Abstract
Banana wilt outbreaks that are attributable to Moko disease-causing strains of the pathogen Ralstonia solanacearum (Rs) remain a social and economic burden for both multinational corporations and subsistence farmers. All known Moko strains belong to the phylotype II lineage, which has been previously recognized for its broad genetic basis. Moko strains are paraphyletic and are distributed among seven related but distinct phylogenetic clusters (sequevars) that are potentially major threats to Musaceae, Solanaceae, and ornamental crops in many countries. Although clustered within the Moko IIB-4 sequevar, strains of the epidemiologically variant IIB-4NPB do not cause wilt on Cavendish or plantain bananas; instead, they establish a latent infection in the vascular tissues of plantains and demonstrate an expanded host range and high aggressiveness toward Solanaceae and Cucurbitaceae. Although most molecular diagnostic methods focus on strains that wilt Solanaceae (particularly potato), no relevant protocol has been described that universally detects strains of the Musaceae-infecting Rs phylotype II. Thus, a duplex PCR assay targeting Moko and IIB-4NPB variant strains was developed, and its performance was assessed using an extensive collection of 111 strains representing the known diversity of Rs Moko-related strains and IIB-4NPB variant strains along with certain related strains and families. The proposed diagnostic protocol demonstrated both high accuracy (inclusivity and exclusivity) and high repeatability, detected targets on either pure culture or spiked plant extracts. Although they did not belong to the Moko clusters described at the time of the study, recently discovered banana-infecting strains from Brazil were also detected. According to our comprehensive evaluation, this duplex PCR assay appears suitable for both research and diagnostic laboratories and provides reliable detection of phylotype II Rs strains that infect Musaceae.
Introduction
The Cavendish banana and plantain (cooking banana) (Musa spp.) are among the most economically important crops, and they also represent staple foods in developing countries. However, various pathogens affect their production, and the most epidemiologically active pathogens are Fusarium oxysporum f.sp. cubense race 4 [1], banana bunchy top virus [2], and two bacteria that cause bacterial wilt, namely Xanthomonas vasicola pv. musacearum [3] and Ralstonia solanacearum (Rs) [4]. Bacterial wilt caused by Rs on both bananas and plantains continues to be a major constraint on the production of these crops, both for multi-national corporations and for subsistence farmers [5].
Recognized as a species complex (Rssc) [6], Rs is phylogenetically classified into four groups, called phylotypes, that take into account the phylogeography and evolutionary histories of the various strains [6,7]. The four groups are Asian phylotype I; American phylotype II, which encompasses the Moko and 4NPB strains; African phylotype III; and Indonesian phylotype IV, which encompasses the closely related species R. syzygii (Sumatra disease of clove trees) and the blood disease bacterium (BDB) [8,9]. A recently proposed taxonomic revision divides the Rssc into three genomic species [10]. While phylotype II is classified as a separate genomic species, its name remains R. solanacearum. The Rssc comprises strains that are capable of causing wilt in Musaceae plants and that cluster into two distant phylogenetic groups: (i) Moko disease-causing strains reported from Latin America, Asia, and the Philippines (Moko is recognized as Bugtok disease [11,12]) and (ii) the BDB originating in Indonesia and Malaysia. Systemic vascular infection by Rs induces symptoms that begin with the yellowing of leaves and tissue necrosis and that lead to a general collapse of the plant. The fruits are inedible and exhibit internal vascular discoloration. Specific symptoms can be observed, particularly with BDB, which produces a reddish coloration of the vascular ring in the fruit [13]. Bugtok disease only affects the floral bud, leading to hardening of the fruit (stone fruit) [14].
Phylotype II harbors the largest number of epidemiologically active ecotypes, such as Brown rot, Moko, NPB, and Granville wilt. As a working definition, the phylotypes are further subdivided into sequevars [6]. The Moko disease-causing strains are paraphyletic and have historically clustered into four sequevars: IIA-6, IIA-24, IIB-3, and IIB-4. The pathological variant IIB-4NPB was first reported in diseased anthurium (Anthurium andreanum) in Martinique [15] and was phylogenetically assigned to the Moko lineage IIB-4. These strains are variants that are not pathogenic to bananas (NPB) but that demonstrate a host range that expands to Cucurbitaceae.
The Moko-associated strains, in addition to being soil-borne and transmitted through wounds and cuttings, can also be actively transmitted by insects through the bud [16]; the pathogen then migrates down into the plant, leading to symptoms that start with fruit decay and end in plant collapse. In addition to these two groups, the epidemiological 4NPB lineage variant, which is grouped into the Moko sequevar IIB-4 lineage, does not cause wilt on Cavendish or plantain bananas; instead, this variant establishes itself and moves within the vascular tissues of plantains, even via soil-borne contamination, as it establishes a latent infection through the root system [15].
