SUMMARY
CENP-A is a centromere-specific histone H3 variant that epigenetically determines centromere identity to ensure kinetochore assembly and proper chromo-some segregation, but the precise mechanism of its specific localization within centromeric heterochromatin remains obscure. We have discovered that CUL4A-RBX1-COPS8 E3 ligase activity is required for CENP-A ubiquitylation on lysine 124 (K124) and CENP-A centromere localization. A mutation of CENP-A, K124R, reduces interaction with HJURP (a CENP-A-specific histone chaperone) and abrogates localization of CENP-A to the centromere. Addition of monoubiquitin is sufficient to restore CENP-A K124R to centromeres and the interaction with HJURP, indicating that “signaling” ubiquitylation is required for CENP-A loading at centromeres. The CUL4A-RBX1 complex is required for loading newly synthesized CENP-A and maintaining preassembled CENP-A at centromeres. Thus, CENP-A K124R ubiquitylation, mediated by the CUL4A-RBX1-COPS8 complex, is essential for CENP-A deposition at the centromere.
INTRODUCTION
The centromere plays an essential role in accurate chromosome segregation, and defects in its function lead to aneuploidy and thereby cancer. In most eukaryotes, the centromere has no defined DNA sequence but consists of large arrays of repetitive DNA; in humans, this sequence is a 171-bp α-satellite DNA, although several other sequence types are found in this region. Except in the budding yeast, centromere identity relies not on the DNA sequence but on the presence of a special nucleosome that contains the histone H3 variant CenH3 (CENtromere Protein A [CENP-A] in humans). Therefore, CENP-A is proposed to be the epigenetic mark of the centromere (Karpen and Allshire, 1997), and recently this mark was demonstrated to act through a two-step mechanism to identify, maintain, and propagate centromere function indefinitely using gene targeting in human cells and fission yeast (Fachinetti et al., 2013). CENP-A-containing nucleosomes are formed with canonical histones H2A, H2B, and H4 at the active centromeres, but their structure remains controversial (Black and Cleveland, 2011). CENP-A nucleosomes localize to the inner plate of mammalian kinetochores (Warburton et al., 1997) and bind to the 171-bp α-satellite DNA. Active centromeres require CENP-A nucleosomes to direct the recruitment of a constitutive centromere-associated network (CCAN) and the kinetochore proteins, which together orchestrate the attachment of chromosomes to the mitotic spindle and regulate cycle progression through the spindle checkpoint. CENP-A contains a short centromere targeting domain (CATD) within the histone fold region (Black et al., 2004). Replacement of the corresponding region of H3 with the CATD is sufficient to direct H3 to the centromere (Black et al., 2004), and this chimeric histone can rescue the viability of CENP-A-depleted cells (Black et al., 2004, 2007).
Previous studies in human cells show that newly synthesized CENP-A is recruited only during a brief interval in G1 immediately following mitosis (Hemmerich et al., 2008; Jansen et al., 2007). The assembly of new centromeric nucleosomes depends on the Holliday junction recognition protein (HJURP), which is a CENP-A-specific chromatin assembly factor (Bernad et al., 2011; Dunleavy et al., 2009; Foltz et al., 2009). Like CENP-A, HJURP is also recruited during early G1 (Dunleavy et al., 2009; Foltz et al., 2009; Jansen et al., 2007; Schuh et al., 2007). Primary structural analysis has revealed that human HJURP is a distant counterpart of Scm3, which is required to deposit centromeric nucleosomes in yeast (Sanchez-Pulido et al., 2009). CENP-A interacts with HJURP as a soluble pre-nucleosomal complex, and HJURP recruitment to centromeres depends on the activity of the Mis18 complex (Barnhart et al., 2011; Moree et al., 2011), which influences the histone modification and DNA methylation status of centromeres (Fujita et al., 2007; Kim et al., 2012). The human proteins hMis18 and M18BP1/KNL2 are recruited to the centromere at telophase G1, suggesting that the hMis18 complex and RbAp46 and RbAp48 (homologs of Mis16) prime the centromere for CENP-A localization (Fujita et al., 2007; Maddox et al., 2007). Recently, McKinley et al. have shown that faithful CENP-A deposition requires integrated signals from Plk1 and cyclin-dependent kinase (CDK), with Plk1 promoting the localization of the Mis18 complex, and CDK inhibiting Mis18 complex assembly (McKinley and Cheeseman, 2014). In addition, the remodeling and spacing factor (RSF) complex actively supports the assembly of CENP-A chromatin (Perpelescu et al., 2009), and the CENP-A licensing factor M18BP1/KNL2 and the small GTPases-activating protein MgcRacGAP cooperate to promote the stability of newly loaded CENP-A at centromeres (Lagana et al., 2010; Prendergast and Sullivan, 2010).
Although CENP-A is thought to be regulated epigenetically, the function of post-translational modification (PTM) in centro-mere assembly remains obscure. The phosphorylation of CENP-A serine 7 (S7) in human cells by Aurora B in the final stages of cytokinesis has been described (Zeitlin et al., 2001), and the phosphorylation of CenH3 serine 50 (S50) in maize has been characterized during chromosome segregation (Zhang et al., 2005). Studies on the CENP-A S7A mutant show that the phosphorylated human CENP-A nucleosomes might be “bridged” to CENP-C via the phospho-binding 14-3-3 proteins in mitosis (Goutte-Gattat et al., 2013). Bailey et al. reported 3 PTMs on human CENP-A (trimethylation of Gly1 and phosphor-ylation of Ser16 and Ser18), suggesting that the major modifications on the N-terminal tail of CENP-A alter the physical properties of the chromatin fiber at the centromere (Bailey et al., 2013). In addition, yeast Cse4 (CENP-A homolog) is methylated on arginine 37 (R37), and this methylation regulates the recruitment of kinetochore components to centromeric sequences (Samel et al., 2012). However, the molecular mechanisms of these PTMs of CENP-A on recruitment to centromeres have not yet been elucidated.
In this study, we report that PTM of CENP-A via ubiquitylation at lysine 124 (K124) regulates CENP-A localization. CUL4ARBX1-COPS8 E3 ligase activity is required for CENP-A ubiquity-lation at K124, and this modification is important to localize CENP-A to centromeres.
RESULTS
CUL4A E3 Ligase Is Required to Localize CENP-A to Centromeres
The interphase-centromere complex (ICEN) has been isolated by anti-CENP-A native chromatin immunoprecipitation (Ando et al., 2002; Izuta et al., 2006; Obuse et al., 2004). We hypothesized that some of the ICEN proteins may play a role in localizing CENP-A to centromeres. Therefore, we performed small interfering RNA (siRNA) knockdown to screen for proteins whose deletion induced the delocalization of CENP-A at centromeres (Table S1). The Cullin 4A (CUL4A) siRNA significantly induced a reduction of CENP-A at centromeres (Figures 1A–1C). Ectopic expression of CUL4A-Flag rescues the reduction of CENP-A at centromeres when CUL4A siRNA targets 3’ UTR (Figures S1A–S1C), excluding the possibility of off-target effects of siRNA. By confirming that the protein levels of CENP-A in total cell lysates were similar to CENP-A levels in lysates from luciferase (Luc) siRNA-treated cells under the same culture conditions (Figure S1G), we eliminated the possibility that CUL4A depletion causes CENP-A protein degradation (see also Figures S1H–S1M).
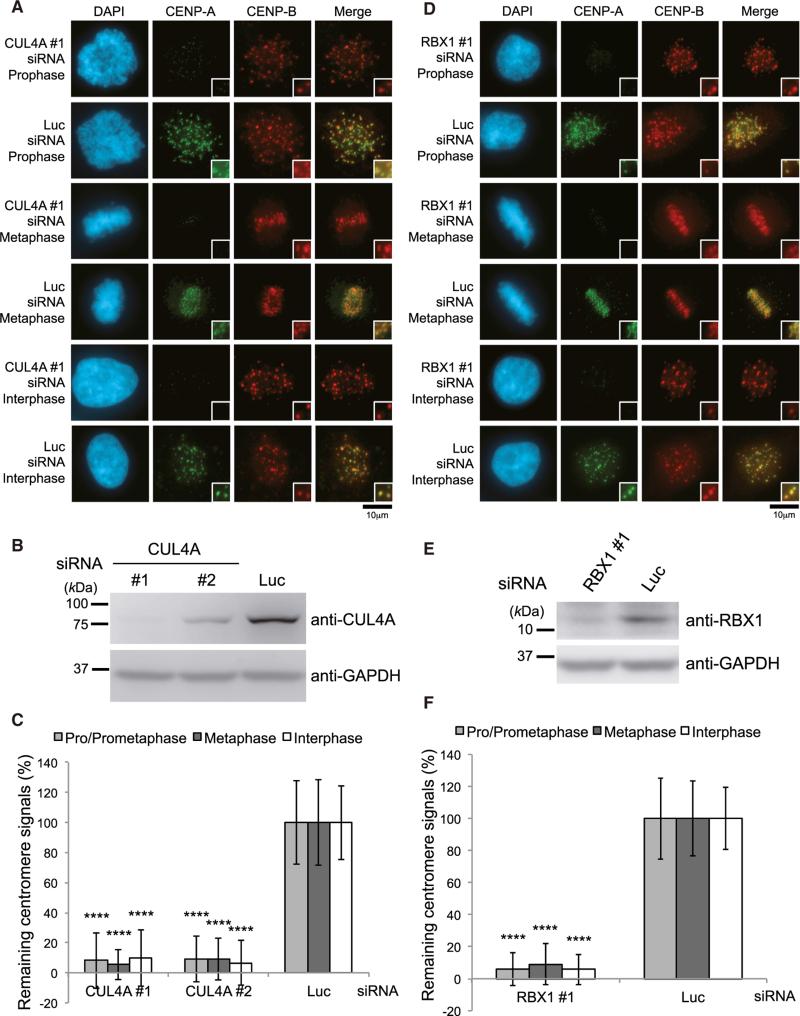
Figure 1. The CUL4A Complex Is Required for CENP-A Localization to Centromeres.
