Abstract
Acute liver failure is a severe, but rare, outcome of hepatitis A virus infection. Unusual presentations of prevalent infections have often been attributed to pathogen-specific immune deficits that exhibit Mendelian inheritance. Genome-wide resequencing of unrelated cases has proven to be a powerful approach for identifying highly penetrant risk alleles that underlie such syndromes. Rare mutations likely to affect protein expression or function can be identified from sequence data, and their association with a similarly rare phenotype rests on their existence in multiple affected individuals. A rare or novel sequence variant that is enriched to a significant degree in a genetically diverse cohort suggests a candidate susceptibility allele. Whole genome sequencing of ten individuals from ethnically diverse backgrounds with HAV-associated acute liver failure was performed. A set of rational filtering criteria was used to identify genetic variants that are rare in the population, but enriched in this cohort. Single nucleotide polymorphisms, insertions, and deletions were considered and autosomal dominant, autosomal recessive, and polygenic models were applied. Analysis of the protein-coding exome identified no single gene with putatively deleterious mutations shared by multiple individuals, arguing against a simple Mendelian model of inheritance. A number of rare variants were significantly enriched in this cohort, consistent with a complex and genetically heterogeneous trait. Several of the variants identified in this genome-wide study lie within genes important to hepatic pathophysiology and are candidate susceptibility alleles for hepatitis A virus infection.
Keywords: acute liver failure, genome-wide, hepatitis A, host genetics, immunity, viral hepatitis
Introduction
Hepatitis A virus (HAV) is a significant cause of liver-related morbidity and mortality with an annual incidence of up to 1.4 million cases worldwide [Anon 2013]. The clinical spectrum of HAV disease is broad, ranging from asymptomatic seroconversion to acute liver failure and death. Approximately one third of infections are clinically inapparent, and 30–50% of those with symptoms require hospitalization [Yang et al. 1988]. Severe infection leading to acute liver failure is extremely rare, affecting <2% of hospitalized patients and 0.015–0.3% of individuals in large common-source epidemics [Cooksley 2000; Taylor et al. 2006].
Risk factors for HAV acute liver failure remain poorly defined. Disease severity is highly correlated with the degree of hepatocellular necrosis in histological specimens. Extreme presentations could therefore result from either over-exuberant immune responses or loss of initial virologic control, leading to greater numbers of infected hepatocytes. The distribution of clinical outcomes in common-source epidemics suggests that the genotype and virulence of the infecting strain play only a minor role, and most infections in the United States are due to genotype IA [Ajmera et al. 2011]. While advanced age, chronic hepatitis B infection, and preexisting liver disease are correlated with severe disease, many patients with acute liver failure are young and without significant comorbidities [Willner et al. 1998; Cooksley 2000]. Genetic polymorphisms in tumor necrosis factor alpha and beta loci have been associated with fulminant hepatitis, and familial clusters of acute liver failure suggest that host genetic factors are a significant contributor to the clinical spectrum of HAV disease [Tsuchiya et al. 2004; Yalniz et al. 2005; Ajmera et al. 2011]. Indeed, a case control study of individuals with HAV associated acute liver failure identified a 6 amino acid insertion within the virus’ cellular receptor, TIM1/HAVCR1, as a susceptibility allele [Kim et al. 2011].
Recent work indicates that severe outcomes of prevalent infections are often due to Mendelian deficits in immunity [Alcaïs et al. 2009]. Risk alleles have typically been identified in children and often in the setting of consanguinity. Healthy individuals are known to carry many loss-of-function mutations, and the role of these rare, high impact variants on more common infections in adult populations has not been extensively explored [Tennessen et al. 2012]. Genome-wide resequencing of unrelated cases with extreme clinical presentations can be an effective method for identifying these alleles [Ng et al. 2010]. Rare mutations likely to affect protein expression or function can be identified from sequence data, and their association with a similarly rare phenotype rests on their existence in multiple affected individuals. A rare or novel sequence variant that is enriched to a significant degree in a genetically diverse cohort suggests a candidate susceptibility allele. In this study, this approach was used to discover genetic risk factors for severe outcomes of HAV infection.
Materials and methods
Patients and Samples
Individuals with HAV acute liver failure were identified through the Acute Liver Failure Study Group (ALFSG), a multicenter research collaboration. The ALFSG database consists of over 2000 adult patients meeting pre-determined consensus criteria for acute liver failure, defined as jaundice or illness <26 weeks prior to admission and mental status changes with coagulopathy (INR >1.5), without known chronic liver disease. HAV infection was confirmed serologically by the clinical laboratory at each study site at the time of enrollment (e.g. anti-HAV IgM positivity) and deemed by the treating medical team to be the primary cause of either acute liver injury or acute liver failure in 39 patients [Taylor et al. 2006]. The ALFSG provided genomic DNA for 8 subjects. Genomic DNA was also provided by three additional subjects initially enrolled at the University of California, San Francisco (UCSF) site. Informed, written consent was provided by all subjects or their next of kin. The ALFSG study protocols were reviewed and approved by the Institutional Review Board (IRB) at each of the participating sites, and this study was approved by the IRBs of the UCSF and the University of Michigan. This study was performed according to the World Medical Association Declaration of Helsinki http://www.wma.net/e/policy/b3.htm.
