosome activation.
Introduction
Nuclear pre-mRNA splicing, in which introns are removed from exonic sequences, is an essential step for the expression of many eukaryotic genes. The splicing process, which occurs by two consecutive transesterifications, is carried out by the spliceosome, a large and highly dynamic complex of the U1, U2, U4/U6 and U5 small nuclear ribonucleoproteins (snRNPs) and non-snRNP proteins (Smith et al., 2008; Wahl et al., 2009). After recognition of the splice sites and before the splicing process, a large spliceosomal complex is formed on the pre-mRNA that is known as the B complex, which contains all five snRNPs and many non-snRNP proteins. Subsequently, substantial structural and compositional alterations, including the exit of the U1 and U4 snRNAs and the exchange of protein components, lead to spliceosome activation. The first transesterification generates spliceosomal complex C which, after additional rearrangements, carries out the second splicing step. While it has recently been established that RNA catalyzes the splicing reaction (Fica et al., 2013), there are more than 200 spliceosome-associated proteins known in the human system several of which are indispensable for the splicing process (Wahl et al., 2009; Hegele et al., 2012). Among those is PRP19, a highly conserved splicing factor that is essential for the activation of the spliceosome (Ohi and Gould, 2002; Chan et al., 2003; Makarova et al., 2004). PRP19’s role in RNA splicing is through a heteromeric protein complex known as the Nineteen Complex (NTC) in the budding yeast Saccharomyces cerevisiae (Tarn et al., 1994) and the PRP19/CDC5L complex in humans (Makarova et al., 2004; Grote et al., 2010). Although the protein composition of the human and yeast complexes differ, they share the subunits PRP19, CDC5L/Cef1p (human/yeast nomenclature), SPF27/Snt309, and PRL1/Prp46. In the human system, these subunits were shown to form a salt stable core complex (Grote et al., 2010). Additionally, there are 3–4 non-homologous proteins in each system that are considered bona fide subunits of these complexes (Chanarat and Strasser, 2013) which, from here on, will be generally referred to as ‘PRP19 complexes’. Moreover, several other spliceosomal proteins were found to consistently co-purify with the PRP19 complex, although their association appears to be less stable than that of the bona fide subunits (Makarov et al., 2002; Ohi et al., 2002). In the human system, these proteins have been termed ‘PRP19-related’ proteins, and they include factors such as SKIP, SYF1, CRN, ISY1, etc. (Wahl et al., 2009).
Pre-mRNA splicing has been a focus in the protozoan parasites Trypanosoma brucei, Trypanosoma cruzi, and Leishmania spp., which cause devastating human diseases, because maturation of each and every mRNA in these organisms requires spliced leader (SL) trans splicing (Günzl, 2010; Michaeli, 2011; Preuβer et al., 2012). Protein coding genes in these trypanosomatids are arranged in long tandem arrays that are polycistronically transcribed. Individual mRNAs are then processed from precursor RNA by SL trans splicing and polyadenylation. In trans splicing, the SL is derived from the 5′ end of the small nuclear SL RNA and spliced onto the 5′ end of each mRNA. The SL carries a so-called cap4 structure, which comprises a standard m7G cap nucleotide and methylations of the first four SL nucleotides (Bangs et al., 1992). Cap4 formation appears to be a pre-requisite for trans splicing (Ullu and Tschudi, 1991; McNally and Agabian, 1992) although individual methylations are dispensable for the process (Arhin et al., 2006a,b; Zamudio et al., 2006; 2007). SL trans splicing is achieved by the same two transesterification reactions as in ‘cis splicing’ (Murphy et al., 1986; Sutton and Boothroyd, 1986). Furthermore, dependence of trans splicing on U snRNAs (Tschudi and Ullu, 1990) and on orthologs of various spliceosomal factors (Liang et al., 2006; Tkacz et al., 2008; 2010; Luz Ambrósio et al., 2009) demonstrated that trans splicing in trypanosomes is carried out by a spliceosomal complex that is similar to its human and yeast counterparts. However, characterization of the trypanosomal spliceosome has been difficult because, so far, a larger spliceosomal complex could not be quantitatively purified from trypanosomatids. Furthermore, trypanosome protein sequences are highly divergent from their human and yeast counterparts, making bioinformatic annotation of proteins and their genes challenging in some cases. Nevertheless, tandem affinity purification of the T. brucei snRNP proteins SmD1 (Luz Ambrósio et al., 2009), SmB (Palfi et al., 2009), and the Leishmania tarentolae snRNP proteins SmD3, LSm3 and U1A (Tkacz et al., 2010) led to the identification of several new splicing factors increasing the trypanosomatid repertoire of known spliceosomal proteins to ~ 60 (Günzl, 2010; Preuβer et al., 2012). Most of these proteins are snRNP proteins and relatively few non-snRNP proteins have been identified so far, suggesting that mainly individual snRNPs were purified and that many non-snRNP proteins remained undetected in these purifications (Günzl, 2010). PRP19 is such a spliceosomal non-snRNP protein that was identified by sequence homology and shown to be essential for cell viability, trans splicing and removal of the PAP (poly A polymerase) intron in T. brucei (Tkacz et al., 2010). In addition, PRP19 silencing led to hypomethylation of the SL RNA cap. Little is known, however, about PRP19 complex subunits. While CDC5 (Günzl, 2010) and PRL1 (Tkacz et al., 2010) co-purified with splicing complexes and were then identified bioinformatically, an SPF27 ortholog has been elusive. Furthermore, only a few homologs of PRP19-related proteins have been identified so far (Günzl, 2010). (Please note that since T. brucei is a parasite of humans, when possible, we prefer to use the nomenclature of the human system for trypanosome splicing factors).
Here we have tandem affinity-purified T. brucei PRP19 and mass spectrometrically identified 47 co-purifying proteins. Among those were 35 spliceosomal orthologs, which included the three newly annotated proteins SPF27, SKIP, and PPIL1, as well as six proteins that appear to be conserved only among trypanosomatids. A sedimentation analysis revealed a stable PRP19 complex of seven subunits that, although containing the four core subunits, deviated in protein composition from its human and yeast counterparts. As expected, PRP19 complex subunits were primarily associated with the activated spliceosome. Finally, silencing of SPF27 affected SL RNA cap methylation similarly to the previous PRP19 knockdown. However, as our data indicate, cap4 methylation defects may be a general phenomenon of blocking spliceosome activation in trypanosomes.
Results
Tandem affinity purification of PRP19 and identification of co-purified proteins
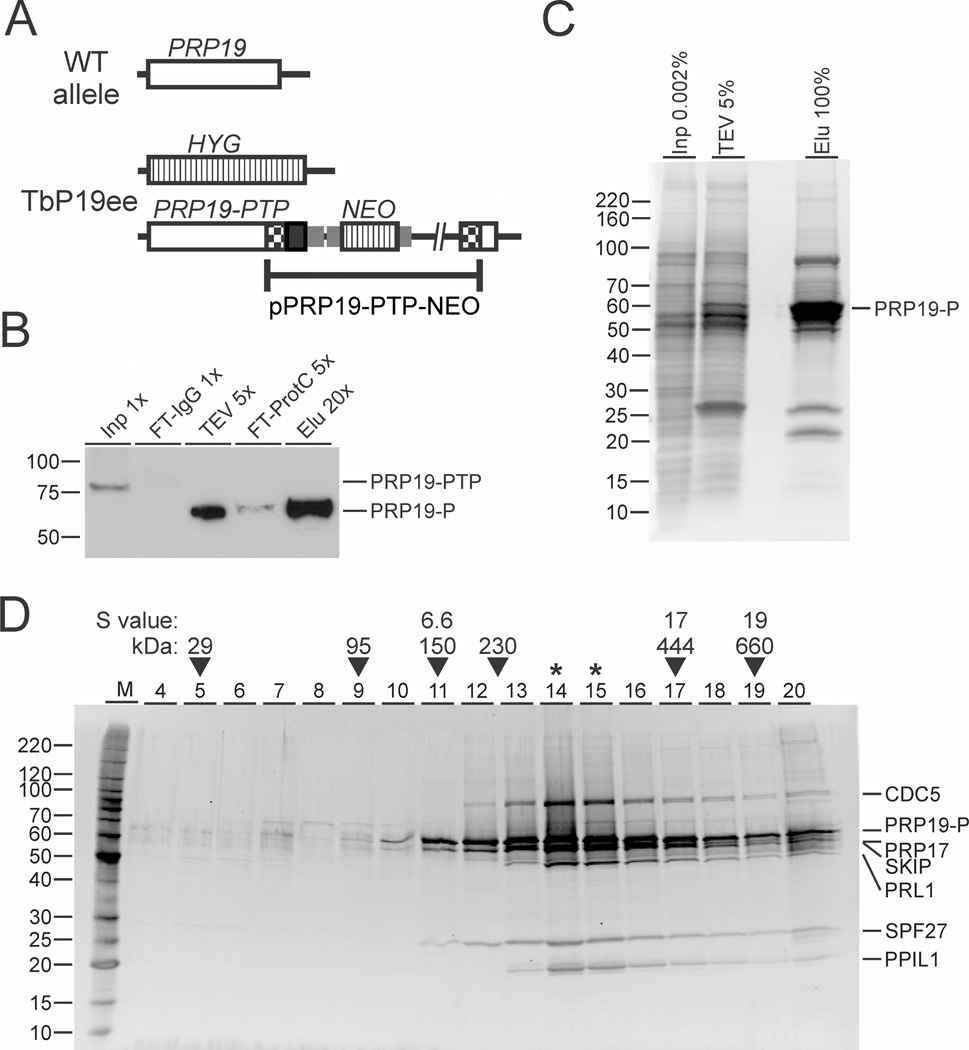
For tandem affinity purification (TAP) of PRP19 (accession number Tb927.2.5240 at www.GeneDB.org or www.TriTrypDB.org) we generated the insect-stage, procyclic T. brucei cell line TbP19ee, which expressed PRP19 with a C-terminal PTP tag (PRP19-PTP) and no untagged PRP19. The PTP tag is a composite tag consisting of a protein C epitope (ProtC) followed by a tobacco etch virus (TEV) protease cleavage site and tandem protein A domains (ProtA) (Schimanski et al., 2005b). The cell line was obtained by replacing one PRP19 allele of wild-type procyclics with the coding region of the selectable marker hygromycin phosphotransferase (HYG) and by targeted integration of plasmid PRP19-PTP-NEO into the remaining PRP19 allele, thereby fusing the PTP sequence to the PRP19 gene (Fig. 1A). Since TbP19ee cell cultures did not exhibit a growth defect (data not shown) and PRP19 is an essential gene in T. brucei (Tkacz et al., 2010), we inferred that the tag did not disrupt PRP19 function. We then prepared extract from 5 × 1010 trypanosomes and performed PTP tandem affinity purification of PRP19-PTP. While the expected size of PRP19-PTP is 74 kDa, anti-ProtC immunoblotting of extract revealed a single band of ~ 80 kDa (Fig. 1B, lane 1). PTP-based TAP involves IgG affinity chromatography through ProtA, release of the tagged protein by TEV protease cleavage, and recapture of the tagged protein by anti-ProtC immunoaffinity chromatography (Schimanski et al., 2005b). Immunoblot monitoring of the TAP procedure showed that both chromatography steps led to efficient depletion of the tagged protein from extract and from TEV protease eluate (lanes 2 and 4) and that the final eluate contained a substantial amount of purified PRP19 (lane 5). Note, that removal of ProtA decreased the molecular mass of PRP19-P by ~ 15 kDa. The final eluate was then separated on a SDS-polyacrylamide gel and stained with Coomassie blue (Fig. 1C). The profile of protein bands exhibited a massive protein enrichment in the 55–65 kDa range, which included PRP19-P, and three major protein bands with apparent sizes of ~ 85, 26 and 22 kDa. In addition, many bands of minor abundance and different size were detected, which could be components of the spliceosome.