Recently, unanticipated Moko disease-related strains from Brazil were reported [17] that clustered (i) into the previously described Solanaceae-related sequevars IIA-41 and IIB-25, which are not related to historic Moko lineages, and (ii) into a newly proposed sequevar, IIA-53. This finding highlights the fact that the Moko ecotype benefits from a broad genetic basis and harbors far more genetic diversity than anticipated [17,18]. The current Moko strain-specific molecular diagnostic method consists of a Musa multiplex PCR (Mmx-PCR) [19,20] that targets the historically known sequevars IIA-6, IIA-24, IIB-3, and IIB-4 by producing a size-specific amplification band for each Moko sequevar. IIB-4 Moko strains also produce another specific amplification band that is not observed with IIB-4NPB strains. However, this protocol was unable to specifically detect the Moko disease-related strains reported in Brazil by Albuquerque et al. [17], as it relies on the characterization of historically known sequevars. Therefore, it appears that there is no official diagnostic method suitable for the specific detection of phylotype II strains that infect bananas. There is a strong need for such a method when conducting territory and border surveillance, for basic material in vitro, and for plantlet banana production, as these processes must be free of quarantine-propagative pathogens, including Rssc strains. In addition to representing a major threat to the banana trade and to sustainable production, these strains may also threaten Solanaceae production, as most of them are pathogenic to potato (Solanum tuberosum) or tomato (S. lycopersicum) under artificial conditions [21].
Based on the whole-genome sequences of Moko strains, we developed a robust, simple, and affordable duplex PCR assay that is specific for phylotype II Rssc strains that can be retrieved from banana and plantain tissues (Moko disease-causing strains and IIB-4NPB pathological variants). Here, we present an extensive characterization of the performance of the duplex PCR assay.
Materials and Methods
Bacterial strains and viruses
A set of 111 reference strains was selected to cover the known genetic diversity among Rssc strains and strains related to the Rssc (Table 1). Within the Rssc, a total of 40 bacterial strains were selected as targets, while 45 bacterial strains were selected as non-targets. Additionally, 21 non-target bacterial strains that are phylogenetically close to the Rssc or related to banana diseases were selected. Finally, 5 viruses were selected as being related to banana diseases. The bacterial strains were obtained from the Centre de coopération Internationale en Recherche Agronomique pour le Développement (CIRAD—Saint Pierre, Reunion Island) and were stored at -80°C on cryobeads (Microbank, Pro-labs Diagnostics, Toronto, Canada). The bacteria were cultured overnight in Luria-Bertani broth (LB) at 28°C with 250 rpm agitation, streaked on modified Sequeira semi-selective solid medium containing agar (18 g/L), yeast extract (1 g/L), peptone (11 g/L), glycerol (6.3 g/L), crystal violet (2 mg/L), polymyxin-β-sulfate (10 mg/L), tyrothricine (20 mg/L), chloramphenicol (5 mg/L), 2,3,5-triphenyltetrazolium chloride (11 mg/L), Tilt (Propiconazole; Syngenta, Bâle, Switzerland; 0.004%), and penicillin (20 U) and incubated for 48h at 28°C. Calibrated bacterial suspensions were generated in 0.1 M Tris-HCl (pH 7.1) (Sigma-Aldrich, Saint-Louis, MO, USA) adjusted initially to 108 CFU/mL, as determined by measuring an optical density of 0.1 at 650 nm (Biomate 3, Thermo Scientific, Boston, MA, USA). Successive dilutions were prepared using molecular biology-grade water and were quantified on modified Sequeira semi-selective solid medium.
Table 1. Accuracy assessment on pure culture of target and non-target strains related to the Ralstonia solanacearum species complex and other related families.
