Abstract
Tuberculosis, a pandemic disease is caused by Mycobacterium tuberculosis (Mtb). DNA polymerase III encoded by DnaE2 of Mtb is specifically required for its survival in vivo, and hence can be considered to be a potential drug target. Amino acid sequence analysis of the MtbDnaE2 and its human counterpart does not show any significant similarity. Therefore, a 3D model of the MtbDnaE2 was generated using Modeller 9v10 with the template structure of E. Coli DNA polymerase III alpha subunit (2HNH_A). The generated models were validated using a number of programmes such as RAMPAGE/PROCHECK, VERIFY_3D, and ProSA. MtbDnaE2 has few conserved residues and four conserved domains similar to that present in DNA polymerase III of E. coli. In silico screening was performed with bioactive anti-tuberculosis compounds and 6-AU (a known inhibitor of DNA polymerase III of Bacillus subtilis) and its analogues against the modeled MtbDnaE2 structure. Docking was performed using GOLD v5.2 software which resulted in the identification of top ten compounds with high GOLD fitness scores and binding affinity (X-Score). To further evaluate the efficacy of these compounds, in silico ADMET analysis was performed using MedChem Designer v3. Given their high binding affinity to the targeted MtbDnaE2, which is essential for DNA replication in the Mtb and good ADMET properties, these compounds are promising candidates for further evaluation and development as anti-tubercular agents.
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis (Mtb), is a leading cause of death globally. According to the World Health Organisation (WHO), almost 95% of reported cases occur in developing countries and 25% of affected adults die from the infection [1]. The recent emergence of multidrug resistant strains of M. tuberculosis has further complicated the challenge of curing tuberculosis [2, 3]. Therefore, understanding the molecular mechanisms of the Mtb infection can help in the development of new drugs that may be more effective than traditional therapies. Analysing the genome sequence of the Mtb and human allows one to identify unique enzymes/proteins that are present only in the pathogen’s metabolic pathway, and not in the host’s [4]. Such unique proteins exclusively present in the pathogen can thus be targeted as potential drug targets [5].
DNA polymerase III (DnaE2) is one such enzyme that barely shares any similarity with the proteins involved in the host’s DNA replication machinery. DnaE2 belongs to the Y family of error prone DNA polymerases that has been reported to be responsible for pathogen survival and drug resistance [6]. Hence, its inactivation would impede Mtb’s survival within the host [7, 8]. DNA polymerase III is strongly conserved in a broad group gram-positive pathogens such as Staphylococcus, Streptococcus, Enterococcus, and Mycoplasma [9], and has been considered to be a drug target [10]. Many deoxyribonucleotide analogues act as inhibitors or a substrates for DNA polymerase and can inhibit proliferation [11]. An analogue of dGTP, 6-anilino-1H-pyrimidine-2, 4-dione (6-AU) is one of the most common drugs that target DNA polymerase III of gram positive bacteria [12, 13].
In the present study, we have evaluated the therapeutic potential of a large number of compounds against the DNA polymerase III alpha subunit of Mtb (MtbDnaE2). The DNA polymerase III alpha subunit of M. tuberculosis (H 37 Rv) which was selected, did not show any significant sequence identity (BLASTP threshold e value < 0.005) with the human proteins. Since the crystal structure of the MtbDnaE2 has not been determined yet, comparative model of the MtbDnaE2 was generated using DNA polymerase III (2HNH_A) of E.coli (DNA polIIIα) as the template. The best models were validated by various structure verification programs. Its conserved residues and domains were analyzed in order to predict action mechanisms. MtbDnaE2 model was used for structure based drug designing. After docking, lead compounds that bind to MtbDnaE2 were identified as potential inhibitors of Mtb. We have also analyzed the efficacy of top compounds through Adsorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) studies.
Materials and Methods
Sequence analysis
The sequence of MtbDnaE2 (Accession Number NP_217887.2) was obtained from the NCBI database (www.ncbi.nlm.nih.gov) [14]. BlastP was used to identify the suitable template in the Protein Data Bank (PDB) for modeling of the target sequence. The identified top hits included replicative enzymes PDB entries 2HPI_A, 2HNH_A, 3HQA, 3EOD, and 4JOM_A [15–19]. The PDB entries 3HQA and 3EOD were not considered as crystal structures of these templates have only 73 and 130 amino acid (aa) residues respectively. Pairwise amino acid sequence alignment of the selected template and the target sequence was performed using ClustalW for identification of conserved residues in the target [20].
Comparative modeling and validation, and conserved domains prediction
Though the crystal structure of the E. coli DNApolIIIα (PDB entry 2HNH_A), retrieved from the protein data bank (http://www.rcsb.org/pdb/home/home.do) [21] does not have the C- terminal region (911–1160 aa), it was considered to be the best template with 33% sequence identity, based on its higher sequence coverage and resolution (2.30 Ǻ, R value = 0.190,R free 0.258) in comparison to that of other available structures (S1 Table). Models were generated using Modeller 9v10 software [22]. The final model of the MtbDnaE2 was visualized using PyMOL v1.7 viewer [23]. MtbDnaE2 models were validated using verification programs such as RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) [24]/ PROCHECK and VERIFY_3D available through NIH (http://nihserver.mbi.ucla.edu/SAVES) [25]. RAMPAGE/PROCHECK provide positional information of each amino acid residues between phi (ϕ) and psi (ψ) angles in a Ramachandran plot [26]. VERIFY_3D is a unique type of verification program that takes into account the location and environment of amino acids and verifies the compatibility of the model from 3D to 1D [27]. ProSA (Protein Structure Analysis) program that analyzes the interaction energy of each amino acid residue in the model, was used to confirm model stability and accuracy on the basis of energy and Z score [28]. Conserved domains were predicted using Pfam server (http://pfam.sanger.ac.uk/)[29].
In silico screening of anti-tuberculosis (bioactive) compounds and, 6-AU and its analogues against the MtbDnaE2
In silico screening of anti-tuberculosis (bioactive) compounds was performed against the MtbDnaE2 comparative modeled structure. Bioactive compounds (49413) were obtained from ChEMBL database [30]. A known inhibitor of DNA polymerase III of Bacillus subtilis, 6-AU, and its 20 analogues with 95% structural similarity were screened from PubChem Similarity Search [31]. Three dimensional (3D) conformations of all compounds were generated using the CORINA program v3.2 [32].