Fig. 1. Trypanosoma brucei has a stable PRP19 complex of seven subunits.
A. Schematic (not to scale) of a PRP19 wild-type allele and the PRP19 locus in procyclic TbP19ee cells that exclusively express PRP19-PTP and no untagged PRP19. PRP19, HYG/NEO coding regions and PTP tag are indicated by open, striped and black boxes respectively. Gene flanks for RNA processing signals are drawn as smaller gray boxes. Checkered boxes indicate plasmid-derived portions of the PRP19 coding region.
B. Immunoblot monitoring of the PRP19-PTP tandem affinity purification. PRP19-PTP and PRP19-P (after removal of the ProtA domains) were detected with the monoclonal anti-ProtC antibody HPC4 in crude extract (Inp), flowthrough of the IgG column (FT-IgG), TEV protease eluate (TEV), flowthrough of the anti-ProtC column (FT-ProtC) and the final eluate (Elu). x-Values indicate relative amounts loaded.
C. Aliquots of extract, the TEV protease eluate and the final eluate were separated on a 10–20% SDS-polyacrylamide gradient gel and stained with Coomassie blue. Percentages indicate relative amounts analyzed.
D. The eluate of a PRP19-PTP tandem affinity purification was sedimented through a 10–40% linear sucrose gradient by ultracentrifugation and 20 fractions were taken from top to bottom. Proteins from each fraction were separated on a 10–20% SDS-polyacrylamide gradient gel and stained with Sypro Ruby. Proteins specified on the right were identified by excision of individual protein bands and LC/MS/MS analysis. For comparison, sedimentations of Taq DNA polymerase (95 kDa), IgG (150 kDa, 6.6S), apoferritin (444 kDa, 17S) and thyroglobin (660 kDa, 19S) were analyzed in parallel gradients (arrowheads). In addition, sedimentation peaks from previous, comparable experiments of TEV protease (29 kDa) and the SNAPc/TRF4/TFIIA transcription factor complex (230 kDa) are indicated as well. Asterisks indicate the sedimentation peak.
To identify co-purified proteins, we performed liquid chromatographytandem mass spectrometry (LC/MS/MS) with the final TAP eluate that was not separated electro-phoretically. By limiting the analysis to proteins that were identified by at least two unique peptides and excluding recurrent PTP purification contaminants, we identified 47 proteins (Table 1). Of those, 35 proteins were homologs of known spliceosomal proteins, two of the six proteins without annotation (conserved hypothetical) were previously co-purified with trypanosomatid splicing complexes (Luz Ambrósio et al., 2009; Palfi et al., 2009), and only six of the annotated proteins did not have a known splicing function. Moreover, the list contains several orthologs of human and yeast PRP19 complex subunits as well as PRP-related proteins (see below) confirming that the tagged PRP19 was functionally incorporated into RNA splicing complexes and indicating that the PRP19 purification was highly specific.
Table 1.
Mass spectrometric identification of PRP19 co-purified proteins.
| Annotationa | Accession # | Mr (kDa) | Protein score |
# unique pept. |
% coverage | # pept. |
|---|---|---|---|---|---|---|
| PRP19 | Tb927.2.5240 | 54.2 | 65,296 | 32 | 75.1 | 39 |
| CDC5/Cef1 | Tb927.5.2060 | 80.1 | 31,713 | 57 | 82.0 | 64 |
| CDC40/PRP17 | Tb927.3.1930 | 52.8 | 9,959 | 33 | 83.7 | 37 |
| SKIP/Prp45b,c | Tb927.9.5880 | 50.4 | 8,862 | 25 | 50.3 | 30 |
| PRL1/Prp46 | Tb927.10.10170 | 48.8 | 6,349 | 23 | 68.2 | 23 |
| SPF27/Snt309b,c | Tb927.11.14150 | 24.5 | 5,775 | 16 | 74.8 | 23 |
| PPIL1b | Tb927.8.2090 | 21.7 | 5,615 | 11 | 67.0 | 13 |
| DHX38/Prp16 | Tb927.10.7280 | 121.2 | 4,398 | 63 | 66.6 | 63 |
| U5-200K/Brr2 | Tb927.5.2290 | 249.3 | 3,936 | 67 | 37.2 | 67 |
| [U5-]PRP8 | Tb927.9.11110 | 276.8 | 3,811 | 79 | 36.0 | 79 |
| HSP73 (HSP70) | Tb927.11.11330 | 76.0 | 3,523 | 36 | 65.1 | 40 |
| CRN/Clf1, Syf3 | Tb927.10.9660 | 87.6 | 2,654 | 25 | 45.8 | 25 |
| CACTINc | Tb927.11.11610 | 64.6 | 2,303 | 24 | 57.2 | 24 |
| U5-116K/Snu114 | Tb927.11.15430 | 105.4 | 2,165 | 33 | 43.8 | 33 |
| SYF1 | Tb927.5.1340 | 92.1 | 1,846 | 21 | 35.9 | 21 |
| KIAA1604/Cwc22 | Tb927.11.10750 | 66.8 | 1,737 | 20 | 35.3 | 20 |
| Conserved hypoth.c | Tb927.8.5650 | 103.2 | 1,233 | 22 | 34.8 | 22 |
| Conserved hypoth. | Tb927.2.3400 | 36.6 | 852 | 10 | 32.9 | 10 |
| U5-CWC21 | Tb927.9.3480 | 20.4 | 693 | 10 | 55.2 | 10 |
| U2A′ (U2-40K) | Tb927.10.14360 | 36.5 | 662 | 15 | 65.3 | 15 |
| Conserved hypoth.c | Tb927.10.11230 | 34.7 | 567 | 11 | 42.7 | 11 |
| SmD1 | Tb927.7.3120 | 11.7 | 528 | 4 | 45.3 | 4 |
| LSm2 | Tb927.8.5180 | 13.3 | 507 | 5 | 44.8 | 5 |
| LSm8 | Tb927.3.1780 | 14.0 | 435 | 6 | 43.4 | 6 |
| VSG pseudogene | Tb927.5.4980 | 48.5 | 428 | 5 | 20.0 | 5 |
| Aminopeptidase | Tb927.11.3570 | 98.0 | 408 | 9 | 15.5 | 9 |
| AD002/CWC15 | Tb927.10.11950 | 22.4 | 403 | 6 | 33.2 | 6 |
| U5-40K | Tb927.11.11150 | 35.0 | 398 | 5 | 18.7 | 5 |
| SmF | Tb927.9.10250 | 8.4 | 394 | 4 | 56.0 | 4 |
| PRP31 | Tb927.10.10700 | 39.6 | 371 | 4 | 13.2 | 4 |
| SSm2-1 (Sm15K) | Tb927.6.4340 | 12.8 | 342 | 3 | 33.3 | 3 |
| ISY1 | Tb927.8.1930 | 31.7 | 313 | 5 | 29.0 | 5 |
| SmD2 | Tb927.2.5850 | 12.5 | 282 | 4 | 35.1 | 4 |
| PABP2 (PABP1) | Tb927.9.10770 | 62.2 | 282 | 7 | 15.5 | 7 |
| Conserved hypoth. | Tb927.11.2960 | 12.0 | 280 | 5 | 40.7 | 5 |
| Actin A | Tb927.9.8850 | 41.9 | 266 | 3 | 11.4 | 3 |
| SSm2-2 (Sm16.5K) | Tb927.10.4950 | 14.7 | 255 | 4 | 31.3 | 4 |
| SmE | Tb927.6.2700 | 9.7 | 217 | 3 | 37.2 | 3 |
| SmB | Tb927.2.4540 | 12.3 | 169 | 5 | 40.4 | 5 |
| U2B″ | Tb927.3.3480 | 13.6 | 167 | 5 | 27.4 | 5 |
| Argonaute AGO1 | Tb927.10.10850 | 99.2 | 162 | 3 | 3.7 | 3 |
| ALBA3 | Tb927.4.2040 | 20.8 | 153 | 2 | 10.5 | 2 |
| Conserved hypoth.c | Tb927.4.3540 | 41.2 | 136 | 3 | 12.9 | 3 |
| Pumilo PUF2 | Tb927.10.12660 | 92.2 | 126 | 2 | 4.5 | 2 |
| LSm4 | Tb927.11.13960 | 14.2 | 122 | 2 | 15.0 | 2 |
| Conserved hypoth.c | Tb927.8.5840 | 21.5 | 118 | 2 | 10.7 | 2 |
| Cyclophilin (PPlase) | Tb927.9.11740 | 20.3 | 103 | 3 | 24.0 | 3 |
Annotation is according to the human/yeast systems unless there is a trypanosome-specific name. Names in parentheses refer to alternative names published for T. brucei.
Proteins that were annotated in this study.
[putative] Spliceosomal proteins that co-purified in a trypanosome splicing complex for the first time.
PRP19 co-purifying proteins, identified by LC/MS/MS, are ranked according to protein score. Listed proteins have a score of > 100 and were identified by 2 or more unique peptides. Proteins that cosedimented with PRP19 in linear sucrose gradients are highlighted in yellow, proteins denoted as PRP19 subunits or PRP19-related proteins in other systems in green, un-annotated proteins in blue, and [putative] spliceosome components in gray. Annotated proteins without a known role in RNA splicing were not highlighted. In addition, standard contaminants of trypanosome TAPs such as α/β tubulin, translation elongation factor, retrotransposon hot spot proteins, histones, low scoring ribosomal proteins, and chaperones were eliminated from this list, although it should be noted that prefoldin chaperones were particularly enriched in these purifications.