| Strain | Description | 93F/93R 1 | 5F/5R 1 | ||||
|---|---|---|---|---|---|---|---|
| Alternative ID | Isolation Host | Country | Phylotype | Sequevar | |||
| 9–1 | Bluggoe banana | Grenada | IIA | 6 | ++-/++- | —-/—- | |
| A3909 | Heliconia rostrata | USA | IIA | 6 | +++/+++ | —-/—- | |
| GMI8044 | BA7 | Cavendish banana | Grenada | IIA | 6 | +++/+++ | —-/—- |
| GRE T11L1 | Solanum lycopersicum | Grenada | IIA | 6 | +++/-++ | —-/—- | |
| GUY B06E2 | Cavendish banana | French Guiana | IIA | 6 | -++/-+- | —-/—- | |
| UQRS457 | Heliconia rostrata | Hawaii | IIA | 6 | +++/++- | —-/—- | |
| UW588 | Cavendish banana | Guatemala | IIA | 6 | +-+/+— | —-/—- | |
| B26 | Musa spp. | Brazil | IIA | 24 | +—/++- | —-/—- | |
| B34 | Musa spp. | Brazil | IIA | 24 | +++/+++ | —-/—- | |
| B43 | Musa spp. | Brazil | IIA | 24 | +++/+++ | —-/—- | |
| B50 | Musa spp. | Brazil | IIA | 24 | +++/+++ | —-/—- | |
| B91 | Musa spp. | Brazil | IIA | 24 | +++/+++ | —-/—- | |
| IBSBF1900 | Musa spp. | Brazil | IIA | 24 | +++/+++ | —-/—- | |
| CFBP1183 | UQRS35, JS793; 07–027 | Heliconia rostrata | Costa Rica | IIB | 3 | +++/+++ | —-/—- |
| CFBP1416 | K138; JS746; JS748 | Plantain banana | Costa Rica | IIB | 3 | +++/+++ | —-/—- |
| CIP414 | Bug14 | Musa spp. | Philippines | IIB | 3 | +++/+-+ | —-/—- |
| CIP417 | Bug2 | Musa spp. | Philippines | IIB | 3 | +-+/+-+ | —-/—- |
| JT644 | UW9; S147 | Heliconia rostrata | Costa Rica | IIB | 3 | +++/+++ | —-/—- |
| UW11 | S167; K207 | Heliconia rostrata | Costa Rica | IIB | 3 | +-+/-++ | —-/—- |
| UW166 | UQRS18, CIP27 | Plantain banana | Costa Rica | IIB | 3 | +++/+++ | —-/—- |
| UW2 | UQRS17, R377, CIP2 | Musa spp. | Costa Rica | IIB | 3 | +++/+++ | —-/—- |
| UW28 | Solanum tuberosum | Cyprus | IIB | 3 | +++/+++ | —-/—- | |
| CFBP1415 | JS740; K82 | Solanum tuberosum | Colombia | IIB | 4 | +++/+++ | —-/—- |
| CFBP1418 | JS790 | Heliconia rostrata | Costa Rica | IIB | 4 | ++-/+++ | —-/—- |
| LNPV31.10 | 2006 539 | Musa spp. | French Guiana | IIB | 4 | -++/-++ | —-/—- |
| SVG B09B1 | Musa spp. | Saint Vincent | IIB | 4 | ++-/+++ | —-/—- | |
| UW160 | GMI8138; S253 | Plantain banana | Peru | IIB | 4 | +++/+-+ | —-/—- |
| UW163 | S256; GMI8235 | Plantain banana | Peru | IIB | 4 | +++/-++ | —-/—- |
| UW170 | Heliconia rostrata | Colombia | IIB | 4 | +++/+++ | —-/—- | |
| UW179 | R368; CIP30; K254 | Plantain banana | Colombia | IIB | 4 | +++/+++ | —-/—- |
| ANT80 | CIR02–080 | Anthurium andreanum | Martinique | IIB | 4NPB | +++/+-+ | +++/+++ |
| CFBP6780 | 02–143–1; 8283 | Solanum lycopersicum | Martinique | IIB | 4NPB | +++/+++ | +++/+++ |
| CFBP6783 | ANT75; CIR02–075 | Heliconia caribea | Martinique | IIB | 4NPB | +++/+++ | +++/+++ |
| CFBP6797 | PV8; SPV02–30308; 8280 | Solanum americanum | Martinique | IIB | 4NPB | +++/+++ | +++/+++ |
| CFBP7014 | 06–024; 8291 | Anthurium andreanum | Trinidad | IIB | 4NPB | +++/+-+ | +++/+-+ |
| IBSBF1454 | Cucurbita pepo | Brazil | IIB | 4NPB | +-+/+-+ | +++/+++ | |
| IBSBF1503 | Cucumis sativus | Brazil | IIB | 4NPB | +++/+++ | -++/+++ | |
| LNPV24.25 | 2001 868 | Solanum lycopersicum | France | IIB | 4NPB | ++-/+— | +++/+++ |