(A) CUL4A siRNA causes delocalization of CENP-A from centromeres. HeLa cells were transfected for 48 hr with CUL4A or luciferase (Luc) siRNAs. Scale bar, 10 μm.
(B) Western blot analysis of total cell lysates of HeLa cells harvested 48 hr after transfection with CUL4A siRNA (Table S4) or Luc siRNA. GAPDH protein was used as a loading control.
(C) CENP-A signals at centromeres given in (A) were quantified. Signals were normalized with Luc siRNA-treated cells, and the mean percentages (±SD) are shown. ****p < 0.0001 compared with Luc siRNA-treated cells (Student's t test).
(D) RBX1 siRNA causes delocalization of CENP-A from centromeres. HeLa cells were transfected for 72 hr with RBX1 or luciferase (Luc) siRNAs. Scale bar, 10 μm.
(E) Western blot analysis of HeLa cells transfected with RBX1 or Luc siRNA(s) for 72 hr.
(F) CENP-A signals at centromeres given in (D) were quantified. Signals were normalized with Luc siRNA-plus vector-transfected cells, and the mean percentages (±SD) are shown. ****p < 0.0001 compared with Luc siRNA-treated cells (Student's t test).
Cullin-RING E3 ubiquitin ligases (CRLs) are the most prominent class of ubiquitin ligases (Merlet et al., 2009). CRLs contain 3 major elements: a cullin scaffold, a RING finger protein (RBX1 or RBX2) that recruits a ubiquitin-charged E2 enzyme, and a substrate adaptor that places substrates in proximity to the E2 enzyme to facilitate ubiquitin transfer (Bennett et al., 2010). RBX1 siRNAs induced a significant reduction of CENP-A at centromeres (Figures 1D–1F) under the conditions in which degradation of CENP-A protein was not observed either by RBX1 depletion or by the combination of CUL4A and RBX1 depletion (Figure S1G). Ectopic expression of Flag-RBX1 rescues the reduction of CENP-A at centromeres when RBX1 siRNA targets 3’ UTR (Figures S1D–S1F), which rules out off-target effects of siRNA. These findings indicate that CUL4A-RBX1 E3 ligase is specifically required to localize CENP-A to centromeres.
The CUL4A-RBX1 Complex Contributes to CENP-A Ubiquitylation In Vivo
Wang et al. reported that the CUL4-DDB1-RBX1/ROC1 complex functions as a histone ubiquitin ligase and that ubiquitylation of histones H3 and H4 occurs in the cellular response to DNA damage (Wang et al., 2006). On the basis of the high CENP-A homology to histone H3, we hypothesized that CENP-A ubiquitylation by the CUL4A complex might affect CENP-A loading to centromeres. To test this hypothesis, we performed an in vivo ubiquity-lation assay using HeLa Tet-Off cells harboring pcDNA3-HAUbiquitin plus pTRM4-CENP-A-Flag or pTRM4 vector only in the absence of tetracycline or doxycycline (Figure S2A). CENPA-Flag proteins were ubiquitylated in HeLa cells, and putative mono- and diubiquitylated proteins were identified by the appearance of bands of appropriate molecular sizes (Figure 2A). CUL4A or RBX1 single siRNA led to a significant reduction in ubiquitylation (Figures 2B and S2B, second lane from right). These results suggest that CENP-A ubiquitylation by CUL4ARBX1 E3 ligase is important for CENP-A loading to centromeres.
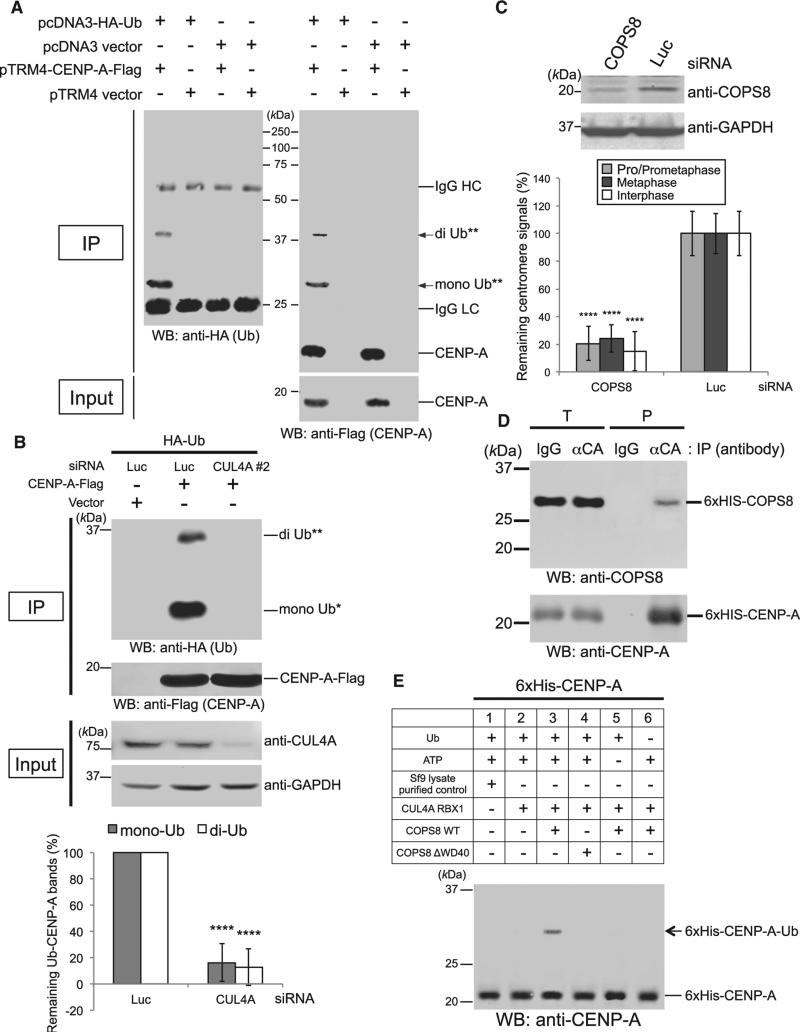
Figure 2. The CUL4A-RBX1 Complex Contributes to CENP-A Monoubiquitylation and Diubiquitylation through the Adaptor Protein COPS8.
(A) Representative images of the in vivo ubiquitylation assay (see Experimental Procedures). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) were detected by western blot (WB) analysis using the indicated antibodies. Bands of putative di-Ub-CENP-A-Flag (**) and putative mono-Ub-CENP-A-Flag (*) are indicated with arrows.
(B) Representative images of the in vivo ubiquitylation assay with the combination of CUL4A or Luc siRNA. For western blot analysis, GAPDH protein was used as a loading control. The histogram shows quantified putative mono- and diubiquitylated-CENP-A-Flag bands (ratio of the ubiquitylated band signal to Flag band signal was normalized with pTRM4-CENP-A-Flag plus Luc siRNA-transfected cells [left column]). Experiments were repeated (n ≥ 3 experiments), and the mean percentages (±SD) are shown. ****p < 0.0001 compared with pTRM4-CENP-A-Flag plus Luc siRNA-transfected cells (left column, Student's t test).
(C) (Top) Western blot analysis of HeLa cell total lysates harvested 72 hr after transfection with COPS8 or Luc siRNA(s). (Bottom) CENP-A signals at centromeres in HeLa cells were quantified 72 hr after transfection with COPS8 or Luc siRNA(s). Signals were normalized with Luc siRNA-treated cells, and the mean percentages (±SD) are shown. ****p < 0.0001 compared with Luc siRNA-treated cells (Student's t test).
(D) Purified 6xHis-CENP-A directly interacts with purified 6xHis-COPS8. Proteins in 3% of the total reaction (T) and immunoprecipitated (P) were detected. Proteins were immunoprecipitated with anti-CENP-A antibody (αCA) or immunoglobulin G control (IgG) (Table S3; see Figures S2F–S2H, for verification of purified recombinant proteins). Note that 6xHis-CENP-A migrates slower than Flag-tagged CENP-A on SDS-PAGE gels.
