Abstract
Orientia bacterium is the agent of the scrub typhus, a seriously neglected life-threatening disease in Asia. Here, we report the detection of DNA of Orientia in rodents from Europe and Africa. These findings have important implications for public health. Surveillance outside Asia, where the disease is not expected by sanitary services, needs to be improved.
Keywords: Scrub typhus, Zoonoses, Emerging disease, Rodent-borne disease, Metagenomics
Findings
Orientia tsutsugamushi is the only known species belonging to the Orientia bacterial genus. The bacterium causes scrub typhus in humans. It is an obligate intracytosolic bacterium that is transmitted during feeding by larval trombiculid mites, and is hosted by rodents [1]. In Asia, approximately one million cases of scrub typhus occur annually, where it is probably one of the most underdiagnosed and underreported febrile illnesses requiring hospitalization [2], with an estimated 10% fatality rate unless treated appropriately. Formerly thought to be geographically restricted to Asia [3], Orientia was recently identified in sick patients from the Arabian Peninsula [4] and Chile [5]. Miscellaneous reports of scrub typhus-like illness have previously questioned the presence of the bacterium in the Congo [6] and Cameroon [7].
Methods
In order to generate a global picture of zoonotic bacteria that are likely to be harboured by rodents, we applied a metagenomic approach using spleen samples of 1334 rodents from France (Ardennes region), Senegal (along the Senegal River) and Thailand (northern and north-eastern provinces of Loei, Nan and Buriram). Rodents were trapped in both natural habitats and villages within rural landscapes. They were euthanized by cervical dislocation and dissected. In order to prevent cross contamination during dissection, we systematically alternated the use of two sets of dissecting instruments. After dissecting a rodent, the set used was immersed in bleach then water and let in alcohol, while we dissected another rodent with the other set [8]. Spleens were placed in RNAlater® storage solution (Sigma-Aldrich, Saint Louis, MO, USA) then stored at -20°C until further analysis. Genomic DNA was then extracted from the spleen using the DNeasy® 96 Tissue Kit (Qiagen, Germany). Spleen DNA samples were screened for the presence of bacteria using universal primers targeting the hyper variable region V4 of the 16S rRNA gene (251 bp) via Illumina MiSeq (Illumina) sequencing. The V4 region has been proven to have excellent taxonomic resolution at the genus level [9]. A multiplexing strategy enabled the identification of bacterial genera in each individual sample. We followed the method described in [10] to perform PCR amplification, indexing, pooling, multiplexing and de-multiplexing and finally taxonomic identification using the SILVA SSU Ref NR 119 database as a reference (http://www.arb-silva.de/projects/ssu-ref-nr/). In total we performed four different MiSeq runs, two with rodents from France (N = 557), one with rodents from Asia (N = 423) and one with rodents from Africa (N = 354). For each run, we systematically used negative controls (of DNA extraction and PCR) and none were positive for Orientia. Though we did not use positive controls for Orientia because the bacterium was not expected in European and African samples. We used positive controls for other bacterial genus like Leptospira, Borrelia, Bartonella and Mycoplasma, and all were found positive for the expected bacterial genera.
Results and discussion
From over a total of 1334 rodents tested, 110 were found positive for Orientia (Table 1). As expected, Orientia was detected in five sampled rodent species from Thailand: Rattus tanezumi (5 positives/67 tested), Rattus exulans (1/81), Bandicota savilei (2/26), Berylmys bowersi (1/17), and Leopoldamys edwardsi (1/10). More surprisingly, the bacterium was also detected in numerous rodents collected from both France and Senegal. In Senegal, Orientia sequences were only identified in the exotic house mouse (Mus musculus domesticus) (48 positives/207 tested), while the sympatric endemic multimammate rats (Mastomys erythroleucus) were all found to be negative (0/147), suggesting introduction of the bacterium via the exotic rodent. In France, Orientia sp. was detected in three rodent species: Myodes glareolus (44/302), Arvicola scherman (2/64) and Microtus arvalis (6/49). Positive for Orientia were trapped inside human houses in Asia and Africa, and in close proximity to human dwellings in Asia and France.
Table 1.