| LNPV30.75 | 2006 0261 | Capsicum annuum | French Guiana | IIB | 4NPB | +-+/+— | +++/+++ |
| PV1 | SPV02–60196 | Solanum melongena | Martinique | IIB | 4NPB | +-+/-++ | +++/+++ |
| R288 | CFBP6442; UW373 | Morus alba | China | I | 12 | —-/—- | —-/—- |
| CFBP7058 | CMR134 | Vaccinium membranaceum | Cameroon | I | 13 | —-/—- | —-/—- |
| PSS4 | Solanum lycopersicum | Taiwan | I | 15 | —-/—- | —-/—- | |
| ACH92 | CFBP6425 | Zingiber officinale | Australia | I | 16 | —-/—- | —-/—- |
| IBSBF1882 | Begonia semperflorens | Brazil | I | 17 | —-/—- | —-/—- | |
| GMI1000 | JS753 | Solanum lycopersicum | French Guiana | I | 18 | —-/—- | —-/—- |
| JT519 | Pelargonium asperum | Reunion | I | 31 | —-/—- | —-/—- | |
| CIP365 | WP144 | Solanum tuberosum | Philippines | I | 45 | —-/—- | —-/—- |
| MAD-017 | Capsicum annuum | Madagascar | I | 46 | —-/—- | —-/—- | |
| GMI8254 | TO1 | Solanum lycopersicum | Indonesia | I | 47 | —-/—- | —-/—- |
| K60 | CFBP2047; LMG2299T | Solanum lycopersicum | USA | IIA | 7 | —-/—- | —-/—- |
| RF32 | Solanum lycopersicum | Trinidad | IIA | 7 | —-/—- | —-/—- | |
| CIV30 | Solanum lycopersicum | IvoryCoast | IIA | 35 | —-/—- | —-/—- | |
| CFBP2957 | MT5 | Solanum lycopersicum | Martinique | IIA | 36 | —-/—- | —-/—- |
| CFBP2958 | GT4 | Solanum lycopersicum | Guadeloupe | IIA | 39 | —-/—- | —-/—- |
| JQ1143 | Solanum tuberosum | Reunion | IIA | 39 | —-/—- | —-/—- | |
| CIP239 | UW469 | Solanum tuberosum | Brazil | IIA | 40 | —-/—- | —-/—- |
| AP31H | Solanum tuberosum | Uruguay | IIB | 1 | —-/—- | —-/—- | |
| CFBP1417 | Solanum tuberosum | Australia | IIB | 1 | —-/—- | —-/—- | |
| CFBP3858 | JS907; PD2763 | Solanum tuberosum | Netherlands | IIB | 1 | —-/—- | —-/—- |
| IPO1609 | Solanum tuberosum | Netherlands | IIB | 1 | —-/—- | —-/—- | |
| JT516 | Solanum tuberosum | Reunion | IIB | 1 | —-/—- | —-/—- | |
| LNPV19.66 | Solanum tuberosum | France | IIB | 1 | —-/—- | —-/—- | |
| UW551 | Pelargonium asperum | Kenya | IIB | 1 | —-/—- | —-/—- | |
| CFBP3879 | PD1958; CFBP1414 | Solanum tuberosum | Colombia | IIB | 2 | —-/—- | —-/—- |
| DGBBC1138 | Solanum tuberosum | Guinea | III | 43 | —-/—- | —-/—- | |
| CMR33 | Solanum lycopersicum | Cameroon | III | 20 | —-/—- | —-/—- | |
| CFBP6942 | CMR32 | Vaccinium membranaceum | Cameroon | III | 29 | —-/—- | —-/—- |
| CMR15 | CFBP6941 | Solanum lycopersicum | Cameroon | III | 29 | —-/—- | —-/—- |
| CMR20 | Solanum lycopersicum | Cameroon | III | 29 | —-/—- | —-/—- | |
| CFBP3059 | JS904 | Solanum melongena | Burkina Faso | III | 23 | —-/—- | —-/—- |
| CFBP7038 | CMR66 | Vaccinium membranaceum | Cameroon | III | 49 | —-/—- | —-/—- |
| JT525 | Pelargonium asperum | Reunion | III | 19 | —-/—- | —-/—- | |
| NCPPB1029 | B509 | Pelargonium asperum | Reunion | III | 19 | —-/—- | —-/—- |
| NCPPB1018 | JS950 | Solanum tuberosum | Angola | III | 21 | —-/—- | —-/—- |
| MAFF301558 | 06–042 | Solanum tuberosum | Japan | IV | 8 | —-/—- | —-/—- |
| JT663 | R008 | Syzygium aromaticum | Indonesia | IV | 9 | —-/—- | —-/—- |
| R24 | UQRS466 | Syzygium aromaticum | Indonesia | IV | 9 | —-/—- | —-/—- |
| R28 | Syzygium aromaticum | Indonesia | IV | 9 | —-/—- | —-/—- | |