Sequencing and Variant Identification
Genomic DNA libraries were prepared and sequenced on the Illumina HiSeq platform using the manufacturer’s reagents and protocols. Sequence reads were aligned to the GRCh37 reference human genome sequence using the Burrows-Wheeler Alignment Tool (BWA) paired end alignment algorithm (BWT-SW). Picard and SAMtools were used to convert the SAM alignment files into BAM format, generate BAM indices, mark PCR duplicates, and merge and divide sample-level BAM files into a multisample, study-level BAM for each chromosome. For each multi-sample, chromosomal BAM, local realignment around InDels and base quality score recalibration was performed with the Genome Analysis Toolkit (GATK) version v2.2-16 as described in the Broad Institute’s “Best Practice Variant Detection with the GATK v4” guidelines. Multi-sample variant calling was performed using the GATK UnifiedGenotyper. Variant call files (VCFs) were annotated with snpEff using the GRCh37.64 ENSEMBL annotation database. The GATK Variant Quality Score Recalibration feature was used to identify false-positive variant calls, targeting quality control thresholds that excluded fewer than 1% and 5% of known, high confidence SNPs (HapMap3) and InDels (Mills Devine dataset), respectively. To identify stereotypical sequence errors and mis-calls, whole genome sequences from three Center for the Study of Human Polymorphism (CEPH) individuals (provided by Illumina) were analyzed using an identical pipeline.
Filtering and Secondary Analysis
VCFs were imported into a MySQL database pre-populated with tables containing reference genomic data from both public and in-house sources. This database included: estimates of allele frequency in various populations (derived from 1000 Genomes data, dbSNP, NHLBI exomes, Complete Genomics Public Genomes, in-house exomes, and three control genomes provided by Illumina), lists of known pseudogenes and immune genes, coordinates of repetitive and redundant regions of the genome, and published lists of genes with high loss-of-function rates. Further details on the reference genomic data used and their sources are provided as Supplementary Methods. Variant calls were linked to these reference tables by chromosome, position number, reference allele, and alternate allele. Candidate genetic variants were identified by applying a series of rational filters to this linked dataset based on estimates of population allele frequency, predicted biologic effect, genomic context, and various modes of inheritance.
The appropriate allele frequency thresholds for autosomal recessive and autosomal dominant models of inheritance were calculated based on Hardy-Weinberg equilibrium with a conservatively estimated trait prevalence of < 2%. Unless otherwise specified, predicted biologic effect was determined in a conservative manner with consideration of all variants likely to affect the translated polypeptide (e.g. non-synonymous SNP, InDel, splice site variant, or start/stop gain/loss). Only autosomal variants sequenced to an average depth of ≥10 reads and passing the VQSR threshold were considered. Single nucleotide polymorphisms lying within 3 base pairs of a repetitive region or segmental duplication were excluded, as were InDels that extended to within 10 base pairs of such features. Pseudogenes, genes reported to have high predicted loss of function rates in the general population, and genes known to commonly exist as false positives in next generation sequencing datasets were excluded. Variants passing these filters were then ranked by the proportion of individuals harboring the variant. For noncoding variants, putative regulatory function was based on an ENCODE Project RegulomeDB score ≥5 and evolutionary conservation was based on genome evolutionary rate profiling (GERP) score. Genetic models applied (i.e. autosomal recessive, single gene, single variant vs. autosomal dominant, multiple genes, multiple variants) are described in the results below and further details of each database query are provided as Supplementary Methods.
Results
Thirty-nine individuals with HAV acute liver failure were identified from a multicenter registry and genomic DNA was obtained from eleven. Interim sequencing results identified one sample with an abnormally high number of heterozygous genotype calls, suggesting either contamination or chimerism. Further examination of study records revealed that, because of clinical circumstances, DNA for this subject had been collected post-liver transplant. This sample was excluded, and the characteristics of the final sequenced cohort are described in Table 1. All ten individuals were anti-HAV IgM positive, and nine met criteria for acute liver failure. Case 2 was not encephalopathic at admission, and therefore, did not meet strict criteria for acute liver failure. Data from this individual were included in the analysis as the severity of his disease was similar to the other cases (see Supplementary Table 1). With the exception of case 8, who was also infected with HIV, none had significant medical comorbidity. Four of the individuals were infected with HAV genotype IA [Ajmera et al. 2011]. While viral genotype information was not available on the remaining individuals, previous work from this cohort suggests that nearly all cases of acute liver failure at the participating centers are due to genotype IA [Ajmera et al. 2011]. The race/ethnicity of the cohort was diverse with five non-Latino of European descent, three Asians, and one Latino. Notably, the cohort included a pair of first cousins (Cases 1 and 2) who presented in close succession.