Identification of a stable T. brucei PRP19 complex of seven subunits
To determine the proteins that form a stable PRP19 complex, we sedimented PRP19-PTP-purified proteins through a linear sucrose gradient by ultracentrifugation. We took 20 fractions of the gradient from top to bottom, collected the proteins from each fraction with a hydrophobic resin and stained the proteins after electrophoretic separation in the gel with sypro ruby (Fig. 1D). The major protein bands of the purification co-sedimented with peak signals in fractions 14 and 15, which is between the ~ 230 kDa-large T. brucei SNAPc/TRF4/TFIIA transcription factor complex (Schimanski et al., 2005a) and the 444 kDa-large apoferritin marker. Furthermore, in fraction 20, major and minor bands were co-detected indicating that the purified material contained larger complexes that sedimented to the bottom of the gradient.
To identify the proteins in the bands seen in fractions 14 and 15, gel slices were excised and their protein content analyzed by LC/MS/MS. We identified seven proteins that coincided with the seven proteins that obtained the highest protein score listed in Table 1. The largest protein was identified as trypanosome CDC5. The strong bands of 65 and 55 kD contained tagged PRP19-P and PRP17 (Palfi et al., 2009) respectively. The strong PRP19 signal is consistent with PRP19 forming a tetramer within a single PRP19 complex (Ohi et al., 2005). In contrast, PRP17 was not a component of previously characterized human and S. cerevisiae PRP19 complexes and, therefore, has not been considered a bona fide component of the PRP19 complex (Wahl et al., 2009; Chanarat and Strasser, 2013). On the other hand, systematic yeast-2-hybrid screens have identified PRP17 as a direct interactor of the PRP19 WD40 domain (Fig. 2A) in human and yeast systems (Ren et al., 2011; Hegele et al., 2012). Moreover, in Schizosaccharomyces pombe, PRP17 was consistently detected in tandem affinity purifications of PRP19 complexes, and it was determined that there are four PRP17 molecules per complex (Ren et al., 2011). Our results corroborate these findings. In T. brucei, PRP17 co-purified and co-sedimented with PRP19 identifying it as a bona fide subunit of the trypanosome PRP19 complex. Moreover, the strong signals of the 65 and 55 kDa bands suggest that there is more than one PRP17 molecule present in the trypanosome complex. However, the strong 55 kDa signal appears to be not confined to PRP17 alone because we detected a second protein in this band. This protein is encoded in gene Tb927.9.5880, which has been annotated as a ‘conserved, hypothetical protein’ indicating that its sequence is conserved only among kinetoplastids. A standard BLAST search identified an internal SKIP/SNW domain strongly indicating that this protein is the ortholog of the spliceosomal Ski-binding protein (SKIP) and PRP45p of mammals and S. cerevisiae respectively (Fig. 2A and B). Interestingly, in the human and yeast systems, SKIP/PRP45p is not considered to be a PRP19 complex subunit but a PRP19-related/associated protein (Makarova et al., 2004; Fabrizio et al., 2009).
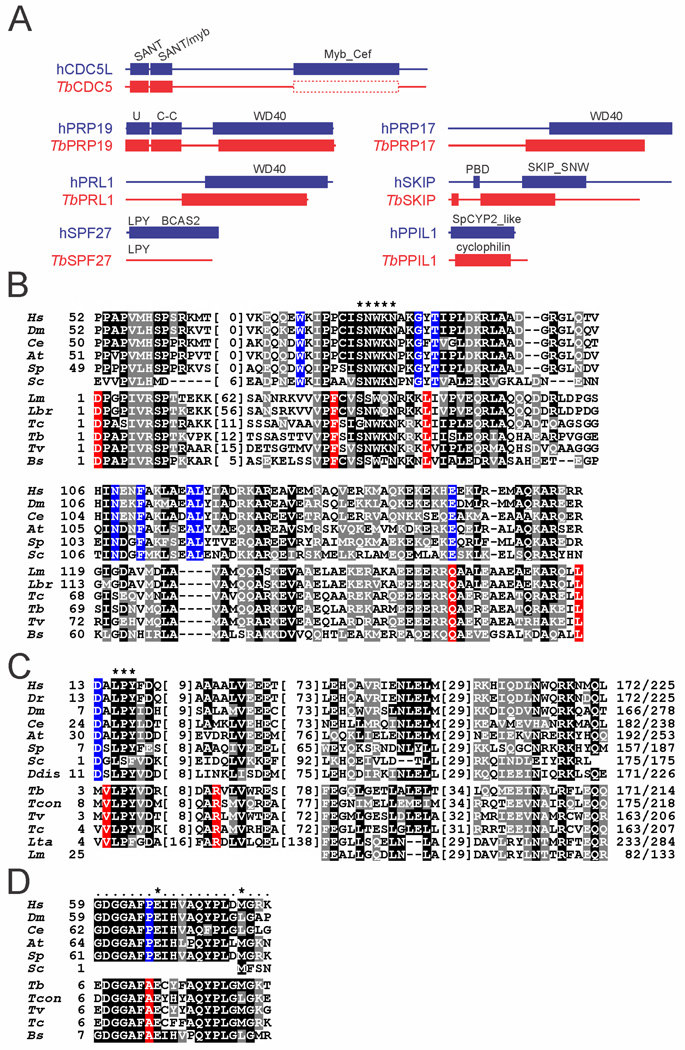
Fig. 2. Domain structure and sequence conservation in PRP19 complex subunits.
A. Domain structure drawn to scale of trypanosome PRP19 complex subunits and their human orthologs. The protein domains were identified by NCBI blastp searches of the Conserved Domain Database (Marchler-Bauer et al., 2011) (E-values are specified for T. brucei protein domains): CDC5: SANT domain (cd00167), E = 1.03e−08; SANT/myb-like domain (cd11659), E = 8.07e−10; the very limited sequence similarity between the human Myb_Cef domain (pfam11831) and the corresponding region of trypanosomatid CDC5 sequences is indicated by the dashed box. PRP19: U-Box (smart00504), E = 1.06e−14; PRP19 domain (coiled-coiled [C-C] protein interaction domain, pfam08606), E = 4.87e−20; WD40 (cd00200), E = 5.77e−18. PRL1: WD40 (cd00200), E = 1.4e−75. SPF27: while the LPY motif is present in T. brucei SPF27, the human BCAS2 domain (pfam05700) was not recognized. PRP17: WD40 (cd00200), E = 2e−37. SKIP: PPIL1-binding domain (PBD); SKIP_SNW domain (pfam02731), E = 7.51e−08. PPIL1: cyclophilin domain [cd00317], E = 2.18e-27 (note that while the trypanosome sequence returned the general cyclophilin domain, the more specific SpCYP2_like cyclophilin domain (cd01922) was identified in the human ortholog.
B. Multiple sequence alignment of the SKIP/SNW domain of Homo sapiens (Hs, accession number NP_036377), Drosophila melanogaster (Dm, AGB95213), Caenorhabditis elegans (Ce, CAA98552), Arabidopsis thaliana (At, AEE35947), Schizosaccharomyces pombe (Sp, CAB41231), Saccharomyces cerevisiae (Sc, P28004), and the kinetoplastids Leishmania major (Lm, LmjF.15.1030), Leishmania braziliensis (Lbr, LbrM.15.1070), Trypanosoma cruzi (Tc, TcCLB.509445.20), T. brucei(Tb, Tb927.9.5880), Trypanosoma vivax(Tv, TvY486_0902160), and Bodo saltans (Bs, ACI16065). Since the N-terminal ~ 50 amino acids of the domain are only weakly conserved among kinetoplastids, they were omitted from the alignment. Numbers indicate the position within the SKIP/SNW domain. Positions with more than 50% similarity or identity are shaded in gray and black respectively. Identical positions in model organisms without any conservation in kinetoplastids are shaded blue and identical position in kinetoplastids without conservation in model organisms are shaded in red. A hyphen indicates lack of an amino acid at this position. Numbers in parentheses specify lengths of non-conserved insertions. The highly conserved SNWKN motif is indicated by asterisks.
C. Corresponding alignment of four short conserved domains in SPF27 orthologs. Asterisks mark the highly conserved LPY motif, which, among Leishmania species, is only present in L. tarentolae as LPF. Accession numbers: Hs, NP_005863; Dr(Danio rerio), NP_001007775; Dm, NP_651596; Ce, NP_498360; At, NP_566599; Sp, CAB57933; Sc, EEU07188; Ddis (Dictyostelium discoideum), XP_640072; Tb, Tb927.11.14150; Tcon (Trypanosoma congolense), TcIL3000.11.14450; Tv, TvY486_1114990; Tc, TcCLB.511727.110; Lta (Leishmania tarentolae), LtaP32.0970; Lm, LmjF.32.0900.
D. Multiple sequence alignment of the 21 amino acid-long PPIL1 binding domain in SKIP orthologs. Numbers indicate positions relative to the starting methionine. The two key residues for PPIL1 binding are marked by asterisks.
The band of ~ 50 kDa has a lower strength signal and contained only a single protein, namely the trypanosome ortholog of human PRL1 and yeast PRP46p. It is considered a core component of the yeast and human PRP19 complexes (Makarova et al., 2004; Fabrizio et al., 2009). Trypanosomatid PRL1 had been identified previously, co-purifying with LSm3 in Leishmania tarentolae (Tkacz et al., 2010). Accordingly, T. brucei PRL1 shares clear sequence homology to human PRL1 (E = e−80) including a C-terminal WD40 protein-protein interaction domain (Fig. 2A).
The sixth protein, with an apparent size of 25 kDa, was found to be encoded by gene Tb927.11.14150. The corresponding amino acid sequence was conserved only among kinetoplastid organisms and, in contrast to the SKIP identification, did not reveal a conserved sequence motif. However, a key subunit of similar size of PRP19 complexes across systems is SPF27. To analyze whether this trypanosomatid protein could be a SPF27 ortholog, we performed a multiple sequence alignment of SPF27 orthologs from model organisms and of the kinetoplastid orthologs of Tb927.11.14150. As shown in Fig. 2C, we found four short domains of sequence conservation between the two orthologous groups including a nearly invariant LPY motif near the N-terminus of the protein. Although this degree of sequence conservation is not strong, the findings that SPF27 is a bona fide PRP19 complex subunit in all systems and that Tb927.11.14150 was a major protein in the PRP19 purification cosedimenting with the PRP19 complex subunits argue that this protein is trypanosome SPF27.
