Abstract
Potato has a variety of reproductive uniquenesses besides its clonal propagation by tubers. These traits are controlled by a different kind of genetic control. The reproductive information has been applied to enable interspecific hybridization to enhance valuable traits, such as disease and pest resistances, from the tuber-bearing Solanum gene pool. While progress has been made in potato breeding, many resources have been invested due to the requirements of large populations and long time frame. This is not only due to the general pitfalls in plant breeding, but also due to the complexity of polyploid genetics. Tetraploid genetics is the most prominent aspect associated with potato breeding. Genetic maps and markers have contributed to potato breeding, and genome information further elucidates questions in potato evolution and supports comprehensive potato breeding. Challenges yet remain on recognizing intellectual property rights to breeding and germplasm, and also on regulatory aspects to incorporate modern biotechnology for increasing genetic variation in potato breeding.
Keywords: polyploidy, tetrasomic inheritance, reproductive biology, potato genome
I. Potato reproductive biology and genetics
1. Reproductive uniqueness
The diversity of potato species was discussed in a special issue by Machida-Hirano (2015) with respect to geographic distribution and species variation. Common cultivated potato varieties include tetraploid (2n = 4x = 48) with a basic chromosome number of 12, while there are cultivated species at the diploid (2n = 2x = 24) to pentaploid (2n =5x = 60) levels. The triploid and pentaploid cultivated species are grown only on highland plateaus and slopes of the Andes, but diploid cultivated species are grown more widely and also used for breeding tetraploid varieties. Wild and cultivated potato genetic resources provide a variety of reproductive and genetic features associated with species differentiation and breeding applications (Table 1). The number of species varies among the taxonomic classifications and around 200 wild species have been identified (Hawkes and Jackson 1992, Hijmans and Spooner 2001, also refer to Machida-Hirano 2015). The majority of the 200 wild species are diploid, and many of these diploids species can be crossed with tetraploids using 2n gametes, which can be easily found in potato genetic resources both in cultivated and wild species (Iwanaga and Peloquin 1982, Watanabe and Peloquin 1989, 1991, Watanabe et al. 1991).
Table 1.
Reproductive features associated with potato breeding using tuber-bearing Solanum spp.
| Feature | Association with breeding | Key references |
|---|---|---|
| Tetraploid | Complex and large segregating population | Allard (1960), Bradshaw (1994) |
| Odd ploidies (3x, 5x) | Ending up with further sexual hybridization | Marks (1966) |
| 2n gametes | Facilitating use of diploid gene pool in tetraploids by interploidy crosses | den Nijs and Peloquin (1977) |
| Dihaploid | Assisting capturing diversity and selection at diploid level | Hermsen and Verdenius (1973), Ortiz and Peloquin (1994) |
| Endosperm Balance Number (EBN) | Providing prediction of hybridization success | Johnston et al. (1980), Johnston and Hanneman (1982) |
| Poor flowering in common cultivars | No cross hybridization | Ross (1986) |
| Male sterility in common cultivars | Unilateral hybridization | Iwanaga et al. (1991a), Salaman (1910) |
| Self-incompatibility at diploids | Limitation of crossing counterparts with the same incompatibility allele | Kaufmann et al. (1991), Pal and Nath (1942) |
Propagation
Potato can be propagated vegetatively by tubers, tissue culture, or even by cuttings (Harris 1978). Usually “seed” implies tuber propagules in the potato community, and true potato seed means botanical seed. Potato species can also produce true potato seeds. Potato fruit looks like a small green tomato. A fertile fruit can contain over 200 true potato seeds in tetraploid cultivars. While diploid wild species are often self-incompatible (Cipar et al. 1964, Pal and Nath 1942), the tetraploid cultivars and polyploidy species are self-compatible. Moreover, many polyploidy species are disomic polyploids with a selfing nature, but they have the potential to produce selfed true seed and tubers (Watanabe and Orrillo 1994).
Endosperm Balance Number
Species are sexually separated principally by their ploidy and Endosperm Balance Number (EBN; Hawkes and Jackson 1992). The EBN was hypothesized as a 2 maternal : 1 paternal EBN genic ratio in endosperm and embryo, and a gene dose of the EBN is a prerequisite for successful interspecific crossability based on normal endosperm development to support embryo survival (Johnston et al. 1980). The EBN is controlled by a few genes with a variety of alleles (Bamberg 1994). The concept of endosperm development and hybrid embryo success was independently developed originally by two groups: one by the potato group at the University of Wisconsin as the EBN (Johnston et al. 1980) and the other by Nishiyama and Yabuno (1978) as polar-nuclei activation indexed by activation/response values (AVs/RVs) on Avena and Ipomoea. Assignment of EBNs and AVs/RVs depends on plumpness, size, and germinability of hybrid seeds (Katsiotis et al. 1995). The applicability of the concept has been proven in different species, such as Brassica (Nishiyama and Inomata 1966, Nishiyama et al. 1991), Impatiens (Arisumi 1982), and Trifolium (Hussain and Williams 2008), and the concept can be applied to many different taxa in relation to interspecific hybridization.
2n gametes
The common occurrence of 2n gametes is a unique feature of tuber-bearing Solanum (Iwanaga and Peloquin 1982, Watanabe and Peloquin 1989, 1991, Watanabe et al. 1991). The 2n gametes are gametes with a somatic chromosome number, often mixed with unreduced gametes. Unreduced gametes are gametes without genetic recombination during the process of gamete formation, which implies no genetic reduction and no reduction of chromosome number. The 2n gametes contribute to polyploidization and enable introgression from diploid to polyploidy taxa (Carputo et al. 2003, den Nijs and Peloquin 1977). Ploidy manipulation can be systematically performed with knowledge of the EBN and use of 2n gametes to utilize the wild species gene pool in cultivated potato breeding (Hanneman 1994, Johnston and Hanneman 1982). The 2n gametes occur widely in angiosperm and they are likely a common mechanism of polyploidization (Harlan and De Wet 1975). Furthermore, 2n gametes have been used for ploidy manipulation in breeding in many taxa of higher plants (Dewitte et al. 2012).
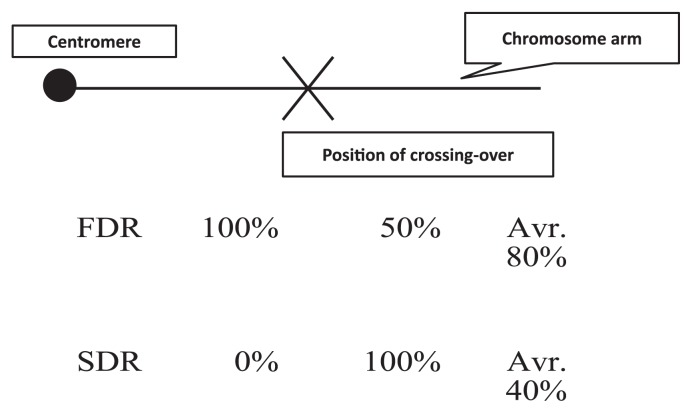
The 2n gamete formation is genetically controlled by simple genetic mechanisms, such as parallel spindles (ps) and premature cytokinesis (pc) in microsporogenesis, which are recessive. The ps gene results in meiotic variation at the second metaphase with parallel orientation of spindle fibers of two chromosome division plates; consequently, the two microspores end up with an unreduced number of chromosomes after cytokinesis instead of a four microspore formation, which is genetically equivalent to first division restitution (FDR) in meiosis (Mok and Peloquin 1975a, 1975b). The pc gene induces premature cytokinesis right after the first division, which is genetically equivalent to second division restitution (SDR). The frequency of 2n pollen varies in different environments, but selection of diploid breeding lines with stable 2n pollen production is feasible (Watanabe et al. 1994, 1995b, 1996a, 1996b). Female 2n gametes have been reported and they exhibit simple inheritance (Iwanaga 1984, Werner and Peloquin 1991).
The genetic significance of 2n gametes is that they are crucial for potato germplasm enhancement. Heterozygosity and allelic interactions are important for many traits, so it is essential to support conservation transmission of parental heterozygosity to the progeny (Carputo et al. 2003). FDR 2n gametes transmit 100% of parental heterozygosity between a centromere and first crossing-over and 50% from the crossing-over to the distal end of a chromosome (Fig. 1). Likewise, SDR 2n gametes transmit 0% of parental heterozygosity between a centromere and first crossing-over and 100% from the crossing-over to the distal end of a chromosome. Overall, FDR 2n gametes, a result of the ps gene, average 80% of the parental heterozygosity, and the SDR gametes, a result of the pc gene, average 40% of the parental hybridity (Mok and Peloquin 1975c).
Fig. 1.
Percentage of genetic constituents which can be transmitted by alternative genetic modes of 2n gametes. FDR and SDR indicate first division restitution and second division restitution, respectively. Based on one cross-over occurrence at a chromosome arm on potato, FDR 2n gametes can transmit an average of 80% and SDR results in an average 40% of parental genetic constituent.
Haploids
Dihaploids from tetraploids, which are diploid, occur naturally (Hougas and Pelqouin 1958) and also can be introduced by pseudogametic parthenogenesis using special pollinators (Hermsen and Verdeniu 1973). Haploid production efficiency varies by pollinator (Peloquin et al. 1996). Haploids from tetraploids often do not flower and are also male-sterile due to inbreeding during the processes of haploidization. Haploidization can also be induced by another culture, but often the derived haploids do not have vigor compared with the ones by pseudogametic parthenogenesis (Ortiz and Peloquin 1994). However, those tissue culture-based haploids facilitated genetics study at the diploid level (Veilleux 1999) and germplasm enhancement for disease and pest resistances (Wenzel and Uhrig 1981).
Selection of haploids can lead to diploid breeding lines to obtain wild diploid genetic variation (Carputo et al. 2003, Hougas and Peloquin 1958) and F1 hybrids between the haploids, and wild species can be used as bridging genotypes to transfer useful traits from diploids to the tetraploid cultivated gene pool (Watanabe et al. 1995b). In addition, this type of haploid has been used to study the segregation of traits at the tetraploid level if many haploids are produced from one tetraploid genotype.
Self-incompatibility
Self-incompatibility occurs commonly in tuber-bearing Solanum species, especially at the diploid level. Self-incompatibility of diploid wild species results in genetic variation within a taxon, and allows outcrossing sexual reproduction with heterozygosity and vegetative propagation (tubers) under variable climatic conditions (Cipar et al. 1964). The locus S with multiple alleles controls gametophytic self-incompatibility (reviewed by Xu et al. 1990). The genetic structure of the S gene is rather simple: the S2 allele consists of two exons and one intron of 117bps with a total size of 786 bps (Kaufmann et al. 1991). The S locus was identified on chromosome I (Gebhardt et al. 1991). A rare self-compatibility-inducing mutant gene, Sli, has also been identified, and this unique mutant gene can enable pure genetic lines to be established to study the breeding value of heterozygosity in potatoes (Hosaka and Hanneman 1998). The Sli gene was mapped at the distal end of chromosome XII; thus, the Sli gene is independent of the S locus.