Table 1.
Clinical characteristics of sequenced cohort
| Case 1a | Case 2a | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 26 | 20 | 60 | 51 | 40 | 57 | 30 | 41 | 21 | 47 |
| Gender | M | M | F | F | M | M | F | M | F | M |
| Race, ethnicity | Asian | Asian | White | White | Latino | White | White | White | Asian | White |
| Days from onset to enrollment | 6 | 8 | 24 | 7 | 9 | 8 | 50 | 13 | 9 | 11 |
| History of liver disease | no | no | no | no | no | no | no | no | no | no |
| Anti-HAV IgM | + | + | + | + | + | + | + | + | + | + |
| INR > 1.5 | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Altered mental status | yes | no | yes | yes | yes | yes | yes | yes | yes | yes |
| Peak encephalopathy grade | III | 0b | IV | IV | IV | IV | I | II | III | III |
| Outcome | Transplant | Spontaneous survival | Transplant | Spontaneous survival | Spontaneous survival | Transplant | Transplant | Transplant | Spontaneous survival | Transplant |
nd = no data
first cousins
not encephalopathic (acute liver injury)
One hundred thirty-three gigabases of sequence were generated per individual (Table 2). On average, 95.5% of reference bases were covered at a read-depth of 10x or greater, with a mean coverage of 93.9% across RefSeq exons. In total, approximately three million SNPs (17,000–18,000 in coding regions) were identified per individual, 1–2% of which were novel. Between 600,000 and 700,000 InDels were observed per individual (300–500 in coding regions), 5–12% of which were novel. The observed transition to transversion ratio was 3.29–3.37 across coding regions and 2.12–2.13 across the genome as a whole. Concordance between sequence variant calls and SNP genotype calls was 99.94–99.97% in the seven samples for which array-based genotypes were available. These values are similar to those reported by others using comparable pipelines [Pelak et al. 2010].
Table 2.
Summary statistics on whole genome sequence data
|
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | |||
|
| ||||||||||||
| Coverage Statistics | All Sites | Raw Yield (Gb) | 142.3 | 133.6 | 127.0 | 136.3 | 136.5 | 127.5 | 128.2 | 132.8 | 117.7 | 146.1 |
| Aligned Reads (Gb) | 123.5 | 115.9 | 111.6 | 120.2 | 120.3 | 112.0 | 111.8 | 115.7 | 103.2 | 129.4 | ||
| Coverage ≥10x | 95.4% | 95.2% | 95.7% | 96.0% | 95.1% | 94.9% | 95.7% | 95.3% | 96.0% | 96.1% | ||
|
| ||||||||||||
| Codinga | Coverage ≥10x | 93.3% | 92.8% | 92.8% | 93.8% | 92.4% | 92.3% | 93.0% | 94.4% | 96.9% | 97.0% | |
|
| ||||||||||||
| SNP Statistics | All Sites | SNPs | 3,188,561 | 3,181,072 | 3,151,179 | 3,158,296 | 3,172,525 | 3,113,583 | 3,148,716 | 3,106,186 | 3,199,635 | 3,092,950 |
| % Novelb | 2.49% | 2.44% | 0.90% | 1.06% | 0.98% | 0.97% | 0.97% | 0.88% | 1.96% | 0.86% | ||
| Ti/Tv | 2.12 | 2.12 | 2.12 | 2.12 | 2.12 | 2.12 | 2.12 | 2.13 | 2.12 | 2.13 | ||
|
| ||||||||||||
| Codinga | SNPs | 19,019 | 18,922 | 18,490 | 18,839 | 18,873 | 18,397 | 18,616 | 18,665 | 18,748 | 18,399 | |
| % Novelb | 2.73% | 2.71% | 0.89% | 1.05% | 1.25% | 0.88% | 0.95% | 0.81% | 2.09% | 0.71% | ||
| Ti/Tv | 3.34 | 3.33 | 3.35 | 3.36 | 3.33 | 3.29 | 3.37 | 3.31 | 3.3 | 3.37 | ||
|
| ||||||||||||
| InDel Statistics | All Sites | InDels | 705,851 | 703,504 | 701,111 | 705,163 | 705,434 | 696,876 | 701,599 | 689,105 | 689,441 | 672,362 |
| % Novelb | 8.04% | 7.90% | 7.50% | 7.62% | 7.50% | 7.51% | 7.43% | 7.37% | 7.43% | 7.03% | ||
|
| ||||||||||||
| Codinga | InDels | 411 | 395 | 398 | 393 | 392 | 390 | 384 | 420 | 466 | 453 | |
| % Novelb | 7.79% | 9.37% | 5.28% | 4.83% | 6.89% | 4.36% | 4.69% | 7.38% | 11.80% | 8.39% | ||
RefSeq
dbSNP 137
Filtering by allele frequency and functional impact proved an efficient approach to identifying candidate variants. By looking for variant loci shared among these unrelated and ethnically diverse individuals, a subset of rare, potentially deleterious polymorphisms were prioritized from a total of eight million variants. Autosomal recessive and autosomal dominant models were considered separately, with the allele frequency cut-offs for each based on an estimated trait frequency of 2% and Hardy-Weinberg equilibrium [Cooksley 2000; Taylor et al. 2006]. The filtering parameters are detailed in Supplementary Methods, and the complete, annotated outputs for each model are provided in Supplementary Table 2.