Analysis of the smallest band of 21 kDa revealed a protein that is encoded by gene Tb927.8.2090 and had been annotated as a cyclophilin-type peptidyl-prolyl cis-trans isomerase in the genome data base because it harbors the conserved domain of this enzyme class (Fig. 2A). The protein was identified previously co-purifying with SmB and annotated as cyclophilin CYP2 (Palfi et al., 2009). However, a search for the closest homolog of this protein in the human genome returned the PRP19-related factor PPIL1. A multiple sequence alignment of the trypanosomatid orthologs of this protein and PPIL1 sequences from different model organisms showed sequence conservation along the whole protein (Supporting Information Fig. S1). Moreover, human PPIL1 binds to a 21 amino acid-long region in the N-terminal part of SKIP (Wang et al., 2010). This region is highly conserved in trypanosome SKIP sequences including two key residues (Fig. 2D). Further support for Tb927.8.2090 being a PPIL1 ortholog comes from a comparison with yeast genome databases. While S. pombe harbors PPIL1 and full-length SKIP orthologs, S. cerevisiae does not have a PPIL1 ortholog (spliceosome database; Cvitkovic and Jurica, 2013) and its SKIP protein lacks the N-terminal domain including the PPIL1 binding site (Wang et al., 2010). Accordingly, a blastp search of the S. pombe genome database with the Tb927.8.2090-encoded amino acid sequence identified the PPIL1 ortholog Cyp1 as the most similar sequence with a low Expect Value (E = 2e−25) whereas the same search of the S. cerevisiae database returned the mitochondrial peptidylprolyl isomerase CPR3 as the most similar sequence with a considerably higher Expect Value (E = 9e−14). We therefore concluded that gene Tb927.8.2090 encodes T. brucei PPIL1.
Finally, to confirm the characterization of the PRP19 complex, we generated a cell line that expressed CDC5 with a C-terminal PTP tag. Tandem affinity-purification of CDC5-PTP and sedimentation analysis revealed a protein complex with similar banding pattern and sedimentation property as the PRP19-purified complex (Supporting Information Fig. S2). Accordingly, mass spectrometry of the excised protein bands revealed the same seven proteins as in the PRP19-PTP-purified complex. Thus, we have identified a stable PRP19 complex in T. brucei consisting of seven subunits.
SPF27 silencing affects trans and cis splicing of pre-mRNA
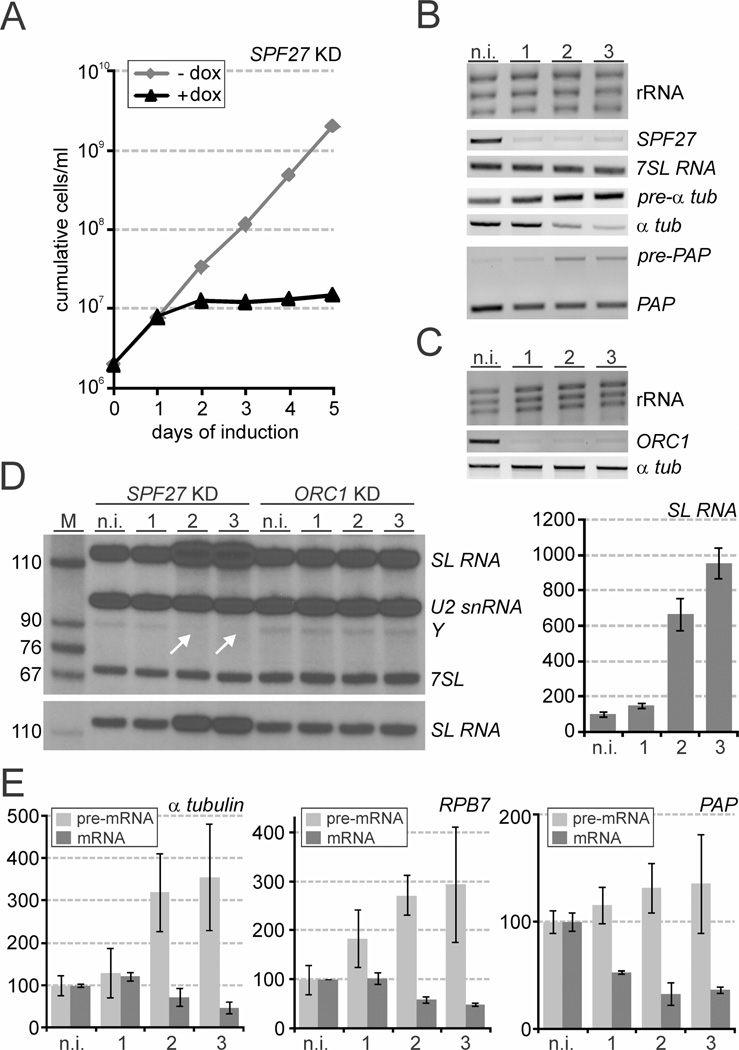
Next, we wanted to verify for one co-purified subunit whether its depletion in cells had effects on cell viability, RNA splicing and SL cap4 formation, which corresponded with the previous PRP19 silencing experiment (Tkacz et al., 2010). We chose SPF27 since it was the least conserved subunit of the PRP19 complex. By stable integration of a tetracycline repressor-regulated SPF27 stem-loop construct into procyclic 29-13 cells, which constitutively express the tetracycline repressor and T7 RNA polymerase (Wirtz et al., 1999), we generated a cell line for conditional SPF27 silencing. Induction of the SPF27 knockdown with the more stable tetracycline derivative doxycycline, which was added to the medium, stopped culture growth between one and two days of induction (Fig. 3A). Subsequently, the cell density remained nearly constant until day 5 of the experiment strongly indicating that SPF27 is an essential gene (Fig. 3A). Interestingly, while the previous PRP19 knockdown generated a similar growth defect (Tkacz et al., 2010), depletion of the essential snRNP proteins LSm2 and U5-Cwc21 led to rapidly declining cell numbers in our hands (Luz Ambrósio et al., 2009), suggesting that disabling the PRP19 complex affected trypanosomes differently than depletion of snRNP proteins. This difference appears not to be a consequence of inefficient gene knockdown because a semi-quantitative RT-PCR analysis showed that SPF27 mRNA was rapidly and strongly reduced in the presence of doxycycline (Fig. 3B). Analyzing total RNA prepared from non-induced cells and from cells in which SPF27 was silenced for 1 to 3 days clearly revealed pre-mRNA splicing defects. While the amounts of the control RNAs, rRNA and the non-spliceosomal 7SLRNA, which is the RNA component of the signal recognition particle (Michaeli et al., 1992), did not change in these RNA preparations, semi-quantitative RT-PCR analysis clearly indicated that the abundance of mature α tubulin mRNA decreased upon SPF27 silencing whereas, in random hexamer-derived cDNA of the same RNA preparations, the signal of unspliced α tubulin pre-mRNA, lacking the SL, clearly increased (Fig. 3B). Similarly, a competitive PCR of cDNA across the PAP intron showed a relative increase and decrease of unspliced and mature PAP RNA respectively (Fig. 3B). Together, these results revealed a splicing defect both in SL trans splicing (α tubulin mRNA) and in cis splicing (PAP mRNA).
Fig. 3. SPF27 silencing halted trypanosome proliferation and inhibited splicing of pre-mRNA.
A. Procyclic trypanosome culture growth in the absence and presence of doxycycline, which induced SPF27 silencing.
B. Analysis of total RNA prepared from non-induced cells (n.i.) and cells in which SPF27 was silenced for 1, 2 or 3 days. rRNA was detected by ethidium bromide staining, 7SL RNA and [pre-]mRNA of α tubulin (αtub) by semi-quantitative PCR and [pre-]mRNA of poly-A polymerase (PAP) by competitive PCR.
C. Corresponding analysis of rRNA, ORC1 mRNA and α tubulin mRNA in ORC1-silenced cells.
D. Left panels, primer extension of total RNA prepared from SPF27- and ORC1-silenced cells with three 32P-5′-endlabeled oligonucleotides detecting SL RNA, U2 snRNA, the SL intron after the first step of SL trans splicing in the Y structure, and the 7SL RNA by denaturing PAGE and autoradiography. Arrows indicate the loss of the Y structure intermediate in SPF27-silenced cells. The lower panel is a lower exposure the SL RNA-specific product that better shows the increase of the SL RNA signal upon SPF27 silencing. Right panel, RT-qPCR analysis of SL RNA abundance in SPF27-silenced cells. The amounts are relative to that of non-induced cells, which was arbitrarily set to 100. Averages were derived from three independently conducted gene knockdown experiments.
E. Corresponding RT-qPCR analyses of pre-mRNA and of mature mRNA of α tubulin, RPB7 and PAP. For pre-mRNA analyses, random hexamer-derived cDNA was amplified with a sense oligonucleotide that hybridized upstream of the SL trans splice site (α tubulin and RPB7) or with an antisense oligonucleotide that hybridized within the PA P intron.
To confirm that these changes in RNA abundances are specific to SPF27 silencing and not due to pathways of death, we co-analyzed the knockdown of a non-spliceosomal gene that affected trypanosome culture growth similar to SPF27 silencing. We chose the ORC1 gene encoding subunit 1 of the origin recognition complex (Godoy et al., 2009) because, in our hands, the ORC1 knockdown arrested culture growth after the second day of induction (Supporting Information Fig. S3). The RNA analysis showed that, while ORC1 mRNA abundance was very strongly reduced after 1 day of induction, the level of α tubulin mRNA was unaffected even after 3 days of ORC1 silencing (Fig. 3C). Together, these results confirmed that SPF27 is a spliceosomal protein.
Since the PRP19 complex is known to be important for spliceosome activation, we expected that SPF27 silencing would affect the first step of splicing. In trans splicing such a defect should lead to an increase of the SL RNA splicing substrate and a decline of the Y structure intermediate that is formed after the first transesterification (Luz Ambrósio et al., 2009; Tkacz et al., 2010). To test this we conducted a primer extension assay with three radiolabeled oligonucleotides that specifically hybridize to the intronic sequence of the SL RNA and, as controls, to the U2 snRNA and the 7SL RNA. In the Y structure, the 5′ end of the intron is covalently linked to the branch site that forms a barrier for reverse transcriptase, leading to an intron-specific primer extension product. Indeed, in SPF27-silenced cells, primer extension showed a strong increase of SL RNA abundance on day 2 and 3 of induction and a concomitant reduction of the Y structure signal in these total RNA preparations, a phenotype that was not observed in ORC1-silenced cells (Fig. 3D, left panel).