GOLD v5.2 (Genetic Optimization for Ligand Docking) molecular docking program was used with default parameters for in silico screening of all the above compounds against the modeled MtbDnaE2 structure to identify potential inhibitors as it performs flexible docking using genetic algorithm, thus allowing better binding of ligand at a specific site of receptor [33]. The binding pocket residues for MtbDnaE2 model were predicted using pocket finder (http://www.modeling.leeds.ac.uk/pocketfinder) [34]. The predicted residues were cross-checked with the template (2HNH_A). Protein preparation with binding pocket information and ligand library preparation were carried out using the docking wizard of GOLD program. A few independent parameters were applied for fitness function (hydrogen bond energies, atom radii and polariabilities, torsion potentials, etc.), taken from the GOLD parameter file. Best docked complexes were ranked based on their GOLD fitness score and non-bonded interaction analysis. Binding affinity was calculated using X-Score v1.2.1 [35]. Hydrogen bond contacts, lipophilic interactions and non-bonded contacts were calculated using LIGPLOT v.4.5.3 [36]
Evaluation of ADMET of top compounds
MedChem Designer v.3 was used to evaluate drug permeability (S+logp), distribution (S+logD), topological polar surface area (TPSA), Moriguchi octanol-water partition coefficient (MLogP), molecular weight (Mwt), and the total number of nitrogen and oxygen atoms[37] Such analysis for any molecule provides information about its adsorption, distribution, metabolism, excretion, and toxicity (ADMET). These features can be used to estimate drug efficacy and suitability.
Results and Discussion
Sequence analysis
The MtbDnaE2 (Acc No. NP_217887.3) was selected as a target because it shares very little sequence similarity with the human DNA polymerase alpha subunit (Swissprot ID P09884). PSI-BLAST analysis of MtbDnaE2 and human DNA polymerase alpha subunit showed very little sequence similarity (only 20% similarity spanning only 107 amino acid residues regions of the human (877–983) and MtbDnaE2 (371–469 aa). Pfam database showed no similar functional domains between the two. Therefore, the atomic architecture of the binding site of the targeted ligands with MtbDnaE2 is not likely to be present in human proteins, and thus the ligands are likely to be highly specific towards Mtb. The MtbDnaE2 was therefore modeled using a suitable template for molecular docking studies and to identify its inhibitors.
Amino acid sequence alignment of the MtbDnaE2 with E. coli DNApolIIIα showed that few amino acid residues involved in the catalytic reaction of E. coli DNApolIIIα [16] were also conserved in the MtbDnaE2 (S1 Fig.). The MtbDnaE2 sequence has conserved amino acids residues comprising P-D-X-D-X-D from 380–385, which are also correspondingly present (400–405 aa) in E. coli DNApolIIIα. Three acidic residues—D381, D383, and D437 of MtbDnaE2, aligned with E.coli DNApolIIIα sequences (D401, D403 and D457). The two aspartate residues (D401, D403) have been reported to be involved in phosphotransferase activity with two Mg2+ ions [38]. The third aspartate amino acid residue plays a major role in the nucleophilic reaction, during the interaction of incoming nucleotides [39]. As observed in E. coli DNApolIIIα (G363, S364, and K543), equivalent amino acid residues (G344, S345 and K509) were also highly conserved in MtbDnaE2. The residues G363 and S364 formed a binding pocket for the incoming nucleotide while K543 participated in salt bridge creation with the phosphate group of the terminal 3´ base [40]. Amino acid residues R342 and R370 from the palm domain (corresponding equivalent residues R362 and R390 in E. coli DNApolIIIα) and R666, R667 from the finger domain of MtbDnaE2 (equivalent to R709 and R710 in E. coli DNApolIIIα) generally interact with dATP [16]. Thus, the analysis revealed the presence of a number of conserved active residues in MtbDnaE2, as present in E. coli DNApolIIIα. Hence MtbDnaE2 can catalyze the reaction in a similar manner as reported for E. coli DNApolIIIα.
The amino acid sequences of the three templates (2HPI_A, 2HNH_A and 4JOM_A) showed similar identity (33%) with the MtbDnaE2. While amino acid sequences of all the three templates showed comparable sequence coverage (excluding the C-terminal region in 2HNH_A structure), 2HNH_A structure is solved at a better resolution (2.30 Ǻ, R value = 0.190, R free = 0.258) when compared to 2HPI_A (3.00 Ǻ, R value = 0.225, R free = 0.275) and 4JOM_A (2.9 Ǻ, R value = 0.197, R free = 0.245) and was therefore selected as the template for comparative modeling.
Comparative modeling of the MtbDnaE2
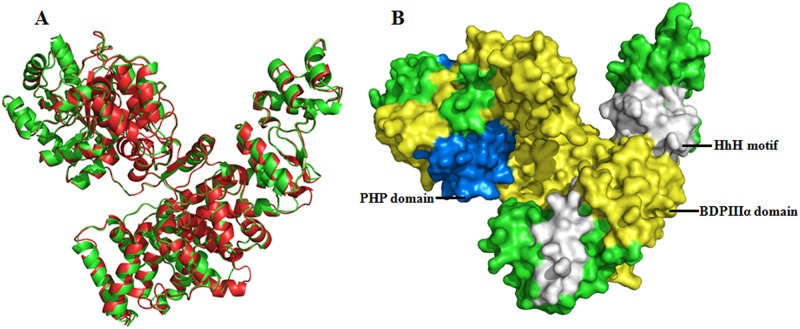
For homology modeling, template (2HNH_A) was selected on the basis of resolution,(R = 2.3Å), E-value (6e-134), Max Score (431), Query cover (84%), identity (33%), and positives (50%). The conserved domains analysis also revealed that the template (2HNH_A) has similar domains as in MtbDnaE2 except OB fold (due to the absence of C-terminal region encompassing 911–1160 aa). Based on these analyses, a comparative model with a DOPE score of -96276.38 was generated using the crystal structure of E coli DNApolIIIα (2HNH_A) as the template (Fig. 1A). A Ramachandran plot of the best MtbDnaE2 model generated using RAMPAGE showed 91.8% amino acid residues in favorable regions, 7% amino acid residues in allowed regions, and 1.3% amino acid residues in disallowed regions (S2 Fig.). Similar to the template, most of the amino acid residues in the model were arranged in favourable regions that comprised phi (ϕ) and psi (ψ) angles of Ramachandran plot, demonstrating that the modeled MtbDnaE2 structure was reasonably good. Verify-3D showed that 98% of the amino acid residues had an averaged 3D-1D score > 0.2 in the modeled MtbDnaE2 structure (S3A Fig.). Using ProSA, the Z-score of the modeled MtbDnaE2 was determined to be -13.9, which is comparable to that of the template E. coli DNApolIIIα (-16.19) (S3B Fig.). ProSA analysis revealed that most of the residues in the modeled MtbDnaE2 had negative interaction energy, and only a small number of residues displayed positive interaction energy (S3C Fig.). The modeled MtbDnaE2 structure was superposed onto the template (2HNH_A) and calculated RMSD is 1.40 Ǻ, indicating the model to be of good quality (Fig. 1A). These results clearly suggest that the comparative modeled structure of MtbDnaE2 is of good quality, and can be used for further studies in the absence of experimentally driven structure.
Fig 1. Superimposed Comparative Ribbon Model Structure and Conserved Domains of MtbDnaE2.