(E) In vitro ubiquitylation assay using purified components (see Experimental Procedures). Purified 6xHis-tagged components (CUL4A, RBX1, and COPS8 WT or COPS8 ΔWD40) (see Figure S3F) were used and shown in the upper table. The band of putative 6 × His -CENP-A-Ub is indicated with an arrow. Overview images are shown in Figure S3E.
The CUL4A Complex Uses Adaptor COPS8/CSN8 to Target CENP-A
CUL4 assembles multiple different ligases to target different substrates (Bennett et al., 2010; Jackson and Xiong, 2009; Merlet et al., 2009). However, we did not detect significant CENP-A signal reduction at centromeres by using siRNAs against other CUL4-binding linkers and adaptors (DDB1, DDB2, WDR5, and EED) (He et al., 2006) (Figures S2C and S2D; Table S1). To identify the specific CUL4A adaptor protein that targets CENP-A, CUL4A and associated proteins were immunopurified from extracts of HeLa cells that transiently expressed myc-tagged CUL4A (myc3-CUL4A). Subsequently, liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed by the linear ion-trap method. Five proteins (CAND1, DDB1, COPS4/ CSN4, COPS3/CSN3, and COPS8/CSN8) were identified in myc-CUL4A immunoprecipitates by LC-MS/MS analysis as significant peptide hits, based on the criteria that they were identified with more than two peptides (after subtraction of nonspecific peptides) having an expected score of 10e−6 or less (Table S2). We performed siRNA knockdown experiments to examine whether these candidate proteins have a role in recruiting CENP-A to centromeres. COPS8 siRNAs significantly induced the delocalization of CENP-A at centromeres (Figure 2C), but DDB1 or COPS4 siRNAs did not (Figures S2C and S2E; Table S1). Depletion of COPS8 protein induced a significant reduction in CENP-A ubiquitylation (Figure S2B, last lane from left). Recombinant purified COPS8 specifically bound purified CENP-A (Figures 2D and S2F–S2H), and this interaction was confirmed in different systems (Figures S2I and S2J; in the yeast two-hybrid analysis, CAND1 did not interact with CENP-A, and CENP-A showed dimerization). These data support that the interaction between COPS8 and CENP-A is specific and direct (see also Figures S3A–S3C and Table S6).
The CUL4A-RBX1-COPS8 Complex Ubiquitylates CENPA In Vitro
To test whether the E3 activity of the CUL4A-RBX1-COPS8 complex could ubiquitylate CENP-A in vitro, a ubiquitylation assay was performed using Sf9 cell lysates expressing 6xHis-CUL4A, RBX1, and 6xHis-COPS8 and in-vitro-transcribed and -translated 35S-labeled GST-CENP-A (Figure S3D). We also used purified 6xHis-tagged proteins (CUL4A, RBX1, COPS8, and CENP-A) that were expressed in Sf9 cells (Figures 2E, S3E, and S3F). In both systems, putative monoubiquitylation bands of CENP-A were found only in the sample with a mixture of three factors (CUL4A, RBX1, and COPS8) but not in other control samples (Figures 2E, S3D, and S3E). Addition of COPS8 ΔWD40 instead of WT did not induce proper ubiquitylation in both systems (Figures 2E and S3E, compare lanes 3 and 4; Figure S3G, compare lanes 3 and 4). These results confirm that, only in the presence of the specific adaptor COPS8, the recombinant CUL4A-RBX1 complex can ubiquitylate CENP-A in vitro.
CENP-A K124 Ubiquitylation Is Essential to Localize CENP-A to Centromeres and Important for Binding to HJURP
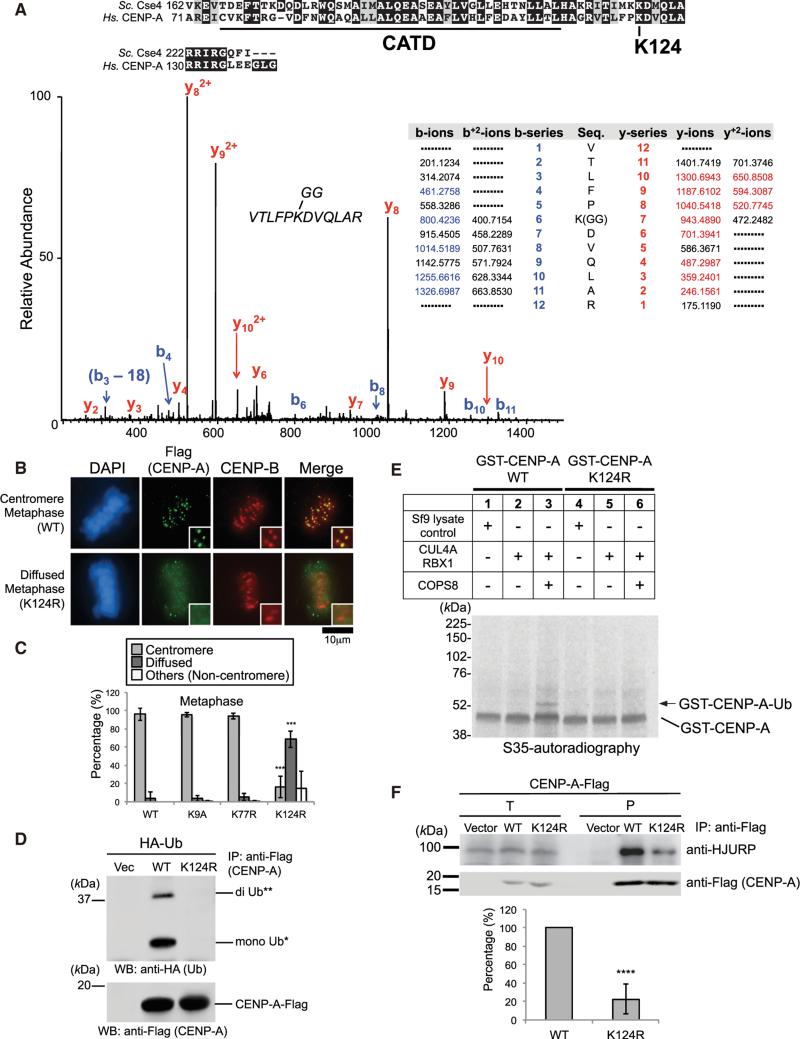
Our immunoprecipitation mass spectrometry analysis revealed that CENP-A-Flag is ubiquitylated on lysine 124 (K124) in HeLa cells (Figure 3A; Table S7). The crystal structure of the CENP-A nucleosome revealed that the K124 site resides in the CENP-A α3 helix, although the site is not within the CATD region (Figures 3A, top, and S4A). Interestingly, K124 is conserved among mammals, birds, lizards, plants, and a group of fungi (i.e., budding yeast) (Figures 3A, top, and S4B). CENP-A lysine mutants (Figure S4C; see also Figure S7F) were constructed and their centromeric localization was tested. The K124R mutation substantially abrogated the centromeric localization of exogenous CENP-A, giving diffused signals in both mitotic and interphase HeLa cells (Figures 3B and 3C; representative data of other cell-cycle stages shown in Figures S4D and S4E). Neither K9A (which corresponds to histone H3 K9 methylation) nor K77R (a unique lysine site in CATD) mutations significantly affected centromere delocalization of CENP-A (Figures 3C and S4E).
Figure 3. CENP-A K124 Ubiquitylation Is Required for CENP-A Localization at Centromeres.
(A) (Top) C-terminal conserved region between S. cerevisiae Cse4 and human CENP-A. The ubiquitylation site (lysine 124) on human CENP-A is indicated, and the CATD is underlined. (Bottom) Lysine 124 on CENP-A is a ubiquitylation site in vivo. Collision-induced dissociation analysis of the VTLFPKGGDVQLAR (m/z 500.94) peptide is displayed (coverage 55%). The b (blue) and y (red) ions detected during fragmentation are highlighted in the table. The y-7 ion (m/z 943.4890) confirms the modification of K124 by the di-glycine motif (see Table S7).
(B) The CENP-A K124R mutant delocalizes from centromeres (see Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red) as a centromere location control were visualized. Note that diffused signals appear in the exogenous CENP-A-Flag over-expression, presumably because its expression level is approximately 1.0–1.4 orders of magnitude (10- to 25-fold) higher than endogenous CENP-A (data not shown). Representative images of other cell-cycle stages are shown in Figure S4D. Scale bar, 10 μm.
(C) Histograms summarizing the localization patterns shown in (B). More than 50 metaphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. “Others (Non-centromere)” indicates mostly damaged cells, dead cells, or cells with nucleolar localization (only in inter-phase) presumably due to transfection or other treatments. Histograms of other cell-cycle stages are shown in Figure S4E. ***p < 0.001 compared with CENP-A WT-Flag (Student's t test).
(D) The CENP-A K124R mutation abrogates ubiquitylation of CENP-A in vivo (see Experimental Procedures). WT, CENP-A (WT)-Flag; K124R, CENP-A (K124R)-Flag.
(E) The CENP-A K124R mutation abrogates ubiquitylation of CENP-A in vitro (see Experimental Procedures). 6xHis-tagged components (CLU4A, RBX1, and COPS8) are shown in the upper table. Band of putative GST-CENP-A-Ub is indicated with arrow.