Numbers of rodent tested and found positive for Orientia sp. for the different rodent species sampled in France, Senegal and Asia
| Geographic area | Rodent species | Number tested | Number positive |
|---|---|---|---|
| France | Myodes glareolus | 302 | 44 |
| Arvicola scherman | 64 | 2 | |
| Microtus arvalis | 49 | 6 | |
| Microtus agrestis | 7 | 0 | |
| Microtus subterraneus | 4 | 0 | |
| Apodemus sylvaticus | 67 | 0 | |
| Apodemus flavicolis | 34 | 0 | |
| Rattus norvegicus | 30 | 0 | |
| Senegal | Mus musculus | 207 | 48 |
| Mastomys erythroleucus | 147 | 0 | |
| Asia | Bandicota indica | 20 | 0 |
| Bandicota savilei | 26 | 2 | |
| Berylmys berdmorei | 19 | 0 | |
| Berylmys bowersi | 17 | 1 | |
| Chiropodomys gliroides | 2 | 0 | |
| Hapalomys delacouri | 1 | 0 | |
| Leopoldamys edwardsi | 10 | 1 | |
| Leopoldamys sabanus | 1 | 0 | |
| Maxomys surifer | 15 | 0 | |
| Menetes berdmorei | 1 | 0 | |
| Mus caroli | 14 | 0 | |
| Mus cervicolor | 17 | 0 | |
| Mus cookii | 11 | 0 | |
| Mus fragilicauda | 1 | 0 | |
| Niviventer fulvescens | 17 | 0 | |
| Rattus argentiventer | 1 | 0 | |
| Rattus exulans | 81 | 1 | |
| Rattus losea | 32 | 0 | |
| Rattus nitidus | 1 | 0 | |
| Rattus tanezumi | 67 | 5 | |
| Total | 1334 | 110 |
In rodents, blood, spleen and other organs are routinely used for Orientia PCR detection [11], although one should notice that such assay is limited to the time window of rickettsemia, i.e. when the bacteria are infecting macrophages in peripheral blood. In our experiments we targeted the spleen because this organ is known to act primarily as a blood filter and then appears appropriate for detecting bacteria infecting blood cells. However, although there are many lines of evidence that Orientia may chronically infect humans and rodents, the persistence of the bacteria in organs, and spleen in particular, is currently poorly known [12]. Thus we cannot discard the possibility of false negatives in our assays.
Sequence analyses revealed that the Orientia sequences identified in this study were between 100 to 94.4% identical to GenBank-published Orientia sequences isolated from humans, mites and rodents in Asia. All sequences shared only 90.8 to 86.5% identity with GenBank-published Rickettsia, the closest bacterial genus to Orientia, thus consolidating our finding on the presence of Orientia in Europe and Africa. We also performed phylogenetic analyses of both the haplotypes identified in this study, and those from GenBank databases, using the neighbor-joining method [13]. Bootstrap analysis was performed on 1,000 replicates. Haplotypes from this study clustered strongly within the Orientia phylogroup and were clearly separated from the Rickettsia phylogroup. Asian haplotypes were distributed amongst Genbank haplotypes from Asia and Arabian peninsula (Chuto haplotype), African haplotypes fell with the Chuto haplotype, whereas European haplotypes clustered into a new basal phylogroup (Figure 1).
Figure 1.
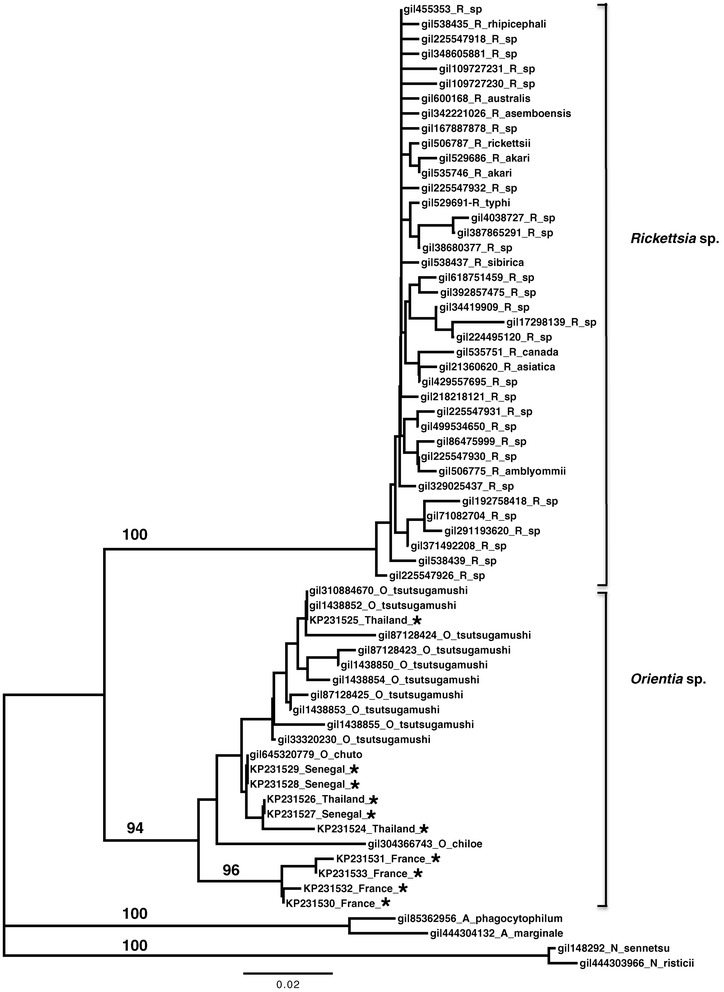
Phylogenetic tree based on the V4 region of the 16S rRNA gene. GenBank accession numbers are indicated. Only different haplotypes were shown. A complete list of sequences uploaded to GenBank can be provided upon request. Numbers beside branches indicate bootstrap values (>80). O: Orientia; R: Rickettsia; N: Neorickettsia; A: Anaplasma. The tree was rooted with the phylogenetically closest genus Anaplasma and Neorickettsia. Scale bar indicates evolutionary distances. Samples sequenced in the present study are marked with _*.