(A) Superimposed comparative model of MtbDnaE2 (green) on the 2HNH_A (red) template with RMSD value (1.4Å). This model was generated by Modeller9v10. The model’s ribbon structure was visualized by PyMoL Viewer (B) Connolly surface form of MtbDnaE2 model shows three out of the four conserved domains (identified by domain search)—PHP domain at N terminal, BDPIIIα domain, and HhH motif visualized by PyMOL viewer. The OB fold domain identified in the C-terminal region of MtbDnaE2 can not be seen, due the absence of C-terminal region in the template structure.
Domain analysis using Pfam database shows that the MtbDnaE2 sequence contains four structurally conserved domains—(i) Polymerase and histidinol phosphatases (PHP) domain at the N-terminal (1–166 aa), which exhibits pyrophosphatase activity that hydrolyzes the pyrophosphate by product of DNA synthesis[41], (ii) bacterial DNA polymerase III alpha subunit domain spanning aa 280–691, involved in pathogen replication system, (iii) Helix-hairpin-helix motif (HhH) located at aa767-856, which interacts with DNA with non-sequence specific manner, and (iv) Oligonucleotide/oligosaccharide binding fold domain (OB-fold) at the C-terminal (aa 934–1005), which interacts with the ssDNA [16]. Three of these domains are also present in the template structure (2HNH_A). The OB-fold domain being part of the C-terminal region is not present in the template as well as in the modeled structure (Fig. 1B).
In silico screening of anti-tuberculosis (bioactive) compounds on the modeled MtbDnaE2
To screen for potential inhibitor(s) of the MtbDnaE2, 49413 compounds (with reported anti-tuberculosis activity on the basis of cellular assays, not directed against the MtbDnaE2) were retrieved from the ChEMBL and docked against the modeled MtbDnaE2 structure. Active residues in the modeled structure identified by pocket finder were cross checked with the template (2HNH_A). The GOLD fitness score for the top ranking eight compounds ranged from 77.62 to 84.62 and the predicted binding energy ranged from -7.90 to -9.71 kcal/mol (calculated using the X-Score) (Table 1). These eight ligands have also been reported to be inhibitors of Mycobacterium tuberculosis by both in vitro and in vivo cell based assays [42–44] and not directed specifically against the MtbDnaE2. High GOLD fitness score and binding affinity to the modeled MtbDnaE2 of these compounds suggest that MtbDnaE2 could be the possible site of action of these compounds (Table 1).
Table 1. Molecular Docking Results and Interaction Analysis for Identified putative anti-mycobacterium Inhibitors against MtbDnaE2.
| S.No | ChEMBL/ Assay IDs | Compound Name | Hydrogen Bonds | Lipophilic Interactions | Non-bonded Interactions | GOLD Fitness Score | X-Score (kcal/mol) | IC50Ref |
|---|---|---|---|---|---|---|---|---|
| C1 | CHEMBL119236 | 1-[[ethoxy(tetradecyl)phosphoryl] oxymethyl]-3-phenoxybenzene | 1; Tyr 799 | 8; Glu 361,Arg 362, Arg 403, Ile 407, Cys 409, Gly 425, Ala 428, His 602 | 29 | 84.62 | -8.86 | 862.29μM42 |
| SID 103345338 | ||||||||
| AID 38730 | ||||||||
| C2 | CHEMBL326268 | 2-(1,3-dioxoisoindol-2-yl)ethoxy-heptylphosphinic acid | 4; Cys 402, Arg 403, Asn 405, Lys 605 | 9; Asp 406, Leu 408, Arg 412, Leu 598, Ser 599, His 602, Leu 618, Ser 792, Leu 796 | 21 | 81.13 | -8 | 1.47μM42 |
| SID 104036885 | ||||||||
| AID38730 | ||||||||
| C3 | CHEMBL325149 | 1-[[ethoxy(nonyl)phosphoryl]oxymethyl]-3-phenoxybenzene | NIL | 13; Arg 403, Ile 407, Leu 408, Cys 409, Arg 412, Thr 427, Ala 428, Val 429, Leu 598, His 602, Ser 792, Ser 795, Leu 796 | 30 | 80.22 | -9.12 | 14.83μM42 |
| SID 103346167 | ||||||||
| AID 38730 | ||||||||
| C4 | CHEMBL175147 | 1-cyclopropyl-6-fluoro-7-[4-[2-[[5-(5-methyl-2,4-dioxopyrimidin-1-yl)-2,5-dihydrofuran-2-yl]methoxy]-2-oxoethyl]piperazin-1-yl]-4-oxoquinoline-3-carboxylic acid | 7; Ile 407, Ser 599, His 602, Lys 605, Asp 606, Ser 795, Tyr 799 | 9; Cys 402, Asp 406, Gln 410, Arg 412, Ala 428, Tyr 603, Leu 618, Ser 792, Leu 796, | 38 | 79.27 | -9.7 | 6.25μg/ml43 |
| SID 103437119 | ||||||||
| AID 143439 | ||||||||
| C5 | CHEMBL332399 | 2-[[ethoxy(nonyl)phosphoryl]oxymethyl]isoindole-1,3-dione | 2; Arg 403, His 602 | 8; Asp 406, Ile 407, Leu 408, Arg 412, Leu 598, Ser 792, Leu 796, Tyr 799 | 27 | 78.93 | -8.54 | 87.09μM42 |
| SID 103345226 | ||||||||
| AID 38730 | ||||||||
| C6 | CHEMBL118269 | 2-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]isoindole-1,3-dione | 1; Tyr 799 | 11; Arg 403, Leu 408, Arg 412, Gly 425, Ala 428, Glu 457, Gln 460, Leu 598, His 602, Ala 619, Leu 796 | 31 | 77.87 | -8.52 | 40.990μM42 |
| SID 103345259 | ||||||||
| AID 38730 | ||||||||
| C7 | CHEMBL177774 | benzyl 2-(6-decylsulfanylpurin-9-yl)acetate | 2; His 602, Tyr 799 | 14; Arg 403, Asp 406, Ile 407, Leu 408, Cys 409, Arg 412, Ala 428, Val 429, Gly 594, Leu 595, Leu 598, Ser 792, Ser 795, Leu 796 | 34 | 77.74 | -8.59 | Not determined44 |
| SID 103438626 | ||||||||
| AID 143119 | ||||||||
| C8 | CHEMBL418868 | 2-(1,3-dioxoisoindol-2-yl)ethoxy-hexylphosphinic acid | 3; Arg 403, Ile 407, Lys 605 | 7; Cys 402, Asp 406, Leu 408, Arg 412, Leu 598, His 602, Leu 796 | 18 | 77.62 | -7.9 | 4.39μM42 |
| SID 103345288 | ||||||||
| AID 38730 |
ChemBL compound ID (CHEMBLID); PubChem Substance accession identifier (SID), Assay ID (AID) for the identified compounds namely C1-C8 are given.
