Abstract
The maturation of naive CD8+ T cells into effector CTLs is a critical feature of a functional adaptive immune system. Development of CTLs depends, in part, upon the expression of the transcriptional regulator eomesodermin (EOMES), which is thought to regulate expression of two key effector molecules, perforin and granzyme B. Although EOMES is important for effector CTL development, the precise mechanisms regulating CD8+ effector cell maturation remains poorly understood. In this study, we show that Notch1 regulates the expression of EOMES, perforin, and granzyme B through direct binding to the promoters of these crucial effector molecules. By abrogating Notch signaling, both biochemically as well as genetically, we conclude that Notch activity mediates CTL activity through direct regulation of EOMES, perforin, and granzyme B.
A functional CD8+ T cell response is an essential component of the adaptive immune response to many bacterial and viral pathogens (1). Upon engagement with Ag, naive CD8+ T cells rapidly expand and differentiate into CD8+ effectors, producing cytokines such as IFN-γ and the effector molecules perforin and granzyme B. Effector CTLs play a key role in the defense against intracellular bacteria and viruses, using at least two distinct mechanisms to mediate direct killing of infected target cells CTLs can lyse targets by perforin-mediated release of granzyme B, a serine protease that induces apoptosis (2). CTLs also express FAS ligand (FasL)3 and can engage FAS on a target cell resulting in apoptosis (3).
The differentiation of a naive CD8+ T cell into a functional CTL is driven, in part, by the T-box transcription factor eomesodermin (EOMES). EOMES belongs to the family of T-box transcription factors, and shares 74% homology with T-bet (4). EOMES plays a critical role during vertebrate development and EOMES deficiency in mice results in embryonic death (5). Dominant negative EOMES expression in CD8+ T cells results in loss-of-function of CD8+ T cells, whereas ectopic expression of EOMES was shown to induce expression of IFN-γ, perforin, and granzyme B, suggesting this transcription factor is important in differentiation of naive CD8+ T cells into effector CTLs (4).
Previously, we showed that signaling through the TCR in both CD4+ and CD8+ T cells induces the activation of Notch1 (6). Notch proteins are single-pass transmembrane receptors that require multiple enzymatic cleavages to produce the full-length heterodimer displayed on the cell surface (7). In mice and humans, four Notch genes (Notch1–4) and at least five ligands (Delta-like DL1, DL3, and DL4 and Jagged1 and Jagged2) have been identified (8). Upon ligand binding, Notch undergoes final processing by gamma-secretase (9, 10), and activation of all Notch isoforms can be effectively prevented using pharmacological inhibitors of gamma-secretase (GSI) (11). This final gamma-secretase-induced cleavage generates the mature, intracellular signaling peptide NICD (Notch intracellular domain), which translocates to the nucleus, and potentiates the transcriptional activity associated with Notch. In the nucleus, NICD associates with the DNA binding protein CSL (CBF-1/RBP-Jκ for mammals, Suppressor of Hairless for Drosophila, Lag-1 for C. elegans), resulting in the Notch-mediated conversion of CSL from a repressor into a transcriptional activator.
Earlier data from our laboratory indicated Notch regulates the expression of the T-box protein, T-bet, in Th1 cells (12). T-bet and the closely related protein, EOMES, both are implicated in the development of effector CD8+ T cells (4, 13, 14). These observations, as well as our earlier data demonstrating TCR signaling in CD8+ T cells induces the activation of Notch, led us to ask whether Notch signaling plays a role in the development or function of effector CTLs. In this report, we describe an intrinsic role in CD8+ T cells for gamma-secretase in the regulation of EOMES, perforin, and granzyme B. We also present genetic data from Notch1 antisense (AS) mice showing a reduction in EOMES, perforin, and granzyme B. Lastly we show that reintroduction of activated Notch into GSI-treated CD8+ T cells partially rescues EOMES expression. These data lead us to conclude Notch is the major target of GSI and that Notch signaling is an important component of cytolytic effector function in CD8+ T cells.
Materials and Methods
Mice and preparation of CD8+ T cells
C57BL/6 and Notch1 AS mice were housed in the animal care facility at the University of Massachusetts (Amherst, MA) in accordance with Institutional Animal Care and Use Committee guidelines. Transgenic Notch1 AS mice originated from backcrossing of (C57BL/6H2b × SJLH2s) with Notch1 AS transgene controlled by the mouse mammary tumor virus long terminal repeat promoter (15). Splenocytes or CD8+ T cells (2–3 × 106/ ml) from 8- to 12-wk-old mice were stimulated to proliferate with soluble anti-mouse CD3ε and anti-mouse CD28 (BD Pharmingen) for indicated time periods. For experiments using GSI in vitro, cells were pretreated for 30 min at 37°C with 50 μM IL-CHO, as previously described (6), or with DMSO as vehicle control, before stimulation. For in vivo GSI experiments, LY-411,575 was formulated in rodent chow to deliver 5 mg/kg/day, based on average consumption, and administered to mice for 13 days. CD8+ T cells (>95% purity) were isolated from unstimulated splenocytes using the IMag bead separation system (BD Pharmingen).
Semiquantitative RT-PCR and quantitative real-time PCR
Total RNA was isolated from CD8+ T cells using RNA-BEE (Tel-Test). After DNase treatment (Promega), reverse transcription of 1–2 μg of RNA was performed with oligo(dT) primers (Invitrogen), and cDNAs were analyzed by semiquantitative PCR or real-time quantitative PCR. For semi-quantitative PCR, three different cycles described below were tested and PCR were performed with the following primers: EOMES (forward) 5′-ATGCAGTTGGGAGAGCAGCTCCTG-3′, (reverse) 5′-GTTGCACAG GTAGACGTG-3′; perforin (forward) 5′-CAAGCAGAAGCACAAGT TCGT-3′, (reverse) 5′-CGTGATAAAGTGCGTGCCATA-3′; granzyme B (forward) 5′-ACTTTCGATCAAGGATCAGCA-3′, (reverse) 5′-ACT GTCAGCTCAACCTCTTGT-3′; and GAPDH (forward) 5′-ACTTTCGA TCAAGGATCAGCA-3′, (reverse) 5′-ACGGAAGGCCATGCCAGTGA GCTT-3′. Conditions were as follows: the shortest cycle, 94°C 2 min, 94°C 30 s, 55°C 30 s, 68°C 30 s (20 cycles), no extension; the optimal cycle, 94°C 2 min, 94°C 30 s, 55°C 30 s, 68°C 30 s (25 cycles), 68°C 5 min; and the longest cycle, 94°C 2 min, 94°C 30 s, 55°C 30 s, 68°C 30 s (30 cycles), 68°C 5 min. For real-time quantitative PCR, semiquantitative PCR products of perforin and granzyme B were subcloned into pCR2.1 (Invitrogen) as cDNA control plasmids. Specific primers from PrimerBank of the Center for Computational and Integrative Biology (Harvard Medical School, Boston, MA) were used: perforin (PrimerBank ID no. 6755042a3) (forward) 5′-GCTCCCACTCCAAGGTAGC-3′, (reverse) 5′-TTTGTACC AGGCGAAAACTGT-3′; and granzyme B (ID no. 7305123a3) (forward) 5′-TGCTGCTAAAGCTGAAGAGTAAG-3′, (reverse) 5′-CGTGTTT GAGTATTTGCCCATTG-3′. The control plasmids of each target sequence were diluted to generate standard curves. Real-time quantitative PCR amplification was performed on a Bio-Rad iCycler by using SYBR Green PCR Core Reagents (PE Applied Biosystems). To quantify the amount of cDNA for an individual transcript, SYBR Green fluorescence was measured at the end of each cycle. The cycle threshold (Ct), the cycle at which exponential growth of the PCR product was first detected, was determined for known concentrations of plasmid DNA (pCR2.1 Perforin, pCR2.1 granzyme B, and pGEM 18 S rRNA), and then a standard curve was created. Template copy numbers were calculated for each sample by interpolating the Ct values on the standard curve, by using the iCycler software. All samples and standards were run in triplicate for any given experiments. The values of perforin and granzyme B normalized to 18 S rRNA as the average copy number. Specific products were verified by melt-curve analysis and gel electrophoresis. Real-time quantitative PCR conditions were the following: the first cycle, 95°C 8 min 30 s; the second cycle, 95°C 30 s, 53°C 30 s, 72°C 30 s (40 cycles); the third cycle, 95°C 1 min; the fourth cycle, 53°C 10 s and increased temperature after cycle 2 by 0.5°C (80 cycles) for collecting melt-curve data. Specific primers for 18 S rRNA were (forward) 5′-TGGTGGAGCGATTTGTCTGG-3′ and (reverse) 5′-TCAATCTCGGGTGGCTGAAC-3′.