To substantiate these findings, we carried out quantitative RT-PCR analyses with three independently derived RNA preparations for each time point of SPF27 silencing. On days two and three after induction, the amount of SL RNA increased ~ 6- to ~ 9-fold over the un-induced level respectively (Fig. 3D, right panel). These numbers were in accordance with those of a previous study in which silencing the gene of CDC2-related kinase 9 (CRK9) also resulted in a block of trans splicing before the first step (Badjatia et al., 2013). In agreement with such a block, we found a ~ 3-fold increase of α tubulin pre-mRNA abundance in SPF27-silenced cells, which coincided with a ~ 50% reduction of the mature mRNA level on day three of induction (Fig. 3E, left panel; note that our analysis of mature mRNA did not discriminate between a correct mRNA and a polyadenylated, unspliced mRNA). Comparable results were obtained when we analyzed a second mRNA encoding the RPB7 subunit of RNA polymerase II (RNAPII; Fig. 3E, middle panel). Finally, while the PAP mRNA level strongly decreased upon SPF27 silencing as expected, a PAP intron-dependent qPCR detected only a small, insignificant increase of PAP pre-mRNA in induced cells (Fig. 3E, right panel). Possibly, unspliced PAP mRNA is highly unstable, preventing an accumulation of this RNA in splicing-impaired cells.
In summary, the increase of SL RNA and unprocessed pre-mRNA levels paralleled by the concomitant decrease of the Y structure intermediate and mature mRNA abundances demonstrated that SPF27 depletion caused a block of the first trans splicing step, further indicating a role of the PRP19 complex for spliceosome activation in trypanosomes and corroborating SPF27 as a bona fide subunit of this complex.
PRP19 and SPF27 co-localize in the nucleus
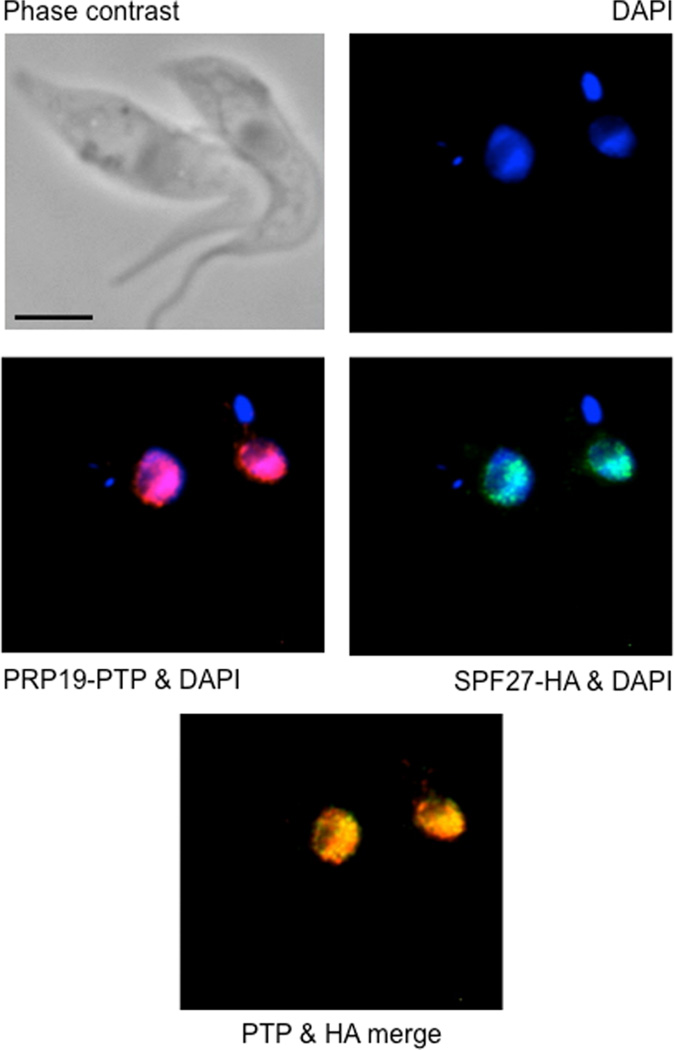
To further validate SPF27 as a component of the PRP19 complex, we examined if SPF27 co-localizes with PRP19 in trypanosomes. We generated a procyclic cell line in which PRP19 and SPF27 were C-terminally tagged with PTP and HA respectively. Detection of both proteins by indirect immunofluorescence microscopy revealed that both proteins localized to the nucleus, exhibiting a speckle-like distribution that is characteristic of spliceosomal proteins, including trypanosome and human PRP19 (Fig. 4) (Tkacz et al., 2010; Spector and Lamond, 2011; Marechal et al., 2014). Consistently, both proteins colocalized to a large extent (Pearson coefficient of 0.73, n = 53), supporting our findings that both proteins are subunits of the same spliceosomal protein complex. In comparison, PRP19 localization was previously found to be not as widespread in the nucleus as in our analysis but more focused at the so-called SL RNP factory in which SL RNA is synthesized (Tkacz et al., 2010). This discrepancy could be due to using different tags (PTP versus YFP), tagging different ends (C- versus N-terminus) or detecting PRP19 by indirect versus direct immunofluorescence microscopy.
Fig. 4.
PRP19 and SPF27 co-localize predominantly in nuclear speckles. PRP19-PTP (red) was co-localized with SPF27-HA (green) in procyclic trypanosomes by indirect immunofluorescence. DNA detected by DAPI staining revealed the large nucleus and the smaller kinetoplast of trypanosomes. The black bar represents 10 µm.
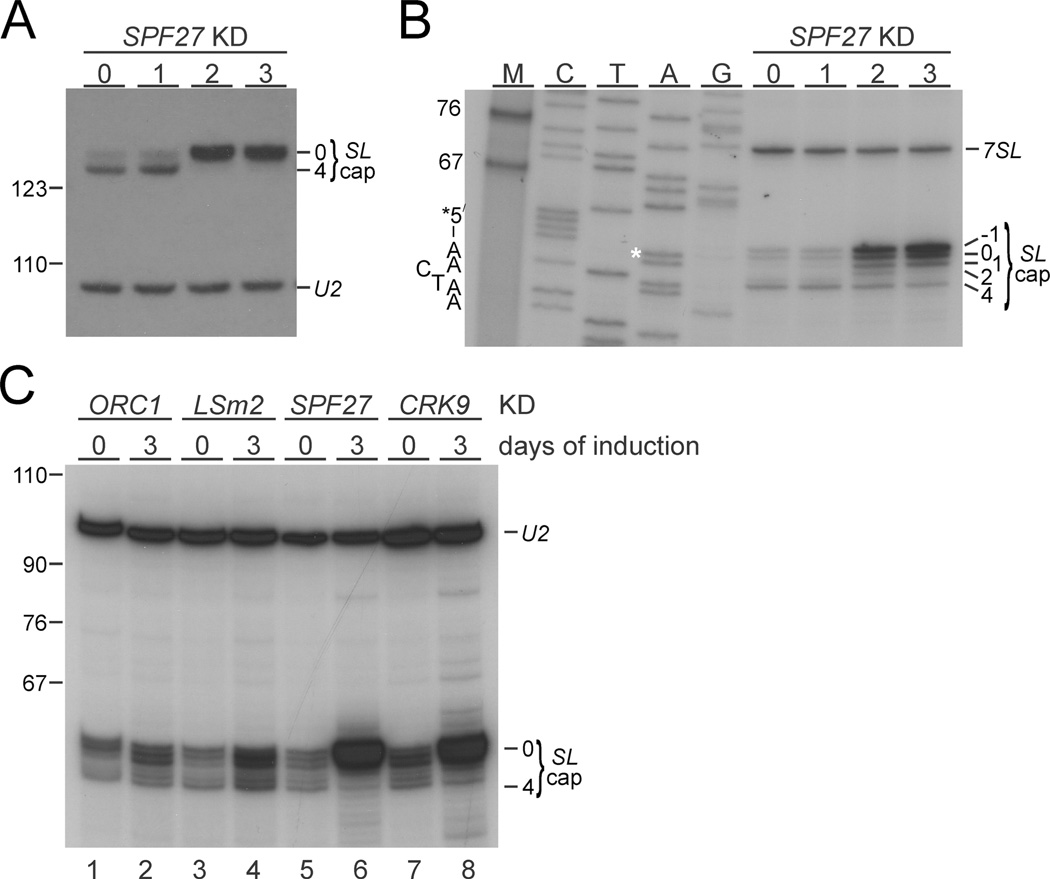
SPF27 silencing leads to SL cap hypomethylation
SL cap4 methylations at positions 1, 2 and 3/4 are carried out by the methyl transferases MTR1 (Zamudio et al., 2007), MTR2 (also known as MT47 or COM1) (Arhin et al., 2006b; Hall and Ho, 2006) and MTR3 (also known as MT57) (Arhin et al., 2006a; Zamudio et al., 2006) respectively, and they occur sequentially from position 1 to 4 (Mair et al., 2000; recently reviewed by Sturm et al., 2012). Previously, Tkacz et al. (2010) reported that PRP19 silencing led to a loss of cap3/4 and an accumulation of SL RNAs with cap2 indicating a functional link between PRP19 and MTR3. To test whether SPF27 silencing led to a comparable defect, we performed a modified primer extension assay with MMLV reverse tran-scriptase and reduced ribonucleotide concentrations, both of which promote polymerization stops at methylated residues. We first extended oligonucleotide SL_PE, which hybridizes to the single-stranded Sm binding region of the SL RNA (positions 111 to 130) and which we had used previously to show that CRK9 silencing caused the loss of all cap4 methylations (Badjatia et al., 2013). As shown in Fig. 5A, we found that the shorter extension product, indicative of SL RNA with a fully methylated cap4 structure, disappeared after 2 days of SPF27 silencing and that the accumulated SL RNA signal was found almost exclusively in the top band instead, suggesting the predominant presence of SL RNA with a hypomethylated cap. Since a nearly congruent primer extension pattern was obtained when CRK9 was silenced (Badjatia et al., 2013), this result suggested a complete loss of cap4 methylations instead of a specific cap3/4 loss upon SPF27 silencing. However, Tkacz et al. had used an oligonucleotide that hybridized further upstream of the SL RNA, precisely up to the GU splice site nucleotides of the SL RNA intron. Since it was possible that the use of different primers led to different extension results, we repeated the primer extension assay with oligonucleotide SL40 (Zamudio et al., 2006) that also hybridized up to GU splice site. Although the same assay conditions and RNA preparations were used, primer extension of SL40, in contrast to that of SL_PE, was different revealing cap4 products on days 2 and 3, and a cap2-specific product that was clearly detectable on accumulated SL RNA 2 days into the SPF27 knockdown (Fig. 5B). Possibly, the shorter extension of SL40 made reverse transcription more sensitive to methylated residues, resulting in a better detection of methylated and partially methylated SL RNA caps. Nonetheless, consistent with the SL_PE extension results, the cap4 product diminished over the time course, the cap2 product disappeared on day 3, and the accumulating SL RNA predominantly had a cap0. Thus, these findings did not reveal a specific cap3/4 defect but they clearly indicated that SPF27 silencing led to an accumulation of SL RNA that is progressively hypomethylated with most SL RNA having a cap0 structure.