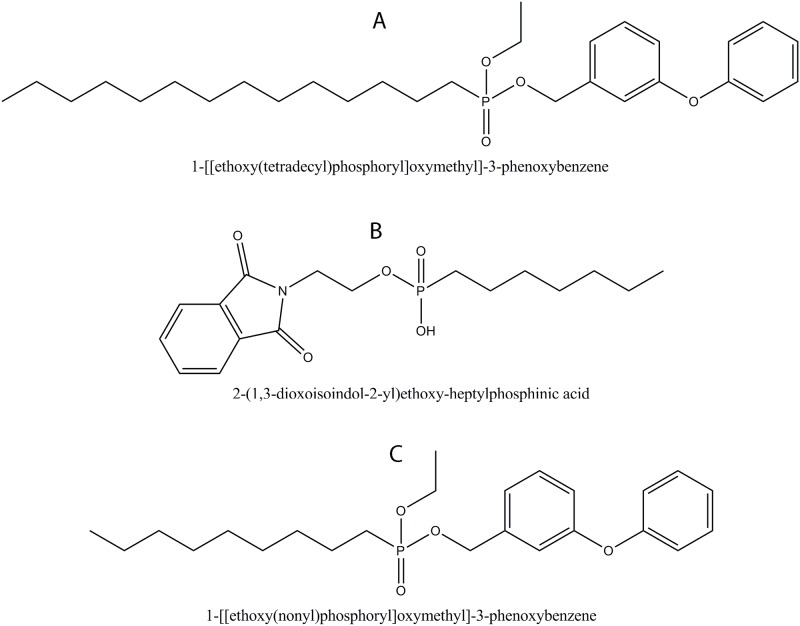
The top compounds (i) 1-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]-3-phenoxybenzene, (ii) 2-(1,3-dioxoisoindol-2-yl)ethoxy-heptylphosphinic acid and (iii) 1-[[ethoxy(nonyl)phosphoryl]oxymethyl]-3-phenoxybenzene, designated as C1, C2, and C3 are shown in Fig. 2A–2C, respectively.
Fig 2. Structure of Top Extracted Compounds by in silico Screening.
(A) 1-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C1),(B) 2-(1,3-dioxoisoindol-2-yl)ethoxy-heptylphosphinic acid (C2), (C) 1-[[ethoxy(nonyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C3).
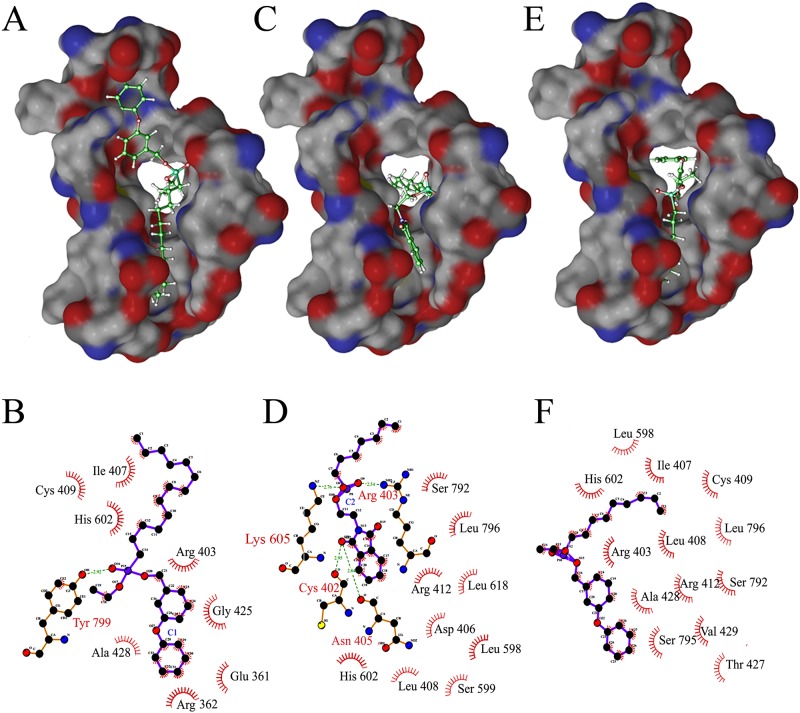
C1 and C2 only have 1 and 4 hydrogen bonding interactions with MtbDnaE2, respectively while C3 did not have any hydrogen bonding interaction with MtbDnaE2. C1 has 8 lipophilic interactions and 29 non-bonded interactions (Fig. 3A &B). Compounds C2 and C3 has 9 and 13 lipophilic interactions and 21and 30 non-bonded interactions, respectively (Fig. 3C-F). High lipophilic interactions of the C3 may accountable for better binding with MtbDnaE2 at -9.12 kcal/mol X-Score (Table 1). Predicted binding energy for C1 and C2 is -8.86 kcal/mol and -8.00 kcal/mol, respectively (Table 1). Residue His602 of MtbDnaE2 were common in top 3 lipophilic interactions, whereas Arg403, Ile 407, Cys409, and Ala428 were common in lipophilic interactions of C1 and C3 with MtbDnaE2.
Fig 3. Docked Complex of MtbDnaE2 with ChEMBL Database Compounds.
Panels A, C, and E show the electrostatic surface potential of modeled MtbDnaE2 with bound ligands (stick models, shown in atom colors). (A) 1-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C1), (C) 2-(1,3-dioxoisoindol-2-yl)ethoxy-heptylphosphinic acid (C2), (E) 1-[[ethoxy(nonyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C3) respectively. In ligplot, Panels B, D, and F show 2D representation of ligand C1, C2, and C3 with the interacting amino acid residues of MtbDnaE2. Green dashed lines indicate hydrogen bonds, with the numbers indicating the interatomic distances (Å). The labeled arcs with radial spokes that point toward the ligand atoms show the hydrophobic interactions with the corresponding amino acid residues.