ELISA and Western blot
For ELISA, supernatants from CD8+ T cells prepared as described were harvested after incubation at 37°C for times indicated and analyzed as previously described (12). For immunoblotting, 25 μg of total protein was resolved as previously described (12). Sequentially stripped blots were reprobed with anti-EOMES (Abcam), anti-Notch1 (Santa Cruz Biotechnology), anti-Notch1ICD (N1ICD; Cell Signaling Technology), anti-T-bet (Santa Cruz Biotechnology), anti-FasL (Santa Cruz Biotechnology), anti-heat shock protein 70 (Affinity BioReagents), and anti-GAPDH (Chemicon International).
Retroviral infection
Briefly, 1 day before transfection, Phoenix-Eco cells (2 × 105/ml), used for producing retroviral supernatants, were cultured with fresh medium (1:1 of DMEM to RPMI 1640, 10% FBS, 20 μg/ml gentamicin, and 2 mM l-glutamine (all Life Technologies)) in 100-mm culture dishes. Cells were transfected with 4 μg each of DNA construct (N1ICD and EOMES in pMX-IRES-hCD8 vector or pMX-IRES-hCD8 empty vector) and added to Phoenix-Eco cells. Retroviral supernatants were collected after 3 days and added to CD8+ T cells from splenocytes stimulated for 1 day. Infected cells were cultured at 37°C with 5% CO2 for 3 days. Infection efficiency was monitored by human CD8+ expression using anti-human CD8-PE (BD Pharmingen), an LSRII flow cytometer, and DIVA software (BD Immunocytometry Systems).
Intracellular staining and cell surface staining
Prepared splenocytes or CD8+ T cells were stained as previously described (6). Cells were stained with anti-mouse CD8a-PE, -FITC, or -allophycocyanin (BD Pharmingen), anti-mouse CD4-FITC (BD Pharmingen), anti-mouse CD25-FITC (BD Pharmingen), anti-mouse CD44-FITC (BD Pharmingen), anti-mouse CD69-FITC (BD Pharmingen), anti-mouse IFN-γ-PE (BD Pharmingen), anti-mouse/human Notch1-PE (eBioscience); anti-mouse IgG1κ isotype control PE or FITC (eBioscience), anti-mouse perforin PE or FITC (eBioscience), anti-rat IgG2a isotype control PE or FITC (eBioscience), anti-mouse/human granzyme B PE or FITC (eBioscience), anti-mouse IgG1κ isotype control PE (eBioscience), anti-mouse CD178 (FasL) PE (BD Pharmingen), with 5 μl of anti-hamster IgG1κ isotype control PE (BD Pharmingen). Stained cells were analyzed using an LSRII flow cytometer and DIVA or FlowJo software.
Chromatin immunoprecipitation (ChIP) assay
ChIP analysis was performed using 1 × 106 CD8+ T cells from GSI- or DMSO-pretreated splenocytes stimulated for 1 day, as described, using the ChIP Assay kit (Upstate Cell Signaling Solutions). The following primers were used for PCR: mouse EOMES primer set1 (472 bp) (forward) 5′-AGTTTCCCGTGTGATCGCATTGG-3′, (reverse) 5′-AGGCCGTCAC TTTCATTACTCAG-3′; mouse EOMES primer set2 (369 bp) (forward) 5′-GGTAGACCATGTTCGCAGACTTCA-3′, (reverse) 5′-CATTTAG CAACCAGCCATTTCCTC-3′; mouse perforin primer (forward) 5′-CTCA GAAGCAGGGAGCAGTC-3′, (reverse) 5′-TGCGATCTATCCCCAGGC AG-3′; and mouse granzyme B primer (forward) 5′-AGCTTGGGTTTC TGGGACTCTGA-3′, (reverse) 5′-TATGAAAACTCCTGCCCTACTG CC-3′. Abs used were rabbit anti-Notch1, rabbit anti-RBP-Jκ, goat anti-p50, normal rabbit IgG, and normal goat IgG (all Santa Cruz Biotechnology). Conditions were 94°C 2 min, 94°C 30 s, 55°C 30 s, 68°C 30 s (35 cycles), 68°C 5 min.
Ag-specific CTL functional assay
The E.G7-OVA and EL4 lines were purchased from American Type Culture Collection (ATCC CRL-2113 and ATCC TIB-39, respectively). For CTL priming, C57BL/6 mice were immunized with 5 × 106 E.G7-OVA (or EL4, as control) cells/500 μl PBS. For in vivo GSI administration, LY-411,575 mouse chow (5 mg/kg/day) was orally administered to C57BL/6 mice for a total of 19 days, beginning 3 days before immunization and continuing for 16 days, at which time mice were sacrificed. Primed splenocytes were restimulated in vitro with gamma-irradiated (30,000 rad) 1.5 × 106 E.G7-OVA or EL4 cells. After 5 days, live CD8+ T cells were isolated. CTL analysis was performed in black flat-bottom plates (Costar), using DELFIA EuTDA cytotoxicity reagents (PerkinElmer) and a time-resolved fluorometer (SpectraMax M5 plus SOFTmax PRO), following the manufacturer's instructions.