Male sterility
Male sterility occurs often in tetraploid cultivars and related taxa (Grun et al. 1962). It is the consequence of nuclear-cytoplasm interactions; the dominant Ms gene interacts with the cytoplasm of S. tuberosum Group Tuberosum cultivars to cause male sterility and the dominant Rt gene restores fertility (Iwanaga et al. 1991a). For example, the diploid hybrids between S. tuberosum Group Tuberosum haploids × Group Phureja yield all or almost all male sterile progeny with the dominant Ms from Group Tuberosum influencing Tuberosum cytoplasm but the reciprocal cross results in fertile filial lines. The occurrence of male sterility often results in difficulties for potato breeders, as the choice of parental lines can restrict the introgression of traits. However, once information on the genotypes associated with male-sterility is organized, prediction can be made easily to choose parental lines for germplasm enhancement.
2. Tetraploid genetics
While progress has been made in potato breeding, many resources have to be invested due to the requirements of large populations and long time frame. This is not only due to the general pitfalls in plant breeding, but also due to the complexity of polyploid genetics. Tetraploid genetics is the most prominent aspect associated with potato breeding (Ortiz and Watanabe 2004, Table 2). Often, a trait controlled by a single gene can be quantitatively scored by phenotype due to its specificity in segregation (Fig. 2). Here, the term “polysomic polyploidy” from Mackey (1970) is used instead of the word “autopolyploid” used by Grant (1971), Stebbins (1950), and other botanists. Tetrasomic behavior of chromosomes and corresponding genetic segregation have been reported (Matsubayashi 1991, Swaminathan and Magoon 1961). However, variation was observed in frequency of quadrivalent formation in tetraploid cultivars.
Table 2.
Comparison of inheritance in tetraploid potatoes and polyploidy wild Solanum species
| Feature | Species example | Segregation | Multivalent in meiosis | Chromatid segregation | Fertility | Key references | |
|---|---|---|---|---|---|---|---|
| Tetrasomic cultivars | tetraploid | S. tuberosum | Tetrasomic and variable | Yes | Possible | Low to medium |
Allard (1960) Bradshaw (1994) Ortiz and Watanabe (2004) |
| Disomic species (4x, 6x) | polyploid | 4x: S. acaule, S. stoloniferum, 6x: S. demissum | Disomic | No | No | Very high | Watanabe and Orrillo (1994) |
Fig. 2.
One variation and complexity of zygotic outputs in polysomic polyploids. Assuming a diallelic model with complete dominant A and recessive a at a locus, genotypic and phenotypic segregation are displayed in the crosses from heterozygous parental genotypes. Ovals indicate genotypic segregation and squares show phenotypic segregation. Increase of ploidy requires far larger progeny population size for specific phenotypic selection.
Inter-locus interaction (epistasis) and heterozygosity are important factors in potato breeding, while additive components also contribute to quantitative traits. Identification of individual chromosomes is very difficult by conventional cytogenetic methods, while monitoring recombination and introgression is also difficult by phenotypic evaluation at the tetraploid level. Theoretical prediction of segregation has been supported by cytogenetic and genetic research, and these findings were tested and applied in potato breeding (Ross 1986).
There are four unique aspects of polyploids: 1) genotypic variation at a locus, 2) possibility of multiple alleles in a locus of the same genotype, 3) possibility of the occurrence of allelic interactions with multiple alleles, and 4) occurrence of chromatid segregation as well as chromosome segregation (Allard 1960, Bradshaw 1994).
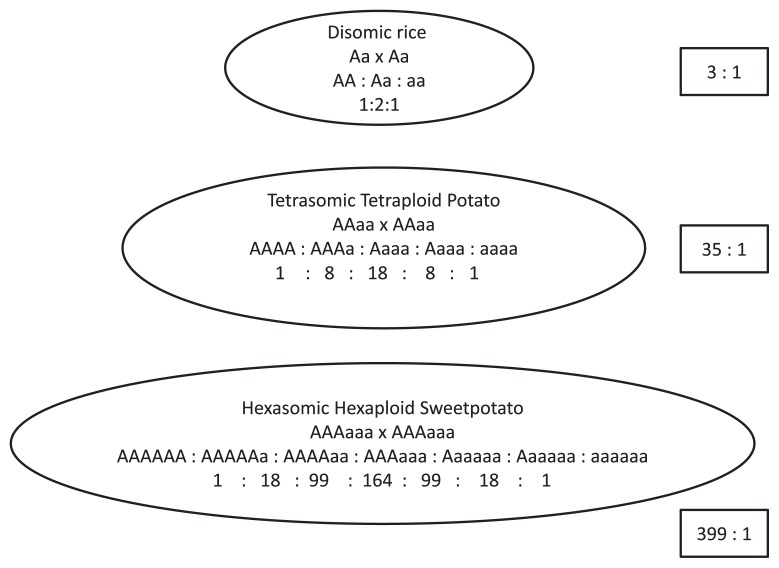
Due to tetrasomic conditions, one locus accommodates four alleles, and with a diallelic locus, A and a, five classes of genotype are possible: AAAA, AAAa, AAaa, Aaaa, and aaaa. If an additive effect of an allele occurs, then one locus can result in considerable quantitative segregation (Fig. 2).
A diversity of multiple alleles is possible: each allele can have a different function, such as a1, a2, a3, and a4.
These different alleles can interact, resulting in various allelic interactions: diallelic, a1a2, a1a3, a1a4, a2a3, a2a4, a3a4; triallelic, a1a2a3, a1a3a4, a2a3a4; or tetrallelic a1a2a3a4. Thus, heterosis is not the only result of interaction between two alleles, but there a wide range of possibilities.
A locus distal to the centromere with a chance of crossing-over can be involved in chromatid segregation. For example, a locus with a diallelic condition and genotype Aaaa produces gametic output at a 1 : 1 ratio to Aa : aa by chromosome segregation and 15AA : 12Aa : 1aa by chromatid segregation, assuming random assortment of the homologous chromosomes and chromatids. Thus, a rare gametic genotype and zygotic progeny phenotype are possible. Another example is AAaa genotype which produces AA : aa = 1 : 1 with chromosome segregation and AA : Aa : aa = 3 : 8 : 3 ratio with chromatid segregation and larger phenotypic ratio could be expected as 208 : 1 if AAaa genotypes are inter-mated.
Even with chromosome segregation, tetrasomic tetraploid genetics is far more complicated than the diploid situation; moreover, the outcrossing nature and high heterozygosity of tetraploid potato cultivars cause more complications in segregation. Potato breeding objectives often involve quantitative traits which are highly influenced by the environment. Considering a wide range of phenotype variation with tetrasomic inheritance and environmental effects, a large true seed population is needed in crosses and subsequent phenotypic selection for successful potato breeding.
3. Interspecific hybridization
The genetic base of the potato cultivars is narrow, and could be very susceptible to new strains of pests and diseases, such as potato late blight, which resulted in the Great Irish Famine. While there are many wild relatives of potato, due to the variety of reproductive mechanisms resulting in genetic isolation from cultivated potato, germplasm enhancement has been developed with a variety of methods using the knowledge of reproductive biology and genetics (Jansky 2000). Further challenges in potato breeding are associated with the use of wild relatives with different reproductive and genetic characteristics (Table 1). Differences in ploidy can be overcome with the selection and use of 2n gametes, alleviating the elements listed in Table 1. However, technical trials are needed for successful interspecific sexual hybridization among distantly related genera. Past efforts are discussed below on the attempts to generate hybrids between non-tuber-bearing and tuber-bearing genera.
Accumulated knowledge of reproductive biology and genetics of Solanum species, including some non-tuber-bearing species, has made it possible to use disomic tetraploid species (2EBN) (Iwanaga et al. 1991b, Watanabe et al. 1992, 1995c) and distantly related diploid wild species (1EBN), which often furnish high levels of important resistance. These species cannot be crossed directly with cultivated potato lines, as hybrids result in odd ploidies which do not allow for further introgression.
Embryo rescue combined with rescue pollination (second compatible pollination) can break down major cross incompatibility barriers in many wild Solanum species that contain resistance (R) genes of potential value, which cultivated potatoes do not have (Iwanaga et al. 1991b, Watanabe et al. 1992, 1994, 1995c). Yield performance also can be high due to introduction of unique alleles from a wild gene pool (Fig. 2). The sexual filial progeny have a use in interspecific hybridization. Previous approaches, including somatic hybrids, required complicated ploidy manipulation and/or bridging to introgress the valuable genes from these species, while the current new strategy uses simpler method(s). Developments in enhancement methods indicate the feasibility of introgression of tetraploid wild species, such as S. acaule and S. stoloniferum (Watanabe et al. 1992), and diploid non-tuber-bearing wild species, such as S. brevidens (Watanabe et al. 1995c), while generalization of these methods can be exploited to achieve genotype-independent success. Alleviating the problems in obtaining successful filial generations using different wild species also provides an opportunity for comparison of genetic efficiency with somatic fusion-derived hybrids using such species as S. brevidens, which furnishes many relevant resistances for potato breeding, such as resistance to potato leaf roll virus, tuber soft rot, and early blight.
This approach has been used by the potato breeding community for germplasm enhancement with wild species, as can be found easily in various papers. Protoplast fusion is another approach for using distantly related taxa. However, there has not been significant cultivar developed via this approach (Millam et al. 1995).
4. Germplasm enhancement at the diploid level using haploids and 2n gametes and high heritability of traits from 2x to 4x
Potato genetic improvement has been achieved by establishing systematic germplasm enhancement methods and breeding approaches. The International Potato Center (CIP) has contributed to international collaboration by establishing research networks with broad participation (Watanabe et al. 1995a). Germplasm enhancement with diploid tuber-bearing Solanum species, including some diploid cultivated taxa, has been performed in many potato breeding programs (Watanabe et al. 1995b, 1996a, 1996b, 1999a, 1999b, 1999c). The methods alleviate some of the bottlenecks that occur in conventional breeding at the tetraploid level. These involve two major biological tools: 1) haploids from 4x cultivars and 2) 2n gametes. Haploids from tetraploids can be obtained easily by pseudogametic parthenogenesis using a haploid inducing pollinator (Hermsen and Verdenius 1973, Hougas and Peloquin 1958), and are used to capture useful genes from wild or exotic germplasm. Breeding at the diploid level with disomic inheritance facilitates simultaneous selection for target resistance traits as well as some agronomic characteristics (Ortiz and Peloquin 1994). Transmission of useful traits from diploid breeding lines has been successfully achieved by the use of 2n gametes, especially using FDR 2n pollen, which occurs widely and frequently in the diploid tuber-bearing Solanum species. Furthermore, the concept of an EBN greatly assisted prediction of success in interspecific and/or interploidy crosses (Carputo et al. 2003, Hanneman 1994).
Using the above strategy, many potato cultivars have been established with successful quantitative disease and insect pest resistances (Watanabe et al. 1999a, 1999b, 2005). These resistance sources from wild species were transmitted to tetraploid breeding lines in a few years via germplasm enhancement methods (Watanabe and Watanabe 2000). Bacterial wilt, early blight, potato tuber moth, and root-knot nematode have been successfully introgressed into potato breeding lines. Consequently, multiple resistant diplopid clones have been generated and used for tetraploid potato breeding.