| PSI07 | CFBP7288 | Solanum lycopersicum | Indonesia | IV | 10 | —-/—- | —-/—- |
| R229 | NCPPB3726 | Musa spp. | Indonesia | IV | 10 | —-/—- | —-/—- |
| UQRS283 | T38 | Solanum lycopersicum | Indonesia | IV | 10 | —-/—- | —-/—- |
| UQRS627 | 1712075KkB-PWR | Musa spp. | Indonesia | IV | 10 | —-/—- | —-/—- |
| UQRS633 | 240808RB-MND | Musa spp. | Indonesia | IV | 10 | —-/—- | —-/—- |
| ACH732 | CIP357 | Solanum lycopersicum | Australia | IV | 11 | —-/—- | —-/—- |
| LMG5942T | Ralstonia pickettii | —-/—- | —-/—- | ||||
| LMG21421 | Ralstonia insidiosa | —-/—- | —-/—- | ||||
| LMG6866T | Ralstonia mannitolytica | —-/—- | —-/—- | ||||
| LMG1199T | Ralstonia eutropha | —-/—- | —-/—- | ||||
| LMG2172T | Pseudomonas corrugata | —-/—- | —-/—- | ||||
| LMG5093 | Pseudomonas syringae pv. tomato | —-/—- | —-/—- | ||||
| LMG2162T | Pseudomonas cichorii | —-/—- | —-/—- | ||||
| LMG16206 | Pseudomonas putida | —-/—- | —-/—- | ||||
| LMG1794T | Pseudomonas fluorescens | —-/—- | —-/—- | ||||
| LMG2404T | Pectobacterium corotovorum subsp. carotovorum | —-/—- | —-/—- | ||||
| LMG2129T | Burkholderia andropogonis | —-/—- | —-/—- | ||||
| LMG1222T | Burkholderia cepacia | —-/—- | —-/—- | ||||
| LMG2804T | Erwinia chrysanthemi | —-/—- | —-/—- | ||||
| LMG2894 | Clavibacter michiganensis subsp. sepedonicus | —-/—- | —-/—- | ||||
| LMG7333 | Clavibacter michiganensis subsp. michiganensis | —-/—- | —-/—- | ||||
| NCPPB881T | Xanthomonas gardneri | —-/—- | —-/—- | ||||
| NCPPB4321T | Xanthomonas perforans | —-/—- | —-/—- | ||||
| LMG911T | Xanthomonas vesicatoria | —-/—- | —-/—- | ||||
| NCPPB2968T | Xanthomonas euvesicatoria | —-/—- | —-/—- | ||||
| CFBP5827 | Xanthomonas campestris pv. raphani | —-/—- | —-/—- | ||||
| CFBP7171 | Xanthomonas vasicola pv. musacearum | —-/—- | —-/—- | ||||
| BBTV_NvllCal | Banana bunchy top virus 3 | —-/—- | —-/—- | ||||
| CMV011 | Cucumber mosaic virus 3 | —-/—- | —-/—- | ||||
| BSV-001 | Banana streak virus 3 | —-/—- | —-/—- | ||||
| BBrMV008 | Banana bract mosaic virus 3 | —-/—- | —-/—- | ||||
| BanMMV001 | Banana mild mosaic virus 3 | —-/—- | —-/—- | ||||
| Inclusivity 2 | 93% [0.86–0.97] | 100% [0.88–1] | |||||
| Exclusivity 2 | 100% [0.98–1] | 100% [0.99–1] | |||||
| Accuracy 2 | 97%[0.95–0.99] | 100%[0.99–1] | |||||
1 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated three times (separated by a slash)
2 calculated according to the ratio of agreements relatively to the total repetitions performed within the experiment (agreements and disagreements); numbers in brackets indicates the binomial exact proportion confidence interval
3 viruses were tested on plant extracts. Codification for international collection are as follow: IBSBF: Biological Institute Culture Collection of Phytopathogenic Bacteria (Brazil); CFBP: Collection Française de Bactéries Phytopathogènes (French Collection of Phytopathogenic Bacteria) (France); BCCM/LMG: Belgian Co-ordinated Collections of Micro-organisms/ Laboratory of Microbiology of Ghent (Belgium); NCPPB: National Collection of Plant Pathogenic Bacteria (UK); MAFF: Ministry of Agriculture, Forestry and Fisheries Genebank (Japan).