Results
Notch signaling regulates the expression of perforin and granzyme B in developing effector CTLs
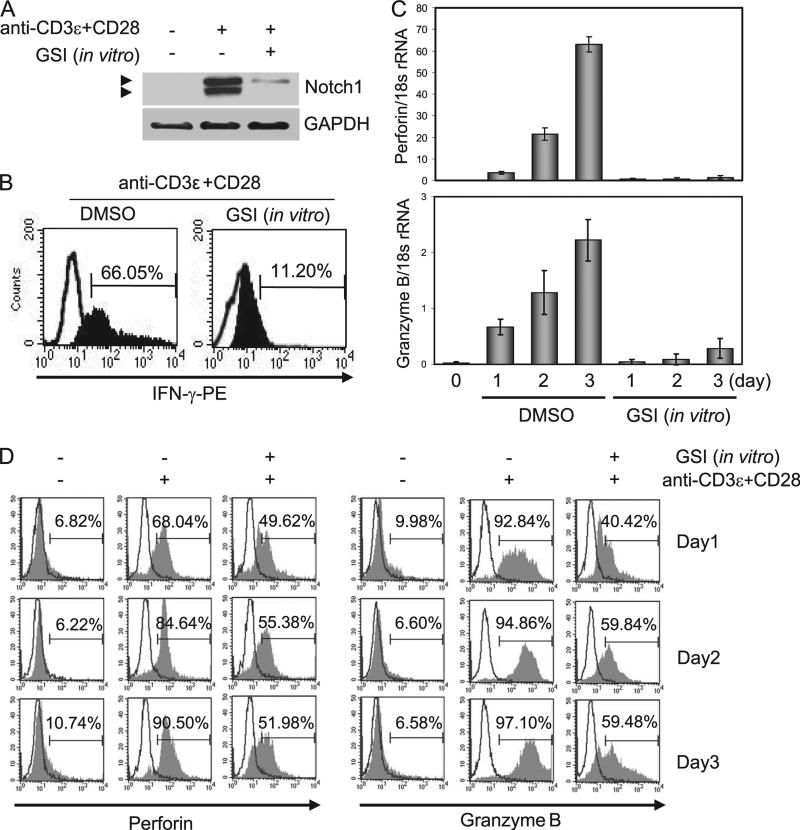
To address what role Notch may have in the development of effector CTLs, splenocytes from C57BL/6 mice were pretreated with DMSO, as vehicle control, or with a GSI to block Notch activation then stimulated with Abs against CD3ε and CD28 for 2 days. CD8+ T cells were isolated and whole cell lysates were immuno-blotted with anti-Notch1 (Fig. 1A). In control cells, we observed increasing expression of Notch1 intracellular domain (N1ICD) up to 3 days following stimulation (data not shown); however, in vitro treatment with GSI significantly abrogated Notch1 expression, consistent with previous reports (6, 12). Additionally CD8+ T cells were analyzed for IFN-γ expressions by intracellular staining (Fig. 1B). Compared with DMSO control, we noted substantial IFN-γ reduction by in vitro treatment with GSI. We next analyzed mRNA transcripts and protein expression of perforin and granzyme B, two key mediators of CTL effector function. In DMSO-treated cells, stimulation for 2 days with anti-CD3ε and anti-CD28 resulted in up-regulation both of perforin and granzyme B mRNA. In contrast, in vitro treatment with GSI markedly diminished mRNA transcripts of perforin, whereas granzyme B was less affected (Fig. 1C). Using intracellular staining and flow cytometric analysis of CD8+ T cells, we assessed the level of protein expression of perforin and granzyme B up to 3 days poststimulation. Consistent with the mRNA expression profile, protein expression both of perforin and granzyme B increased poststimulation, and in vitro treatment with GSI significantly reduced expression of both proteins, compared with DMSO controls (Fig. 1D).
FIGURE 1.
Stimulating splenocytes with anti-CD3ε plus anti-CD28 induces Notch-dependent perforin and granzyme B expression in CD8+ T cells. A, CD8+ T cells were isolated from splenocytes that were stimulated for 2 days and whole cell lysates were analyzed for Notch1 expression by immuno-blotting. Extracellular and transmembrane portions of Notch1 (top bands) and N1ICD (bottom bands) are shown. B, Isolated CD8+ T cells as in A were used for analysis of IFN-γ expression by intracellular staining and flow cytometry. The indicated days are stimulation periods with anti-CD3ε and anti-CD28. C, Parallel samples were used to determine the expression of perforin and granzyme B by real-time quantitative PCR and by intracellular staining (D) followed by flow cytometric analysis. For these experiments, stimulations were conducted for 1–3 days. Open histograms represent isotype controls. Data shown represent one of three independent replicates.
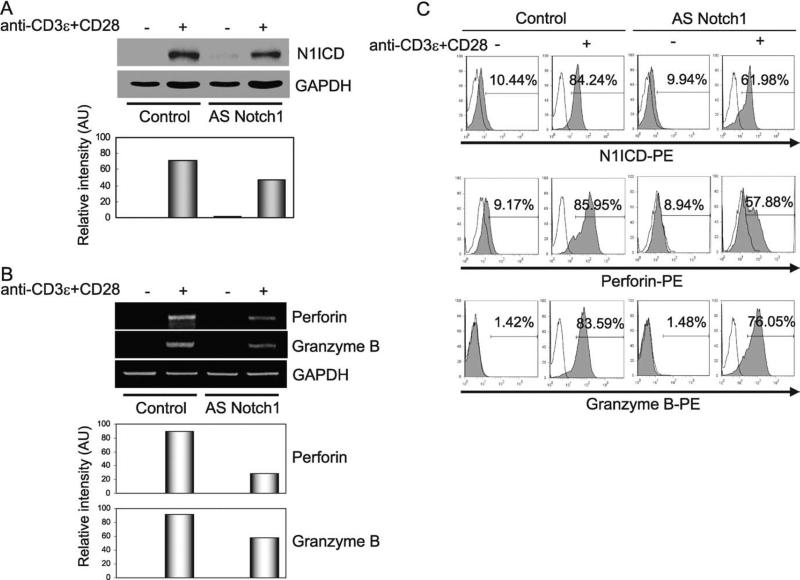
To further investigate, in vivo, the effects of blocking Notch signaling, we used cells from Notch1 AS mice that have been engineered to express reduced levels of Notch1 in lymphocytes, as well as in other cell lineages (6). Results obtained using cells from these animals are more modest, nonetheless, CD8+ T cells from Notch1 AS mice exhibit reduced expression of N1ICD, compared with littermate controls (Fig. 2A), decreased mRNA transcripts of perforin and granzyme B (Fig. 2B), as well as reduced protein expression, as determined by intracellular staining and flow cyto-metric analysis (Fig. 2C). Thus, using pharmacological and genetic means to block Notch activation in CD8+ T cells, we show that inhibiting Notch signaling also decreases expression of the principal mediators of CTL activity in CD8+ T cells, perforin and granzyme B, both at the transcript and protein levels.
FIGURE 2.
Notch1 AS mice express reduced levels of perforin and granzyme B in CD8+ T cells. A, Splenocytes from Notch1 AS mice were stimulated with anti-CD3ε plus anti-CD28 for 2 days, CD8+ T cells were isolated and we used immunoblotting to analyze whole cell lysates for N1ICD expression (upper bands). Graph (lower) represents band intensity normalized against GAPDH. B, RT-PCR was used to assess perforin and granzyme B mRNA transcript expression in CD8+ T cells from Notch1 AS mice (upper bands). Quantifications of the relative expression of perforin and granzyme B transcripts are represented (lower panels). C, Splenocytes from Notch1 AS mice were isolated and stimulated with anti-CD3ε plus anti-CD28 for 2 days, and intracellular staining was conducted to determine the percentage of CD8+ T cells expressing N1ICD, perforin, and granzyme B; blank peaks indicate isotype control. Data shown represent one of at least three independent replicates.