Fig. 5. Accumulated SL RNA in SPF27-silenced cells is hypomethylated.
A. SL RNA and, as a loading control, U2 snRNA primer extension products of total RNA, prepared from non-induced (n.i.) cells and cells in which SPF27 was silenced for 1 to 3 days, were analyzed on a sequencing gel that separates SL RNA cap4–derived signals from signals of hypomethylated SL RNA caps (SL cap0). On the left, marker bands are indicated.
B. Corresponding primer extension analysis of the same RNA with oligonucleotide SL40, which hybridizes just downstream of the 5′ splice site in the SL RNA (position 40 to position 59 relative to SL RNA 5′ end). For comparison, a sequencing ladder, which was obtained by linear amplification sequencing with the same oligonucleotide, was co-analyzed. Each lane is marked with the complementary base of the chain-terminating nucleotide used, so that the SL RNA sequence can be directly inferred from top to bottom. The starting A residue is marked with an asterisk. M, marker. On the left, the SL starting sequence 5′-AACTAA-3′ is indicated while specific cap products and the signal of the 7SL RNA, which was co-analyzed as a loading control, are specified on the right. Note that primer extension of SL40 generated a product with an extra nucleotide (position −1) that has been observed before and may be due to terminal transferase activity of reverse transcriptase (Zamudio et al., 2009).
C. SL RNA primer extension analysis, using primer SL40, of total RNA, which was prepared from uninduced cells or from cells in which ORC1, LSm2, SPF27, or CRK9 was silenced for three days. Co-detection of U2 snRNA served as a loading control.
Since SPF27 and CRK9 silencing led to similarly strong accumulations of SL RNA and, according to the SL_PE primer extension results, to a similar loss of cap4 methylation, we co-analyzed total RNA from both gene knockdowns with primer SL40. In addition, we analyzed RNA from the ORC1 knockdown as a negative control and repeated an LSm2 silencing experiment, which was shown previously to also affect the first step of trans splicing (Luz Ambrósio et al., 2009). As shown in Fig. 5C, depletion of the U6 snRNP-specific LSm2 protein caused only a mild increase of SL RNA that did not exhibit any methylation defect. This result is not due to ineffective gene silencing because LSm2 silencing rapidly and strongly affected trypanosome proliferation in culture (Luz Ambrósio et al., 2009; data not shown). In contrast, the extent of SL RNA accumulation and the very strong cap0 signal were nearly identical in SPF27 and CRK9 knockdowns. The similarity of these phenotypes, despite the fact that SPF27 silencing merely halted culture growth (Fig. 1A) while CRK9 silencing led to a strong decline of cell numbers (Badjatia et al., 2013), suggests that SPF27 and CRK9 function in the same step of RNA splicing. LSm2 silencing led to a loss of U6 snRNA (Tkacz et al., 2008; Luz Ambrósio et al., 2009), which should affect the formation of the B complex before spliceosome activation. Therefore, strong SL RNA accumulation and cap hypomethylation may be specific consequences of interference with spliceosome activation.
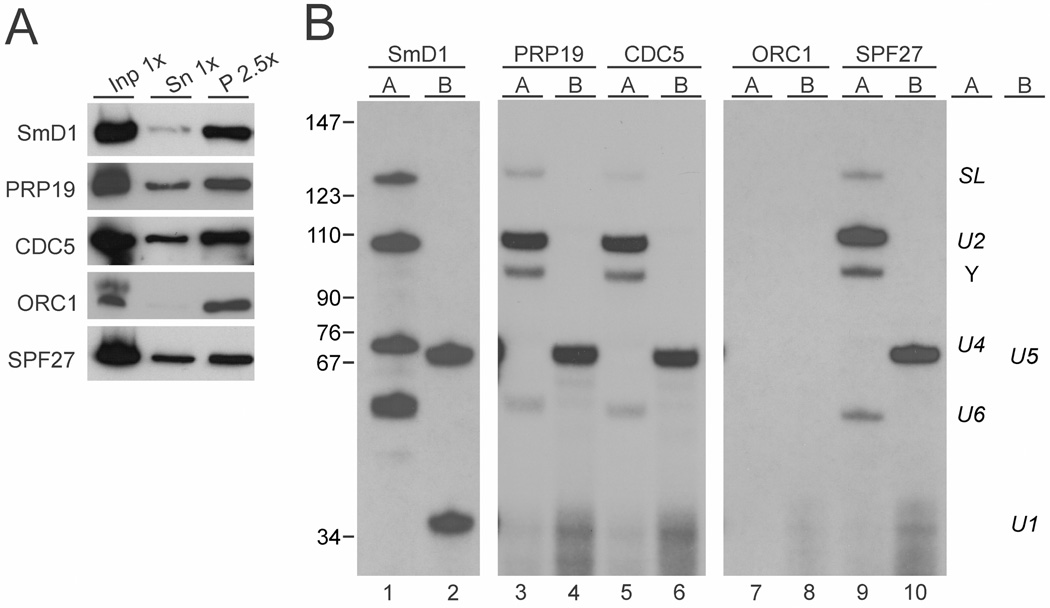
The trypanosome PRP19 complex is associated with U2, U5 and U6 snRNAs
In a final step, we wanted to see whether the trypanosome PRP19 complex is associated with a complex comparable to the activated spliceosome in the yeast and human systems, which is characterized by the presence of U2, U5 and U6 snRNAs and the absence of U4 and U1 snRNAs. For this analysis, we prepared extract from TbP19ee cells, and efficiently pulled down PRP19-PTP by IgG beads (Fig. 6A). Total RNA, prepared from the precipitated material, was then analyzed by two distinct primer extension assays. In assay A, four oligonucleotides were used to detect SL RNA and the Y structure intermediate as well as U2, U4 and U6 snRNA, whereas assay B contained two oligonucleotides complementary to U1 and U5 snRNAs (Fig. 6B). In a positive control precipitation of the PTP-tagged SmD1 core snRNP protein (Luz Ambrósio et al., 2009) all six snRNAs were efficiently detected by correctly sized primer extension products (lanes 1 and 2) while in a negative control precipitation of PTP-tagged ORC1 none of these RNAs co-precipitated (lanes 7 and 8). Since all PTP-mediated pull-downs were of similar efficiency (Fig. 6A), these results confirmed the specificity of the primer extension assays. In comparison, the pull-down of PRP19 mainly co-precipitated U2 and U5 snRNAas well as the Y structure intermediate (Fig. 6B, lanes 3 and 4). In addition, full length SL RNA and U6 snRNA were co-precipitated although with a reduction in relative signal strength whereas U4 and U1-specific products were hardly detectable. This snRNA pattern shows that the PRP19 complex is predominantly associated with a complex that resembles an activated spliceosome and it suggests that the trypanosome PRP19 complex assembles into the spliceosome after the discard of U1 and U4 snRNA has been initiated. Moreover, the clear detection of the Y structure in PRP19-precipitated material confirms that the trypanosome PRP19 remains associated with the spliceosome after the first step of splicing. Since SmD1 binds directly to the SL RNA (Palfi et al., 2000) and PRP19 is associated with snRNAs only through the spliceosome, the reduction of SL RNA signal versus the positive control is expected. Similarly, the reduction of the U6 signal is likely due to the presence of free U4/U6 snRNP in extract that is not assembled into the spliceosome and, therefore, is precipitated with SmD1 but not with a non-snRNP protein. When we repeated this experiment with two different cell lines that expressed either PTP-tagged CDC5 (lanes 6 and 7) or PTP-tagged SPF27 (lanes 9 and 10) the results were congruent with those for PRP19, further validating CDC5 and SPF27 as bona fide subunits of the trypanosome PRP19 complex. Overall, we found that only minor amounts of snRNPs co-precipitated with the PRP19 complex because, in contrast to the SmD1 precipitation in which snRNAs were efficiently co-depleted from the supernatant, the pull down of SPF27 did not detectably reduce the abundance of snRNAs in extract (Supporting Information Fig. S4). This result is consistent with the finding that in trypanosomal extracts the majority of snRNAs are assembled in individual snRNPs rather than in large spliceosomes (Cross et al., 1991).
Fig. 6. The trypanosome PRP19 complex specifically associates with U2, U5 and U6 snRNAs.
A. Extract was prepared from procyclic cell lines that express SmD1, PRP19, CDC5, ORC1 or SPF27 with a functional, C-terminal PTP tag. PTP-tagged proteins were precipitated by IgG beads that bind the protein A domain of PTP and were detected with the peroxidase anti-peroxidase complex that is bound by protein A. Inp, input; Sn, supernatant; P, precipitate. x-Values indicate relative amounts analyzed.
B. Total RNA was prepared from the precipitates and analyzed by two primer extension reactions. In reaction A, oligonucleotides were used that are specific to SL RNA, U2 snRNA, U4 snRNA and U6 snRNA, whereas in reaction B, a U5- and a U1-specfic oligonucleotide were combined. The SmD1 and ORC1 pull-downs served as positive and negative controls respectively. Note that the exposure time of the SmD1 analysis was much shorter than those of the other assays.
Finally, depletion of Sm and LSm proteins as well as of other snRNP proteins in trypanosomes led to a loss of the cognate U snRNA (Liu et al., 2004; Tkacz et al., 2008; 2010; Luz Ambrósio et al., 2009). In contrast, SPF27 silencing for three days did not alter the abundance of spliceosomal snRNAs, verifying the non-snRNP nature of the trypanosome PRP19 complex (Supporting Information Fig. S5). Thus, the effective co-precipitation of the U snRNAs with PRP19 complex components suggests that trypanosomes form splicing complexes that are comparable to the B and C complexes described in other organisms and that these complexes remained stable in our extracts.