Under a recessive model, no single coding polymorphism meeting all filtering criteria was identified in more than one affected individual (Table 3). The racial and ethnic diversity of the cohort increased the likelihood that individuals might harbor distinct risk alleles in a common disease gene. Therefore, in a second model, variants were grouped by transcript and their combined effects examined at the gene level. This analysis also identified no rare mutations at the gene level that were shared among a majority of cases. Finally, a polygenic model for HAV acute liver failure was considered, with individual risk alleles existing in a limited number of disease genes that are not often shared among individuals. Using this model, four candidate genes were identified that had a single, homozygous non-synonymous SNP in one individual each: natural killer cell cytotoxicity receptor 3 ligand 1 (NCR3LG1), reelin (RELN), syntaxin binding protein 1 (STXBP1), and tetratricopeptide repeat domain 40 (TTC40).
Table 3.
Candidate genes by genetic model
| Model Type a | Autosomal Recessive b | Autosomal Dominant b |
|---|---|---|
| Single, Coding Variant | none | none |
| Single Gene, Multiple Coding Variants | none | none |
| Multiple Genes, Multiple Coding Variants | NCR3LG1 RELN STXBP1 TTC40 |
GPC1 MST1R DNAH12 NFATC4 |
| Single, Noncoding Variant | 45 variants near 25 genes | 735 variants near 551 genes |
| Regulatory or evolutionary conserved region | 13 variants near 7 genes | 358 variants near 291 genes |
| (Regulatory OR conserved) AND near immune, inflammatory, apoptotic, or hepatitis gene | none | 13 variants near 12 genes |
See Supplementary Methods for details of filtering criteria
See Supplementary Table 2 for complete, annotated lists of variants by model. Data are for SNPs not found in 3 control CEPH genomes. The total number of SNPs identified is reported in the text.
A parallel series of filters were applied to look at dominant effects within the protein-coding exome. No single coding variant was observed in more than 20% of cases (Table 3). Grouping variants by gene, no candidate gene affecting more than 40% of cases was identified. Because of the large number of heterozygous variants shared among individuals, particularly the first-cousin pair, an additional filter was imposed when considering polygenic as opposed to single-gene effects. This filter required that at least two unrelated subjects were affected and that at least one contributing variant was observed in multiple individuals. Four candidate genes met these criteria. Four individuals were heterozygous for one of three rare missense mutations in glypican 1 (GPC1). Three individuals were heterozygous for one of two rare missense mutations in macrophage stimulating 1 receptor (MST1R). A similar distribution was observed in dynein axonemal heavy chain 12 (DNAH12). Finally, two unrelated individuals were heterozygous for a 9-base pair insertion in nuclear factor of activated T-cells, calcineurin-dependent 4 (NFATC4). None of these alleles were identified in dbSNP137, the 1000 genomes project integrated phase 1 release, or the 54 unrelated genomes in the Complete Genomics public database. Screenshots of the primary sequencing data supporting the variant calls at each of the coding loci are shown in Supplementary Figure 1.
A six amino acid insertion within TIM1/HAVCR, the gene encoding the HAV receptor, has been previously identified as a risk factor for acute liver failure in a case control study [Kim et al. 2011]. This insertion, 157insMTTTVP, is now annotated as the reference allele in build 37 of the human genome. None of our ten cases were homozygous for this risk allele, and six individuals were heterozygous.