Previous data from our laboratory suggested Notch activation is associated with the proliferation of T cells as well as the activation of NF-κB and the production of the effector cytokine IFN-γ (6). CD25 and CD69 was partly reduced by the in vitro treatment of splenic T cells with GSI, suggesting that both TCR signals as well as Notch activation are required for sustained and maximal expression of these markers of early T cell activation. Additionally Adler et al. (16) showed that Notch signaling enhances CD25 expression on CD4+ T cells. To determine whether purified CD8+ T cells display altered cell surface expression of these activation markers in the absence or presence of GSI in vitro, CD8+ T cells were isolated from splenocytes of age-matched C57BL/6 mice and then incubated with anti-CD3ε and anti-CD28 for 2 days. As shown in supporting data, GSI treatment had little to no effect on the expression of CD25, CD44, and CD69 (see supplemental Fig. S1).4
Notch signaling plays an intrinsic role in CD8+ T cells to regulate the expression of perforin and granzyme B
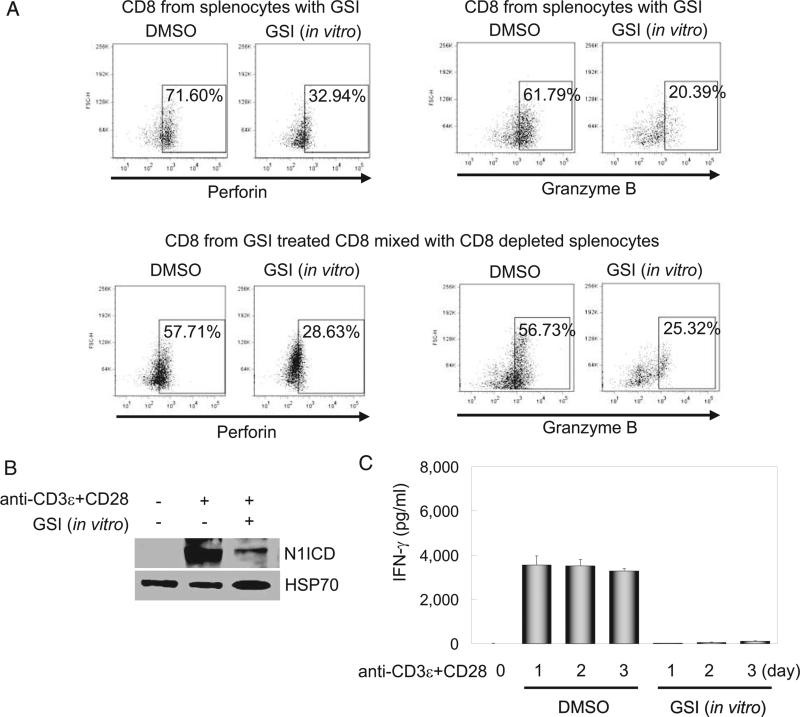
In the experiments shown in Fig. 1, splenocytes containing both CD4+ and CD8+ T cells were activated with anti-CD3ε and anti-CD28, followed by analysis of perforin and granzyme B in highly purified CD8+ T cells. It is well established that CD4+ T cells provide cytokines, including IFN-γ, that drive the maturation of CD8+ effector cells, and we have previously reported a role for Notch1 in mediating IFN-γ production (17). Therefore, it is possible that reduced Notch signaling in the CD4+ population contributed, at least partially, to the diminished expression of perforin and granzyme B observed in Fig. 1. To address whether Notch signaling plays an intrinsic role in the differentiation of CD8+ T cells into CTLs, purified CD8+ T cells (see supplemental Fig. S2, A)4 were treated in vitro with GSI or DMSO, mixed with CD8-depleted splenocytes, and stimulated for 2 days. As controls, total splenocytes were treated in vitro with GSI or DMSO, then stimulated for 2 days. At the end of the culture period, cells were stained intracellularly for perforin or granzyme B and analyzed by flow cytometry; whole cell lysates were prepared from parallel cultures to monitor Notch inhibition. Fig. 3A, top row, shows results from control experiments in which total splenocytes were stimulated for 2 days in vitro with either GSI or DMSO. After 2 days of culture, CD8+ cells were gated and intracellular expression of either perforin (Fig. 3, top left) or granzyme B (Fig. 3, top right) was determined. As expected from data presented in Fig. 1, these control experiments demonstrated that GSI effectively blocks the appearance of both intracellular perforin and granzyme B. To address the question whether GSI works intrinsically on CD8+ T cells, we isolated CD8+ T cells and treated them with GSI and washed them to remove extracellular GSI. This treated CD8+ population was then mixed with CD8-depleted splenocytes and stimulated as described for 2 days. As described for the control experiment, at the end of the 2-day incubation, CD8+ cells were gated and intracellular protein levels of perforin and granzyme B determined (Fig. 3A, bottom row). Again, we found that GSI effectively blocks the level of intracellular perforin and granzyme B, suggesting Notch functions intrinsically in CD8+ T cells to regulate effector function. As a control, to ensure GSI functioned to block Notch processing in these experiments, CD8+ T cells from Fig. 3A were analyzed for the presence of N1ICD, the activated form of Notch1. As shown in Fig. 3B (and supported in supplemental material Fig. S2, B),4 GSI treatment significantly blocks the appearance of N1ICD. Additionally, supernatants were harvested from these cultures and assayed for the expression of IFN-γ. As shown in Fig. 3C, supernatants from DMSO-treated controls expressed high levels of IFN-γ, whereas supernatants from GSI-treated CD8+ T cells did not secrete appreciable levels of this cytokine (see supplemental Fig. S2, C).4
FIGURE 3.
Intrinsic Notch1 signaling directly regulates perforin and granzyme B expression in CD8+ T cells. A, Purified CD8+ T cells from un-stimulated splenocytes were pretreated in vitro with GSI, washed two times, mixed with CD8-depleted cells, and stimulated with anti-CD3ε plus anti-CD28. As a control, similar to parallel samples analyzed by real-time quantitative PCR and by intracellular staining as in Fig. 1D, stimulated bulk splenocytes in the absence or presence of GSI in vitro were compared (top panels). Using intracellular staining and flow cytometry, the percentage of CD8+ T cells expressing perforin and granzyme B after 2 days was determined. All perforin- or granzyme B-positive plots are gated on CD8+. B, CD8+ T cells were isolated from splenocytes, pretreated in vitro with GSI then stimulated with anti-CD3ε plus anti-CD28 for 2 days. We verified N1ICD inhibition by immunoblot (top band); heat shock protein HSP70 was used as a loading control (bottom band). C, CD8+ T cells prepared as in A were stimulated for 1–3 days, supernatants were harvested, and IFN-γ determined by ELISA. Data represent the mean ± SD of three individual replicates. All experiments were repeated three times.
Notch binds to both perforin and granzyme B promoters
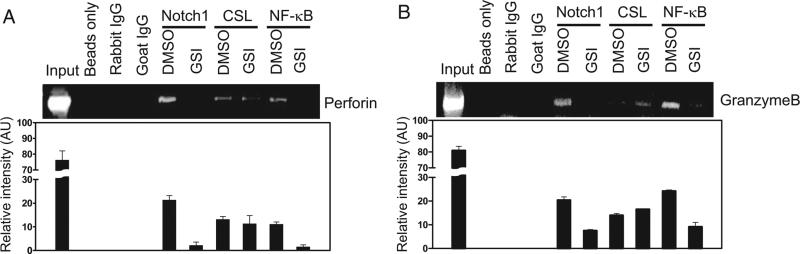
To investigate whether Notch1 directly regulates transcription of CTL effector molecules, we examined mouse genomic DNA, ~1.8 and 0.96 kb upstream of the perforin and granzyme B promoter start sites, respectively (see supplemental Fig. S3).4 Within these regions, we identified multiple putative CSL and NF-κB binding sites. To determine whether N1ICD forms a complex with CSL on these promoters, we performed ChIP analysis both on the perforin and granzyme B promoters. We observed that N1ICD binds to both promoters and GSI treatment abrogates N1ICD binding. We also detected CSL bound to both promoters; however, treatment with GSI showed no effect on CSL binding. The CSL binding site overlaps with that of the transcription factor NF-κB (18–20). Previously, we reported that N1ICD also can interact with NF-κB to directly regulate IFN-γ (17). Therefore, we questioned whether NF-κB was also found bound to the perforin and granzyme B promoters. ChIP analysis demonstrates that Notch and NF-κB occupy sites both in the perforin and the granzyme B promoter regions analyzed, and GSI treatment abrogates recruitment of NF-κB to these binding sites (Fig. 4). Taken together, these data suggest that NF-κB and CSL both are recruited to the perforin and granzyme B promoters and, in the case of NF-κB, interaction with nuclear complexes that require Notch1 directly regulates these CTL effector molecules.