Discussion
In this study we have characterized the spliceosomal non-snRNP PRP19 complex of T. brucei. Since trypanosomes and the model organisms from yeast to humans belong to different phylogenetic lineages, e.g. the Excavata and Opisthokonta respectively (Katz, 2012), a comparison of these complexes can distinguish between properties that are highly conserved over extended evolutionary time and those that vary between species. Accordingly, the subunits PRP19, CDC5, PRL1 and SPF27 are invariably present in the human, yeast and trypanosome complexes indicating that these four proteins form the conserved core of the complex (Table 2).
Table 2.
PRP19 complex subunits in H. sapiens, T. brucei and S. cerevisiae.
| H. sapiens | T. brucei | S. cerevisiae |
|---|---|---|
| PRP19 | PRP19 | Prp19 |
| CDC5L | CDC5 | Cef1 |
| PRL1 | PRL1 | Prp46 |
| SPF27 | SPF27 | Snt309 |
| AD002 | assoc. | assoc. (Cwc15) |
| CTNNB1 | n.k.h. | n.k.h. |
| HSP73 | assoc. | – |
| – | PRP17 | assoc. |
| assoc. | SKIP | assoc. |
| assoc. | PPIL1 | n.k.h. |
| assoc. | assoc. | Isy1 |
| assoc. | assoc. | Syf1 |
| assoc. | n.k.h. | Syf2 |
| assoc. | assoc. | Clf1 (Syf3) |
assoc., associated; proteins that were co-purified with PRP19 complexes but not considered bona fide subunits. In humans they are referred to as ‘PRP19-related’ (Wahl et al., 2009; Chanarat and Strasser, 2013); n.k.h., no known homolog; –, protein was not co-purified.
PRP17 is not a subunit of the human PRP19 complex and it is considered to be associated with the S. cerevisiae PRP19 complex. Trypanosome PRP17, however, stably co-purified and co-sedimented with the PRP19 complex independent of whether the complex was purified through PRP19 or CDC5 (Fig. 1D). This finding confirmed the association of S. pombe PRP17 and PRP19 (Ren et al., 2011), and it strongly suggested that PRP17 functions in the spliceosome through association with PRP19 across eukaryotes. In the model of the S. pombe PRP19 complex, a PRP19 tetramer binds four PRP17 molecules through interaction of the PRP17 N-terminus with the PRP19 WD40 domain (Ren et al., 2011). These domains are conserved in the trypanosome orthologs (Fig. 2A) and the strong protein bands that contained PRP19 and PRP17 (Fig. 1D) may indicate that this model does fit the trypanosome complex. However, our sedimentation analysis revealed a complex of ~ 350 kDa, which is substantially smaller than the expected 675 kDa of a complex containing a PRP19 tetramer, four PRP17 molecules and monomers of each of the other subunits. Possibly, tandem purification and sedimentation led to a partial disruption of the PRP19 complex. Alternatively, we cannot rule out that there are two distinct PRP19 complexes of different composition that were not separated during sedimentation.
An interesting deviation from the yeast and human system was the finding that SKIP and PPIL1 were stably associated with trypanosome PRP19. Identification of these two proteins in trypanosomes implies an early evolutionary origin of the SKIP-PPIL1 sub-complex (Wang et al., 2010). Furthermore, although previous work demonstrated a direct interaction between the yeast SKIP ortholog and PRP46p (PRL1) in vitro (Albers et al., 2003), these proteins have not been considered bona fide subunits of the PRP19 complex and were classified as PRP19-related proteins (Wahl et al., 2009; Chanarat and Strasser, 2013). Since organization of PRP19-related proteins is in general not well understood, our finding suggests that the SKIP/PPIL1 sub-complex functions within the PRP19 complex independent of other PRP19-related proteins. This seems to be at least true for trypanosomes since we recently succeeded in characterizing a distinct complex of PRP19-related proteins that does not contain PRP19 complex subunits including SKIP and PPIL1 (DL Ambrósio and A Günzl, unpublished data).
Another important aspect of our study was the discovery that depletion of SPF27 led to a strong accumulation of SL RNA with a hypomethylated cap. The extent of SL RNA accumulation and the pattern of cap hypomethylation closely resembled the effects seen upon CRK9 silencing but not those of the LSm2 knockdown, which also affected trans splicing (Fig. 5C) (Luz Ambrósio et al., 2009; Badjatia et al., 2013). Thus, these findings suggested that CRK9 functions in spliceosome activation. Originally, our interest in CRK9 stemmed from its apparent role in the phosphorylation of RPB1, the largest subunit of RNAPII. In other systems, RPB1 phosphorylation is critically important for RNAPII transcription initiation and elongation (Buratowski, 2009). However, in trypanosomes the loss of RPB1 phosphorylation upon CRK9 silencing did not lead to an RNAPII-specific transcription block but, instead, coincided with SL RNA cap hypomethylation and the trans splicing block. Since cap4 formation was thought to be co-transcriptional (Mair et al., 2000) and cap4 methylations a pre-requisite for SL trans splicing (Ullu and Tschudi, 1991; McNally and Agabian, 1992), we had speculated that CRK9’s primary substrate is RPB1, and that cap methylation and splicing defects are downstream consequences of RPB1 dephosphorylation (Badjatia et al., 2013). However, the possibility that CRK9 functions directly in spliceosome activation may point to a regulatory loop in trypanosomes in which the status of RNAPII phosphorylation is directly linked to the trans splicing mechanism. Accordingly, it will be interesting to analyze whether blocking spliceosome activation independent of CRK9 affects RNAPII phosphorylation.
Overall, this is the first characterization of a spliceosomal non-snRNP protein complex in trypanosomes. It is an important step towards understanding the spliceosome of T. brucei and related parasites. The human spliceosome is a validated anti-cancer drug with the first inhibitor in clinical trial (van Alphen et al., 2009; Eskens et al., 2013). Given the special use of this RNA-protein machinery in trypanosome SL trans splicing, and considering the great evolutionary distance between trypanosomes and humans, a trypanosome-specific splicing inhibitor may wait to be discovered.
Experimental procedures
DNAs
pPRP19-PTP-NEO was generated by inserting 627 bp of the C-terminal PRP19 coding region (position 901 to position 1527 relative to the translation initiation codon) into the pC-PTP-NEO tagging construct (Schimanski et al., 2005b) using the ApaI and NotI restriction sites. pCDC5-PTP-NEO, pSPF27-PTP-NEO, and pORC1-PTP-NEO were obtained analogously by cloning 519 bp (position 1663 to position 2181), 695 bp position −53 to position 642) and 326 bp (position 983 to position 1308) of the CDC5, SPF27 and ORC1 genes into pC-PTP-NEO respectively. For pSPF27-HA-BLA, the 695 bp DNA fragment was cloned into pC-HA-BLA (Nguyen et al., 2007), again using the ApaI and NotI restriction sites. The conditional knockdown constructs pT7-SPF27-stl and pT7-ORC1-stl were obtained by cloning respectively, 571 bp (position 7 to position 577) and 526 bp (position 7 to position 532) of the SPF27 and ORC1 coding regions in a stem-loop arrangement into the vector pT7-stl (Brandenburg et al., 2007). The constructs for conditional knockdown of LSm2 and CRK9 have been described previously (Luz Ambrósio et al., 2009; Badjatia et al., 2013).
Cells
Cell culturing and stable transfection of procyclic T. brucei brucei strain 427 and of cell line 29-13 (Wirtz et al., 1999) was carried out as described previously (Laufer et al., 1999). After each transfection, cell lines were cloned by limiting dilution. Cell line TbP19ee was generated by inserting BmgBI-linearized pPRP19-PTP-NEO into one PRP19 allele and deleting the remaining wild-type allele with a PCR product in which 100 bp of the PRP195′ and 3′ gene flanks were fused to the hygromycin phosphotransferase (HYG) coding region. Cell lines that express CDC5-PTP, SPF27-PTP or ORC1-PTP were generated by transfecting SnaBI-linearized pCDC5-PTP-NEO, BsmI-linearized pSPF27-PTP-NEO, and XhoI-linearized pORC1-PTP-NEO respectively. Cells that express both PRP19-PTP and SPF27-HA were obtained by two consecutive transfections, first with linearized pPRP19-PTP-NEO and then with BsmI-linearized pSPF27-HA-BLA. Cell lines for conditional gene knockdowns were generated by targeting the T7-stl constructs to the RRNA spacer region. For each transfection correct integration of DNA was verified by PCR with at least one oligonucleotide hybridizing outside the transfected DNA sequence. After selection, 427 cells with antibiotic resistance markers were grown in 40 µg ml−1 hygromycin, 40 µg ml−1 G418, and/or 10 µg ml−1 blasticidin whereas 29-13 cell lines were kept in 50 µg ml−1 hygromycin, 15 µgml−1 G418, and 4 µg ml−1 phleomycin. Conditional gene silencing experiments were induced by adding doxycycline to a final concentration of 1 µg ml−1. Cells were counted and diluted to a density of 2 × 106 cells/ml daily.
Protein analysis
PTP-tagged proteins on immunoblots were detected either with the peroxidase anti-peroxidase reagent (Sigma) or a monoclonal anti-protein C epitope antibody (Roche). Extract preparation and tandem affinity purification of PTP-tagged proteins were carried out according to the standard protocol (Schimanski et al., 2005b) Purified proteins were separated on SDS–10 to 20% polyacrylamide gradient gels (BioRad) and stained either with sypro ruby (BioRad) or with Coomassie blue (Gelcode Coomassie stain; Thermo Fisher Scientific). For the sedimentation analysis of PRP19 and CDC5 complexes, the final eluate of a standard tandem affinity purification (~ 1.8 ml) was vacuum concentrated to ~ 100 µl, dialyzed overnight against E-80 buffer (150 mM sucrose, 20 mM HEPES-KOH pH 7.7, 20 mM potassium glutamate, 80 mM potassium chloride, 3 mM magnesium chloride, 0.2 mM EDTA, 0.5 mM EGTA), loaded onto a 4 ml linear 10–40% sucrose gradient in E-80 buffer, and ultracentrifuged at 41,000 r.p.m. in a Beckman SW55Ti rotor for 19 h at 4°C. Subsequently, the gradient was fractionated from top to bottom in 20 aliquots. Proteins in each aliquot were collected by a hydrophobic resin (StrataClean, Stratgene), and resus-pended in SDS loading buffer for electrophoresis. Proteins that co-purified with PRP19-P were analyzed from a gel lane of the final eluate by the Proteomic Core facility of the Rockefeller University. The gel lane was cut into multiple pieces and peptides, derived from trypsin-digested proteins, were separated by liquid chromatography and analyzed by tandem mass spectrometry (LC/MS/MS) using an Ultimate 3000 HPLC system (Dionex) and a nanospray LTQ Orbitrap XL (Thermo Scientific) mass spectrometer. Individual protein bands obtained after sucrose gradient sedimentation were analyzed equivalently by the Keck facility (Yale University). Proteins were identified using Mascot and NCBI non-redundant protein sequence database of eukaryotes, with carbamidomethyl (C) as static and oxidation (M) as variable modifications. Peptide Mass tolerance was set to ± 25 ppm and fragment mass tolerance to ± 0.8 Da. Peptides were considered as identified when their score was ≥22 and expectation values were ≤ 0.05.