(F) (Top) The CENP-A K124R mutation reduces the interaction with HJURP in vivo (see Immunoprecipitation Assay in Supplemental Experimental Procedures; see also Discussion and Figures S5E and S7E). Proteins in 2% of the total cell lysates (T) and immunoprecipitates (P) were detected. (Bottom) Histogram shows quantified coprecipitated endogenous HJURP bands represented in (top) (ratio to Flag band signal normalized with pTRM4-CENP-A WT-Flag-transfected cells [left column]). Experiments were repeated (n ≥ 4 experiments), and mean percentages (±SD) are shown. ****p < 0.0001 compared with pTRM4-CENP-A WT-Flag-transfected cells (left column, Student's t test).
These results raised two possibilities regarding the in vivo function of CENP-A ubiquitylation on K124: (1) K124 ubiquitylation is involved in ubiquitin-mediated proteolysis to eliminate overexpressed and/or mislocalized CENP-A to euchromatin, or (2) CENP-A ubiquitylation on K124 is involved in loading CENP-A to centromeres. To test the first possibility, we compared the stabilities of CENP-A-Flag WT and the K124R mutant by monitoring protein levels. Both CENP-A-Flag WT and the K124R mutant showed similar protein stabilities after new protein synthesis was inhibited by adding doxycycline to cell cultures (Figure S4F). These data suggest that ubiquitylation on K124 is likely not involved in ubiquitin-mediated proteolysis to eliminate overexpressed and/or mislocalized CENP-A. Furthermore, we monitored protein stability of endogenous CENP-A after cycloheximide (CHX) treatment in CUL4A- or RBX1-depleted cells. Neither CUL4A nor RBX1 depletion stabilized endogenous CENP-A (Figure S4G), which is consistent with our observation in the asynchronous or Taxol-treated condition (Figure S1G). These data indicate that neither CUL4A nor RBX1 is involved in ubiquitin-mediated proteolysis of CENP-A, suggesting that the CUL4A-RBX1 complex contributes to “signaling” ubiquitylation, which is required for CENP-A localization at centromeres (see Discussion).
To confirm that the K124R mutation abrogates putative monoand diubiquitylation bands, both in vivo and in vitro ubiquitylation assays were performed using the K124R mutant. Presence of the K124R mutation abrogated the appearance of ubiquitylated forms (Figures 3D, 3E, S3G, and S5A–S5D; see also Discussion and Figures S7A–S7D). The K124R mutation, CUL4A, or RBX1 siRNA significantly reduced the interaction of CENP-A with endogenous HJURP in HeLa cells (Figures 3F and S5E; see also Discussion and Figure S7E). Collectively, these data suggest that CENP-A K124 is monoubiquitylated or diubiquitylated by the CUL4A-RBX1 complex and that CENP-A K124 ubiquitylation is required for efficient interaction with HJURP to properly localize CENP-A at centromeres.
Monoubiquitin Fusion Is Sufficient to Load CENP-A K124R at Centromeres
On the basis of the results above, we hypothesized that a covalently linked monoubiquitin serves as a signal to load CENP-A at centromeres. To test this hypothesis, we constructed an N-terminal Flag-tagged and C-terminal ubiquitin-fused WT and K124R mutant CENP-A (Figures 4A and 4B). To prevent ubiquitin-fused CENP-A protein from potential polyubiquitylation, we also used the monoubiquitin mutant Ub (K48R), which has lost a major site for polyubiquitylation (Antonelli et al., 1999; Codomo et al., 2014; Thrower et al., 2000). These constructs allowed us to study the role of CENP-A monoubiquitylation. We tested the centromeric localization of constructs by capturing anti-Flag immunofluorescent signals. Both Flag-CENP-A (WT) and Flag-CENP-A (WT)-Ub (WT) maintained their centromere localization, whereas Flag-CENP-A (K124) substantially abrogated centromere localization of CENP-A (Figures 4C and 4D, column 6; representative data of other cell-cycle stages shown in Figures S5F and S5G). The Flag-CENP-A (K124R)-Ub (K48R) protein, which presumably mimics monoubiquitylated CENP-A, substantially restored the localization to centromeres (Figures 4D and S5G, compare columns 5 and 6) more efficiently than did CENP-A (K124R)-Ub (WT) (Figures 4D and S5G, compare columns 4–6; see also Discussion).
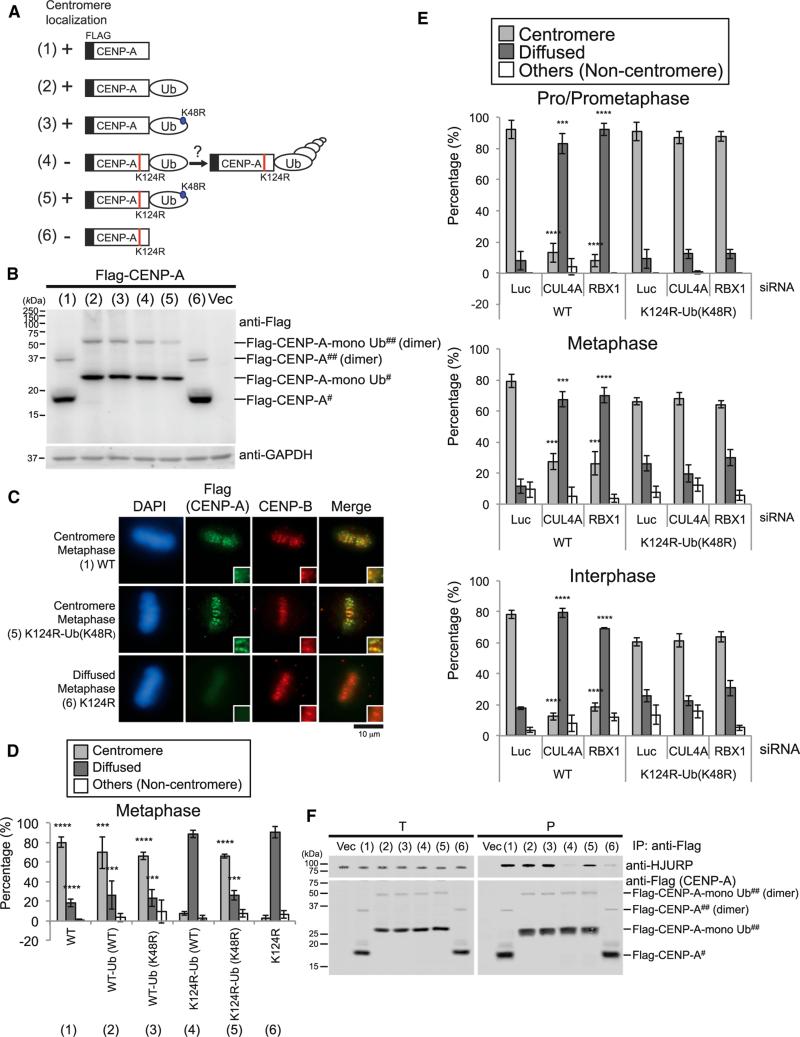
Figure 4. CENP-A Monoubiquitin Fusion Rescues Delocalization of the CENP-A K124R Mutant from Centromeres.
(A) Schematic figures of each construct used in this study. The K124R mutation site on CENP-A (red) and the K48R mutation site on monoubiquitin (blue) are indicated. (1) WT, non-fused CENP-A (WT); (2) WT-Ub (WT), CENP-A (WT)-Ub (WT); (3) WT-Ub (K48R), CENP-A (WT)-Ub (K48R); (4) K124R-Ub (WT), CENP-A (K124R)-Ub (WT); (5) K124R-Ub (K48R), CENP-A (K124R)-Ub (K48R); (6) K124R, non-fused CENP-A (K124R).
(B) Western blot analysis of HeLa Tet-Off cell total lysates. Cells were cultured without tetracycline/doxycycline and harvested 48 hr after transfection with each construct indicated in (A). Putative dimer (##) and monomer (#) of each construct are also indicated. Note that ubiquitylated CENP-A bands do not appear in this membrane because of non-HA-Ub coexpression, MG132-untreated, and non-immunoprecipitated samples.
(C) CENP-A monoubiquitin fusion rescues delocalization of the CENP-A K124R mutant from centromeres (see Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red) as a centromere location control were visualized. Representative images of other cell-cycle stages are shown in Figure S5F. Scale bar, 10 μm.
(D) Histograms summarizing the localization patterns shown in (C). More than 50 metaphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. Representative histograms of other cell-cycle stages are shown in Figure S5G. ****p < 0.0001 and ***p < 0.001 compared with non-fused Flag-CENP-A K124R [column (6)] (Student's t test).
(E) CENP-A monoubiquitin fusion bypasses the requirement for the CUL4A complex. Histograms summarizing the localization patterns of non-fused Flag-CENPA (WT) and monoubiquitin-fused Flag-CENP-A (K124R)-Ub (K48R) mutant. More than 50 pro/prometaphase and metaphase cells and more than 200 interphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. ****p < 0.0001 and ***p < 0.001 compared with non-fused Flag-CENP-A WT plus Luc siRNA-transfected cells (Student's t test).