Prediction of heterozygosity transmission by 2n gametes, which is associated with effective transmission of quantitative traits, is shown in Table 3. The estimation of transmission rate of quantitative traits from diploids to tetraploids via different genetic modes of 2n gametes is given in Table 3. A simple genetic model is presented as percentage of 2n gametes with desirable allelic combination for QTLs, assuming complete dominance at each locus and dominant alleles are responsible for the trait, an additive effect over loci and P value, and probability of crossing-over between a locus of interest and the centromere. The expected outcome was the percentage of progenies with high frequencies of alleles/loci responsible for a quantitative trait. By comparison with 4x × 4x mating, 4x × 2x crosses are favored when high allelic frequency for the resistance do not exist in the tetraploids. On the other hand, when a high allelic frequency occurs in the tetraploids, 4x × 4x is favored over 4x × 2x. The overall outcome from the simulation showed that 4x × 2x with FDR was generally more desirable. The results can be used in conjunction with the existing genetic map information to determine an optimum population size in 4x × 2x progenies to make efficient selection in plant breeding. This theoretical information enabled the generation of multiple resistances to be transferred to 4x × 2x progenies (Watanabe et al. 1999a, 1999b, Watanabe and Watanabe 2000).
Table 3.
Transmission estimation of multiple loci with heterozygosity by 2n gametes. FDR is first division restitution and SDR is second division restitution. The p indicates the frequency of a crossing-over between the centromere and a locus
| # loci | FDR | SDR | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| p = 0 | p = 0.5 | p = 1.0 | p = 0 | p = 0.5 | p = 1.0 | |
| 1 | 100 | 87.5 | 75.0 | 50 | 75.0 | 100 |
| 2 | 100 | 76.6 | 56.3 | 50 | 56.3 | 100 |
| 3 | 100 | 67.0 | 42.2 | 50 | 42.2 | 100 |
| 4 | 100 | 58.6 | 31.6 | 50 | 31.6 | 100 |
| 5 | 100 | 51.3 | 23.7 | 50 | 23.7 | 100 |
| 6 | 100 | 44.9 | 17.8 | 50 | 17.8 | 100 |
| 7 | 100 | 39.3 | 13.3 | 50 | 13.3 | 100 |
| 8 | 100 | 34.4 | 10.0 | 50 | 10.0 | 100 |
| 9 | 100 | 30.1 | 7.5 | 50 | 7.5 | 100 |
| 10 | 100 | 26.3 | 5.6 | 50 | 5.6 | 100 |
II. Molecular markers and Genome research
1. Genome research
The potato genetic and genome research communities established research resource databases together with other plant science groups. The SOL Genomics Network (SGN) (http://solgenomics.net/, Mueller et al. 2005) was derived from the Tomato Genetics Cooperative (TGC) but now supports all Solanaceae species. The SGN provides information on maps & markers, genes, genomes & sequences, breeding tools, pathways, and phenotypes all in one site.
Potato genome research has been effectively supported by tomato genome information due to the high synteny between their genomes and their close molecular evolutionary relationship (Bonierbale et al. 1988). Tomato genome information has been employed for marker exploitation for potatoes, such as COS (Conserved Ortholog Set) (http://solgenomics.net/markers/cos_markers.pl) (Fulton et al. 2002) and COSII (http://solgenomics.net/markers/cosii_markers.pl) (Wu et al. 2006). The COS was developed originally by screening an extensive tomato expression sequence tag (EST) database against the Arabidopsis genomic sequence. The first COS contained 1,025 genes that are single or low copy in both genomes. The COSII was the result of extensive expansion of comparative assessment of the genome database. Lindqvist-Kreuze et al. (2013) applied the COS to 300 genes using eight accessions of tuber-bearing Solanum species. These gene sequences were mapped with a BLASTN algorithm. They found that the majority of the genes are single copy and corresponded well with the tomato genome, but some showed repetitiveness with unexpected location in the potato genome. The effectiveness of COS markers has been proven by different taxa beyond Solanum, such as Lycium (Levin et al. 2009), Manihot esculentum (Castelblanco and Fregene 2006), Rosaeae (Cabrera et al. 2009), and Theobroma cacao (Khun et al. 2012).
The Potato Genome Sequence Consortium (PGSC) was established (http://www.potatogenome.net/index.php/Main_Page) and all of the genome information has been released (PGSC 2011). The PoMaMo database (Meyer et al. 2005) once supported genome research, but now it is integrated into the potato genome under the PGSC. The database is available at http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml (Sharma et al. 2013).
Diploid potato has an estimated genome size of 844 Mb (Bennett et al. 1997, PGSC 2011). The PGSC has sequenced two diploid potato genotypes: the heterozygous diploid S. tuberosum Group Tuberosum genetics line RH89-039-16 (RH) and the doubled monoploid S. tuberosum Group Phureja clone DM1–3 (DM). The major achievements are as follows:
Sequence information was assembled with 86% of the whole genome of the doubled-monoploid potato clone, and 39,031 protein-coding genes were predicted.
A palaeopolyploid origin was also proposed with two major gene duplication events.
Presence of potato-specific genes was indicated for 2,642 genes.
Sequence of the heterozygous diploid clone indicated that gene presence/absence variants and other potentially deleterious mutations occur frequently and are a likely cause of inbreeding depression.
Tuber development evolved from gene family expansion, tissue-specific expression, and recruitment of genes to new metabolic pathways.
Potato research has benefitted from overall advances in the plant sciences and biotechnology. Here are some examples: carbohydrate metabolism, plant hormone interaction with tuberization, tissue culture for germplasm conservation and clean seed tuber production, pathogen detection methods, and transgenic approaches (Watanabe et al. 1997). Tuberization is controlled by day length (Rodríguez-Falcón et al. 2006, Seabrook et al. 1991) and plant hormones, such as gibberellin and jasmonic acid (Amador et al. 2001, Pruski et al. 2001). Each potato genotype tuberizes under only the given day-length requirement with specific physiological conditions. While simplified in vitro studies have been conducted, so far no elucidation of the genetic system of tuberization has been reported and it is considered a complicated genetic system (Brown 2007, Struik et al. 1999).
A novel finding in the plant science community has opened a new arena for studying tuberization. The mobile FLOWERING LOCUS T (FT) protein is a main component of long-range ‘florigen’, or flowering hormone, which was originally identified in Arabidopsis thaliana (Kobayashi et al. 1999). Trangenic short-day tuberizing potato genotypes with FT induce tuberization under long-day conditions (Navarro et al. 2011). Using the genome sequence (PGSC 2011) and information on FT, Kloosterman et al. (2013) identified the potato genomic element StSP6A, which induces mobile tuberization signal. The StSP6A signal resulted in the induction of tuber development at the stolon termini. They have postulated that diverse allelic variation of the gene is closely associated with the domestication of potatoes to diverse latitudes with corresponding different day lengths.
2. Potato genetic map
The first potato genetic map was reported in 1988 using tomato RFLP (restriction fragment length poylmorphism) markers (Bonierbale et al. 1988). Since potatoes are highly heterozygous, F1 mapping can be conducted using two heterozygous genotypes. Two linkage maps were obtained from a cross between a diploid clone of S. tuberosum Group Phureja and a diploid hybrid line from S. tuberosum Group Tuberosum × (S. tuberosum Group Phureja × S. chacoense). The alignment of the RFLP loci shows a high level of similarity to that of the tomato map and the major differences were paracentric inversions on three chromosomes.
Generation and uses of molecular markers in potato genome studies are listed in Table 4. With advances in molecular marker knowledge in general, the potato genetics community also has applied different markers. Many maps have been constructed and used for identifying specific loci, and markers have been used in breeding for specific traits.
Table 4.
Representative potato genetic markers and maps
| Marker type | Number of loci or markers | cM | References | Comments |
|---|---|---|---|---|
| RFLP (restriction fragment length polymorphism) | 135 | 1189 | Bonierbale et al. (1988) | First potato map |
| RFLP | 141 | 690 | Gebhardt et al. (1989) | High heterozygosity observed in diploid genetics lines |
| RFLP | 304 | 1034 | Gebhardt et al. (1991) | Increase of mapped loci |
| RFLP | 1030 (average 1.2 cM interval) | 1276 | Tanksley et al. (1992) | Highly saturated map for comparison with tomato |
| RFLP, transposons, isozymes | 175 | 1120 | Jacobs et al. (1995) | Integration of known loci and new markers |
| AFLP (amplified fragment length polymorphism) | 770 | – |
Vos et al. (1995) van Eck et al. (1995) Rouppe van der Voort et al. (1998) |
AFLP used on potatoes and high resolution is possible |
| RGL (resistance gene-like fragment) | Concept generation for landmark | – | Leister et al. (1996) | Applicability for disease and pest resistances |
| SSR | 89 | – | Milbourne et al. (1998) | Fingerprinting |
| ISSR (inter simple sequence repeat) | 4 | – | Prevost and Wilkinson (1996) | Validation for fingerprinting |
| RAPD (random amplified polymorphism), RFLP | 100 | 606 | Hosaka and Hanneman (1998) | An example of target-trait-specific rapid generation of mapping |
| COS (Conserved Ortholog Set) | 1025 | – | Fulton et al. (2002) | Further validation of tomato-potato orthology |
| SSR | 156 | – | Ghislain et al. (2004) | 15 SSR marker sets selected as highly informative |
| SSR (simple sequence repeat, microsatellite) | 61 | – | Feingold et al. (2005) | EST-based SSR |
| Multiple gene family homologues (P450) | 15 primer pairs produced 27 loci | – | Yamanaka et al. (2003, 2005) | Simple approach to make a marker and map |
| AFLP | 10,305 (10,365 with markers such on CAPS, SCAR) | 751 maternal/773 paternal | Van Os et al. (2006) | Small number of progeny individuals with 130 and 381 AFLP primer combinations made over 10,000 markers |
| NB-LRR disease resistance gene homologues | 738 RGLs | 47 R genes physically mapped | Bakker et al. (2011) | BAC-based physical map of R genes |
| NB-LRR R genes | 438 | 370 R genes physically mapped | Jupe et al. (2012) | |
| BAC physical map | 2800 contigs | 1.64 times the coverage of the genome | de Boer et al. (2011) | Foundation for genome sequencing |
| SNP (single nucleotide polymorphism), SSR, AFLP, DArT (diversity array technology) | 2469 | 936 | Sharma et al. (2013) | Integration of genetic and physical maps |
AFLP (amplified fragment length polymorphism) markers are highly transferable between potato populations (Rouppe van der Voort et al. 1997, 1998). However, Ishidore et al. (2003) examined Linkage I with 1,260 loci, and there was a change in PstI-site methylation patterns within a population. The map indicated highly variable recombination frequencies. Although the map indicated that centromeric regions were highly saturated and showed extensive marker clustering, some areas lacked AFLP markers. Currently, there is no miracle marker set to serve all purposes.
Microsatellite markers (also known as simple sequence repeats—SSRs) have assisted progress in mapping the potato genome (Provan et al. 1996), and are valuable for fingerprinting closely related genotypes (Milbourne et al. 1998). AFLP showed greater merit than multilocus SSRs for fingerprinting in a comparative study of RAPD (random amplified polymporphic DNA), ISSR (inter simple sequence repeat), AFLP, and SSR (McGregor et al. 2000) These can be made from the EST database (Feingold et al. 2005) and made more systematical by potato genome information (PGSC 2011).