Molecular docking of 6-AU and its analogues against MtbDnaE2 comparative model
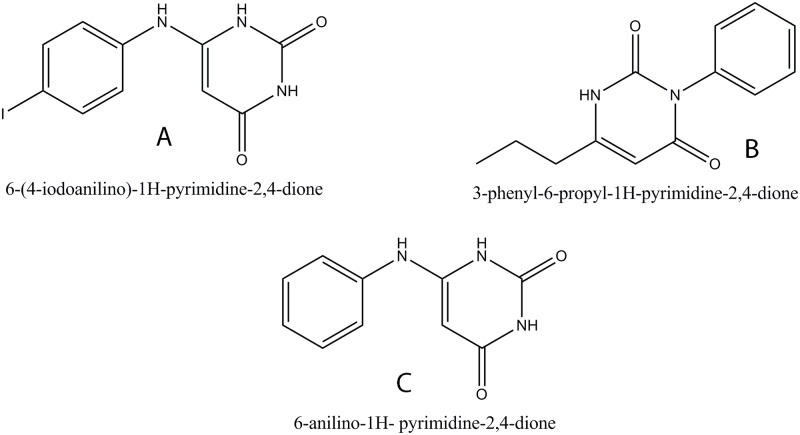
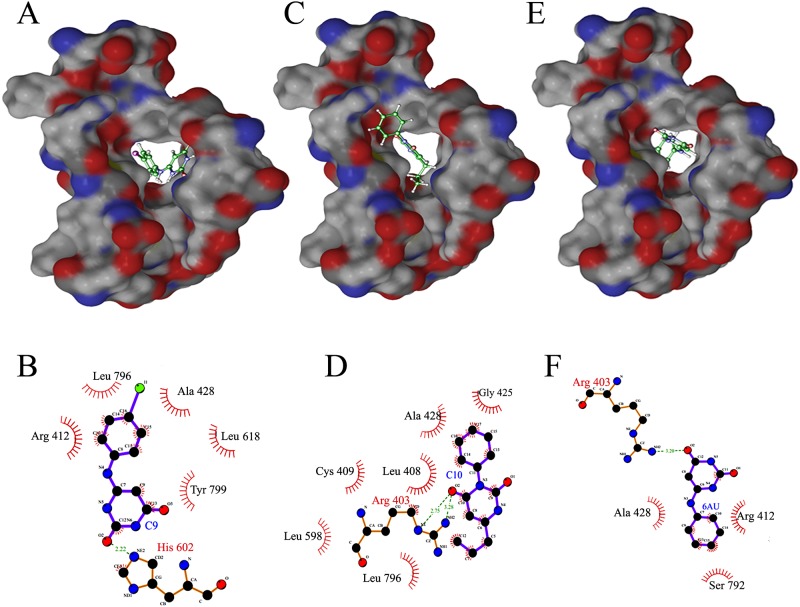
6-AU is a known inhibitor of the DNA polymerase III alpha subunit in a number of gram positive bacteria, including B. subtilis with reported IC50 of 4.7μM [45]. The enzyme interacts with 6-AU compounds through a guanine-like base pairing domain and an enzyme specific aryl domain. The action of these compounds is competitive with dGTP as they are able to form Watson- Crick like hydrogen bonds with an unopposed cytosine residue in the template strand just distal to the DNA primer terminus. The aryl group of these compounds binds near the enzyme’s active site, thus resulting in the formation of an inactive ternary complex [46]. However, 6-AU and its analogues have not been evaluated for their interaction with MtbDnaE2. Therefore, 6-AU and its analogues were identified by a structural similarity search using the PubChem database and were analyzed by docking on the modeled MtbDnaE2 structure. Two analogues of 6-AU—C9 (6-(4-iodoanilino)-1H-pyrimidine-2,4-dione) and C10 (3-phenyl-6-propyl-1H-pyrimidine-2,4-dione, selected on the basis of their GOLD fitness scores (35.81 to 41.56) and X-Score (-7.19 to -7.57 kcal/mol),were analyzed using 6-AU as a positive control (Fig. 4A-C, Table 2). No activity information is available for the identified 6-AU analogues, C9 and C10 [47, 48 ]. Docking studies revealed that the Arg 403 of the MtbDnaE2 was involved in hydrogen bonding with C10 and 6-AU whereas His 602 hydrogen bonded only with C9 (Table 2).
Fig 4. Structure of 6-AU and its Top Analogues.
(A) 6-(4-iodoanilino)-1H-pyrimidine-2,4-dione (C9), (B) 3-phenyl-6-propyl-1H-pyrimidine-2,4-dione (C10), (C) 6-anilino-1H- pyrimidine-2, 4-dione (6-AU) (control).
Table 2. Molecular Docking Results and Interaction Analysis for Identified 6-AU and its analogous against MtbDnaE2.
| S.No | Compound ID | Compound Name | Hydrogen Bonds | Lipophilic Interactions | Non-bonded Interactions | GOLD Fitness Score | X-Score (kcal/mol) | IC50Ref |
|---|---|---|---|---|---|---|---|---|
| C9 | CID 737913 SID 8052167 AID 372 | 6-(4-iodoanilino)-1H-pyrimidine-2,4-dione | 1; His 602 | 5; Arg 412, Ala 428, Leu 618, Leu 796, Tyr 799 | 11 | 41.56 | -7.57 | Anti-Mtb activity not reported47 |
| C10 | CID 22075432 SID 37716788 | 3-phenyl-6-propyl-1H-pyrimidine-2,4-dione | 1; Arg 403 | 6; Leu 408, Cys 409, Gly 425, Ala 428, Leu 598, Leu 796 | 20 | 40.05 | -7.44 | Anti-Mtb activity not reported48 |
| 6-AU (Control) | CID 309801 SID 103355885 AID 57364 | 6-anilino-1H- pyrimidine-2,4-dione | 1; Arg 403 | 3; Arg 412, Ala 428, Ser 792 | 09 | 35.81 | -7.19 | 4.7μM45 |
Compound ID (CID); PubChem Substance accession identifier (SID), Assay ID (AID) for the identified compounds are given.
The compounds C9 and C10 showed 5 and 6 lipophilic interactions while 6-AU showed 3 lipophilic interactions with the modeled MtbDnaE2 structure (Fig. 5). The compounds C9 and C10 showed 11 and 20 non-bonded interactions whereas the 6-AU showed only 9 non-bonded interactions with the modeled MtbDnaE2 structure (C9, Fig. 5A&B; C10, Fig. 5C &D and 6-AU, Fig. 5E &F). Residue Ala 428 residues of the MtbDnaE2 were commonly involved in lipophilic interactions with all three compounds, whereas Arg 412 showed lipophilic interaction with only C9 and 6-AU (Fig. 5B&F). Ser 792 showed lipophilic interaction only with the 6-AU (Fig. 5F, Table 2). Since these residues (G363, S364, and K543) are involved in nucleotide binding and in salt bridge formation with the phosphate group of the terminal 3′base [16], the selected compounds’ interaction with the equivalent residues in MtbDnaE2 is expected to inhibit its polymerization activity. The binding energy calculated using X-Score of C9 and C10 are -7.57 kcal/mol and -7.44 kcal/mol respectively, which were slightly lower than that determined for 6-AU (-7.19 kcal/mol) (Table 2). This interaction study suggested that two analogues of 6-AU, C9 and C10, showed better GOLD fitness score and binding energy than 6-AU, possibly due to the presence of different functional groups.
Fig 5. Docked Complex of MtbDnaE2 with 6-AU and its Analogues.