(F) CENP-A monoubiquitin fusion rescues the interaction with HJURP. The in vivo immunoprecipitation assay was performed using N-terminal Flag-tagged constructs shown in (A) (see Experimental Procedures). Proteins in 3% of the total cell lysates (T) and immunoprecipitates (P) were detected.
To determine the specific function of CUL4-RBX1 E3 activity on K124 monoubiquitylation, we examined the localization of Flag-CENP-A (K124R)-Ub (K48R) in CUL4A- or RBX1-depleted cells. Flag-CENP-A (K124R)-Ub (K48R) was localized at centromeres, whereas Flag-CENP-A was significantly delocalized from centromeres in CUL4A- or RBX1-depleted cells (Figure 4E), indicating that CUL4A and RBX1 are required for loading CENP-A to centromeres via ubiquitylation of K124R.
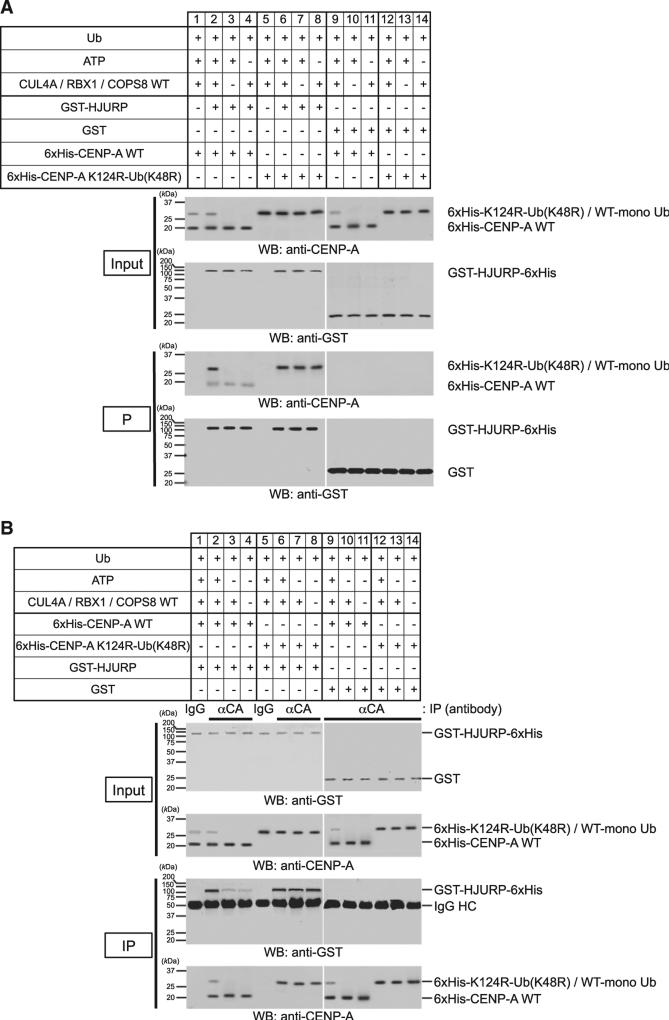
Importantly addition of monoubiquitin rescued the interaction of the CENP-A K124R mutant with HJURP in vivo (Figure 4F, sample 5) and in vitro (Figures 5A and 5B, compare lanes 3 and 4 with lanes 7 and 8; verification of protein purification was shown in Figures S3F and S5H; see also Figure S5I). In vitro, the fusion of monoubiquitin bypassed the requirement of CUL4A, RBX1, and COPS8 for interaction of CENP-A and HJURP (Figures 5A and 5B, lanes 7 and 8, respectively). Taken together, these data demonstrate that monoubiquitylation is an essential signaling modification required for efficient interaction with HJURP and subsequent recruitment of CENP-A to centromeres.
Figure 5. Ubiquitylation of CENP-A by the CUL4A-RBX1 Complex Enhances the Binding to HJURP In Vitro.
(A) In vitro GST pull-down assay was performed using purified proteins after CENP-A ubiquitylation in vitro. In vitro ubiquitylation was performed as in Figure 2E and subsequently purified GST-HJURP-6xHis or GST was added to perform in vitro GST pull-down assay. Proteins in 3% of the total reaction (Input) and pulled down with glutathione Sepharose (P) were detected (see Protein Purification and In Vitro GST Pull-Down Assay in Supplemental Experimental Procedures).
(B) In vitro immunoprecipitation was performed using purified proteins after CENP-A ubiquitylation in vitro. In vitro ubiquitylation was performed as Figure 2E and subsequently purified GST-HJURP-6xHis or GST was added to perform immunoprecipitation. Proteins in 3% of the total reaction (Input) and immunoprecipitates (IP) obtained were detected (see Immunoprecipitation Assay in Supplemental Experimental Procedures).
The CUL4A-RBX1 Complex Contributes to Localization of Newly Synthesized CENP-A to Centromeres and to Maintenance of Old CENP-A at Centromeres
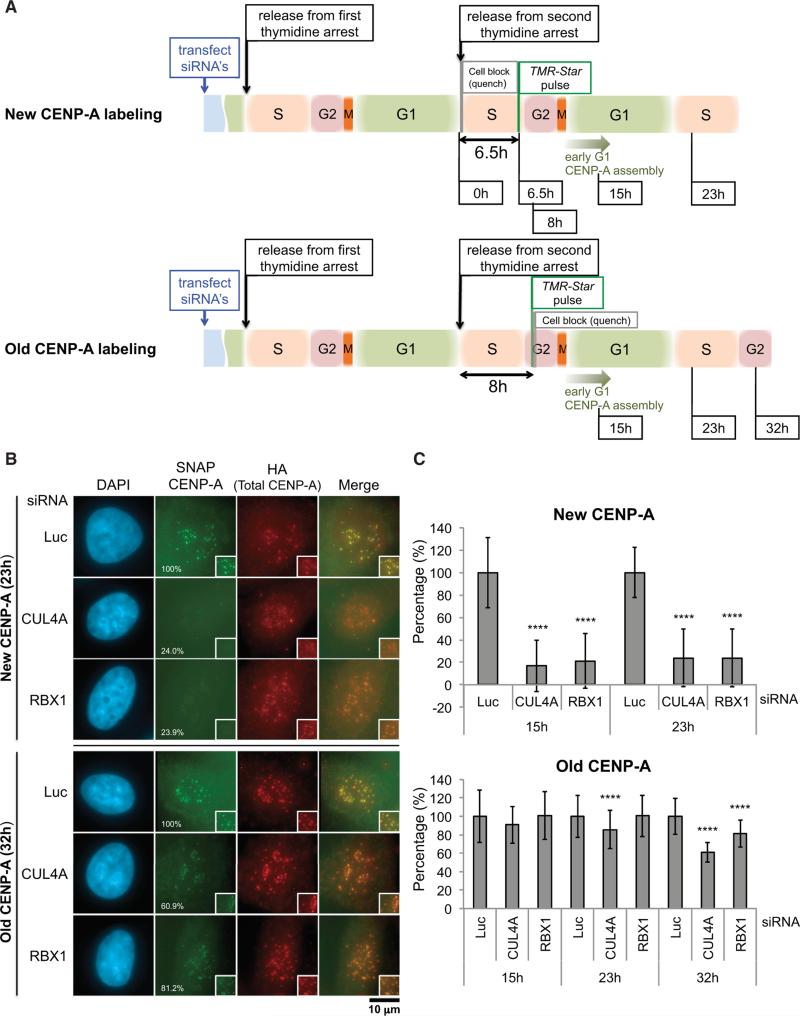
To determine the role of the CUL4A-RBX1 complex in CENP-A localization at centromeres, we monitored the effects on both loading of newly synthesized CENP-A (“new CENP-A”) and maintenance of pre-existing CENP-A (“old CENP-A”) using the SNAP-tag pulse-chase system (Carroll et al., 2009; Foltz et al., 2009; Jansen et al., 2007; Lagana et al., 2010). The SNAP protein tag can catalyze the formation of a covalent bond to a benzylguanine moiety coupled to different fluorescent or non-fluores-cent membrane-permeable reagents (Keppler et al., 2003). This tag allows pulse-chase experiments to be conducted at the level of a single protein. We applied quench-chase-pulse labeling with this technique to follow the fate of newly synthesized CENP-A (“New CENP-A labeling,” Figure 6A, top) as well as pulse labeling to determine CENP-A turnover specifically at centromeres (“Old CENP-A labeling,” Figure 6A, bottom) (Jansen et al., 2007; Lagana et al., 2010). CUL4A or RBX1 siRNAs were transfected during the double-thymidine block-and-release experiment, using established HeLa cell lines stably expressing centromere-localized CENP-A-SNAP at near endogenous levels (Jansen et al., 2007; Lagana et al., 2010). First, we confirmed that CUL4A or RBX1 depletion did not significantly affect cell-cycle progression both in asynchronous cells (Figure S6A) and in double-thymidine blocked or released cells (Figure S6B). Jansen et al. reported that for approximately 11 hr after release from thymidine the initially synchronized cell population does not load substantial levels of CENP-A; loading occurs concomitantly with entry into G1, and loading of newly synthesized CENP-A begins in telophase (Jansen et al., 2007). Consistent with these findings, we did not observe substantial levels of CENP-A loading at 0, 6, and 8.5 hr (data not shown), and the majority of cells had newly incorporated pulse-labeled CENP-A at 15 hr after thymidine release (Figure 6C, top; data not shown). CENP-A assembly was saturated at 23 hr post-thymidine release, corresponding to the next G1-S in the Luc control sample (Figure 6C, top; data not shown). However, CUL4A- or RBX1-depleted cells had substantially impaired assembly of newly synthesized CENP-A (Figures 6B and 6C, top). Preassembled old CENP-A was also reduced slightly but significantly in CUL4A siRNA (23 and 32 hr) or RBX1 siRNA (32 hr) -treated cells (Figures 6B and 6C, bottom), suggesting that the CUL4A-RBX1 complex also has a role in maintaining “old” CENP-A at centromeres.