The phylotype multiplex PCR (Pmx-PCR) developed by Fegan and Prior [6] was used to confirm that the strains belonged to the Rssc and to determine their phylotypes. Partial egl sequencing was performed to determine the sequevars [22]. The Moko lineage strains were typed using the Moko multiplex PCR (Mmx-PCR) designed to identify Moko strains in the historic phylotypes IIA-6, IIB-3, IIB-4, IIB-4NPB [19], and IIA-24 [20].
Viruses were obtained from lyophilized reference samples in the Plant Health Laboratory (Anses-Reunion Island).
No DNA extraction was required during the workflow.
Duplex PCR assay development
Marker selection was performed by conducting genome comparisons using the Gene Phyloprofile Exploration tool (default parameters) of the MicroScope platform [23] (Genoscope, Evry, France) of both the Moko and IIB-4NPB strains versus the non-target strains from the Rssc and non-Rssc databases (available online: https://www.genoscope.cns.fr/agc//microscope/about/microscopeprojects.php?P_id=67). The fully sequenced and annotated genomes of Moko-related strains included the following: Grenada 9–1 (IIA-6), UW181 (IIA-6), B50 (IIA-24), IBSBF1900 (IIA-24), CFBP1416 (IIB-3), CIP417 (IIB-3), Molk2 (IIB-3), UW163 (IIB-4), and UW179 (IIB-4). Genomes from IIB-4NPB strains included CFBP6783, IBSBF1503, and CFBP7014. Primers were designed using Primer3 [24,25] running under Geneious v6.1.8 (Biomatters, available at http://www.geneious.com/).
The optimized 25-μL (total volume) PCR reaction mixture contained 1x PCR GoTaq Green Buffer Mix and 0.625 U of GoTaq Hot Start polymerase (Promega, Madison, WI, USA), 1.5 mM MgCl2, 0.2 mM dNTPs, 2 μM each of the 4 primers, and 2 μL of the analysis strain suspended in molecular biology-grade water. The PCR amplification was performed using a Veriti thermal cycler (Applied Biosystems, Carlsbad, CA, USA) with the following parameters: 5 min at 96°C, followed by 35 cycles of 94°C for 15 s, 70°C for 30 s, and 72°C for 30 s, with a final extension step for 10 min at 72°C.
Performance assessment
Accuracy (analytical specificity)
The performance of the two new sets of primers to detect Moko disease-causing strains and 4NPB variant strains was fully evaluated, as required in the ISO 17025 standard [26] (general requirements for the competence of testing and calibration laboratories), by following European and Mediterranean Plant Protection Organization (EPPO) protocols [27,28], thereby ensuring the highest confidence in method development and validation. The assessment first involved the evaluation of the accuracy, referred to as “analytical specificity” in the EPPO standard [27], which relies on the qualitative detection capacity of the method and that includes two criteria, namely inclusivity and exclusivity. Inclusivity is the ability to avoid false negatives, while exclusivity is the ability to avoid false positives. The accuracy evaluation was performed using the duplex PCR assay with 111 strains (Table 1; n = 40 target strains; n = 71 non-target strains) at a concentration of 108 CFU/mL in molecular biology-grade water. The inclusivity and the exclusivity were assessed for three replicates and were calculated using the ratio of agreement (true positives) relative to the total repetitions performed within the experiment (agreements and disagreements). Accuracy was calculated following the same exact methods as for the inclusivity and exclusivity (Table 1). Binomial exact proportion confidence interval was calculated for all the three parameters, namely inclusivity, exclusivity, and accuracy scores (R statistical softwarev3.1.2 [29], package “STATS” function “binom.test”). PCR was performed on pure cultured strains.
Detectability (analytical sensitivity) and repeatability
Second, the assessment involved the evaluation of the detectability, referred to as “analytical sensitivity” in the EPPO standard [27], and repeatability (EPPO standard), which relies on the quantitative detection capacity of target strains. Detectability refers to the smallest amount of target that can be reliably detected, and repeatability characterizes the level of agreement among replicates of a sample tested under the same conditions. The detectability and repeatability evaluation was performed using the primer sets 93F/93R and 5F/5R in simplex PCR (Table 2, n = 15, and Table 3, n = 10) and in duplex PCR (Table 4, n = 22). To simulate routine laboratory conditions, this test was performed using strains calibrated at 108 CFU/mL in molecular biology-grade water that were successively diluted to 6 different concentrations, ranging from 106 to 101 CFU/mL, with banana pseudo-stem extracts (2.0 g of pseudo-stems were ground in 5 mL of molecular biology-grade water). As above, the detectability and repeatability were assessed for three replicates and were calculated according to the ratio of agreement (true positives) to the total repetitions performed within the experiment (agreements and disagreements). PCR was performed on spiked plant extracts.