Panels A, C, and E show the electrostatic surface potential of modeled MtbDnaE2 with bound ligands (stick models, shown in atom colors). (A) 6-(4-iodoanilino)-1H-pyrimidine-2,4-dione (C9),(C) 3-phenyl-6-propyl-1H-pyrimidine-2,4-dione (C10), (E) 6-anilino-1H- pyrimidine-2,4-dione (control). Panels B, D, and F show 2D representations of ligand C9, C10, and 6-AU with interacting amino acid residues of MtbDnaE2 respectively. Green dashed lines indicate hydrogen bonds, with the numbers indicating the interatomic distances (Å). The labeled arcs with radial spokes that point toward the ligand atoms show the hydrophobic interactions with the corresponding amino acid residues.
ADMET analysis of the identified compounds
MedChem Designer was used for ADMET analysis of the identified compounds [37]. Generally, lipophilicity is the logarithm value of the partition coefficient P (logP) between octanol and water (buffer), which explains the partition of the unionized (neutral) form of the compound, whereas logD describes the total partition of both the ionized and the unionized forms of the compound. Compounds C1 and C3 identified from ChEMBL showed log p value more than 5 indicating their lipophilic proprieties, whereas compound C2, 6-AU and its analogues C9 and C10 showed low logP scores of 1.59, 0.155, 0.847 and 2.0, respectively, indicating their hydrophilic nature. When compounds are ionized at various pH, their logD value differs from that of logP. Acidic compounds show lower logD value [49]. Thus, similar logP and logD values for compounds C1 and C3 (Table 3) suggest that they exist in their unionized forms (Table 3). Compouinds C2, 6-AU, C9, and C10, were found to have lower logD values in comparision to their logP (Table 3), indicating their acidic nature. MlogP (Moriguchi octanol-water partition coefficient) is well known and is traditionally used in QSAR model structure analysis. It describes the lipophilicity of a compound, which indicates the penetration of the compound from aqueous solutions to lipid-rich zones. Moriguchi's logP (MLogP) of greater than 4.15 suggests that the compound would be poorly absorbed [50]. The calculated MLogP of compounds C1 and C3 were found to be 5.625 and 4.618, respectively suggesting that these compounds are likely to be poorly absorbed. On the other hand, compounds C2 6-AU and its analogues (C9 and C10) showed calculated MLogP value significantly less than 4.15, suggesting that these compounds would be easily absorbed.
Table 3. ADMET analysis of top interactive compounds with MtbDnaE2 comparative model.
| Compounds name | S+logP | S+logD | MlogP | TPSA | M_NO | MWt |
|---|---|---|---|---|---|---|
| 1-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C1) | 8.816 | 8.816 | 5.625 | 47.92 | 4 | 502.76 |
| 2-(1,3-dioxoisoindol-2-yl)ethoxy-heptylphosphinic acid (C2) | 1.589 | 1.498 | 2.304 | 93.39 | 6 | 365.45 |
| 1-[[ethoxy(nonyl)phosphoryl]oxymethyl]-3-phenoxybenzene (C3) | 6.574 | 6.574 | 4.618 | 47.92 | 4 | 432.62 |
| 1-cyclopropyl-6-fluoro-7-[4-[2-[[5-(5-methyl-2,4-dioxopyrimidin-1-yl)-2,5-dihydrofuran-2-yl]methoxy]-2-oxoethyl]piperazin-1-yl]-4-oxoquinoline-3-carboxylic acid (C4) | 0.199 | -0.421 | 0.657 | 164.83 | 13 | 617.76 |
| 2-[[ethoxy(nonyl)phosphoryl]oxymethyl]isoindole-1,3-dione (C5) | 3.671 | 3.665 | 3.408 | 82.39 | 6 | 407.53 |
| 2-[[ethoxy(tetradecyl)phosphoryl]oxymethyl]isoindole-1,3-dione (C6) | 5.904 | 5.896 | 4.484 | 82.39 | 6 | 477.66 |
| benzyl 2-(6-decylsulfanylpurin-9-yl)acetate (C7) | 4.792 | 3.881 | 3.464 | 68.79 | 6 | 456.73 |
| 2-(1,3-dioxoisoindol-2-yl)ethoxy-hexylphosphinic acid (C8) | 1.124 | 1.011 | 2.062 | 93.39 | 6 | 351.42 |
| 6-(4-iodoanilino)-1H-pyrimidine-2,4-dione (C9) | 0.847 | -0.038 | 1.793 | 76.55 | 5 | 341.19 |
| 3-phenyl-6-propyl-1H-pyrimidine-2,4-dione (C10) | 2.0 | 1.372 | 2.446 | 55.73 | 4 | 242.36 |
| 6-anilino-1H- pyrimidine-2,4-dione (6-AU, as positive control) | 0.155 | -0.819 | 1.22 | 76.55 | 5 | 215.29 |
S+LogP = Permeability, S+logD = Distribution, MLogP = Moriguchi octanol-water partition coefficient, TPSA = Topological Polar Surface Area, M_NO = total number of nitrogen and oxygen atoms (Ns and Os), Mwt = moleculer weight, C = Compound no.
Topological polar surface area (TPSA, an indicator of the H-bonding potential of the molecule on the receptor) analysis [51] showed that 6-AU and its analogue C9 had a TPSA score of 76.55, which is the second highest and similar to that of compound C2 (93.39) (Table 3). Greater numbers of nitrogen and oxygen atoms are responsible for the formation of more hydrogen bonds. The 6-AU C9, and C2 can form strong and greater numbers of H-bonds with the target due to the presence of a high number of nitrogen and oxygen atoms in these compounds (Table 3). Thus, based on the binding affinity and ADMET properties considered collectively, the present study suggests that compounds C2, 6-AU and its analogues (C9 and C10) were more hydrophilic due to more hydrogen bonding, a high TPSA score, and a logP of <5. The number of nitrogen and oxygen atoms was also high. According to Lipinski’s “rule of five’,” a compound with logP <5 can also be orally administered.
It is of interest to note that of the top three compounds C1,C2 and C3, compound C2 showing the best ADMET properties also has been reported to have minimum IC50 value (1.47 μM) against M. Tuberculosis in cell based assays. The other compounds, C1, and C3-C8 with good ADMET properties have been evaluated against Mtb, however show several fold higher IC50 values when compared to compound C2. Also, compounds C9 and C10 which have not yet been evaluated for anti-Mycobacterial activity, these compounds (C1, C3-C10) can be used for designing novel analogues which may show lower IC50 values and thus would be more effective.
Conclusions
MtbDnaE2 and human DNA polymerase α subunit do not show significant sequence similarity in their primary structure. MtbDnaE2 showed few conserved residues and four conserved domains which were also present in E.coli DNA polymerase III α subunit. Comparative modeling of the MtbDnaE2 was carried out with crystal structure of DNA polymerase III α subunit of E.coli (DNApolIIIα, 2HNH_A) as a template using Modeller 9v10. A good quality model was generated and was verified by RAMPAGE, SAVES and ProSA energy plot. In silico screening of anti-tuberculosis bioactive compounds (total 49413) resulted in the identification of potential novel inhibitors specific to the target MtbDnaE2. The study resulted in the identification of ten putative inhibitors, including two analogues of 6-anilino-1H-pyrimidine-2, 4-dione (6-AU). Docking interaction analysis identified a few common active residues in MtbDnaE2, also present in the template (2HNH_A), that participated in the biochemical reaction. Based on their strong binding to the MtbDnaE2 and ADMET properties, the shortlisted compounds are likely to inhibit the replicative activity of the MtbDnaE2 and can be evaluated as potential anti-tubercle molecules specific to this target.