Figure 6. The CUL4A-RBX1 Complex Is Required for Incorporating Newly Synthesized CENP-A into Centromeres and for Maintaining Old CENP-A at Centromeres.
(A) Schematic of cell synchronization and labeling protocol for newly incorporated CENP-A (top) and preassembled old CENP-A (bottom). Cell-cycle stages are estimates based on time elapsed after release from double-thymidine-induced arrest at G1-S.
(B) Both newly incorporated CENP-A and a significant amount of preassembled CENP-A are lost in the absence of CUL4A or RBX1. The average intensity of signal (percent) at centromeres in the cell, normalized with Luc siRNA-treated cells at each time point, is listed in the lower left corner of panels showing SNAP CENP-A-stained cells. Representative images for each time point after release from thymidine are shown. Scale bar, 10 μm.
(C) Quantification of SNAP-tag-labeled CENP-A signals from the experiment performed in (B). Signals of more than 100 interphase cells were quantified per experiment and the mean percentages (±SD) are shown. ****p < 0.0001 compared with Luc siRNA-treated cells at each time point (Student's t test).
H3-CATD K125 Ubiquitylation Is Required for H3-CATD Centromere Localization
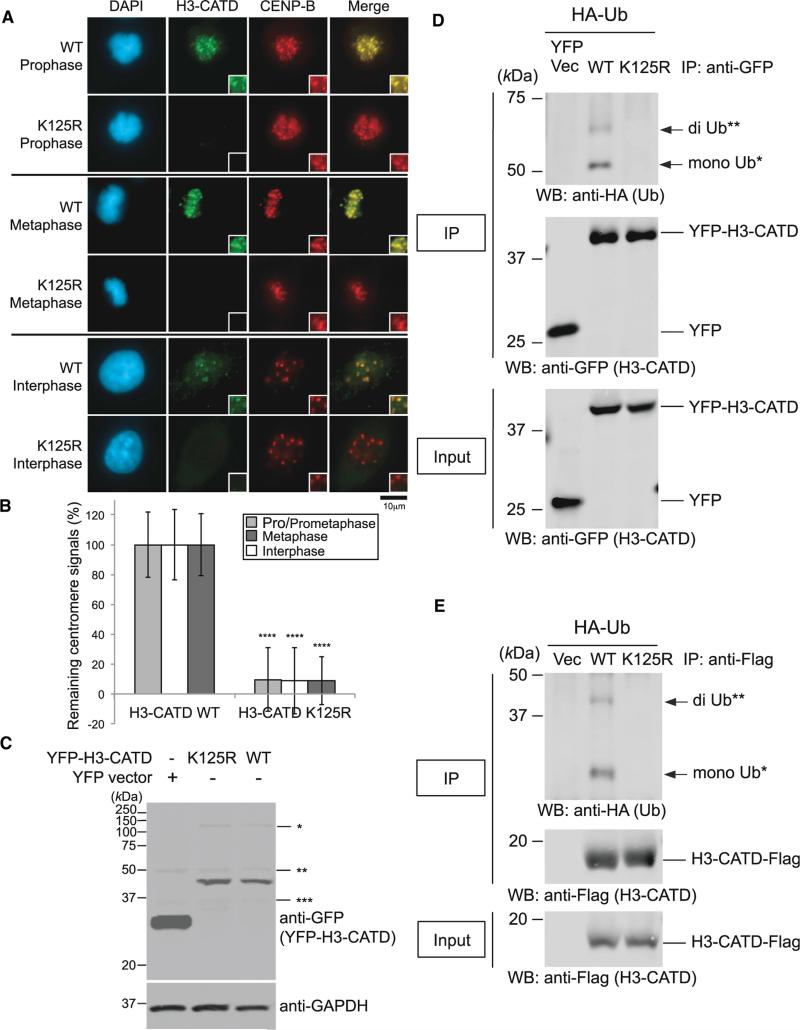
CATD is a cis-acting element within CENP-A required for its assembly at centromeres (Black et al., 2004, 2007), and it comprises the loop 1 and α2 helix of the histone-fold domain. Substitution into histone H3.1 of the CATD is sufficient to promote assembly of chimeric histone H3.1 (H3-CATD) to centromeres (Black et al., 2004). To test whether the ubiquitylation signaling is specific to CENP-A, we studied H3-CATD localization at centromeres with CUL4A and/or RBX1 siRNA(s). A single or a double depletion significantly induced the delocalization of H3-CATD at centromeres (Figures S6E and S6F) without affecting protein levels (Figure S6G) and abrogates ubiquitylation of H3-CATD (Figure S6H). Because the CENP-A K124 site is outside the CATD region, H3-CATD maintains H3-derived lysine K125 (K125 is shifted two amino acid positions from H3 K123 by the chimeric construction). The K125R mutation substantially reduced the centromeric localization of H3-CATD in both mitotic and interphase HeLa cells (Figures 7A and 7B) without affecting expression levels (Figure 7C) and abrogates ubiquitylation of H3-CATD (Figures 7D and 7E). These data indicate a specific role of K124 in collaboration with CATD in centromere localization of CENP-A.
Figure 7. K125R Mutation Abrogates H3-CATD Ubiquitylation and Localization at Centromeres.
(A) Delocalization of the H3-CATD K125R mutant at centromeres. HeLa cells were cultured, transfected, and immunostained (see Immunofluorescence in Supplemental Experimental Procedures). Scale bar, 10 μm.
(B) Histogram quantifying H3-CATD WT and K125R signals at centromeres given in (A). Signals were normalized with those in H3-CATD WT, and mean percentages (±SD) are shown. ****p < 0.0001 compared with H3-CATD WT (Student's t test).
(C) Overexpressed pEYFP-H3-CATD WT and K125R were confirmed, but levels of EYFP-H3-CATD remained unchanged between the WT and K125R mutant. *, **, and *** indicate non-specific bands.
(D) The H3-CATD K125R mutation abrogates ubiquitylation of CENP-A in vivo (pEYFP vector background; see also Experimental Procedures). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) were separated by a 10% SDS-PAGE. YFP Vec, pEYFP vector only; WT, pEYFP-H3-CATD WT; K125R, pEYFP-H3-CATD K125R. Bands of putative di-Ub-YFP-H3-CATD (**) and putative mono-Ub-YFP-H3-CATD (*) are indicated with arrows.
(E) The H3-CATD K125R mutation abrogates ubiquitylation of CENP-A in vivo (pTRM4 vector background; see also Experimental Procedures). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) obtained using ANTI-FLAG M2 Affinity Gel (SIGMA-ALDRICH) were detected. Vec, pTRM4 vector only; WT, pTRM4-H3-CATD WT-Flag; K125R, pTRM4-H3-CATD K125R-Flag. Bands of putative di-Ub-H3-CATD-Flag (**) and putative mono-Ub-H3-CATD-Flag (*) are indicated with arrows.
DISCUSSION
In this study, we found that CENP-A ubiquitylation on lysine 124 (K124) mediated by the CUL4A-RBX1/ROC1 E3 ligase is required for CENP-A recruitment to centromeres. Lysine 124 is the in vivo and in vitro ubiquitylation site on CENP-A. However, the question of how and when one centromeric nucleosome wrapped by DNA accommodates the enlarged size of ubiquity-lated CENP-A (from ~16.0 to 24.5 kDa by monoubiquitylation) remains unanswered. We found ubiquitylation of endogenous CENP-A in chromatin extracts (Figure S7A), and most of the ubiquitylated endogenous CENP-A was found in the chromatin-insoluble fraction (Figures S7B–S7D). Of the total precipitated nucleosomal CENP-A, there were 6%–11% of monoubiquitylated proteins (Figure S7A), suggesting that ubiquitylated CENP-A exists heterogeneically in nucleosomes. Consistent with this, results of the UbFC analysis suggest that K124-ubiquitylated CENP-A exists at centromeres and the nuclear region (Figures S5A–S5D). The latest crystal structure of the human centromeric nucleosome containing CENP-A indicates that the CENP-A nucleosome structure is not compact, as suggested in the free CENP-A-H4 structure (Figure S4A) (Sekulic et al., 2010; Tachiwana et al., 2011). By using single-molecule atomic force microscopy, Bui et al. found that diameters but not heights of CENP-A nucleosomes change over the cell cycle, although they concluded that CENP-A nucleosomes change from tetramers to octamers at the S phase (Bui et al., 2012). Our cell-cycle analysis using chromatin-free extracts revealed that the level of CENP-A ubiquitylation increases in the M and early G1 phases (Figures S6C and S6D), which is consistent with the timing of centromere localization of HJURP and newly synthesized CENP-A (Foltz et al., 2009; Jansen et al., 2007).