FIGURE 4.
Notch1 directly binds the perforin and granzyme B promoters. ChIP analysis was used to determine what nuclear binding partners could be identified on the promoters of perforin (A) and granzyme B (B). PCR primers were designed to amplify putative binding sites for CSL and NF-κB; we performed PCR using 5 μl of DNA eluates and equal volumes of input. Data shown represent one of at least three independent replicates.
Treatment with GSI partially blocks CTL function
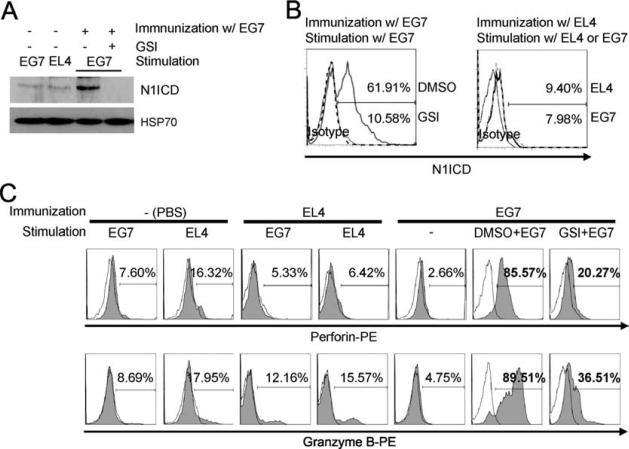
To determine whether GSI influences biological function in CD8+ T cells, we i.p. injected C57BL/6 (H-2b) mice, with E.G7-OVA cells. This syngeneic tumor cell line was constructed by transfecting the parental EL4 cell line with cDNA expressing a MHC class I-restricted peptide of chicken OVA, aa 258–276 (OVA258–276) (21). Sixteen days after immunization, splenocytes from these animals were collected and pretreated in vitro with GSI, or the vehicle control, DMSO, and restimulated in culture for 5 days with gamma-irradiated E.G7-OVA cell or with EL4 cells, as a mock control for stimulation. After 5 days, CD8+ T cells were isolated and N1ICD expression was assessed by immunoblotting and flow cytometric analysis of intracellular staining. We found that N1ICD increased in CD8+ T cells from mice primed with OVA258–276-expressing E.G7-OVA cells, but not in those primed with control EL4 cells, and GSI treatment abrogated expression of N1ICD under E.G7-OVA-immunized conditions (Fig. 5, A and B). Similarly, there was no increase in perforin and granzyme B expression in CD8+ T cells either from E.G7-OVA- or EL4-in vitro-stimulated cultures from mice primed with control EL4 cells (Fig. 5C). In contrast, CD8+ T cells from mice immunized with E.G7-OVA showed robust induction of N1ICD, perforin and granzyme B expression when stimulated in vitro with E.G7-OVA cells (Fig. 5, A–C). In vitro GSI treatment of E.G7-OVA-primed CD8+ T cells significantly reduced perforin and granzyme B expression during stimulation with E.G7-OVA cells (Fig. 5C). These data demonstrate GSI treatment blocks maximal perforin and granzyme B expression and suggest that Notch signaling, induced by MHC class I-restricted OVA peptide in E.G7-OVA-primed CD8+ T cells may play a role in this process.
FIGURE 5.
Specific OVA peptide may increase N1ICD, perforin and granzyme B expression in CD8+ T cells. To study the effects of OVA-specific Ag stimulation on N1ICD expression in the presence of GSI, OVA-specific CD8+ T cells were prepared, as described in Materials and Methods. Briefly, C57BL/6 mice were immunized with PBS, as a negative control, with the parental EL4 cell line as a mock control, or with E.G7-OVA cells, engineered to express an OVA peptide. After 16 days splenocytes were harvested from these mice, pretreated in vitro with GSI then stimulated with the OVA-specific E.G7 cells for 5 days. N1ICD expression was determined by immunoblotting (A) and intracellular staining (B). Dotted or open historams represent the conjugated isotype control. C, Together, intracellular staining and flow cytometric analysis were used to assess the percentage of CD8+ T cells expressing perforin and granzyme B. Data shown are representative of three individual replicates.
To further address the specificity with which GSI regulates CTL effector functions, we asked whether the Fas-FasL pathway, which also plays an important role in CTL-induced cell death, is regulated through targets of GSI. FasL was detected only in CD8+ T cells from mice primed with E.G7-OVA, but not from mice primed with EL4 cells. Additionally, FasL expression was similar whether CD8+ T cells were pretreated in vitro without or with GSI (see supplemental Fig. S4),4 suggesting GSI, likely through its role in blocking Notch signaling, controls CTL activity of CD8+ T cells, specifically, through its direct regulation of perforin and granzyme B transcription, but not through FasL expression.
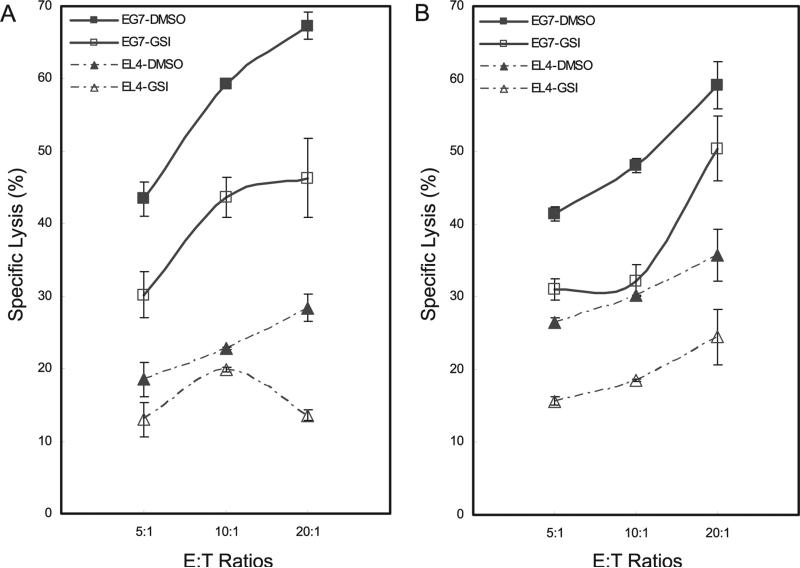
To test the biological impact of GSI in CD8+ CTLs, we used a Europium (Eu) cytotoxicity assay to measure target cell death (22). Live CD8+ T cells from E.G7-OVA-primed mice were mixed either with E.G7-OVA- or EL4-Eu-loaded target cells and cell lysis determined by time-resolved fluorometry (23). CD8+ CTLs were capable of lysing E.G7-OVA cells but not the control EL4 cell line (Fig. 6A, compare triangles). Specific lysis of E.G7-OVA target cells by E.G7-OVA-primed and in vitro-stimulated CD8+ T cells approached 70% (Fig. 6A, filled box symbols) compared with ~25% lysis of control EL4 target cells (Fig. 6A, filled triangles). Consistent with reduced expression of perforin and granzyme B in the absence of Notch signaling, treating E.G7-OVA-primed CD8+ T cells in vitro with GSI blocked target cell lysis, as expected (Fig. 6A, compare open with filled box symbols).