Table 2. Detectability and repeatability assessment in artificially contaminated plant extracts for primers 93F/93R specific to Moko disease-causing strains and IIB-4NPB strains.
| Strain | Phylotype-Sequevar | Concentration (CFU/mL) 1 | |||||
|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | ||
| GMI8044 | IIA-6 | +++/+++ | +++/+++ | +++/—+ | —-/—- | —-/—- | —-/—- |
| 9–1 | IIA-6 | +++/+++ | +++/+++ | +++/+-+ | —+/—+ | —-/—- | —-/—- |
| A3909 | IIA-6 | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| IBSBF1900 | IIA-24 | +++/+++ | +++/+++ | +++/+-+ | —+/—+ | —-/—- | —-/—- |
| B50 | IIA-24 | +++/+++ | +++/+++ | +-+/+-+ | —-/—+ | —-/—- | —-/—- |
| B26 | IIA-24 | +++/+++ | +++/+++ | ++-/++- | -+-/—- | —-/—- | —-/—- |
| CIP417 | IIB-3 | +++/+++ | +++/+++ | +++/-++ | —+/-++ | —-/—+ | —-/—- |
| CFBP1416 | IIB-3 | +++/+++ | +++/+++ | +++/-++ | —+/-++ | —-/—+ | —-/—- |
| MOLK2 | IIB-3 | +++/+++ | +++/+++ | +-+/+++ | —+/+-+ | —-/—- | —-/—- |
| UW160 | IIB-4 | +++/+++ | +++/+++ | —+/-++ | +—/—- | —-/—- | —-/—- |
| UW163 | IIB-4 | +++/+++ | +++/+++ | —+/-++ | —-/—- | —-/—- | —-/—- |
| UW179 | IIB-4 | +++/+++ | +++/+++ | +-+/++- | —-/—- | —-/—- | —-/—- |
| CFBP6780 | IIB-4NPB | +++/+++ | +++/+++ | +—/—- | —-/—- | —-/—- | —-/—- |
| CFBP6783 | IIB-4NPB | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| CFBP7014 | IIB-4NPB | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| Detectability 2 | 100% | 100% | 67% | 24% | 4% | 0% | |
| Repeatability 2 | 100% | 100% | 80% | 80% | 100% | 100% | |
1 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated three times (separated by a slash)
2 calculated according to the ratio of agreements relatively to the total repetitions performed within the experiment (agreements and disagreements).
Table 3. Detectability and repeatability assessment in artificially contaminated plant extracts for primers 5F/5R specific to IIB-4NPB strains.
| Strain | Phylotype-Sequevar | Concentration (CFU/mL) 1 | |||||
|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 10 3 | 10 2 | 10 1 | ||
| ANT80 | IIB-4NPB | +++/+++ | +++/+++ | +-+/+— | —-/-+- | —-/—- | —-/—- |
| CFBP6780 | IIB-4NPB | +++/+++ | +++/+++ | +—/+— | —-/—+ | —-/—- | —-/—- |
| CFBP6783 | IIB-4NPB | +++/+++ | +++/+++ | +—/+— | —+/—+ | —-/—- | —-/—- |
| CFBP6797 | IIB-4NPB | +++/+++ | +++/+++ | +—/-++ | +—/—- | —-/—- | —-/—- |
| CFBP7014 | IIB-4NPB | +++/+++ | +++/+++ | —+/-++ | —-/—- | —-/—- | —-/—- |
| IBSBF1454 | IIB-4NPB | +++/+++ | +++/+++ | +—/—+ | —-/—+ | —-/—- | —-/—- |
| IBSBF1503 | IIB-4NPB | +++/+++ | +++/+++ | +—/+— | —-/—- | —-/—- | —-/—- |
| LNPV24.25 | IIB-4NPB | +++/+++ | +++/+++ | -+-/-+- | —+/—- | —-/—- | —-/—- |
| LNPV30.75 | IIB-4NPB | +++/+++ | +++/+++ | —+/—+ | —-/—- | —-/—- | —-/—- |
| PV1 | IIB-4NPB | +++/+++ | +++/+++ | +—/-+- | —+/—+ | —-/—- | —-/—- |
| Detectability 2 | 100% | 100% | 53% | 23% | 0% | 0% | |
| Repeatability 2 | 100% | 100% | 90% | 83% | 100% | 100% | |
1 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated three times (separated by a slash)
2 calculated according to the ratio of agreements relatively to the total repetitions done within the experiment (agreements and disagreements).