With further advancement of genome research in plants, many relevant technologies can be applied to potato. DArT (diversity array technology) (Wenzl et al. 2004) benefits potato genetics. Finally, new sets of markers have been applied with advances in genome sequencing (PGSC 2011). Single nucleotide polymorphism (SNP) has been used widely for marker generation in many species and many markers have been generated also in potatoes (Sharma et al. 2013).
3. Use of R gene information from potato genome
Potato and its related species furnish a diversity of resistances to diseases and pests. Classical genetics studies have been conducted on inheritance and R genes have specific gene name designations (Valkonen et al. 1996). These R genes have been mapped to different chromosomal locations. The key class of genes that comprise the vast majority of plant R genes contain a nucleotide-binding site (NBS) and leucine-rich repeat (LLR) domain, and they could be used as a genetic marker set (Leister et al. 1996). Mapping information of R genes and their homologues were integrated by Jupe et al. (2012). Many of the R genes for different pests and diseases are independent. The systematics of the R gene structure was postulated using 438 NB-LRR-encoding sequences. Bakker et al. (2011) linked the existing 82 disease and pest resistance loci using 738 NB-LRR disease resistance gene homologues. Further physical localization of 428 R genes has been achieved (Jupe et al. 2012). These studies have supported the implementation of marker-assisted selection (MAS) for potato breeding (Carrasco et al. 2009).
4. Potato virus Y discussion as an example of validation of markers
Extreme resistances (ERs) to potato virus Y (PVY) are available in various Solanum species, including the following: Ryadg, Solanun tuberosum Group ANDIGENA (4x); Rysto, S. stoloniferum (4x); Ryhou, S. hougasii (6x); Ryphu, Group PHUREJA (2x); Rychc, S. chacoense (2x); and Rybrd, S. brevidens (2x). Among potato-related Solanum species, the Ry gene is always dominant and shows ER expression in F1 hybrids produced by direct crossing between Solanum series Tuberosa and non-tuber-bearing S. brevidens (Series Etuberosa) (Valkonen et al. 1995). ER shows no symptoms with extremely low viral accumulation in inoculated seedlings, and cell death confines the virus (Valkonen et al. 1996). Some of these genes have been used in breeding and they have been mapped using these breeding lines and cultivars. Ryadg was mapped on chromosome XI (Hämäläinen et al. 1997). Rysto is on chromosome XII (Song et al. 2005) and Rychc is on chromosome IX (Sato et al. 2006).
The level of ER is the same among these Ry genes, and the resistance is controlled by a single dominant allele. There is no difference in the level of resistance at the simplex and quadriplex levels for the Ry allele. Thus, a Ryryryry genotype at the tetraploid has the same level of resistance as a RyRyRyRy genotype. Moreover, Ry genes have no additive effect by accumulation of the different Ry loci.
In nature, different genotypes or accessions of a species can furnish the same resistance type, but complete genetic identification, including chromosomal location, is required. Valkonen et al. (2008) compared different accessions of S. stoloniferum that confer ER to PVY. They compared the map positions and markers for Rysto identified by Song et al. (2005). Based on sequence analysis, they concluded that the Rysto gene from different accessions of S. stoloniferum can be selected with the same marker sets on chromosome XII.
There are also hypersensitive genes to PVY, designated Ny, which also provide some level of resistance (Valkonen et al. 1994). Ry is epistatic to Ny by prohibiting Ny expression. Ny is also valid for resistance breeding in potatoes, but resistance is lowered by high temperatures. Many Ny were obtained from different species and mapped to different chromosomal locations within the potato genome. Szajko et al. (2014) reported that Ny-2, conferring hypersensitive resistance to PVY, was mapped on chromosome XI in cultivar Romula, but this was not at the same genomic position as Ryadg. The Ny-1 gene was mapped on chromosome IX using the pedigrees from cultivars Albatros and Sekwana.
An R gene is usually identified from a specific potato clone, and pedigree assessment is made on whether the resistance can be identified within the same introgression pedigree for generating valid selection markers. Ryadg, which came from Gp. ANDIGENA and is widely used in the USA and Europe, provides an example. A resistance gene-like DNA fragment was obtained as a PCR product (310 bp) from potatoes. The RFLP clone, designated as ADG2, co-segregated with the RFLP markers that associate highly with Ryadg. ADG2 was characterized as homologous to the corresponding R gene region of several different R genes from a wide range of plant species, including kinase 2 and kinase 3a motifs (Hämäläinen et al. 1998).
Markers for Ry have been generated and validated (Table 5). Progress has been made in selection effectiveness and simpler methodology, with faster time and lower costs, starting from RFLP, CAPS, and then SCAR. With a SCAR, RYSC3, high association with Ryadg was reported (Kasai et al. 2000). This SCAR marker has been widely used in the pedigree programs of North America and Europe. Furthermore, Shiranita et al. (1999) reassessed RFLP markers using ADG2 as a probe, and they have identified general markers for Rys using a wide range of pedigrees and Ry sources from different species consisting of 117 genotypes with 83% correspondence on ER and non-ER, including susceptibility and hypersensitivity. Validation of this marker set (RYSC3) has been confirmed by different potato breeding groups (Ottoman et al. 2009, Sagredo et al. 2009).
Table 5.
Validation history of the selection markers for marker-assisted selection on Ryadg that confers extreme resistance to potato virus Y (PVY) located near the proximal end of chromosome XI. Genotype implies representation of cultivars, breeding lines, and clones of wild species from different origins
| Marker type | Correspondence with the target Ryadg phenotype | References |
|---|---|---|
| RFLP (GP 125) | 30/31 genotypes (96.7%) | Hämäläinen et al. (1997) |
| RFLP (ADG2 probe, 3.5 kb fragment) | 77/77 F1 progeny 112/117 genotypes (95.7%) |
Hämäläinen et al. (1998) Shiranita et al. (1999) |
| CAPS (ADG2 fragment) | 77/77 genotypes (100%) | Sorri et al. (1999) |
| RFLP (ADG2 probe, 10.5 kb fragment) | 97/117 genotypes (83%) for different origins of Ry sources | Shiranita et al. (1999) |
| SCAR (ADG2 fragment with RYSC3 primers) | 103/103 genotypes (100%) | Kasai et al. (2000) |
Likewise, several R genes to potato virus X (PVX) have been reported and they are located at different chromosomal positions (Ritter et al. 1991) but with identical types of resistances, such as ER and hypersensitivity (Tommiska et al. 1998, Valkonen et al. 1996). Furthermore, independence of the virus resistance loci to PVX have been reported (Tiwari et al. 2013).
With the accumulation of independent R gene information and validation of markers, MAS has enabled the establishment of multiple resistance genotypes (Gebhardt et al. 2006). MAS is effective (Ortega and Lopez-Vizcon 2012) but cost-effectiveness varies depending on how a potato breeding program pursues selection rates of target clones (Slater et al. 2013). There is a need for more coordinated collaboration in the potato research community so that various comparative advantages can be pooled for the overall benefit of potato improvement.
5. Example of MAS on quantitative traits
An example of MAS with a combination of molecular markers and graphic genotyping as well as the 2n gamete function is shown in Table 6. Molecular MAS of diploid potato parental lines was tested for quantitative insect resistances conferred by glandular trichomes derived from the diploid wild species S. berthaultii. Diploid potato parental lines were characterized by RFLP, phenotypic trichome expressions, and insect resistances with small insects, such as aphids, potato tuber moths, and mites. Ideotyping of diploid parental lines was performed using the HyperGene computer program (Young and Tanksley 1989) using quantitative trait loci (QTL) information for glandular trichomes (Bonierbale et al. 1994). Progeny testing was conducted on those parents with RFLP-QTL ideotyping by trichome phenotypic observation and insect assays for resistances. Using the selected diploid parents, 2x × 2x and 4x × 2x crosses were made to test whether the QTL could be transmitted efficiently for glandular trichome expression, and if this could confirm insect resistances. The majority of the progeny in the 2x × 2x and 4x × 2x crosses showed Type A trichomes and Type A-mediated insect resistances, but Type B trichome expression was low. Thus, QTL information and the corresponding ideotyping on Type A trichomes are effective enough with the same genetic backgrounds on the glandular trichomes, but QTL for Type B trichomes should be elaborated for higher efficiency of obtaining trichome expression and insect-resistant progenies.
Table 6.
An example of QTL transmission on glandular trichome traits in potato: Frequency of Type A and B glandular trichomes in 4x × 2x potato families
| Family | Female parent (4x) without trichomes (4x) | Male parent (2x) with Type A and B trichomes | A trichomes | Also with B trichomes | No trichomes | Total |
|---|---|---|---|---|---|---|
| 95.201 | Atlantic | M200.38 | 100 | 8 | 2 | 110 |
| 94.202 | Atzimba | M200.38 | 82 | 35 | 6 | 123 |
| 94.204 | Serrana.INTA | M200.38 | 86 | 17 | 3 | 106 |
| 95.205 | Atlantic | 94.104.37 | 30 | 97 | 3 | 130 |
| 95.206 | Atzimba | 94.104.37 | 18 | 99 | 4 | 121 |
| 95.207 | Serrana.INTA | 94.104.37 | 15 | 102 | 5 | 122 |
| 95.208 | Atlantic | 94.106.21 | 16 | 83 | 3 | 102 |
| 95.209 | Atzimba | 94.106.21 | 10 | 100 | 3 | 113 |
| Total | 357 (38.5%) | 541 (58.4%) | 29 (3.1%) | 927 |
III. Partners, Intellectual Property, and New Technology Assessment for Breeding
1. Partners and Industry
Potato is a key crop in global food security. Potato research is supported extensively by two major societies and some regional ones, such as ALAP (La Asociación Latino-americana de la Papa, Latin America Potato Association, http://www.papaslatinas.org) and APA (African Potato Association, http://www.africanpotatoassociation.org). Multisector interaction is facilitated by forums, such as the World Potato Congress (http://www.potatocongress.org), by amalgamating the interest of different communities to promote potato production and commercialization of potato products. The potato industry encompasses a wide range of activities. Potato variety development is critical for the seed potato industry, which produces raw potatoes for the fresh food market, processing for industrial materials, such as starch and alcohol, snack foods, frozen fries, and cooked preserved food.
2. Intellectual properties
The potato industry strongly protects intellectual property rights (IPRs) related to industrial applications, such as processing, product development, and trademarks. In addition, inventions from biotechnology and molecular biology are widely covered by intellectual property protection, especially by patents. Seed-tuber production and quality assurance have a strong association with proprietary technology, such as that on virus eradication, pathogen detection, large scale-production by tissue culture, and technical aspects of breeding, especially those related to selection at large plant breeding stations, that may be covered as trade secrets or even by utility patents (Chapman and Watanabe 2007, Watanabe 2011, Watanabe and Komamine 2004).