FIGURE 6.
Blocking Notch signaling attenuates the CTL activity of OVA-specific CD8+ T cells. A, C57BL/6 mice were immunized with E.G7-OVA cells (box symbols) or EL4 cells, as a mock control (triangles). After 16 days, we harvested splenocytes from these mice, pretreated them in vitro with DMSO (filled symbols) or with GSI (open symbols), and stimulated them with OVA-specific E.G7 cells for 5 days. Live CD8+ T cells were isolated and an Ag-specific cytotoxicity assay, using europium (Eu)-loaded target cells, was used to determine the percentage of specific lysis. B, C57BL/6 mice were fed control chow or chow containing GSI (LY chow) beginning 3 days before immunizing with E.G7-OVA (box symbols) or EL4 (triangles) cells and continuing until splenocytes were harvested 16 days later. Splenocytes were stimulated with OVA-specific E.G7 cells for 5 days, live CD8+ T cells isolated from the splenocytes of control-fed (closed symbols) and GSI-fed (open symbols) mice and the europium cytotoxicity assay was performed as in A. For specific lysis, 5000 target cells (T), OVA-specific E.G7-OVA, or EL4 (as control) were used with 25,000, 50,000, or 100,000 effector (E) CD8+ cells, for E:T ratios of 5:1, 10:1, and 20:1, respectively. Results shown in the cytotoxicity assays represent the mean ± SD of three independent replicates. Data are representative of three independent replicates.
To determine the effects on CTL activity of in vivo treatment with GSI, C57BL/6 mice were fed GSI in rodent chow formulated to deliver 5 mg/kg/day, for 3 days before E.G7-OVA immunization and continuing until animals were sacrificed 16 days later. CTL assays were performed using Europium-primed target cells as described. Similar to results obtained with in vitro GSI treatment, lysis of E.G7-OVA-specific target cells by E.G7-OVA-primed CD8+ T cells was attenuated in cultures of cells from in vivo GSI-administered mice compared with E.G7-OVA-primed controls (Fig. 6B, compare filled with open box symbols). Lysis of EL4 control target cells was reduced compared with E.G7-OVA-specific target cells (Fig. 6B, compare filled box symbols with filled triangles), and further reduced when mice were fed GSI chow (Fig. 6B, compare filled with open triangles). Thus, GSI delivered either in vitro or in vivo effectively alleviated specific target cell lysis mediated by CD8+ CTLs.
Notch signaling regulates EOMES expression through direct binding to the EOMES promoter
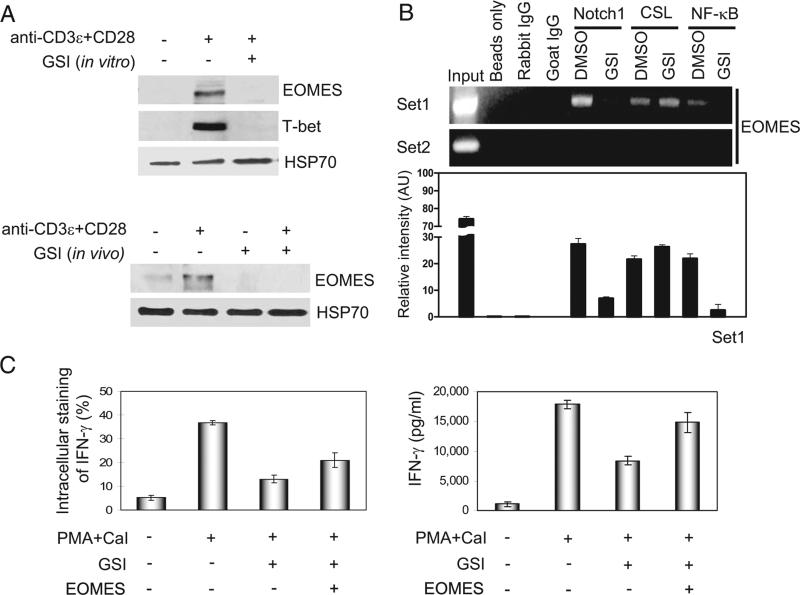
Differentiation of CD8+ T cells is regulated, at least in part, by the T-box transcription factor, EOMES (4, 24). It has been reported that EOMES is significantly up-regulated in effector CD8+ T cells and plays an important role in CTL effector mechanisms, as well as in mediating IFN-γ production (4). To investigate whether Notch1 regulates EOMES, EOMES mRNA and protein expression were analyzed in the presence or absence of Notch signaling. In stimulated CD8+ T cells, inhibiting Notch activation with GSI treatment reduced EOMES mRNA transcript, and this response was confirmed in stimulated CD8+ T cells from Notch1 AS mice (data not shown). Protein expression of EOMES also was reduced in stimulated CD8+ T cells treated in vitro or in vivo with GSI, compared with controls (Fig. 7A). As reported by other studies, T-bet is also expressed in activated CD8+ cells (13, 14). Consistent with these reports, we observed T-bet expression in DMSO-treated cultures. However, as with EOMES expression, we found GSI treatment significantly reduces the expression of T-bet protein (Fig. 7A).
FIGURE 7.
Notch1 signaling directly regulates EOMES expression in GSI-treated CD8+ T cells. A, For in vitro treatment of CD8+ T cells (top blots), splenocytes were harvested from C57BL/6 mice and CD8+ T cells were purified before pretreating them in vitro with DMSO or GSI, stimulating them for 2 days with anti-CD3ε plus anti-CD28, and analyzing whole cell lysates by immunoblotting for EOMES and T-bet expression. For in vivo treatment of CD8+ T cells (bottom blots), control chow, or GSI formulated in rodent chow, was fed to C57BL/6 mice for 13 days. We then harvested splenocytes, purified CD8+ T cells, and stimulated these cells for 2 days with anti-CD3ε plus anti-CD28 before analyzing whole cell lysates by immunoblotting for EOMES expression. B, Two sets of PCR primers (set1 and set2) were designed to include putative binding sites for CSL and NF-κB and ChIP analysis of the EOMES promoter was performed using purified CD8+ T cells pretreated with DMSO or with GSI before stimulation for 2 days with anti-CD3ε plus anti-CD28 (top bands). Whole cell lysates were immunoprecipitated using anti-Notch1, anti-CSL, or anti-p50; rabbit and goat isotype IgG were used as negative controls. For PCR, we used 5 μl of DNA elutes. Results using primer set2 suggest that binding to putative sites within the EOMES promoter is specific. We quantified the relative intensity of the bands at the top, as represented graphically (bottom panel), which represents the mean intensity ± SD of three independent experiments. C, Exogenous EOMES was overexpressed in purified CD8+ T cells, and intracellular staining (left) and ELISA (right) were used to measure the ability of EOMES to induce IFN-γ in CD8+ T cells pretreated with GSI. PMA and calcium ionophore (CaI)-treated (PMA+CaI) and CD8+ T cells retrovirally infected with pMX-EOMES-IRES-hCD8 (EOMES +) or CD8+ T cells retrovirally transduced with empty vector of pMX-IRES-hCD8 as control (EOMES +) are also indicated. Data represent three or four independent replicates.