Conclusion
We have established the presence of Orientia DNA in spleens of rodents from Thailand, as was expected, but also in rodents collected from France and Senegal. In Asia, scrub typhus is considered as a seriously neglected life-threatening disease despite the fact that this ancient disease has been recognized within this region for many years. Our findings, together with those from other recent studies [4,5] suggest that in locales outside of Asia where the disease is not on the public health service radar, surveillance needs to be improved.
Ethical approval
Animals have been treated in accordance with the guidelines of the European Union legislation (Directive 86/609/EEC). The CBGP laboratory has received the approval (no. B 34–169–1) from the regional Head of Veterinary Service (Hérault, France), for the sampling and killing of rodents and the harvesting of their tissues. Dr Cosson has personally received the agreement “certificate d'autorisation d’expérimenter sur animaux vivants” (i.e. “certificate of authorization to experiment on live animals”) (no. C34–105) by the French administration.
Acknowledgments
We thank Mamadou Kane, Christophe A. Diagne, Aliou Sow, Youssou Niang and Mamoudou Diallo for their help during field sampling in Senegal, Hélène Vignes for her assistance with the MiSeq sequencing, and Sylvain Piry, Alexandre Dehne-Garcia and Marie Pagès for their help with the bioinformatic analysis. This study was funded by the INRA metaprogramme PATHO-ID and by the ANR ENEMI (ANR-11-JSV7-0006). Rodents were collected in the course of studies funded by the EU grant FP7-261504 EDENext, the ADEME (APR PREST 2009) and the ANR Biodivhealthsea (ANR-11-CEPL-0002). This study was partially funded by EU grant FP7-261504 EDENext and is catalogued by the EDENext Steering Committee as EDENext289 (http://www.edenext.eu). The contents of this publication are the sole responsibility of the authors and don’t necessarily reflect the views of the European Commission. This work was also supported by the COST Action TD1303 (EurNegVec).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JFC, MG, SM, CB, AD, KB, NC carried out the rodent sampling. MG, MR carried out the molecular genetic studies. MG, MB, EB participated in the sequence analyses. JFC drafted the manuscript and performed the phylogenetic analyses. MG, SM, CB, AD, NC, MVT helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jean François Cosson, Email: cosson@supagro.inra.fr.
Maxime Galan, Email: galan@supagro.inra.fr.
Emilie Bard, Email: ebard@clermont.inra.fr.
Maria Razzauti, Email: maria.razzauti@supagro.inra.fr.
Maria Bernard, Email: mabernard@jouy.inra.fr.
Serge Morand, Email: serge.morand@cirad.fr.
Carine Brouat, Email: carine.brouat@ird.fr.
Ambroise Dalecky, Email: ambroise.dalecky@ird.fr.
Khalilou Bâ, Email: khalilou.Ba@ird.fr.
Nathalie Charbonnel, Email: nathalie.charbonnel@supagro.inra.fr.
Muriel Vayssier-Taussat, Email: mvayssier@vet-alfort.fr.
References
- 1.Paris DH, Shelite TR, Day NP, Walker DH. Unresolved problems related to scrub typhus: a seriously neglected life-threatening disease. Am J Trop Med Hyg. 2013;89:301–7. doi: 10.4269/ajtmh.13-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–36. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;15:203–30. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 4.Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, et al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol. 2010;48:4404–9. doi: 10.1128/JCM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balcells ME, Rabagliati R, Garcia P, Poggi H, Oddo D, Concha M, et al. Endemic scrub typhus-like illness Chile. Emerg Infect Dis. 2011;17:1659–63. doi: 10.3201/eid1709.100960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osuga K, Kimura M, Goto H, Shimada K, Suto T. A case of Tsutsugamushi disease probably contracted in Africa. Eur J Clin Microbiol Infect Dis. 1991;10:95–6. doi: 10.1007/BF01964418. [DOI] [PubMed] [Google Scholar]
- 7.Ghorbani RP, Ghorbani AJ, Jain MK, Walker DH. A case of scrub typhus probably acquired in Africa. Clin Infect Dis. 1997;25:1473–4. doi: 10.1086/516990. [DOI] [PubMed] [Google Scholar]
- 8.Herbreteau V, Jittapalapong S, Rerkamnuaychoke W, Chaval Y, Cosson JF, Morand S. Protocols for field and laboratory rodent studies. Kasetsart University Press; 2011. (freely available on http://www.ceropath.org/research/protocols).
- 9.Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analysing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt G, Kantipong P, Jongsakul K, Watcharapichat P, Phulsuksombati D, Strickman D. Doxycycline and rifampicin for mild scrub-typhus infections in northern Thailand: a randomized trial. Lancet. 2000;356:1057–61. doi: 10.1016/S0140-6736(00)02728-8. [DOI] [PubMed] [Google Scholar]
- 12.Murata M, Sudo K, Suzuki K, Aoyama Y, Nogami S, Tanaka H, et al. Proliferating sites of Riskettsia tsutsugamishi in mice by different routes of inoculation evidenced with immunofluorescence. Jpn J Exp Med. 1985;55:193–9. [PubMed] [Google Scholar]
- 13.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]