Specific examples of patents as representation of IPRs claimed on potato biotechnology and breeding applications are shown in Table 7. There are five specific issues. First, a new cultivar can be protected by a patent rather than by plant variety protection laws under the International Union for the Protection of New Varieties of Plants (UPOV) (UPOV, http://www.upov.int/portal/index.html.en). However, what are patentable items is debatable under the main IPR forums, namely, the WTO’s Trade-Related Aspects of Intellectual Property Rights (TRIPS) (http://www.wto.org/english/tratop_e/trips_e/trips_e.htm) and the World Intellectual Property Organization (WIPO) (http://www.wipo.int/portal/en/index.html), and there is no universal recognition of plant variety as a patentable item; thus, this issue has come under the UPOV scheme. Second, the public sector, including state universities, has filed and owns a variety of patents rather than use the UPOV scheme. Third, the Japanese private sector has been somewhat active in patent filing and use, and some key inventions have been covered, such as those by Kirin Co. Ltd.; however, no proactive research publication has been established by the private sector. In addition, these patents have been considered as inappropriate since they appear to have no further application. Fourth, a patent on genetic markers can be made to allow a diversity of free public users of markers. Such patents have been made by Kyoto University and the University of Tsukuba. Fifth, genes and their uses have been protected under the IPR system, and applications for commercial use often have been hindered due to the difficulty of integrating patent licenses. Financial resources and/or cross-licensing deals need to be considered for IPR coordination.
Table 7.
Examples of patents associated with potato biotechnology and breeding applications sorted by the date of application. Sources are JPO (Japan Patent Office), PTO (USA Patent Office), and EPO (European Patent Office)
| Patent title | Representative patent or PCT | Date of application (dd/mm/yyyy) | Date of publication of application | Priority date | Inventor | Applicant |
|---|---|---|---|---|---|---|
| Potato variety ‘ND1538-1Russ’ | US5434343 A | 02.03.1990 | 18.07.1995 | 02.03.1990 | R.H. Johansen | North Dakota State Univ. |
| Potato alpha-amylase gene | PCT/DK1990/000108 | 24.04.1990 | 12.02.1992 | 24.04.1989 | Kirsten Gausing, Jette D. Kreiberg | Danske Spritfabrikker |
| Lepidopteron insect-resistant transgenic potato plants | US 6100456 A | 16.03.1992 | 08.08.2000 | 16.03.1992 | Sticklen, M.B. and J. Cheng | Board of Trustees Operating Michigan State University |
| A novel SCAR marker for the gene Ryadg conferring extreme PVY resistance | JP3047022 (24.03.00) | 19.04.1999 | 31.10.2000 | 19.04.1999 | Furusawa, I., K. Watanabe and K. Kasai | Kyoto University |
| Gene promoters isolated from potato and use thereof | PCT/US2002/001287 | 18.01.2002 | 01.08.2002 | 23.01.2001 | Z. Dai, B.S. Hooker, L. Shi | Ziyu Dai, Brian S. Hooker, Lifang Shi |
| Novel meloidogyne-resistance gene and utilization thereof | PCT/JP02/12392, JP4320399 | 27.11.2002 | 02.10.2003 | 27.03.2002 | Watanabe, K. and J. Watanabe | Univ. of Tsukuba |
| Comprises transgenic potato which generates starch with modified viscosity and phosphate characteristics; for enhancing quality of paper, cardboard, adhesive, textile, plaster, concrete, fertilizer, medicine, and toothpaste; improving animal feeds | US7897760 B2 | 19.10.2006 | 01.03.2011 | 19.09.1995 | Kossmann, J. and R. Lorberth | BayerBioscience Gmb |
| Protein having glycoalkaloid biosynthase activity and gene encoding same | PCT/JP2011/069643 | 30.08.2011 | 08.03.2012 | 31.08.2010 | Sasaki, K. et al. | Kirin Holdings Ltd. |
On the other hand, some extensive patents have influenced public research, limiting access and activity, such as indicated in Table 8, but many of those patents have now expired. However, private-public cooperation could provide an open arena for initially testing the utility of IPRs before making restrictions and limiting access and use of IPRs (James 2013, Qaim 2009). Key patents have expired or been terminated, making open use possible and helping the research community. This will provide flexibility for the potato breeding community by allowing free IPR platforms for cultivar development.
Table 8.
Examples of patents with an extensive coverage of content on plant biotechnology and breeding applications sorted by the date of application. Sources are PTO (USA Patent Office) and EPO (European Patent Office)
| Patent title | Representative patent, PCT, or notification # | Date of application (dd/mm/yyyy) | Date of publication of application | Priority date | Inventor | Applicant |
|---|---|---|---|---|---|---|
| Vector for coat proteins for potato virus | US 4970168 A | 27.01.1989 | 13.11.1990 | 27.01.1989 | Tumer, N.E. | Monsanto Co. |
| Modification of plant metabolism | EP438904 (GB19890028937) | 21.12.1990 | 11.08.1999 | 21.12.1989 | Burrel, M.M. and K.S. Blundy | Advanced Technologies (Cambridge) Ltd. |
| A method for obtaining plants with reduced susceptibility to plant-parasitic nematodes | WO 1993010251 A1 | 02.11.1992 | 27.05.1993 | 20.11.1991 | Sijmons et al. | Mogen Int. |
3. New technology application to breeding: Transgenic potatoes
Potato research has benefitted from transgenic applications. Since two papers in this potato special issue address related content extensively, this section addresses only safety and application for cultivar development. Field trials of transgenic potatoes are not only conducted in developed countries, but also have been conducted in Mexico for virus resistance, and in India and Bangladesh for late blight resistance.
Historical information on field dissemination of transgenic organisms in general can be obtained at Biotrack, maintained by the Organization for Economic Co-operation and Development (OECD) (http://www.oecd.org/science/biotrack/). This evolved functionally into two databases. FAO GM Foods Platform by the Food and Agriculture Organization of the United Nations (http://www.fao.org/food/food-safety-quality/gm-foods-platform/en/) and new testing information and importation decisions for Living Modified Organisms (LMOs), which is a legal term under the Cartagena Protocol on Biosafety (CPB) for the Convention on Biological Diversity (CBD), may be surveyed in the database of the Biosafety Clearing House Mechanism of CPB (http://bch.cbd.int/about/). LMOs are equivalent to transgenic organisms but with a wider coverage of organisms made by modern biotechnology. LMO is defined as a legal term in the CPB. Commercialization data is reported annually by the ISAAA (International Service for the Acquisition of Agri-biotech Applications) (James 2013) and updated by “Biotradestatus” (http://www.biotradestatus.com), supported by the Global Industry Coalition through CropLife International (http://croplife.org) on the regulatory and market status of agricultural biotechnology products.
Products made by new technology must be assessed for risk and relevant management should be established before dissemination for used. This information is essential for deciding whether the products can be used or not. Use of transgenes by genetic engineering enables the use of genetic variation which could not be incorporated by classical breeding methods. Transgene deployment requires the whole process of plant breeding—from selection of parents to release to farmers. Innovative traits from transgenes cannot be disregarded as a source of insect and disease resistances and abiotic stress tolerance if conventional genetic resources cannot be found (Watanabe et al. 1997, 2011).
Risk assessment and management strategies have been conducted on transgenic crops with respect to environmental and food safety (Watanabe and Komamine 2000). Global field assessment and/or importation approval cases of transgenic potato can be found at the Biosafety Clearing House Mechanism homepage of the CPB (https://bch.cbd.int). Approval of commercial transgenic potatoes can be identified in the GM Approval database (http://www.isaaa.org/gmapprovaldatabase/default.asp) of the ISAAA. Transgenic potato risk assessment procedures can be found in the above databases and also found in the potato biology consensus document (OECD 1997), while geographic location(s) where transgenic potatoes are intended to be used should be examined in those specific locations. Field assessment of transgenic potatoes has also been conducted in the center of origin (Celis et al. 2004). It was reported that there is no harm to many non-target organisms in using nematode-resistant transgenic potato genotypes, but gene flow occurs to wild relatives of potato growing near the transgenic potato crops. The gene flow could increase fitness of wild relatives, but the report suggested that male sterility of the transgenic potatoes drastically reduces the possibility of gene flow out of crop fields and considered it to be a manageable issue.
4. Challenges in the 21st century
Cost-free proprietary technology transfer together with efforts to test new inventions and access their risks has been conducted on potatoes. In the 1990s, the ISAAA mediated between Monsanto and the Mexican public agriculture sector regarding transgenic potato technology on coat-protein virus resistances to PVY and PVX (reviewed in Altman and Watanabe 1995, Watanabe et al. 1997). While commercial varieties were not developed in Mexico, relevant experience on transgenic potatoes with risk assessment and management strategies was established from this case as well as the issue of IPR sharing.
Agricultural knowledge has to be shared for global food security, but in the past two decades, in the context of agriculture knowledge, including indigenous wisdom, intellectual property has been dealt with under different forums, such as the CBD (http://www.cbd.int) and WIPO (http://www.wipo.int). The paradigm has shifted from philanthropic sharing of agricultural knowledge in the global community to sovereignty rights and to individual protection of indigenous group ownership, especially small holders with no governmental assistance. Ownership must be recognized to avoid infringement of IPRS. However, no comprehensive system has been proposed to monitor, advise, mediate, and solve the potential conflicts of agricultural IPRs sustainably. Even with the modern variety protection under the UPOV, there is insufficient surveillance and identification of the appropriate use of the varieties at a global level. While science and technology must adhere to rigorous standards, global food security demands comprehensive systems to enable needed innovations and research products to reach end users both safely while respecting IPRs.
Acknowledgements
This paper was supported in part by a JST/JICA STREPS project, “Diversity Assessment and Development of Sustainable Use of Mexican Genetic Resources” and by Plant Transgenic Design Initiative at Gene Research Center, University of Tsukuba.
Literature Cited
- Allard, R.W. (1960) Chapter 30. Inheritance in autotetraploids. Principles of Plant Breeding. John Wiley & Sons, New York, pp. 385–399. [Google Scholar]
- Altman, D.W. and Watanabe, K.N. (1995) (eds.) Plant Biotechnology Transfer to Developing Countries. R. G. Landes Co., Georgetown, Texas, USA, p. 300. [Google Scholar]
- Amador, V., Bou, J., Martínez-García, J., Monte, E., Rodoríguez-Falcon, M., Russo, E. and Prat, S. (2001) Regulation of potato tuberization by daylength and giberellins. Int. J. Dev. Biol. 45: S37–S38. [Google Scholar]
- Arisumi, T. (1982) Endosperm balance numbers among New Guinea-Indonesian Impatiens species. J. Hered. 73: 240–242. [Google Scholar]
- Bakker, E., Borm, T., Prins, P., van der Vossen, E., Uenk, G., Arens, M., de Boer, J., van Eck, H., Muskens, M., Vossen, J.et al. (2011) A genome-wide genetic map of NB-LRR disease resistance loci in potato. Theor. Appl. Genet. 123: 493–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberg, J.B. (1994) Allelism of endosperm balance number (EBN) in Solanum acaule Bitt. and other wild potato species. Theor. Appl. Genet. 89: 682–686. [DOI] [PubMed] [Google Scholar]
- Bennett, M.D., Cox, A.V. and Leitch, I.J. (1997) http://www.rbgkew.org.uk/cval/database1.html.