Supporting Information
(DOC)
The boxed sequence represents the active residues and more conserved in MtbDnaE2 with respect to 2HNH_A template. The fully conserved residues are represented by (*), strong and weak conservation of amino acids is denoted by (:) and (.), respectively.
(TIF)
In plot, Glycine represents in cross form, proline in triangle form and other residue signify in square form, The most of the favorable and allowed residues cover -98.0 and 2.0% expected range in plot with high density.
(TIF)
A. VERIFY_3D profile (model compatibility from 3D to 1D form) for modeled MtbDnaE2. Scores over 0.2 indicate a high quality model. B &C show ProSA energy plots for the modeled MtbDnaE2 structure. ‘B’ and ‘C’ show overall model quality indicated by Z score and local (knowledge-based energy) quality plots, respectively
(TIF)
Acknowledgments
The authors express their sincere thanks to Dr. Devapriya Choudhury, School of Biotechnology, JNU, New Delhi for critical reading of the manuscript and valuable suggestions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Used infrastructure supported by the Department of Biotechnology (DBT), Government of India, New Delhi at the Centre of Excellence in Bioinformatics at SCIS and SBT, JNU, New Delhi and financial support provided to JNU, New Delhi, under the DBT-BUILDER programme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murray CJL, Styblo K, Rouillon A.Tuberculosis in developing countries: burden, intervention and cost. Bull Int Union Tuberc Lung. Dis.1990; 65: 6–24. [PubMed] [Google Scholar]
- 2. McKinney JD, Jacobs WRJ, Bloom BR. (Persisting problems in Tuberculosis In Emerging Infections (New York: Academic Press; ) 1998; pp: 51–146. [Google Scholar]
- 3. World Health Organisation. WHO⁄ IUATLD Global Project on anti-tuberculosis drug resistance surveillance, Anti-tuberculosis drug resistance in the world: Third global report. Geneva: WHO; 2004. [Google Scholar]
- 4. Cole ST, Brosch R, Parkhill J.Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 5. Anishetty S, Pulimi M, Pennathur G. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput Biol Chem. 2005; 5:368–78. [DOI] [PubMed] [Google Scholar]
- 6. Helena IMB, Michael BR, Clifton EB, Valerie M. DnaE2 Polymerase Contributes to In Vivo Survival and the Emergence of Drug Resistance in Mycobacterium tuberculosis. Cell.2003;113:183–193. [DOI] [PubMed] [Google Scholar]
- 7. Brown NC, Dudycz LW, Wright GE. Rational design of substrate analogues targeted to selectively inhibit replication-specific DNA polymerases. Drugs Exp Clin Re.1986;12:555–564. [PubMed] [Google Scholar]
- 8. Cozzarelli NR. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977; 46:641–668. [DOI] [PubMed] [Google Scholar]
- 9. Barnes MH, Leo C, Brown NC. DNA polymerase III of gram-positive eubacteria is a zinc metalloprotein conserving an essential finger-like domain. Biochemistry.1998;37:15254–15260. [DOI] [PubMed] [Google Scholar]
- 10. Brown NC, Wright GE. DNA polymerase III—A New Target for Antibiotic Development. Curr Opin Anti—Infect Invest Drugs. 1999; 1:45–48. [Google Scholar]
- 11. Wright GE, Brown NC. Deoxyribonucleotide analogs as inhibitors and substrates of DNA polymerases. Pharmacol Ther.1990; 47:447–497. [DOI] [PubMed] [Google Scholar]
- 12. Brown NC, Gambino JJ, Wright GE. Inhibitors of Bacillus subtilis DNA polymerase III 6-(Arylalkylamino) uracils and 6-anilinouracils. J Med Chem.1977;20:1186–1189. [DOI] [PubMed] [Google Scholar]
- 13. Tarantino PMJ, Zhi C, Gambino JJ, Wright GE, Brown NC. 6-Anilinouracil-based inhibitors of Bacillus subtilis DNA polymerase III: antipolymerase and antimicrobial structure-activity relationships based on substitution at uracil N3. J Med Chem.1999; 42: 2035–40. [DOI] [PubMed] [Google Scholar]
- 14. Bethesda MD. The NCBI Handbook [Internet]. 2nd edition National Center for Biotechnology Information; (US: ). 2013(www.ncbi.nlm.nih.gov, Accessed 2014 Oct 24). [Google Scholar]
- 15. Scott B, Richard AW, Thomas AS. The Structure of T. Aquaticus DNA polymerase III is Distinct from Eukaryotic Replicative DNA Polymerases. Cell. 2006;126: 893–904. [DOI] [PubMed] [Google Scholar]
- 16. Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell.2006;5:881–892. [DOI] [PubMed] [Google Scholar]
- 17. Cook WJ, Galakatos N, Boyar WC, Walter RL, Ealick SE. Structure of human desArg-C5a. Acta Crystallogr D Biol Crystallogr.2010; 66:190–197. 10.1107/S0907444909049051 [DOI] [PubMed] [Google Scholar]
- 18. Levchenko I, Grant RA, Sauer RT, Baker TA.The structure of RssB, a ClpX adaptor protein that regulates sigma S.2009. 10.2210/pdb3eod/pdb [DOI] [Google Scholar]
- 19. Barros T, Guenther J, Kelch B, Anaya J, Prabhakar A, O Donnell M, Kuriyan J, et al. A structural role for the PHP domain in E. coli DNA polymerase III. Bmc Struct.Biol. 2013; 13: 8–8. 10.1186/1472-6807-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson JD, Higgins DG, Gibson TJ. "CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice". Nucleic Acids Research. 1994;22: 4673–4680. 10.1093/nar/22.22.4673 (http://embnet.vital-it.ch/software/ClustalW.html, Accessed 2014 Nov 11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyer EF. "The first years of the Protein Data Bank". Protein Science (Cambridge University Press) 1997;7: 1591–1597. 10.1002/pro.5560060724 (http://www.rcsb.org/pdb/home/home.do, Accessed 2014 Nov 16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. David B, Andrej S. Protein Structure Prediction and Structural Genomics. Science. 2001;294: 93 [DOI] [PubMed] [Google Scholar]
- 23. Lill MA, Danielson ML. Computer-aided drug design platform using PyMOL. J Comput Aided MolDes.2011;1:13–9. [DOI] [PubMed] [Google Scholar]
- 24. Simon CL, Ian WD, Arendall BW, Paul I WD B, Michael WJ, Prisant GM, et al. Structure Validation by Cα Geometry: ϕ, ψ and Cβ Deviation PROTEINS: Structure, Function, and Genetics. 2003;50:437–450. [DOI] [PubMed] [Google Scholar]