To verify whether Notch1 directly regulates EOMES expression, we examined ~1.2 kb of genomic DNA upstream of the mouse EOMES start site and again identified multiple putative binding sites for CSL and NF-κB (see supplemental Fig. S5).4 Using ChIP analysis, we determined that Notch1 forms a complex with CSL on the EOMES promoter (Fig. 7B). Treatment with GSI, in vitro, prevented recruitment of Notch1 to the promoter, although CSL binding remained intact, showing binding patterns similar to those observed with the perforin and granzyme B promoters (Fig. 4). Additionally, NF-κB could also be found on the EOMES promoter, and pretreatment with GSI prevented its corecruitment to NF-κB binding sites (Fig. 7B). These data suggest that CSL and NF-κB both are recruited to the EOMES promoter in a nuclear complex with Notch1 to directly regulate EOMES transcription.
EOMES expression is sufficient to drive production of IFN-γ (4), demonstrating IFN-γ is an excellent “readout” of EOMES activity. We previously showed that IFN-γ production in CD8+ T cells requires Notch activation (6) and we present data demonstrating that ectopic expression of N1ICD in CD8+ T cells drives IFN-γ even in the presence of GSI (see supplemental Fig. S6).4 We reasoned whether Notch-induced expression of IFN-γ in CD8+ T cells is mediated through Notch regulation of EOMES, the expression of EOMES should also rescue IFN-γ expression in the presence of GSI. Therefore, we questioned whether ec-topic expression of EOMES is sufficient to drive IFN-γ in CD8+ T cells. Our data show that retrovirally transduced EOMES expression drives IFN-γ in CD8+ T cells even in the presence of GSI, indicating that EOMES functions downstream of Notch signaling to regulate IFN-γ expression (Fig. 7C and see supplemental Fig. S7).4
Discussion
During the course of an infection, two distinct pathways regulate the cytolytic function of both NK and CD8+ T cells; namely granule exocytosis and Fas-FasL interactions (25, 26). The granule exocytosis pathway uses the pore-forming molecule, perforin and the protease, granzymes B, whereas the Fas-FasL pathway facilitates programmed cell death by aggregation of Fas (CD95) on target cells. Notch signaling has been implicated in the development of NK cells (27–31) as well as the development of CD8+ T cells in the thymus, although the latter is still controversial (32–37). However, the regulation of expression of perforin and granzyme B, in NK or CD8+ T cells has not been well studied. In this study, we show that Notch signaling in CD8+ T cells specifically regulates perforin and granzyme B expression but does not influence Fas-FasL interactions. The data presented in this study demonstrate that, in CD8+ T cells, stimulation with anti-CD3ε and anti-CD28, or with OVA258–276-expressing E.G7-OVA cells, increases the levels of perforin and granzyme B expression and blockade of Notch expression results in reduced expression of these molecules, as well as reduced cytolytic function. Furthermore, we show that protein complexes of Notch1, CSL or NF-κB are found on the perforin and granzyme B promoters, suggesting that Notch signaling directly regulates expression of these cytolytic proteins.
Using a pharmacological GSI, in vitro or in vivo, we show that loss of Notch signaling results in down-regulation of perforin and granzyme B expression, in addition to diminished CTL activity, in GSI-treated CD8+ T cells. Because GSI blocks activation of all four Notch proteins, the experiments we describe implicate Notch in the regulation of perforin and granzyme B, but do not determine which unique family member is critical to this process. There also are several other potential targets of GSI in the immune system. In addition to Notch, CD43 and CD44 are thought to be cleaved by gamma-secretase (11). Therefore, the interpretation of data obtained solely through the use of GSI can be confounding. In the experiments reported in this study, we used the use of Notch1 AS mice to confirm and extend our observations using GSI treatment. Notch1 AS mice exhibit 30–40% decreased Notch1 activity compared with control mice; however no differences were observed for Notch2, Notch3, and Notch4 (6). We show that both perforin and granzyme B expression are reduced in activated CD8+ T cells from these animals, suggesting that the effects obtained using GSI treatment are due, at least in part, to blockade of Notch activity. Additionally, EOMES expression in Notch1 AS mice is reduced, again providing genetic evidence for Notch regulation of EOMES.
We suggest the role played by Notch in the development of CTLs is intrinsic to the CD8+ T cell. Although the mechanism is not entirely defined, it is clear that CD4+ T cells play an important role in the CD8+ T cell response (38). In the experiments described in Fig. 1, we stimulated total splenocytes with Abs to CD3ε and CD28 and then used flow cytometric analysis to examine expression of perforin and granzyme B in CD8+ T cells. Therefore, both CD4+ and CD8+ T cells were exposed to these stimulation conditions. This exposure raised the possibility that the role of Notch signaling in perforin and granzyme B expression could be due indirectly to help provided by CD4+ T cells. To rule out this possibility, we prepared highly purified CD8+ T cells and pretreated these cells with GSI for 30 min. These cells were then mixed with CD8-depleted splenocytes, stimulated with anti-CD3ε and anti-CD28 for 2 days, and assayed for intracellular expression of perforin and granzyme B. We found decreased levels of both CTL effector proteins under conditions in which CD8+ T cells were pretreated with GSI. These data provide strong evidence that Notch signaling is an intrinsic requirement for perforin and granzyme B expression in developing CTLs. Because CD8+ T cells were pretreated for only 30 min and then washed, these data also suggest that Notch-dependent events occur within the first several hours following activation of CD8+ T cells. Although it may be surprising that GSI treatment for the first 30 min of activation blocks expression of perforin and granzyme B, we previously have shown that such a treatment protocol also blocks IFN-γ production in CD4+ T cells (12).
In Fig. 6, we provide in vitro and in vivo evidence that GSI partially blocks the development of CTLs. Data presented in Fig. 6A were obtained by immunizing mice with E.G7-OVA cells and restimulating CD8+ T cells in the presence of either GSI or control DMSO. Cells treated with GSI displayed reduced ability to kill E.G7-OVA targets. In Fig. 6B, we show data from CTLs generated in vivo in animals fed GSI in rodent chow. Animals fed GSI display significantly reduced levels of killing as compared with animals fed standard chow. As discussed, these experiments implicate GSI in the development of effector CTLs and only indirectly suggest a function for Notch in this process. However, we also provide evidence that Notch signaling is reduced by both the in vitro and in vivo treatment with GSI. We also show in earlier data that Notch signaling is critical in the expression of EOMES, perforin and granzyme B (Figs. 1 and 2). These three molecules are critical in the development of CTLs, therefore we suggest Notch may play an important role in the development of CTLs. A recent publication from the laboratory of Yasumoto and colleagues (39) demonstrating a cooperative role for Notch2 and CREB1 signaling in T cell cytotoxicity is consistent with our data.
In 2005, Reiner and colleagues (14) reported that effector CD8+ T cell function is controlled, at least in part, by the T-box transcription factor EOMES. These data and our own data demonstrating a role for Notch in the regulation of a related T-box protein T-bet, prompted us to ask whether Notch also regulates EOMES expression. As shown in Fig. 5B, Notch, CSL, and NF-κB all bind the EOMES promoter in a Notch-dependent fashion.