- Bonierbale, M.W., Plaisted, R.L. and Tanksley, S.D. (1988) RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale, M.W., Plaisted, R.L., Pineda, O. and Tanksley, S.D. (1994) QTL analysis of trichome-mediated insect resistance in potato. Theor. Appl. Genet. 87: 973–987. [DOI] [PubMed] [Google Scholar]
- Bradshaw, J.E. (1994) Quantitative genetics theory for tetrasomic inheritance. In: Bradshaw, J.E. and Mackay, G.R. (eds.) Potato Genetics. CAB International, Wallingford, UK, pp. 71–99. [Google Scholar]
- Brown, P.H. (2007) The cannon of potato science: 37. Stolonization, tuber induction and tuberization. Potato Res. 50: 363–365. [Google Scholar]
- Cabrera, A., Kozik, A., Howad, W., Arus, P., Iezzoni, A.F. and van der Knaap, E. (2009) Development and bin mapping of a Rosaceae Conserved Ortholog Set (COS) of markers. BMC Genomics 10: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carputo, D., Frusciante, L. and Peloquin, S.J. (2003) The role of 2n gametes and Endosperm Balance Number in the origin and evolution of polyplopids in the tuber-bearing Solanums. Genetics 163: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, A., Chauvin, J.E., Trognitz, B., Pawlak, A., Rubio-Covarruvias, O. and Zimnoch-Guzowska, E. (2009) Marker-assisted breeding for disease resistance in potato. Potato Res. 52: 245–248. [Google Scholar]
- Castelblanco, W. and Fregene, M. (2006) SSCP-SNP-based conserved ortholog set (COS) markers for comparative genomics in cassava (Manihot esculenta Crantz). Plant Mol. Biol. Report. 24: 229–236. [Google Scholar]
- Celis, C., Scurrah, M., Cowgill, S., Chumbiauca, S., Green, J., Franco, J., Main, G., Kiezebrink, D., Visser, R.G.F. and Atkinson, H.J. (2004) Environmental biosafety and transgenic potato in a centre of diversity for this crop. Nature 432: 222–225. [DOI] [PubMed] [Google Scholar]
- Chapman, J. and Watanabe, K.N. (2007) Chapter 17. 6: Current issues on IP management for health and agriculture in Japan. pp. 1621–1650. In: Krattger, A. et al. (eds.) Mihr-Pipra Handbook of Best Practices for Management of Intellectual Property in Health and Agriculture. Univ. California, Davis, USA. [Google Scholar]
- Cipar, M.S., Peloquin, S.J. and Hougas, R.W. (1964) Variability in the expression of self-incompatibility in tuber-bearing diploid Solanum species. Amer. Potato J. 41: 155–162. [Google Scholar]
- de Boer, J.M., Borm, T.J.A., Jesse, T., Brugman, B., Tang, X., Bryan, G.J., Bakker, J., van Eck, H J. and Visser, R.G.F. (2011) A hybrid BAC physical map of potato: a framework for sequencing a heterozygous genome. BMC Genomics 12: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Nijs, T.P.M. and Peloquin, S.J. (1977) 2n gametes in potato species and their function in sexual polyploidization. Euphytica 26: 585–600. [Google Scholar]
- Dewitte, A., Van Laere, K. and Van Huylenbroeck, J. (2012) Use of 2n Gametes in Plant Breeding. In: Abdurakhmonov, I. (ed.) Plant Breeding. In Tech; Riejeka, Croatia, pp. 59–81. [Google Scholar]
- Feingold, S., Lloyd, J., Norero, N., Bonierbale, M.W. and Lorenzen, J. (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor. Appl. Genet. 111: 456–466. [DOI] [PubMed] [Google Scholar]
- Fulton, T. M., van der Hoeven, R., Eanneta, N.T. and Tanksley, S.D. (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14: 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, C., Ritter, E., Debener, T., Schachtchabel, U., Walkemeier, B., Uhrig, H. and Salamini, F. (1989) RFLP analysis and linkage mapping in Solanum tuberosum. Theor. Appl. Genet. 78: 65–75. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C., Ritter, E., Barone, A., Debener, T., Walkemeier, B., Schachtschabel, U., Kaufmann, H., Thompson, R.D., Bonierbare, M.W., Ganal, M.W.et al. (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83: 49–57. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C., Bellin, D., Henselewski, H., Lehmann, W., Schwarzfischer, J. and Valkonen, J.P.T. (2006) Marker-assisted combination of major genes for pathogen resistance in potato. Theor. Appl. Genet. 112: 1458–1464. [DOI] [PubMed] [Google Scholar]
- Ghislain, M., Spooner, D.M., Rodríguez, F., Villamón, F., Núñez, J., Vásquez, C., Waugh, R. and Bonierbale, M. (2004) Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theor. Appl. Genet. 108: 881–890. [DOI] [PubMed] [Google Scholar]
- Grant, V. (1971) Polyploidy. In: Grant, V. Plant Speciation. Columbia University Press, New York, pp. 283–353. [Google Scholar]
- Grun, P., Aubertin, M. and Radlow, A. (1962) Multiple differentiation of plasmons of diploid species of Solanum. Genetics 47: 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämäläinen, J.H., Watanabe, K.N., Valkonen, J.P.T., Arihara, A., Plaisted, R.L., Pehu, E., Miller, L. and Slack, S.A. (1997) Mapping and marker assisted selection for a gene for extreme resistance to potato virus Y. Theor. Appl. Genet. 94: 192–197. [Google Scholar]
- Hämäläinen, J.H., Sorri, V.A., Watanabe, K.N., Gebhardt, C. and Valkonen, J.P.T. (1998) Molecular examination of a chromosome region that contains resistance to potato virus Y and A potyvirus in potato. Theor. Appl. Genet. 96: 1036–1043. [Google Scholar]
- Hanneman, R.E.Jr (1994) Assignment of Endosperm Balance Numbers to the tuber-bearing Solanums and their close non-tuber-bearing species. Euphytica 74: 19–25. [Google Scholar]
- Harlan, J.R. and De Wet, J.M.J. (1975) On Ö. Winge and a prayer: the origins of polyploids. Bot. Rev. 41: 361–390. [Google Scholar]
- Harris, P.M. (ed.) (1978) The Potato Crop. The scientific basis for improvement. Chapman & Hall, London, p. 730. [Google Scholar]
- Hawkes, J.G. and Jackson, M.T. (1992) Taxonomic and evolutionary implications of the Endosperm Balance Number hypothesis in potatoes. Theor. Appl. Genet. 84: 180–185. [DOI] [PubMed] [Google Scholar]
- Hermsen, J.G. Th. and Verdenius, J. (1973) Selection from Solanum tuberosum group Phureja of genotypes combining high frequency haploid induction with homozygosity for embryo spot. Euphytica 22: 244–259. [Google Scholar]
- Hijimans, R.J. and Spooner, D.M. (2001) Geographic distribution of wild potato species. Am. J. Bot. 88: 2101–2112. [PubMed] [Google Scholar]
- Hosaka, K. and Hanneman, R.E.Jr (1998) Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 2. Localization of an S locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica 103: 265–271. [Google Scholar]
- Hougas, R.W. and Peloquin, S.J. (1958) The potential of haploids in breeding and genetic research. Am. Potato J. 35: 701–707. [Google Scholar]
- Hussain, S.W. and Williams, W.M. (2008) The use of endosperm balance number for predicting gamete selection in complex polyploid interspecific Trifolium repens × T. nigrescens crosses. Plant Breed. 127: 518–523. [Google Scholar]
- Ishidore, E., van Os, H., Andrzejewski, S., Bakker, J., Barrena, I., Bryam, G.J., Caromel, B., van Eck, H., Ghareeb, B., de Jong, W.et al. (2003) Toward a marker-dense meiotic map of the potato genome: lessons from linkage group I. Genetics 165: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga, M. and Peloquin, S.J. (1982) Origin and evolution of cultivated tetraploid potatoes via 2n gametes. Theor. Appl. Genet. 61: 161–169. [DOI] [PubMed] [Google Scholar]
- Iwanaga, M. (1984) Discovery of a synaptic mutant in potato haploids and its usefulness for potato breeding. Theor. Appl. Genet. 68: 87–93. [DOI] [PubMed] [Google Scholar]
- Iwanaga, M., Ortiz, R., Cipar, M.S. and Peloquin, S.J. (1991a) A restorer gene for genetic-cytoplasmic male sterility in cultivated potatoes. Am. Potato J. 68: 19–28. [Google Scholar]
- Iwanaga, M., Freyre, R. and Watanabe, K. (1991b) Breaking the crossability barriers between disomic tetraploid Solanum acaule and tetrasomic tetraploid S. tuberosum. Euphytica 52: 183–191. [Google Scholar]
- Jacobs, J.M., van Eck, H.J., Arens, P., Verkerk-Bakker, B., Te Lintel Hekkert, B., Bastiaanssen, H.J.M., El-Kharbotly, A., Pereira, A., Jacobsen, E. and Stiekema, W.J. (1995) A genetic map of potato (Solanum tuberosum) integrating molecular markers, including transposons, and classical markers. Theor. Appl. Genet. 9: 289–300. [DOI] [PubMed] [Google Scholar]
- James, C. (2013) Global Status of Commercialized Biotech/GM Crops: 2013. ISAAA Brief 46-2013. New York. [Google Scholar]
- Jansky, S. (2000) Breeding for Disease Resistance in Potato. In: Janick, J. (ed.) Plant Breeding Reviews, Volume 19, John Wiley & Sons, Inc., Oxford, UK. [Google Scholar]
- Johnston, S.A., den Nijs, T.P.M., Peloquin, S.J. and Hanneman, R.E.Jr (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57: 5–9. [DOI] [PubMed] [Google Scholar]
- Johnston, S.A. and Hanneman, R.E.Jr (1982) Manipulations of Endosperm Balance Number overcome crossing barriers between diploid Solanum species. Science 217: 446–448. [DOI] [PubMed] [Google Scholar]
- Jupe, F., Pritchard, L., Etherington, G.J., Mackenzie, K., Cock, P.J.A., Wright, F., Sharma, S.K., Bolser, D., Bryan, G.J., Jones, J.D.G.et al. (2012) Identification and localization of the NB-LRR gene family within the potato genome. BMC Genomics 13: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, K., Morikawa, Y., Sorri, V.A., Valkonen, J.P.T., Gebhardt, C. and Watanabe, K.N. (2000) Development of SCAR markers to the PVY resistance gene Ryadg based on a common feature of plant disease resistance genes. Genome 43: 1–8. [PubMed] [Google Scholar]
- Katsiotis, A., Hanneman, R.E.Jr and Forsberg, R.A. (1995) Endosperm Balance Number and the polar nuclei activation hypotheses for endosperm development in interspecific crosses of Solanaceae and Gramineae, respectively. Theor. Appl. Genet. 91: 848–855. [DOI] [PubMed] [Google Scholar]
- Kaufmann, H., Salamini, F. and Thompson, R.D. (1991) Sequence variability and gene structure at the self-incompatibility locus of Solanum tuberosum. Mol. Gen. Genet. 226: 457–466. [DOI] [PubMed] [Google Scholar]