- 25. Lüthy R, James U B, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature.1992; 356: 83–85. 10.1038/356083a0 (http://nihserver.mbi.ucla.edu/SAVES/, Accessed 2014 Nov 11). [DOI] [PubMed] [Google Scholar]
- 26. Ramachandran G N, Sasisekharan V. Advances in Protein Chemistry.1968;23:283 [DOI] [PubMed] [Google Scholar]
- 27. Eisenberg D, Lüthy R, Bowie JU. VERIFY_3D: assessment of protein models with three-dimensional profiles. Methods Enzymol.1997;277:396–404. [DOI] [PubMed] [Google Scholar]
- 28. Wiederstein Sippl. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 2007; 35:W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finn RD, Mistry J, Tate J, Coggill P, Heger A. The Pfam protein families database. Nucleic acids research. 2010;38: D211–22 (http://pfam.sanger.ac.uk/ Accessed 2014 Nov 5). 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrıcia Bento A, Gaulton A, Hersey A, J Bellis L, Chambers J. The ChEMBL bioactivity database:an update Nucleic Acids Research.2013;1–8. 10.1093/nar/gkt1031 (https://www.ebi.ac.uk/chembl/compound, Accessed 2014 Nov 20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolton E, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated Platform of Small Molecules and Biological Activities Chapter 12 IN Annual Reports in Computational Chemistry, 2008;Vol 4, Elsevier: Oxford, UK;, pp. 217–240.https://pubchem.ncbi.nlm.nih.gov/search Accessed 2014 Nov 4. [Google Scholar]
- 32. Schwab CH. Conformations and 3D pharmacophore searching. Drug Discovery Today: Technologies, 2010;Vol 7:e245–e253 (www.molecular-networks.com/online_demos/corina_demo, Accessed 2014 Nov 24). [DOI] [PubMed] [Google Scholar]
- 33. Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with adescription of desolvation. J Mol Biol.1995; 245:43–53. [DOI] [PubMed] [Google Scholar]
- 34. An Ji, Totrov M, Abagyan R. Pocketome via Comprehensive Identification and Classification of Ligand Binding Envelopes. Molecular & Cellular Proteomics. 2005; 4.6:752–761. (http://www.modeling.leeds.ac.uk/pocketfinder,Accessed 2014 June 10). [DOI] [PubMed] [Google Scholar]
- 35. Renxiao W, Luhua L, Shaomeng W. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design. 2002;16: 11–26. [DOI] [PubMed] [Google Scholar]
- 36. Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995; 2: 127–34. [DOI] [PubMed] [Google Scholar]
- 37. Suenderhauf C, Tuffin G, Lorentsen H, Grimm HP, Flament C. Pharmacokinetics of Paracetamol in Göttingen Minipigs: In Vivo Studies and Modeling to Elucidate Physiological Determinants of Absorption. Pharm Res.2014. (http://www.simulations-plus.com/, Accessed 2014 Nov 24). [DOI] [PubMed] [Google Scholar]
- 38. Steitz TA. A mechanism for all polymerases. Nature.1998; 391: 231–232. [DOI] [PubMed] [Google Scholar]
- 39. Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol.1998;1:54–63. [DOI] [PubMed] [Google Scholar]
- 40. Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264: 1930–1935. [DOI] [PubMed] [Google Scholar]
- 41. Aravind L, Koonin EV. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Research, 1998b;Vol. 26:3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanislav G, Ivan P, Janez M, Rosalind AW, Gurdyal SB, Danijel K. Phosphonate inhibitors of antigen 85C, a crucial enzyme involved in the biosynthesis of the Mycobacterium tuberculosis cell wall. Bioorganic & Medicinal Chemistry Letters.2004; 14: 3559–3562. [DOI] [PubMed] [Google Scholar]
- 43. Dharmarajan S, Perumal Y, Narasimharaghavan S, Tanushree R. Synthesis of stavudine amino acid ester prodrugs with broad-spectrum chemotherapeutic properties for the effective treatment of HIV/AIDS. Bioorganic & Medicinal Chemistry Letters.2004;14:1085–1087. [DOI] [PubMed] [Google Scholar]
- 44. Ashish KP, Vibha P, Lainne ES, William JS, and Robert CR. Antimycobacterial Agents. 1. Thio Analogues of Purine. J. Med. Chem 2004;47:273–276. [DOI] [PubMed] [Google Scholar]
- 45. Wright GE, Gambino JJ. Quantitative structure-activity relationships of 6-anilinouracils as inhibitors of Bacillus subtilis DNA polymerase III. J Med Chem.1984;2:181–5. [DOI] [PubMed] [Google Scholar]
- 46. Clements J E, D’Ambrosio J, Brown NC. Inhibition of Bacillus subtilis deoxyribonucleic acid polymerase III by phenylhydrazinopyrimidines. Demonstration of a drug-induced deoxyribonucleicacid-enzyme complex. J. Biol. Chem.1975;250:522–526. [PubMed] [Google Scholar]
- 47. Michael AP, Kyung-Lyum M, Scott RB, Stuart FJ L ZG, John A et al. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Analytical Biochemistry. 2003;322: 33–39. [DOI] [PubMed] [Google Scholar]
- 48.Espacenet Patent search Bibliographic data: EP0589201(A1).1994:03–30.
- 49. Tarun G, Amit K, Bhaskara RJ, Xiaoling L. In silico model of drug permeability across sublingual mucosa. Archives of Oral Biolog.2013;58: 545–551. 10.1016/j.archoralbio.2012.09.020 [DOI] [PubMed] [Google Scholar]
- 50. Todeschini R, Consonni V. Handbook of Molecular Descriptors; Mannhold, Kubinyi H, Timmerman H, Eds.Wiley-VCH: Weinheim, New York, 2000. [Google Scholar]
- 51. David EC. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena.1Prediction of intestinal absorption. J Pharm Sci. 1999;8:807–814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
The boxed sequence represents the active residues and more conserved in MtbDnaE2 with respect to 2HNH_A template. The fully conserved residues are represented by (*), strong and weak conservation of amino acids is denoted by (:) and (.), respectively.
(TIF)
In plot, Glycine represents in cross form, proline in triangle form and other residue signify in square form, The most of the favorable and allowed residues cover -98.0 and 2.0% expected range in plot with high density.
(TIF)
A. VERIFY_3D profile (model compatibility from 3D to 1D form) for modeled MtbDnaE2. Scores over 0.2 indicate a high quality model. B &C show ProSA energy plots for the modeled MtbDnaE2 structure. ‘B’ and ‘C’ show overall model quality indicated by Z score and local (knowledge-based energy) quality plots, respectively
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.