Previously we showed that Notch1 plays a critical role in the expression of IFN-γ in CD4+ cells. The data presented in this study demonstrate a role for Notch in IFN-γ production in CD8+ T cells as well. Because our data demonstrate direct regulation of EOMES expression by Notch occupancy on the EOMES promoter, it is possible that Notch regulates IFN-γ production through EOMES. However a recent report by Mayer et al. (40) demonstrates a critical role for T-bet in in vivo IFN-γ production by CD8+ T cells during infection and suggests that both T-box proteins, EOMES and T-bet, contribute to IFN-γ expression. In this study, we show that inhibition of Notch expression with GSI blocks expression both of EOMES and T-bet. We also present data showing reintroduction of EOMES, as a downstream target of Notch signaling, into GSI-treated cells rescues, at least in part, IFN-γ production. Therefore our data are consistent with the reported roles both for EOMES and T-bet in the regulation of IFN-γ expression in CD8+ T cells and indicate that Notch regulation of EOMES is an important factor in IFN-γ production in CD8+ effector T cells.
We reveal a novel role for Notch in regulating the effector functions of CD8+ CTL cells through the modulation of targeted cell lysis and IFN-γ production. This direct regulation by Notch of perforin, granzyme B, and EOMES may contribute to the effective clearance of pathogens. It may also provide a possible link to the destructive pathology seen in autoimmune diseases such as type 1 diabetes, multiple sclerosis, and rheumatoid arthritis. Identifying this critical function of Notch signaling in CD8+ T cells expands our insight into autoimmune conditions and may also be useful in the design of novel therapies for these diseases.
Supplementary Material
Acknowledgments
We thank Dr. Anthony Capobianco for the gift of Notch1 constructs, Dr. Steven Reiner for EOMES cDNA, and Dr. Yukiko Gotoh for pMX-IRES-hCD8 vector. We also thank Dr. Koji Yasutomo for helpful discussions and sharing of unpublished data.
Footnotes
This work was supported by Grants RO1 AI049361 and PO1 AG 025531 from the National Institutes of Health.
Abbreviations used in this paper
FasL, Fas ligand; EOMES, eomesodermin; GSI, gamma-secretase inhibitor; AS, antisense; ChIP, chromatin immunoprecipitation.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 2.Catalfamo M, Henkart PA. Perforin and the granule exocytosis cytotoxicity pathway. Curr. Opin. Immunol. 2003;15:522–527. doi: 10.1016/s0952-7915(03)00114-6. [DOI] [PubMed] [Google Scholar]
- 3.Nagata S, Suda T. Fas and Fas ligand: lpr and gld mutations. Immunol. Today. 1995;16:39–43. doi: 10.1016/0167-5699(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 4.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 5.Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 6.Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-γ production in peripheral T cells. J. Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- 7.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 8.Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat. Rev. Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 10.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 11.Kopan R, Ilagan MX. Gamma-secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 12.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 13.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 15.Cheng P, Zlobin A, Volgina V, Gottipati S, Osborne B, Simel EJ, Miele L, Gabrilovich DI. Notch-1 regulates NF-κB activity in hemopoietic progenitor cells. J. Immunol. 2001;167:4458–4467. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- 16.Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, Pear WS. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 17.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-κB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang CD, Chang GD, Lee YK, Chen H. A functional composite cis-element for NFκb and RBJκ in the rat pregnancy-specific glycoprotein gene. Biol. Reprod. 2001;65:1437–1443. doi: 10.1095/biolreprod65.5.1437. [DOI] [PubMed] [Google Scholar]
- 19.Beverly LJ, Capobianco AJ. Targeting promiscuous signaling pathways in cancer: another Notch in the bedpost. Trends Mol. Med. 2004;10:591–598. doi: 10.1016/j.molmed.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Shirakata Y, Shuman JD, Coligan JE. Purification of a novel MHC class I element binding activity from thymus nuclear extracts reveals that thymic RBP-Jκ/CBF1 binds to NF-κB-like elements. J. Immunol. 1996;156:4672–4679. [PubMed] [Google Scholar]
- 21.Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 22.Nair S, Wearsch PA, Mitchell DA, Wassenberg JJ, Gilboa E, Nicchitta CV. Calreticulin displays in vivo peptide-binding activity and can elicit CTL responses against bound peptides. J. Immunol. 1999;162:6426–6432. [PubMed] [Google Scholar]
- 23.Nair SK, Snyder D, Rouse BT, Gilboa E. Regression of tumors in mice vaccinated with professional antigen-presenting cells pulsed with tumor extracts. Int. J. Cancer. 1997;70:706–715. doi: 10.1002/(sici)1097-0215(19970317)70:6<706::aid-ijc13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 24.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 25.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 26.Nakazawa T, Agematsu K, Yabuhara A. Later development of Fas ligand-mediated cytotoxicity as compared with granule-mediated cytotoxicity during the maturation of natural killer cells. Immunology. 1997;92:180–187.. doi: 10.1046/j.1365-2567.1997.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoyama K, Delaney C, Varnum-Finney B, Kohn AD, Moon RT, Bernstein ID. The interaction of the Wnt and Notch pathways modulates Nk vs. T cell differentiation. Stem Cells. 2007;25:2488–2497. doi: 10.1634/stemcells.2007-0102. [DOI] [PubMed] [Google Scholar]
- 28.De Smedt M, Taghon T, Van de Walle I, De Smet G, Leclercq G, Plum J. Notch signaling induces cytoplasmatic CD3ε expression in human differentiating NK cells. Blood. 2007;110:2696–2703. doi: 10.1182/blood-2007-03-082206. [DOI] [PubMed] [Google Scholar]
- 29.DeHart SL, Heikens MJ, Tsai S. Jagged2 promotes the development of natural killer cells and the establishment of functional natural killer cell lines. Blood. 2005;105:3521–3527. doi: 10.1182/blood-2004-11-4237. [DOI] [PubMed] [Google Scholar]
- 30.Lehar SM, Dooley J, Farr AG, Bevan MJ. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 2005;105:1440–1447. doi: 10.1182/blood-2004-08-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolink AG, Balciunaite G, Demoliere C, Ceredig R. The potential involvement of Notch signaling in NK cell development. Immunol. Lett. 2006;107:50–57. doi: 10.1016/j.imlet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Robey E, Fowlkes BJ. Selective events in T cell development. Annu. Rev. Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 33.Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 34.Yasutomo K, Doyle C, Miele L, Fuchs C, Germain RN. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 2000;404:506–510. doi: 10.1038/35006664. [DOI] [PubMed] [Google Scholar]
- 35.Doerfler P, Shearman MS, Perlmutter RM. Presenilin-dependent gamma-secretase activity modulates thymocyte development. Proc. Natl. Acad. Sci. USA. 2001;98:9312–9317. doi: 10.1073/pnas.161102498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin. Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 37.Robey E, Schlissel M. Lymphocyte development. Curr. Opin. Immunol. 2003;15:155–157. doi: 10.1016/s0952-7915(03)00017-7. [DOI] [PubMed] [Google Scholar]
- 38.Bevan MJ. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 39.Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 2008;9:1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- 40.Mayer KD, Mohrs K, Reiley W, Wittmer S, Kohlmeier JE, Pearl JE, Cooper AM, Johnson LL, Woodland DL, Mohrs M. Cutting edge: T-bet and IL-27R are critical for in vivo IFN-γ production by CD8 T cells during infection. J. Immunol. 2008;180:693–697. doi: 10.4049/jimmunol.180.2.693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.