- Khun, D., Livingstone, D.III, Main, D., Zheng, P., Sasaki, C., Feltus, F.A., Mockaitis, K., Farmer, A.D., May, G.D., Schnell, R.J.et al. (2012) Identification and mapping of conserved ortholog set (COS) II sequences of cacao and their conversion to SNP markers for marker-assisted selection in Theobroma cacao and comparative genomics studies. Tree Genet. Genomes 8: 97–111. [Google Scholar]
- Kloosterman, B., Abelenda, J., Gomez, M.M., Oortwijn, M., de Boer, J.M., Kowitwanich, K., Horvath, B.M., van Eck, H.J., Smaczniak, C., Prat, S.et al. (2013) Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495: 246–250. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. and Araki, T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Leister, D., Ballvora, A., Salamini, F. and Gebhardt, C. (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat. Genet. 14: 421–429. [DOI] [PubMed] [Google Scholar]
- Levin, R.A., Whelan, A. and Miller, J.S. (2009) The utility of nuclear conserved ortholog set II (COSII) genomic regions for species-level phylogenetic inference in Lycium (Solanaceae). Mol. Phylogenet. Evol. 53: 881–890. [DOI] [PubMed] [Google Scholar]
- Lindqvist-Kreuze, H., Cho, K., Portal, L., Rodríguez, F., Simon, R., Mueller, L.A., Spooner, D.M. and Bonierbale, M. (2013) Linking the potato genome to the conserved ortholog set (COS) markers. BMC Genetics 14: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida-Hirano, R. (2015) Diversity of potato genetic resources. Breed. Sci. 65: 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, J. (1970) Significance of mating systems for chromosome and gametes in polyploids. Hereditas 66: 165–176. [DOI] [PubMed] [Google Scholar]
- Marks, G.E. (1966) The enigma of triploid potatoes. Euphytica 15: 285–290. [Google Scholar]
- Matsubayashi, M. (1991) Phylogenetic relationships in the potato and its relatives. In: Gupta, P.K. and Tsuchiya, T. (eds.) Chromosome Engineering in Plants: Genetics, Breeding, Evolution Part B. Elsevier, pp. 93–118. [Google Scholar]
- McGregor, C.E., Lambert, C.A., Greyling, M.M., Louw, J.H. and Warnich, L. (2000) A comparative assessment of DNA fingerprinting techniques (RAPD, ISSR, AFLP and SSR) in tetraploid potato (Solanum tuberosum L.) germplasm. Euphytica 113: 135–144. [Google Scholar]
- Meyer, S., Nagel, A. and Gebhardt, C. (2005) PoMaMo—a comprehensive database for potato genome data. Nucleic Acids Res. 33: D666–D670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbourne, D., Meyer, R.C., Collins, A.J., Ramsay, L.D., Gebhardt, C. and Waugh, R. (1998) Isolation, characterization and mapping of simple sequence repeat loci in potato. Mol. Gen. Genet. 259: 233–245. [DOI] [PubMed] [Google Scholar]
- Millam, S., Payne, L.A. and Mackay, G.R. (1995) The integration of protoplast fusion-derived material into a potato breeding programme— a review of progress and problem. Euphytica 85: 451–455. [Google Scholar]
- Mok, D.W.S. and Peloquin, S.J. (1975a) Three mechanism of 2n pollen formation in diploid potatoes. Can. J. Genet. Cytol. 17: 217–225. [Google Scholar]
- Mok, D.W.S. and Peloquin, S.J. (1975b) The inheritance of three mechanisms of diploandroid (2n pollen) formation in diploid potatoes. Heredity 35: 295–302. [Google Scholar]
- Mok, D.W.S. and Peloquin, S.J. (1975c) Breeding value of 2n pollen (diplandroids) in tetraploid × diploid crosses in potatoes. Theor. Appl. Genet. 46: 307–314. [DOI] [PubMed] [Google Scholar]
- Mueller, L.A., Solow, T.H., Taylor, N., Skwarecki, B., Buels, R., Binns, J., Li, C., Wright, M.H., Ahrens, R., Wang, Y.et al. (2005) The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol. 138: 1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, C., Abelenda, J.A., Cruz-Oró, E., Cuéller, C.A., Tamaki, S., Silva, J., Shimamoto, K. and Prat, S. (2011) Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122. [DOI] [PubMed] [Google Scholar]
- Nishiyama, I. and Inomata, N. (1966) Embryological studies on cross incompatibility between 2x and 4x in Brassica. Jpn. J. Genet. 41: 27–42. [Google Scholar]
- Nishiyama, I. and Yabuno, T. (1978) Interspecific cross-incompatibility due to disturbed activation of the polar nuclei by the male nucleus. Breed. Sci. (Ikushugaku zasshi) 28: 71–80. [Google Scholar]
- Nishiyama, I., Sarashima, M. and Matsuzawa, Y. (1991) Critical discussion on abortive interspecific crosses in Brassica. Plant Breed. 107: 288–302. [Google Scholar]
- OECD (1997) Series on Harmonization of Regulatory Oversight in Biotechnology No. 8. Consensus Document on the Biology of Solanum tuberosum Subsp. tuberosum (Potato). Environment Directorate, Organisation for Economic Co-operation and Development, Paris: p. 38. [Google Scholar]
- Ortega, F. and Lopez-Vizcon, C. (2012) Application of molecular marker-assisted selection (MAS) for disease resistance in a practical potato breeding programme. Potato Res. 55: 1–13. [Google Scholar]
- Ortiz, R. and Peloquin, S.J. (1994) Use of 24-chromosome potatoes (diploids and dihaploids) for genetical analysis. In: Bradshaw, J.E. and Mackay, G.R. (eds.) Potato Genetics. CAB International, Wallingford, UK, pp. 133–154. [Google Scholar]
- Ortiz, R. and Watanabe, K.N. (2004) Genetic contributions to breeding polyploid crops. Recent Res. Devel. Genet. Breed. 1: 269–286. [Google Scholar]
- Ottoman, R.J., Hane, D.C., Brown, C.R., Yilma, S., James, S.R. and Mosley, A.R. (2009) Validation and implementation of marker-assisted selection (MAS) for PVY resistance (Ryadg gene) in a tetraploid potato breeding program. Am. J. Potato Res. 86: 304–314. [Google Scholar]
- Pal, B.P. and Nath, P. (1942) Genetic nature of self- and cross incompatibility in potatoes. Nature 149: 246–247. [Google Scholar]
- Peloquin, S.J., Gabert, A.C. and Ortiz, R. (1996) Nature of ‘pollinator’ effect in potato (Solanum tuberosum L.) haploid production. Ann. Bot. 77: 539–542. [Google Scholar]
- Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Prevost, A. and Wilkinson, M.J. (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98: 107–112. [Google Scholar]
- Provan, J., Powell, W. and Waugh, R. (1996) Microsatellite analysis of relationships within cultivated potato (Solanum tuberosum). Theor. Appl. Genet. 92: 1078–1084. [DOI] [PubMed] [Google Scholar]
- Pruski, K., Duplirssis, P., Lewis, T., Astatkie, T., Novak, J. and Struik, P.C. (2001) Jasmonate effect on in vitro tuberization of potato (Solanum tuberosum L.) cultivars under light and dark conditions. Potato Res. 44: 315–325. [Google Scholar]
- Qaim, M. (2009) The economics of genetically modified crops. Ann. Rev. Resour. Econ. 1: 665–693. [Google Scholar]
- Ritter, E., Debener, T., Barone, A., Salamini, F. and Gebhardt, C. (1991) RFLP mapping on potato chromosome of two genes controlling extreme resistance to potato virus X (PVX). Mol. Gen. Genet. 227: 81–85. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Falcón, M., Bou, J. and Prat, S. (2006) Seasonal control of tuberization in potato: Conserved elements with the flowering response. Ann. Rev. Plant Biol. 57: 151–180. [DOI] [PubMed] [Google Scholar]
- Ross, H. (1986) Potato breeding—Problems and Perspectives. Verlag Paul Parey, Berlin, p. 132. [Google Scholar]
- Rouppe van der Voort, J.N.A.M., van Zandvoort, P., van Eck, H.J., Folkertsma, R.T., Hutten, R.C.B., Draaistra, J., Gommers, F.J., Jacobsen, E., Helder, J. and Bakker, J. (1997) Use of allele specificity of comigrating AFLP markers to align genetic maps from different potato genotypes. Mol. Gen. Genet. 255: 438–447. [DOI] [PubMed] [Google Scholar]
- Rouppe van der Voort, J.N.A.M., Van Eck, H.J., Draaistra, J., van Zandvoort, P.M., Jacobsen, E. and Bakker, J. (1998) An online catalogue of AFLP markers covering the potato genome. Mol. Breed. 4: 73–77. [Google Scholar]
- Sagredo, B., Mathias, M., Barriento, C., Acuña, I., Kalazich, J. and Santos Rojas, J. (2009) Evaluation of a SCAR RYSC3 marker of the Ryadg gene to select resistance genotypes to potato virus Y (PVY) in the INIA potato breeding program. Chilean J. Agric. Res. 69: 305–315. [Google Scholar]
- Salaman, R.N. (1910) Male sterility in potatoes, a dominant Mendelian character; with remarks on the shapes of the pollen in wild and domestic varieties. J. Linnean Soc. London, Botany 39: 301–312. [Google Scholar]
- Sato, M., Nishikawa, K., Komura, K. and Hosaka, K. (2006) Potato virus Y resistance gene, Rychc, mapped to the distal end of potato chromosome 9. Euphytica 149: 367–372. [Google Scholar]
- Seabrook, J.E.A., Coleman, S. and Levy, D. (1991) Effect of photoperiod on in vitro tuberization of potato (Solanum tubersoum L.). Plant Cell Tissue Organ Cult. 34: 43–51. [Google Scholar]
- Sharma, S.K., Bolser, D., de Boer, J., Sønderkær, M., Amoros, W., Carboni, M.F., D’Ambrosio, J.M., de la Cruz, G., Di Genova, A., Douches, D.S.et al. (2013) Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3 (Bethesda) 3: 2031–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiranita, A., Kasai, K., Hämäläinen, J.H., Valkonen, J.P.T. and Watanabe, K.N. (1999) Applicability of the resistance gene-like fragment ADG2 as an RFLP probe in selection of extreme resistance to potato Y potyvirus (PVY). Plant Biotechnol. 16: 361–369. [Google Scholar]
- Slater, A.T., Cogan, N.O.I. and Forster, J.W. (2013) Cost analysis of the application of marker-assisted selection in potato breeding. Mol. Breed. 32: 299–310. [Google Scholar]
- Song, Y.S., Hepting, L., Schweize, G., Hartl, L., Wenzel, G. and Schwarzfischer, A. (2005) Mapping of extreme resistance to PVY (Rysto) on chromosome XII using anther-culture-derived primary dihaploid potato lines. Theor. Appl. Genet. 111: 879–887. [DOI] [PubMed] [Google Scholar]
- Sorri, V.A., Watanabe, K.N. and Valkonen, J.P.T. (1999) Predicted kinase-3a motif of a resistance gene analogue as a unique marker for virus resistance. Theor. Appl. Genet. 99: 164–170. [Google Scholar]
- Stebbins, G.L.Jr (1950) Variation and Evolution in Plants. Columbia University Press, New York, pp. 298–379. [Google Scholar]
- Struik, P.C., Vreugdenhil, D., van Eck, H.J., Bachem, C.W. and Visser, R.G.F. (1999) Physiological and genetic control of tuber formation. Potato Res. 42: 313–331. [Google Scholar]