Recently, Han et al. reported that lysines 121, 122, and 125 of histone H3 are major ubiquitylation residues in yeast, and the Rtt101Mms1 and Cul4DDB1 E3 ubiquitin ligases regulate nucleo-some assembly in yeast and human cells, respectively (Han et al., 2013). In higher eukaryotes, DDB1 functions as a linker for CUL4. The DDB1 CUL4 Associated Factor (DCAF) proteins are the substrate-specificity factors for Cul4-RING ligases (CRL4s), and mammalian genomes encode more than 50 DCAF proteins, each targeting several different substrates (Angers et al., 2006; Bennett et al., 2010; He et al., 2006; Higa et al., 2006; Jin et al., 2006). He et al. identified the DDB1-binding WD40 protein (DWD) box and demonstrated the binding of 15 DWD proteins with DDB1-CUL4A (He et al., 2006). However, we did not observe DDB1 involvement in CENP-A recruitment to centromeres (Figure S2C). COPS8/CSN8, a subunit of the COP9 signalosome, is a PCI domain protein required for deneddylation in the CRL assemble cycle (Glickman et al., 1998; Merlet et al., 2009; Pierce et al., 2013; Wei and Deng, 2003). Thus, in view of the dogma regarding various recognition subunits of a CRL4 ligase, COPS8 did not seem to be the unique protein providing substrate specificity. However, we have shown that (1) DDB1 and DDB2 are not involved in the CENP-A loading pathway, and thus it is unlikely that DCAF proteins are involved in that pathway; (2) COPS8 depletion reduced CENP-A localization at centromeres; (3) COPS8 interacts directly with CENP-A in vitro; (4) COPS8 bridges CENP-A and CUL4A; (5) COPS8 has a WD40 motif, and deletion of this motif abrogates the interaction with CENP-A and the ability of COPS8 to bridge CUL4A and CENP-A; and (6) deletion of the COPS8 WD40 motif abrogates CENP-A ubiquitylation in vitro. These data suggest that COPS8 itself is the specific adaptor of the CUL4A-RBX1 complex that is required to recognize and ubiquitylate the substrate CENP-A in a non-canonical CRL4 machinery, which does not require DDB1 or DCAF (DDB1-CUL4-associated factors) for substrate recognition (Jackson and Xiong, 2009). Thus, our evidence that COPS8 is required for CENP-A ubiquitylation is striking and redefines the paradigm of CRL4 adaptor proteins. It is possible that COPS8 acts as both a deneddylase and an adaptor for non-canonical CRL4 regulation.
Two independent groups have recently reported that yeast Psh1 (an E3 ubiquitin ligase) controls the cellular level of Cse4 via ubiquitylation and proteolysis in budding yeast (Hewawasam et al., 2010; Ranjitkar et al., 2010). They also showed the toxicity of Cse4 overexpression in the absence of Psh1, suggesting that Psh1 prevents the mislocalization of Cse4 through its proteolytic ubiquitylation. Hewawasam et al. demonstrated the specific ubiquitylation activity of Psh1 toward Cse4 in vitro and mapped the sites of ubiquitylation (Hewawasam et al., 2010). However, the major in vitro ubiquitylation site that they identified (Cse4 lysine 4), which is located in the Cse4 non-homologous N-terminal region, is absent in human CENP-A. In addition, the in vitro ubiquitylation site (Cse4 lysine 163), which has the highest level of ubiquitylation in the histone fold domain targeted by Psh1, is not conserved in human CENP-A (Figure 3A, top). Interestingly, four of the five ubiquitylated lysine sites identified by Hewawasam et al. (2010) are not conserved in human CENP-A, and only Cse4 lysine 155 presumably corresponds to human CENP-A lysine 64 (data not shown). Our study demonstrates that human CENP-A is susceptible to monoubiquitylation or diubiquitylation on lysine 124, whereas Cse4 is polyubiquitylated on multiple lysine sites, most of which are not conserved in human CENP-A (Collins et al., 2004; Hewawasam et al., 2010; Ranjitkar et al., 2010). Psh1 lacks apparent homologs in metazoans (Folco and Desai, 2010). We speculate that the divergence of E3 ligases in the specificity of ubiquitylation sites and biological functions occurred during evolution. Ranjitkar et al. reported that CATD primarily contributes to the interaction with Psh1 and to Cse4 stability via Psh1 ubiquitin ligase (Ranjitkar et al., 2010). Human CENP-A lysine 77 (K77) is the unique lysine site in CATD, but the K77R mutant in our study did not produce significant CENP-A delocalization from centromeres.
Interestingly, very recently Bade et al. demonstrated that CUL3/RDX1 ubiquitylates CENP-A and stabilizes both CENP-A and CAL1 (Bade et al., 2014): a candidate that might fulfill the function of HJURP in Drosophila melanogaster as the CENP-A assembly factor (Chen et al., 2014; Erhardt et al., 2008; Goshima et al., 2007). Similarly, our results do not support the involvement of the CUL4A or RBX1 protein in ubiquitin-mediated proteolysis of CENP-A, because (1) the exogenous K124R mutant (both monoubiquitin fused and non-fused forms) did not show any change in stability compared with wild-type CENP-A in HeLa cell lysates; (2) endogenous CENP-A proteins significantly delocalized after CUL4A or RBX1 depletion, but they did not localize on non-centromere chromosomes or euchromatin, and CUL4A or RBX1 depletion did not affect the stability of endogenous CENP-A; (3) no polyubiquitylation (a typical modification targeting proteins for proteolysis) of CENP-A occurred with MG132 concentration of 5 μM in vivo or 1 mM in vitro, and CENP-A is monoubiquitylated or diubiquitylated both in vitro and in vivo; (4) SNAP-tag pulse-chase analysis revealed that CUL4A-RBX1 plays a role in incorporating newly synthesized CENP-A in centromeres; and (5) monoubiquitylation is not a canonical ubiquitin-based signal for proteasome degradation. Monoubiquitin fusion is sufficient to load CENPA K124R at centromeres, and it restores the affinity of CENPA K124R to HJURP. Therefore, the ubiquitylation alters protein function by mechanisms in which protein degradation is not involved, as in the case of other histones (Sigismund et al., 2004).
There is a discrepancy regarding the experiments using C-terminal ubiquitin fusion constructs (Figure 4): addition of Ub (WT) but not Ub (K48R) can rescue the K124R mutant (Figures 4D and S5G, compare columns 4 and 5). One may presume that addition of Ub (WT) but not Ub (K48R) could induce polyubiquitylation resulting in degradation. However, the protein level of CENP-A (K124R)-Ub (WT) or CENP-A (WT)-Ub (WT) was not reduced substantially (Figure 4B). Thus, we consider that C-terminal tagging Ub (K48R) but not Ub (WT) happens to be able to mimic K124 ubiquitylation structurally. Indeed, CENP-A (K124R)-Ub (WT) did not rescue the interaction of the CENP-A K124R mutant with HJURP in vivo (Figure 4F, sample 4).
Previous studies showed that suppression of HJURP reduced CENP-A levels, presumably affecting the stability of CENP-A (Foltz et al., 2009; Shuaib et al., 2010). However, our results indicate that depletion of CUL4A or RBX1 or K124R mutation does not affect the stability of CENP-A (Figures 4B, S1G, S4C, S4F, S4G, and S5B). The possible difference between the previous data and ours is depletion of CUL4A/RBX1 or K124R mutation abolishes K124 ubiquitylation, but depletion of HJURP still maintains monoubiquitylated CENP-A. However, we did not detect any changes in CENP-A stability between monoubiquitylated form and non-ubiquitylated form (Figure 4B; data not shown). Therefore, a plausible explanation for the inconsistency of CENP-A stability after HJURP dissociation between the previous studies and ours may be because a “basal level” interaction between non-ubiquitylated CENP-A and HJURP is sufficient to stabilize CENP-A.