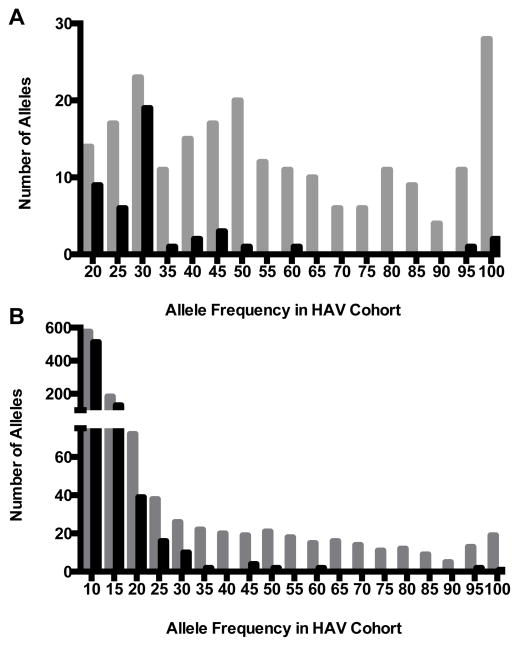
A large number of rare noncoding variants were found within our cohort (Table 2). Because of the large number of variants, a more stringent acute liver failure trait frequency threshold of 0.5% was used in the analysis of these alleles. This adjustment was required to achieve a more actionable list of candidate variants and the selected frequency estimate remains within the range supported by epidemiologic studies. Only SNPs were considered in analyses of noncoding variants, because of pipeline-specific biases related to accurate InDel identification. Despite the use of more restrictive filtering thresholds and in contrast to the results for protein-coding sequences described above, a large number of rare, noncoding variants were shared among subjects. Under a recessive model, 225 SNPs were identified that were shared between two or more individuals in the cohort, 68 of which existed in a homozygous state in more than half of the cases. In an autosomal dominant model, 1124 candidate variants met initial filtering criteria (Figure 1).
Figure 1.
Noncoding variants shared among individuals by mode of inheritance. Shown are number of single nucleotide variants present at each allele frequency assuming either an autosomal recessive (top) or autosomal dominant (bottom) model of inheritance in each of the ten individuals sequenced. Grey bars indicate the number of variants identified using basic frequency cutoffs described in text and Supplementary Methods. Black bars indicate the number of variants remaining after filtering out those present in any of the three control genomes.
The number of SNPs shared by these unrelated individuals was much larger than expected, even for noncoding regions of the genome. Because intergenic sequences of the reference genome can be more stereotyped and have been less well-characterized, current technologies are more prone to systematic errors in alignment and annotation in these regions. Such errors tend to be specific to the particular combination of sequencing platform, reference genome, alignment, and variant calling tools used. To identify systematic sequencing errors in the variant call set, data from three individuals in the CEPH cohort were used. The genomes of these individuals were sequenced in the same laboratory and analyzed in an identical manner.
When presumed “false-positive” susceptibility variants identified among these three randomly selected control genomes were removed, a much smaller number of noncoding SNPs were shared among individuals in our cohort (Figure 1). As anticipated, variant calls from coding regions of the genome were essentially unaffected and none of the candidate alleles identified in coding analyses were observed in the six CEPH chromosomes (Supplementary Table 2). After this final filtering step, 45 noncoding SNPs met basic criteria under an autosomal recessive model, 3 of which (TFCP2, LRP1B, and KRT13) existed in a homozygous state in more than half of the cases (Table 3). In an autosomal dominant model, 735 single nucleotide variants were identified that were shared between two individuals in the cohort. As expected, there was a significant decrease in the number of alleles shared by more than two individuals with just 23 alleles present at a minor allele frequency of 30%.
To further explore those most likely to have biologic effects, the resultant candidate list was compared with reference databases annotated for regulatory elements, evolutionary conservation, and gene function (see Supplementary Methods). Of the 45 noncoding SNPs found in the autosomal recessive analysis, 13 were in regulatory or evolutionarily conserved regions of the genome. In an autosomal dominant model, 358 SNPs were identified, 13 of which lay in or near one of 2,525 genes reported to play a role in hepatitis or other immune, inflammatory and apoptotic processes (see Supplementary Methods). Only one of these 13, an intronic SNP in AKT1, was shared by more than two individuals (Table 3 and Supplementary Table 2).
Discussion
Increased susceptibility to many common infectious diseases may be mediated by pathogen-specific deficits in host immunity [Alcaïs et al. 2009]. Large-scale sequencing efforts indicate that healthy individuals harbor many occult loss-of-function variants [Tennessen et al. 2012], and the phenotypic impact of these rare variants on infectious outcome may only be revealed upon exposure to the appropriate agent. In many cases, candidate gene resequencing across consanguineous pedigrees has identified autosomal recessive alleles responsible for these pathogen-specific deficits. Next generation sequencing of unrelated individuals has accelerated the discovery of similarly penetrant alleles in situations where pedigrees are not available.