- Swaminathan, M.S. and Magoon, M.L. (1961) Origins and cytogenetics of the commercial potato. Advance in Genetics 10: 217–256. [Google Scholar]
- Szajko, K., Strzelczyk-Żyta, D. and Marczewski, W. (2014) Ny-1 and Ny-2 genes conferring hypersensitive response to potato virus Y (PVY) in cultivated potatoes: mapping and marker-assisted selection validation for PVY resistance in potato breeding. Mol. Breed. 34: 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., Ganal, M.W., Prince, J.P., de Vicente, M.C., Bonierbale, M.W., Broun, P., Fulton, T.M., Giovannoni, J.J., Grandillo, S. and Martin, G.B. (1992) High density molecular linkage maps of the tomato and potato. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, J.K., Pandey, S.K., Poonam, P., Chakrabarti, S.K., Gopal, J. and Kumar, V. (2013) Molecular markers of Ryadg gene and serological assay reveal potato virus Y (PVY) resistance in the tetraploid Indian potato (Solanum tuberosum) germplasm. Indian J. Agr. Sci. 83: 397–401. [Google Scholar]
- Tommiska, J., Hämäläinen, J.H., Watanabe, K.N. and Valkonen, J.P.T. (1998) Mapping of the gene Nxphu that controls hypersensitive resistance to potato virus X in Solanum phureja IvP35. Theor. Appl. Genet. 96: 840–843. [Google Scholar]
- Valkonen, J.P.T., Slack, S.A., Plaisted, R.L. and Watanabe, K.N. (1994) Extreme resistance is epistatic to hypersensitivity resistance to potato Virus Yo in a Solanum tuberosum subsp. andigena-derived potato genotype. Plant Disease 78: 1177–1180. [Google Scholar]
- Valkonen, J.P.T., Slack, S.A., Plaisted, R.L., Orrillo, M. and Watanabe, K.N. (1995) Resistances to viruses in F1 hybrids produced by direct crossing between Solanum series Tuberosa and S. brevidens (Series Etuberosa) using S. phureja for rescue pollinations. Plant Breed. 114: 421–426. [Google Scholar]
- Valkonen, J.P.T., Jones, R.A.C., Slack, S.A. and Watanabe, K.N. (1996) Resistance specificities to viruses in potato: standardization of nomenclature. Plant Breed. 115: 433–438. [Google Scholar]
- Valkonen, J.P.T., Wiegmann, K., Hämäläinen, J.H., Marczewski, W. and Watanabe, K.N. (2008) Evidence for utility of the same PCR-based markers for selection of extreme resistance to Potato virus Y controlled by Rysto of Solanum stoloniferum derived from different sources. Ann. Appl. Biol. 152: 121–130. [Google Scholar]
- van Eck, H.J., Rouppe van der Voort, J., Draaistra, J., van Zandvoort, P., van Enckevort, E., Segers, B., Peleman, J., Jacobsen, E., Helder, J. and Bakker, J. (1995) The inheritance and chromosomal localization of AFLP markers in a non-inbred potato offspring. Mol. Breed. 1: 397–410. [Google Scholar]
- Van Os, H., Andrezejewski, S., Bakker, E., Barrera, I., Bryan, G.L., Caromel, B., Ghareeb, B., Isidore, E., de Jong, W., van Koert, P.et al. (2006) Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genome-wide physical map. Genetics 173: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veilleux, R.E. (1999) Anther culture of potato and molecular analysis of anther-derived plants as laboratory exercises for plant breeding courses. Horttechnology 9: 585–588. [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M.et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, J.A., Orrillo, M. and Watanabe, K.N. (1999a) Frequency of multiple quantitative pest resistance traits in 4x × 2x crosses of potato. Breed. Sci. 49: 53–61. [Google Scholar]
- Watanabe, J.A., Orrillo, M. and Watanabe, K.N. (1999b) Resistance to bacterial wilt (Pseudomonas solanacearum) of potato evaluated by survival and yield performance at high temperatures Breed. Sci. 49: 63–68. [Google Scholar]
- Watanabe, J.A., Orrillo, M. and Watanabe, K.N. (1999c) Evaluation of in vitro chromosome-doubled regenerates with resistance to potato tuber moth [Phthorimaea opercullella (Zeller)]. Plant Biotechnol. 16: 225–230. [Google Scholar]
- Watanabe, J.A. and Watanabe, K.N. (2000) Pest resistance traits controlled by quantitative loci and molecular breeding strategies in tuber-bearing Solanum. Plant Biotechnol. 17 : 1–16. [Google Scholar]
- Watanabe, K.N. and Peloquin, S.J. (1989) Occurrence of 2n pollen and ps gene frequencies in cultivated groups and their related wild species in tuber-bearing Solanums. Theor. Appl. Genet. 78: 329–336. [DOI] [PubMed] [Google Scholar]
- Watanabe, K.N. and Peloquin, S.J. (1991) The occurrence and frequency of 2n pollen in 2x, 4x, and 6x wild, tuber-bearing Solanum species from Mexico, and Central and South America. Theor. Appl. Genet. 82: 621–626. [DOI] [PubMed] [Google Scholar]
- Watanabe, K.N., Peloquin, S.J. and Endo, M. (1991) Genetic significance of mode of polyploidization: somatic doubling or 2n gametes? Genome 34: 28–34. [Google Scholar]
- Watanabe, K.N., Arbizu, C. and Schmiediche, P. (1992) Potato germplasm enhancement with disomic tetraploid Solanum acaule. I. Efficiency in introgression. Genome 35: 53–57. [Google Scholar]
- Watanabe, K.N. and Orrillo, M. (1994) Disomic behavior of polyploid tuber-bearing Solanum species. Jpn. J. Genet. 69: 637–643. [Google Scholar]
- Watanabe, K.N., Orrillo, M., Iwanaga, M., Ortiz, R., Freyre, R. and Perez, S. (1994) Diploid potato germplasm derived from wild and landrace genetic resources. Am. Potato J. 71: 599–604. [Google Scholar]
- Watanabe, K.N., Orrillo, M. and Golmirzaie, A.M. (1995a) Potato germplasm enhancement for resistance to biotic stresses at CIP. Conventional and biotechnology-assisted approaches using a wide range of Solanum species. Euphytica 85: 457–464. [Google Scholar]
- Watanabe, K.N., Orrillo, M., Vega, S., Iwanaga, M., Ortiz, R., Freyre, R., Yerk, G., Peloquin, S.J. and Ishiki, K. (1995b) Selection of diploid potato clones from diploid haploid-species F1 families for short day conditions. Breed. Sci. (Ikushugaku zasshi) 45: 341–347. [Google Scholar]
- Watanabe, K.N., Orrillo, M., Vega, S., Hurtado, A., Valkonen, J.P.T., Pehu, E. and Tanksley, S.D. (1995c) Overcoming crossing barriers between nontuber-bearing and tuber-bearing Solanum species: towards potato germplasm enhancement with a broad spectrum of Solanaceous genetic resources. Genome 38: 27–35. [DOI] [PubMed] [Google Scholar]
- Watanabe, K.N., Orrillo, M., Perez, S., Crusado, J. and Watanabe, J.A. (1996a) Testing yield of diploid potato breeding lines for cultivar development. Breed. Sci. (Ikushugaku zasshi) 46: 245–249. [Google Scholar]
- Watanabe, K.N., Orrillo, M., Vega, S., Perez, S., Crusado, J., Golmirzaie, A.M. and Watanabe, J.A. (1996b) Generation of pest resistant, diploid potato germplasm with short day adaptation from diverse range of genetic stocks. Breed. Sci. (Ikushugaku zasshi) 46: 329–336. [Google Scholar]
- Watanabe, K., Golmirzaie, A.M. and Gregory, P. (1997) Chapter 10: Use of Biotechnology Tools in Potato Genetic Resources Management and Breeding. In: Watanabe, K.N. and Pehu, E. (eds.) Plant Biotechnology and Plant Genetic Resources for Sustainability and Productivity. R.G. Landes Co., Georgetown, Texas, USA, pp. 145–154. [Google Scholar]
- Watanabe, K.N. and Komamine, A. (2000) Challenge of Plant and Agricultural Sciences to the Crisis of Biosphere on the Earth in the 21st Century. Landes Bioscience, Austin TX, USA, p. 309. [Google Scholar]
- Watanabe, K.N. and Komamine, A. (2004) Issues on Intellectual Property Rights Associated with Agro-Biotechnology in Japan. In: Erbisch,, F.H. and Maredia, K.M. (eds.) Intellectual Property Rights in Agricultural Biotechnology. 2nd edition Michigan State University, East Lansing and CAB International, Wallingford, UK, pp. 187–200. [Google Scholar]
- Watanabe, K.N., Ortiz, R. and Watanabe, J.A. (2005) Chapter 4: Breeding Potential and Transmission of Traits in 4x -2x Crosses. In: Razdan, M.K. and Mattoo, A.K. (eds.) Genetic Improvement of Solanaceous Crops, Vol. 2: “Potato”, Science Publishers Inc., USA, pp. 83–99. [Google Scholar]
- Watanabe, K.N. (2011) Challenges for Conservation and Utilization of Plant Genetic Resources. pp. 99–107. Proceedings of NIAS-FAO International Symposium on Plant Genetic Resources for Food and Agriculture in Asia and Pacific: Impacts and future directions. NIAS and FAO RAP; Bangkok, RAP Publication 2012/1. p. 118. [Google Scholar]
- Watanabe, K.N., Kikuchi, A., Shimazaki, T. and Asahina, M. (2011) Salt and drought stress tolerances in transgenic potatoes and wild species. Potato Res. 54: 319–324. [Google Scholar]
- Wenzel, G. and Uhrig, H. (1981) Breeding for nematode and virus resistance in potato via anther culture. Theor. Appl. Genet. 59: 333–340. [DOI] [PubMed] [Google Scholar]
- Wenzl, P., Carling, J., Kudrna, D., Jaccoud, D., Huttner, E., Kleinhofs, A. and Killian, A. (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 101: 9915–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.E. and Peloquin, S.J. (1991) Occurrence and mechanisms of 2n egg formation in 2x potato. Genome 34: 975–982. [Google Scholar]
- Wu, F., Muller, L.A., Crouzillat, D., Petiard, V. and Tanksley, S.D. (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euaserid plant clade. Genetics 174: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, B., Grun, P., Kheyr-Pour, A. and Kao, T.-H. (1990) Identification of pistil-specific protein associated with the self-incompatibility alleles in Solanum chacoense. Sex. Plant Reprod. 3: 54–60. [Google Scholar]
- Yamanaka, Y., Suzuki, E., Tanaka, M., Takeda, Y., Watanabe, J.A. and Watanabe, K.N. (2003) Assessment of cytochorome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theor. Appl. Genet. 108: 1–9. [DOI] [PubMed] [Google Scholar]
- Yamanaka, S., Ikeda, S., Imai, A., Luan, Y., Watanabe, J.A. and Watanabe, K.N. (2005) Construction of integrated genetic map between various existing DNA markers and newly developed P450-related PBA markers in diploid potato (Solanum tuberosum). Breed. Sci. 55: 223–230. [Google Scholar]
- Young, N.D. and Tanksley, S.D. (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor. Appl. Genet. 77: 95–101. [DOI] [PubMed] [Google Scholar]