Bui et al. reported that in G1/S-phase-derived cells, CENP-A is acetylated on lysine 124 and histone H4 on lysine 79, suggesting that these covalent modifications occur at the key transition from G1 to S phase (Bui et al., 2012); however, the biological function of the acetylation is not yet clear. Interestingly, Han et al. showed that yeast Rtt101Mms1 (CUL4ADDB1 homolog) preferentially ubiquitylates H3K56ac-H4 over unmodified H3-H4 in vivo and in vitro. Our data insinuate that the CENP-A K124 acetylation, which occurs at the G1/S transition, might “prime” or “block” CENPA K124 for ubiquitylation until the M phase. A recent study in the LacI-LacO system demonstrated that the presence of HJURP is sufficient for forming a functional de novo kinetochore (Barnhart et al., 2011), which is in agreement with our findings that the ubiquitylation of CENP-A is also required for efficient interaction with HJURP to properly localize CENP-A at centromeres (Figures 3F, 4F, 5A, 5B, S5E, and S5I; see also Figures S6C and S6D). A certain level of ubiquitylation in the nucleosome could be still required after the incorporation to maintain “old” CENP-A at the centromere during the G1 phase, which is supported by results of our SNAP assay (Figure 6). Whereas histone ubiquitylation in the cellular response to UV damage is mediated through CUL4-DDB-RBX1/ROC1 (Wang et al., 2006) and the transfer of H3-H4 from the Asf1-H3-H4 complex to other histone chaperones is regulated by the Cul4ADDB1 E3 ligase (Han et al., 2013), the recruitment of CENP-A to centromeres is led by ubiquitylation on CENP-A K124 by the CUL4A-RBX1-COPS8 complex.
Last, but not least, our result is consistent with those from previous studies demonstrating that CATD is important for centro-mere targeting (Figures 7 and S6E–S6H) (Black et al., 2004, 2007): the collaboration of both CATD and ubiquitylation on K124 (K125 of H3-CATD) is required for CENP-A loading at centromeres.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
See Table S3 for antibodies, Table S4 for siRNA sequences, and Table S5 for plasmid vectors used in this study.
Cell Culture and Transfection
HeLa or HeLa Tet-Off cells (Clontech) were cultured in high-glucose DMEM (BioWhittaker) with 10% fetal bovine serum (FBS) (Invitrogen). Cells were grown at 37° C in 5% CO2 in a humidified incubator. Cells were transfected with annealed double-stranded siRNA(s) or mammalian expression plasmids by using Lipofectamine 2000 (Invitrogen), Lipofectamine 3000 (Invitrogen), Lipofectamine LTX (Invitrogen), or Lipofectamine RNAiMAX (Invitrogen). HeLa Tet-Off cells were cultured without tetracycline/doxycycline and transiently transfected with the pTRM4 overexpression vector whose transcription was regulated by the TRE promoter. The double-thymidine block and Taxol treatment were performed as described previously (Jansen et al., 2007; Niikura et al., 2006, 2007, 2010; Tatsumi et al., 2003).
CENP-A In Vivo Ubiquitylation Assay
In short, HeLa or HeLa Tet-Off cells were transfected with the indicated expression vectors and incubated with 5 mM MG132 (Calbiochem) for 24 hr. Cells were then collected and lysed in denaturing buffer A (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% SDS, 1 mM EDTA, 50 mM MG132, and complete EDTA-free protease inhibitor cocktail [Roche]) (Wang et al., 2006) by a sonication and freeze-thaw process. Proteins were immunoprecipitated and the elution was used for western blot analysis with the indicated antibodies. Details are described in Supplemental Experimental Procedures.
CENP-A In Vitro Ubiquitylation Assay
In vitro ubiquitylation assay was performed as described previously (Lorick et al., 2006) with the following minor modifications. In the case that Sf9 cell ly-sates were used, in-vitro-transcribed and -translated 35S-labeled GST-CENPA was incubated with Sf9 cell lysates harboring recombinant 6 × His-components (6 × His-CUL4A, 6 × His-RBX1, and/or 6 × His-COPS8) as indicated. A 25-ml reaction mixture contained 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 μM ATP, 2 mM DTT, 0.2 mM ubiquitin aldehyde, 1 mM MG132, 2 mM LLnL, 100 ng E1 (UBE1), 100 ng E2 (UbcH5c), and 10 μg HA-ubiquitin. GST-proteins were purified on glutathione Sepharose 4 FAST Flow (GE Healthcare), washed four times with STE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) and separated on SDS-PAGE gels. Ubiquitylated 35S signals (GST-CENP-A or GST) were detected by autoradiography.
In the purified system, the indicated 6xHis-tagged proteins (CENP-A, CUL4A, and RBX1) were expressed in Sf9 insect cells and purified with Ni-NTA Agarose (QIAGEN). COPS8 WT and DWD40 proteins were expressed in BL21-Gold (DE3) bacterial cells (Stratagene/Agilent Technologies) and purified with Ni-NTA Agarose (QIAGEN). Then, these purified proteins (CENP-A 310 ng, CUL4A 154 ng, RBX1 184 ng, COPS8 WT 125 ng, and COPS8 ΔWD40 125 ng) were combined to a 12.5-μl reaction mixture contained 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 0.2 μM ubiquitin aldehyde, 1 mM MG132, 2 μM LLnL, 50 ng E1 (UBE1), 50 ng E2 (UbcH5c), and 5 μg ubiquitin. Alternatively, purified Ni-NTA Agarose (QIAGEN) bound to the indicated 6xHis-tagged component(s) was eluted with Native Elution Buffer (see user manual of Ni-NTA Purification System, Invitrogen, 25-0490) and used for ubiquitylation and subsequent assay (see Immunoprecipitation Assay and/or In Vitro GST Pull-Down Assay in Supplemental Experimental Procedures).
Supplementary Material
Highlights.
The CUL4A-RBX1-COPS8 complex ubiquitylates CENP-A K124 in vivo and vitro
CENP-A K124 ubiquitylation is essential to localize CENP-A to centromeres
Ubiquitin fusion restores centromere targeting and HJURP interaction of CENP-A K124R
CUL4A complex is required for loading and maintaining CENP-A at centromeres
ACKNOWLEDGMENTS
The authors thank Yue Xiong, Beezly Groh, Vivien Measday, Iain Cheeseman, Lars E.T. Jansen, Paul S. Maddox, Mitsuhiro Yanagida, Dorota Skowyra, Yoshinori Watanabe, Tatsuo Fukugawa, Akira Kikuchi, Hiroyuki Suzuki, and past and current researchers at The Research Institute at Nationwide Children's Hospital and St. Jude Children's Research Hospital for helpful discussion, experimental guidance, and reagents; Don W. Cleveland, Ben E. Black, William C. Earnshaw, William R. Brinkley, Yue Xiong, Yi Zhang, Hengbin Wang, Pradip Raychaudhuri, Tom K. Kerppola, Yue Xiong, and Stephan Diekmann for their generous gifts of reagents. This study was supported by NIH grants GM68418.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2015.01.024.
REFERENCES
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Crinelli R, Bianchi M, Cerasi A, Gentilini L, Serafini G, Magnani M. Efficient inhibition of macrophage TNF-alpha production upon targeted delivery of K48R ubiquitin. Br. J. Haematol. 1999;104:475–481. doi: 10.1046/j.1365-2141.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- Bade D, Pauleau AL, Wendler A, Erhardt S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell. 2014;28:508–519. doi: 10.1016/j.devcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA. 2013;110:11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENPA assembly. J. Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr., Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codomo CA, Furuyama T, Henikoff S. CENP-A octamers do not confer a reduction in nucleosome height by AFM. Nat. Struct. Mol. Biol. 2014;21:4–5. doi: 10.1038/nsmb.2743. [DOI] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, Cleveland DW. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Desai A. A PSHaver for centromeric histones. Mol. Cell. 2010;40:351–352. doi: 10.1016/j.molcel.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the protea-some regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte-Gattat D, Shuaib M, Ouararhni K, Gautier T, Skoufias DA, Hamiche A, Dimitrov S. Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function. Proc. Natl. Acad. Sci. USA. 2013;110:8579–8584. doi: 10.1073/pnas.1302955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI. Roles of Mis18a in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Yang Y, Jensen JP, Iwai K, Weissman AM. Current Protocols in Cell Biology. John Wiley & Sons; Somerset: 2006. Detection of E3 Activity in Immunoprecipitated Protein. pp. 15.19.25–15.19.26. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell. Mol. Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Ohta S, Vandenbeldt KJ, Abdulle R, McEwen BF, Kitagawa K. 17-AAG, an Hsp90 inhibitor, causes kinetochore defects: a novel mechanism by which 17-AAG inhibits cell proliferation. Oncogene. 2006;25:4133–4146. doi: 10.1038/sj.onc.1209461. [DOI] [PubMed] [Google Scholar]
- Niikura Y, Dixit A, Scott R, Perkins G, Kitagawa K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J. Cell Biol. 2007;178:283–296. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Ogi H, Kikuchi K, Kitagawa K. BUB3 that dissociates from BUB1 activates caspase-independent mitotic death (CIMD). Cell Death Differ. 2010;17:1011–1024. doi: 10.1038/cdd.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L, Sullivan KF. A GTPase switch maintains CENP-A at centromeric chromatin. Nat. Cell Biol. 2010;12:1128–1130. doi: 10.1038/ncb1210-1128. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel A, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromo-some segregation. Proc. Natl. Acad. Sci. USA. 2012;109:9029–9034. doi: 10.1073/pnas.1120968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr. Top. Microbiol. Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li X, Marshall JB, Zhong CX, Dawe RK. Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell. 2005;17:572–583. doi: 10.1105/tpc.104.028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.