This genetic model for infectious disease susceptibility was applied to HAV acute liver failure, a rare outcome of a relatively common viral infection. Given prior results with similarly rare diseases, it was reasonable to hypothesize that the risk of HAV acute liver failure might be explained by a single susceptibility locus across an ethnically diverse collection of patients. However, the results presented in this study suggest that genetic susceptibility to hepatitis A is a heterogeneous and complex trait. While only ten genomes were analyzed, other studies have successfully identified Mendelian risk alleles by sequencing fewer individuals [Ng et al. 2010]. More likely, these results suggest that genetic susceptibility to hepatitis A is a heterogeneous and complex trait. The identification of multiple candidate risk alleles shared by several individuals each suggests that the immunopathogenesis of HAV is modulated by more than one biological pathway. Alternatively, the genetic architecture of HAV susceptibility may differ in adults, as most Mendelian risk alleles for infectious diseases have been identified in children. A notable caveat of this study is that the effects of sex chromosomes, epigenetic factors, or copy number variants were not considered.
Despite the apparent complexity of HAV susceptibility, a handful of candidate variants were identified, several of which exist within biologically plausible loci. Each of these mutations is rare in the general population, and many impact the amino acid sequence of proteins important to hepatic physiology. MST1R is a cell surface receptor tyrosine kinase that regulates hepatic immune responses and is expressed on tissue macrophages and hepatocytes. Its ligand is the hepatocyte growth factor-like (HGFL) protein and MST1R knockout mice are more susceptible to acute liver injury [McDowell et al. 2002]. NCR3LG1 is a ligand for natural killer cell receptors and appears to play an immunomodulatory role in response to TLR and pro-inflammatory cytokine signaling [Matta et al. 2013]. NFATC4, a regulator of T cell and innate immune responses, is highly expressed in the liver, and knockout mice are less susceptible to alcoholic-mediated liver damage [Bukong et al. 2011]. GPC1 is a glycophosphatidylinositol (GPI) linked cell surface protein and member of a large group of heparan sulfate binding proteins that are involved in morphogenesis, adhesion, chemotaxis and inflammation [Fransson 2003; Parish 2006]. Heparan sulfate proteoglycans are key cell surface attachment sites for infectious agents and are thought to play a role in the hepatotropism of hepatitis B virus, hepatitis C virus, and hepatitis D [Barth et al. 2003; Schulze et al. 2007; Leistner et al. 2008; Longarela et al. 2013; Shi et al. 2013]. In contrast, the functions of several of the identified genes do not immediately suggest a role in HAV pathogenesis. Mutations in RELN and STXBP1 have only been associated with neurological disorders, and there are insufficient data to suggest a mechanistic link between DNAH12 and HAV susceptibility. There was little evidence for enrichment of the previously identified HAV risk allele, TIM1-157insMTTTVP, within our cohort [Kim et al. 2011]. The frequency of the allele among our cases was more similar to the controls in this study. This difference could be due to racial or ethnic differences in the study population.
It was surprising to find a significant number of noncoding variants shared by multiple members of a genetically diverse cohort. In many cases, these variants are either very rare (allele frequency < 1%) or not found in dbSNP and the 54 genomes in the Complete Genomics public dataset. Many of these variants, however, were identified in the sequences of three control genomes analyzed with the same bioinformatic pipeline. These data suggest that many noncoding variants annotated as rare or novel in public databases may be more frequent than previously realized. Alternatively, systematic biases in the sequencing platform utilized could result in experiment-specific false-positive variant calls. These results highlight a significant challenge in identifying causative variants in whole genome as opposed to exome sequence. In the analysis of coding regions, a number of false-positive susceptibility alleles were filtered out based on their presence in large public databases of coding sequence as well as exomes sequenced by the authors’ laboratories for other studies. While the cost and availability of whole genome sequence precluded a similarly efficient approach to noncoding variants, analogous control sample filtering strategies should be possible in the near future.
This study is the most comprehensive study of human genetic susceptibility to HAV to date and represents an important step toward defining the biological pathways involved in HAV pathogenesis. The rarity of the identified variants and the corresponding phenotype suggests that validating the results through traditional association studies will be difficult. For example, an adequately powered genome-wide association study would require hundreds or thousands of cases and controls. Rather, one expects that sequencing additional cases will confirm some of the findings and may identify additional risk alleles for this apparently complex trait. The significance of the most plausible candidates can also be evaluated in cell-based assays for HAV infection and small animal models.