RNA analysis
To analyze gene silencing effects on RNA abundances, total RNA was prepared from non-induced or doxycycline-induced cells by the hot phenol method as described previously (Nguyen et al., 2007). rRNA was separated by gel electrophoresis on reliant precast 1.25% SeaKem Gold agarose RNA gels (Lonza) and detected by ethidium bromide staining. For the analysis of mRNA and unspliced pre-mRNA, total RNA was first reverse transcribed with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol using, respectively, oligo(dT) and random hexamers as primers. Secondly, the cDNAs were subjected to semiquantitative and quantitative (q)PCR analysis. Semiquantitative PCR was performed using cycle numbers that were empirically determined to be within the linear amplification range for each oligonucleotide pair. The specificity of oligonucleotide pairs used in qPCR was first verified in analyzing their products by standard agarose gel electrophoresis. To further confirm the validity of the qPCR reactions, a melt curve analysis was included for each qPCR to ensure that only a single product was amplified. In addition, a linear regression analysis of a serial dilution of input material was performed to make sure that the coefficient of determination (r2) was within the 0.98 to 1.0 range.
Spliceosomal U snRNAs, SL RNA and, as a control, 7SL RNA were detected by primer extension assays using 10 µg of total RNA, 5′-32P-end-labeled oligonucleotides and Super-Script II reverse transcriptase. Primers SL_PE and SL40, 7SL_PE, U1_PE, U2f, U4_PE, U5_PE, and U6_PE were used to detect SL RNA, 7SL RNA, U1, U2, U4, U5 and U6 snRNA respectively. Primer extension products were separated on 8 or 15% polycacrylamide – 50% urea gels and visualized by autoradiography. The modified primer extension assay for characterizing the methylation status of the SL RNA cap and generation of the sequencing ladder was conducted with unmodified Moloney’s murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and the Thermo Sequenase cycle sequencing kit (USB), as previously described (Badjatia et al., 2013). For the analysis of PRP19 complex-associated U snRNAs, PTP-tagged proteins were precipitated by immunoglobulin G (IgG) beads (GE Healthcare), and total RNA was prepared and analyzed by primer extension assays as previously published (Luz Ambrósio et al., 2009). DNA oligonucleotides that were used in primer extension assays and in competitive and [semiquantitative reverse transcription (RT-)PCR reactions are specified in Supporting Information Table S1.
Light microscopy
Immunofluorescence microscopy was performed as previously described (Luz Ambrósio et al., 2009). For the detection of PRP19-PTP, a rabbit polyclonal anti-ProtA immune serum (Sigma) followed by an Alexa 594-conjugated antirabbit secondary antibody (Invitrogen) was used, while HA-tagged SPF27 was detected by a rat monoclonal anti-HA antibody (Roche) and an Alexa 488-conjugated anti-rat secondary antibody (Invitrogen). We excluded the possibility that anti-HA antibodies were non-specifically bound by ProtA by analyzing cells that expressed only PRP19-PTP with the antibody mix described above (data not shown). Images were taken on a Zeiss Axiovert 200 microscope equipped with DAPI (4′,6′-diamidino-2-phenylindole), EGFP and Texas-Red filters using a 100× (1.3-numerical-aperture) oil immersion objective. Quantification of the co-localization was done using the Zeiss Axiovision 4.6.3.0 software program for co-localization.
Supplementary Material
Acknowledgements
This work was supported by grant R01 AI073300 to A.G. from the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. National Institutes of Health (NIH). We are grateful to Justin K. Kirkham from our laboratory for critical reading of the manuscript.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Albers M, Diment A, Muraru M, Russell CS, Beggs JD. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA. 2003;9:138–150. doi: 10.1261/rna.2119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen RJ, Wiemer EA, Burger H, Eskens FA. The spliceosome as target for anticancer treatment. Br J Cancer. 2009;100:228–232. doi: 10.1038/sj.bjc.6604801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhin GK, Li H, Ullu E, Tschudi C. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei . RNA. 2006a;12:53–62. doi: 10.1261/rna.2223406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhin GK, Ullu E, Tschudi C. 2′-O-methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol Biochem Parasitol. 2006b;147:137–139. doi: 10.1016/j.molbiopara.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Badjatia N, Ambrósio DL, Lee JH, Günzl A. Trypanosome cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of RNA polymerase II subunit RPB1. Mol Cell Biol. 2013;33:1965–1975. doi: 10.1128/MCB.00156-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- Brandenburg J, Schimanski B, Nogoceke E, Nguyen TN, Padovan JC, Chait BT, et al. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007;26:4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymer-ase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chanarat S, Strasser K. Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta. 2013;1833:2126–2134. doi: 10.1016/j.bbamcr.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Cross M, Günzl A, Palfi Z, Bindereif A. Analysis of small nuclear ribonucleoproteins (RNPs) in Trypanosoma brucei: structural organization and protein components of the spliced leader RNP. Mol Cell Biol. 1991;11:5516–5526. doi: 10.1128/mcb.11.11.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2013;41:D132–D141. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskens FA, Ramos FJ, Burger H, O’Brien JP, Piera A, de Jonge MJ, et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res. 2013;19:6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy PD, Nogueira-Junior LA, Paes LS, Cornejo A, Martins RM, Silber AM, et al. Trypanosome pre-replication machinery contains a single functional orc1/cdc6 protein, which is typical of archaea. Eukaryot Cell. 2009;8:1592–1603. doi: 10.1128/EC.00161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, et al. Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Ho CK. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 2006;34:5594–5602. doi: 10.1093/nar/gkl573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Katz LA. Origin and diversification of eukaryotes. Annu Rev Microbiol. 2012;66:411–427. doi: 10.1146/annurev-micro-090110-102808. [DOI] [PubMed] [Google Scholar]
- Laufer G, Schaaf G, Bollgönn S, Günzl A. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei . Mol Cell Biol. 1999;19:5466–5473. doi: 10.1128/mcb.19.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol Biochem Parasitol. 2006;145:29–39. doi: 10.1016/j.molbiopara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liang XH, Uliel S, Belahcen M, Unger R, Michaeli S. Identification and functional characterization of lsm proteins in Trypanosoma brucei . J Biol Chem. 2004;279:18210–18219. doi: 10.1074/jbc.M400678200. [DOI] [PubMed] [Google Scholar]
- Luz Ambrósio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Günzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol Cell Biol. 1992;12:4844–4851. doi: 10.1128/mcb.12.11.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G, Ullu E, Tschudi C. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J Biol Chem. 2000;275:28994–28999. doi: 10.1074/jbc.M004193200. [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Lührmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Weese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, et al. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- Michaeli S, Podell D, Agabian N, Ullu E. The 7SL RNA homologue of Trypanosoma brucei is closely related to mammalian 7SL RNA. Mol Biochem Parasitol. 1992;51:55–64. doi: 10.1016/0166-6851(92)90200-4. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Watkins KP, Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Schimanski B, Günzl A. Active RNA polymerase I of Trypanosoma brucei harbors a novel subunit essential for transcription. Mol Cell Biol. 2007;27:6254–6263. doi: 10.1128/MCB.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, et al. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol Cell Biol. 2005;25:451–460. doi: 10.1128/MCB.25.1.451-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfi Z, Lücke S, Lahm HW, Lane WS, Kruft V, Bragado-Nilsson E, et al. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc Natl Acad Sci USA. 2000;97:8967–8972. doi: 10.1073/pnas.150236097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfi Z, Jae N, Preuβer C, Kaminska KH, Bujnicki JM, Lee JH, et al. SMN-assisted assembly of snRNP-specific Sm cores in trypanosomes. Genes Dev. 2009;23:1650–1664. doi: 10.1101/gad.526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuβer C, Jae N, Günzl A, Bindereif A. Pre-mRNA splicing in Trypanosoma brucei: factors, mechanisms, and regulation. In: Bindereif A, editor. RNA Metabolism in Trypanosomes. Springer-Verlag Berlin Heidelberg: Springer Press; 2012. pp. 49–76. [Google Scholar]
- Ren L, McLean JR, Hazbun TR, Fields S, Vander KC, Ohi MD, Gould KL. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS ONE. 2011;6:e16719. doi: 10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Günzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei . Mol Cell Biol. 2005a;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Günzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005b;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. ‘Nought may endure but mutability’: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Zamudio JR, Campbell DA. SL RNA biogenesis in kinetoplastids: a long and winding road. In: Bindereif A, editor. RNA Metabolism in Trypanosomes. Springer-Verlag Berlin Heidelberg: Springer Press; 2012. pp. 29–47. [Google Scholar]
- Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz ID, Cohen S, Salmon-Divon M, Michaeli S. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei . Mol Biochem Parasitol. 2008;160:22–31. doi: 10.1016/j.molbiopara.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, et al. Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J Biol Chem. 2010;285:27982–27999. doi: 10.1074/jbc.M109.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc Natl Acad Sci USA. 1991;88:10074–10078. doi: 10.1073/pnas.88.22.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang S, Zhang J, Huang X, Xu C, Wang W, et al. A large intrinsically disordered region in SKIP and its disorder-order transition induced by PPIL1 binding revealed by NMR. J Biol Chem. 2010;285:4951–4963. doi: 10.1074/jbc.M109.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei . Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. Complete cap 4 formation is not required for viability in Trypanosoma brucei . Eukaryot Cell. 2006;5:905–915. doi: 10.1128/EC.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Foldynova-Trantirkova S, Zeiner GM, Lukes J, Bujnicki JM, et al. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei . Mol Cell Biol. 2007;27:6084–6092. doi: 10.1128/MCB.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Campbell DA, Sturm NR. Hypermethylated cap 4 maximizes Trypanosoma brucei translation. Mol Microbiol. 2009;72:1100–1110. doi: 10.1111/j.1365-2958.2009.06696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.