Supplementary Material
Supplementary Figure 1 – Screenshots from the Integrated Genomics Viewer showing evidence for variant calls in Table 3. MS Word Document
Supplementary Methods - Databases and filtering criteria used in this study. MS Word document
Supplementary Table 1 - Additional clinical data on HAV acute liver failure cohort. MS Excel document
Supplementary Table 2 - Raw output of shared variants by genetic model. MS Excel document
Acknowledgments
This work was supported by a pilot grant from the UCSF Liver Center (National Institutes of Health P30 DK026743) and institutional funds from Blood Systems Research Institute. DL was supported by the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the Office of the Director, National Institutes of Health, through UCSF-CTSI Grant Numbers TL1 RR024129 and TL1 TR000144. ASL was supported by National Institutes of Health K08 AI081754. The Acute Liver Failure Study Group was supported by National Institutes of Health U01 DK58369.
We thank Corron Sanders, William M. Lee, and the Acute Liver Failure Study Group for providing clinical data and patient DNA. Members and institutions participating in the Acute Liver Failure Study Group 1998–2012 are as follows: W.M. Lee, M.D. (Principal Investigator); Anne M. Larson, M.D., Iris Liou, M.D., University of Washington, Seattle, WA; Timothy Davern, M.D., University of California, San Francisco, CA (current address: California Pacific Medical Center, San Francisco, CA), Oren Fix, M.D., University of California, San Francisco; Michael Schilsky, M.D., Mount Sinai School of Medicine, New York, NY (current address: Yale University, New Haven, CT); Timothy McCashland, M.D., University of Nebraska, Omaha, NE; J. Eileen Hay, M.B.B.S., Mayo Clinic, Rochester, MN; Natalie Murray, M.D., Baylor University Medical Center, Dallas, TX; A. Obaid S. Shaikh, M.D., University of Pittsburgh, Pittsburgh, PA; Andres Blei, M.D., Northwestern University, Chicago, IL (deceased), Daniel Ganger, M.D., Northwestern University, Chicago, IL; Atif Zaman, M.D., University of Oregon, Portland, OR; Steven H.B. Han, M.D., University of California, Los Angeles, CA; Robert Fontana, M.D., University of Michigan, Ann Arbor, MI; Brendan McGuire, M.D., University of Alabama, Birmingham, AL; Raymond T. Chung, M.D., Massachusetts General Hospital, Boston, MA; Alastair Smith, M.B., Ch.B., Duke University Medical Center, Durham, NC; Robert Brown, M.D., Cornell/Columbia University, New York, NY; Jeffrey Crippin, M.D., Washington University, St Louis, MO; Edwyn Harrison, Mayo Clinic, Scottsdale, AZ; Adrian Reuben, M.B.B.S., Medical University of South Carolina, Charleston, SC; Santiago Munoz, M.D., Albert Einstein Medical Center, Philadelphia, PA; Rajender Reddy, M.D., University of Pennsylvania, Philadelphia, PA; R. Todd Stravitz, M.D., Virginia Commonwealth University, Richmond, VA; Lorenzo Rossaro, M.D., University of California Davis, Sacramento, CA; Raj Satyanarayana, M.D., Mayo Clinic, Jacksonville, FL; and Tarek Hassanein, M.D., University of California, San Diego, CA. The University of Texas Southwestern Administrative Group included Grace Samuel, Ezmina Lalani, Carla Pezzia, Corron Sanders, Ph.D., Nahid Attar, Linda S. Hynan, Ph.D. and Angela Bowling and the Medical University of South Carolina Data Coordination Unit included Valerie Durkalski, Ph.D., Wenle Zhao, Ph.D., Catherine Dillon, Holly Battenhouse, Tomoko Goddard, Lynn Patterson, Jaime Speiser and Caitlyn Nicole Ellerbe
References
- Ajmera V, Xia G, Vaughan G, Forbi JC, Ganova-Raeva LM, Khudyakov Y, Opio CK, Taylor R, Restrepo R, Munoz S, Fontana RJ, Lee WM Acute Liver Failure Study Group. What factors determine the severity of hepatitis A-related acute liver failure? Journal of Viral Hepatitis. 2011;18:e167–74. doi: 10.1111/j.1365-2893.2010.01410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A, Abel L, Casanova J-L. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest. 2009;119:2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. WHO. Hepatitis A Fact Sheet. WHO; 2013. [Google Scholar]
- Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Weizsacker Von F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- Bukong TN, Lo TC, Dolganiuc A. Calcium-dependent NFATc4 signaling contributes to inflammation and steatosis in a mouse model of alcoholic liver disease. Gastroenterology. 2011;140:S983–4. [Google Scholar]
- Cooksley WG. What did we learn from the Shanghai hepatitis A epidemic? Journal of Viral Hepatitis. 2000;7(Suppl 1):1–3. doi: 10.1046/j.1365-2893.2000.00021.x. [DOI] [PubMed] [Google Scholar]
- Fransson L-A. Glypicans. Int J Biochem Cell Biol. 2003;35:125–129. doi: 10.1016/s1357-2725(02)00095-x. [DOI] [PubMed] [Google Scholar]
- Kim HY, Eyheramonho MB, Pichavant M, Gonzalez Cambaceres C, Matangkasombut P, Cervio G, Kuperman S, Moreiro R, Konduru K, Manangeeswaran M, Freeman GJ, Kaplan GG, DeKruyff RH, Umetsu DT, Rosenzweig SD. A polymorphism in TIM1 is associated with susceptibility to severe hepatitis A virus infection in humans. J Clin Invest. 2011;121:1111–1118. doi: 10.1172/JCI44182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leistner CM, Gruen-Bernhard S, Glebe D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell Microbiol. 2008;10:122–133. doi: 10.1111/j.1462-5822.2007.01023.x. [DOI] [PubMed] [Google Scholar]
- Longarela O, Schmidt TT, Schöneweis K, Romeo R, Wedemeyer H, Urban S, Schulze A. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS ONE. 2013;8:e58340. doi: 10.1371/journal.pone.0058340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta J, Baratin M, Chiche L, Forel J-M, Cognet C, Thomas G, Farnarier C, Piperoglou C, Papazian L, Chaussabel D, Ugolini S, Vély F, Vivier E. Induction of B7-H6, a ligand for the natural killer cell-activating receptor NKp30, in inflammatory conditions. Blood. 2013;122:394–404. doi: 10.1182/blood-2013-01-481705. [DOI] [PubMed] [Google Scholar]
- McDowell SA, Mallakin A, Bachurski CJ, Toney-Earley K, Prows DR, Bruno T, Kaestner KH, Witte DP, Melin-Aldana H, Degen SJF, Leikauf GD, Waltz SE. The role of the receptor tyrosine kinase Ron in nickel-induced acute lung injury. Am J Respir Cell Mol Biol. 2002;26:99–104. doi: 10.1165/ajrcmb.26.1.4621. [DOI] [PubMed] [Google Scholar]
- Ng SB, Nickerson DA, Bamshad MJ, Shendure J. Massively parallel sequencing and rare disease. Human Molecular Genetics. 2010;19:R119–24. doi: 10.1093/hmg/ddq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CR. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, Heinzen EL, Need AC, Ruzzo EK, Singh A, Campbell CR, Hong LK, Lornsen KA, McKenzie AM, Sobreira NLM, Hoover-Fong JE, Milner JD, Ottman R, Haynes BF, Goedert JJ, Goldstein DB. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759–1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- Shi Q, Jiang J, Luo G. Syndecan-1 serves as the major receptor for attachment of hepatitis C virus to the surfaces of hepatocytes. J Virol. 2013;87:6866–6875. doi: 10.1128/JVI.03475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Davern T, Munoz S, Han S-H, McGuire B, Larson AM, Hynan L, Lee WM, Fontana RJ US Acute Liver Failure Study Group. Fulminant hepatitis A virus infection in the United States: Incidence, prognosis, and outcomes. Hepatology. 2006;44:1589–1597. doi: 10.1002/hep.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya N, Tokushige K, Yamaguchi N, Hasegawa K, Hashimoto E, Yamauchi K, Shiratori K. Influence of TNF gene polymorphism in patients with acute and fulminant hepatitis. J Gastroenterol. 2004;39:859–866. doi: 10.1007/s00535-004-1402-1. [DOI] [PubMed] [Google Scholar]
- Willner IR, Uhl MD, Howard SC, Williams EQ, Riely CA, Waters B. Serious hepatitis A: an analysis of patients hospitalized during an urban epidemic in the United States. Ann Intern Med. 1998;128:111–114. doi: 10.7326/0003-4819-128-2-199801150-00006. [DOI] [PubMed] [Google Scholar]
- Yalniz M, Ataseven H, Celebi S, Poyrazoğlu OK, Sirma N, Bahçeitoğlu IH. Two siblings with fulminant viral hepatitis A: case report. Acta Medica (Hradec Kralove) 2005;48:173–175. [PubMed] [Google Scholar]
- Yang NY, Yu PH, Mao ZX, Chen NL, Chai SA, Mao JS. Inapparent infection of hepatitis A virus. Am J Epidemiol. 1988;127:599–604. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 – Screenshots from the Integrated Genomics Viewer showing evidence for variant calls in Table 3. MS Word Document
Supplementary Methods - Databases and filtering criteria used in this study. MS Word document
Supplementary Table 1 - Additional clinical data on HAV acute liver failure cohort. MS Excel document
Supplementary Table 2 - Raw output of shared variants by genetic model. MS Excel document