Table 4. Detectability and repeatability assessment in artificially contaminated plant extracts of the duplex-PCR 93F/93R & 5F/5R for Ralstonia solanacearum Moko disease-causing strains and IIB-4NPB strains.
| Strain | Phylotype-Sequevar | Concentration (CFU/mL) 1 | |||||
|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | ||
| GMI8044 | IIA-6 | +++/+++ | +++/+++ | -++/-++ | —-/—+ | —-/—- | —-/—- |
| 9–1 | IIA-6 | +++/+++ | +++/+++ | -++/—+ | +—/—- | —-/—- | —-/—- |
| A3909 | IIA-6 | +++/+++ | +++/+++ | +—/+— | —-/—- | —-/—- | —-/—- |
| IBSBF1900 | IIA-24 | +++/+++ | +++/+++ | -++/-++ | -+-/—- | —-/—- | —-/—- |
| B50 | IIA-24 | +++/+++ | +++/+++ | +-+/+— | +—/—- | —-/—- | —-/—- |
| B26 | IIA-24 | +++/+++ | +++/+++ | ++-/++- | —-/—- | —-/—- | —-/—- |
| CIP417 | IIB-3 | +++/+++ | +++/+++ | —+/-++ | —+/—+ | —-/—- | —-/—- |
| CFBP1416 | IIB-3 | +++/+++ | +++/+++ | -+-/-+- | —-/—- | —-/—- | —-/—- |
| MOLK2 | IIB-3 | +++/+++ | +++/+++ | +—/+— | +—/—- | —-/—- | —-/—- |
| UW160 | IIB-4 | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| UW163 | IIB-4 | +++/+++ | +++/+++ | +—/+— | —-/—- | —-/—- | —-/—- |
| UW179 | IIB-4 | +++/+++ | +++/+++ | -++/-++ | —-/—- | —-/—- | —-/—- |
| ANT80 | IIB-4NPB | +++/+++ | +++/+++ | -+-/-+- | -+-/-+- | —-/—- | —-/—- |
| CFBP6780 | IIB-4NPB | +++/+++ | +++/+++ | —+/—+ | —-/—- | —-/—- | —-/—- |
| CFBP6783 | IIB-4NPB | +++/+++ | +++/+++ | +—/+— | -+-/-+- | —-/—- | —-/—- |
| CFBP6797 | IIB-4NPB | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| CFBP7014 | IIB-4NPB | +++/+++ | +++/+++ | ++-/++- | ++-/++- | —-/—- | —-/—- |
| IBSBF1454 | IIB-4NPB | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| IBSBF1503 | IIB-4NPB | +++/+++ | +++/+++ | +—/+— | +—/+— | —-/—- | —-/—- |
| LNPV24.25 | IIB-4NPB | +++/+++ | +++/+++ | —+/—+ | —+/—+ | —-/—- | —-/—- |
| LNPV30.75 | IIB-4NPB | +++/+++ | +++/+++ | -++/-++ | —-/—- | —-/—- | —-/—- |
| PV1 | IIB-4NPB | +++/+++ | +++/+++ | —-/—- | —-/—- | —-/—- | —-/—- |
| Detectability 2 | 100% | 100% | 41% | 18% | 0% | 0% | |
| Repeatability 2 | 100% | 100% | 95% | 92% | 100% | 100% | |
1 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated three times (separated by a slash)
2 calculated according to the ratio of agreements relatively to the total repetitions performed within the experiment (agreements and disagreements).
Control
Several controls were used throughout the evaluation of the protocol, including a negative process control (non-target strain), a positive process control (strains UW28 (IIB-4) and CFPB7014 (IIB-4NPB)), and a negative PCR control (water). The banana plants used (Musa acuminata cv. 911) (AAA-Cavendish) were obtained in vitro from Vitropic (Saint-Mathieu-de-Tréviers, France) and grown in nurseries until the 5 true leaf stage. The plants were previously typed as R. solanacearum-free; no symptoms of any disease were observed.
Selectivity
Several healthy banana cultivars obtained from Vitropic were tested to assess any cross-reactions; they included Williams (AAA-Cavendish), Flhorban 925 (AAA-Cavendish synthetic hybrid), 902 (AAA-Cavendish), Brimbo 1 (AAB-plantain), Corne (AAB-plantain), Creole French (AAB-French Horn), and Big Ebanga (AAB-plantain).
Full-length protocol test
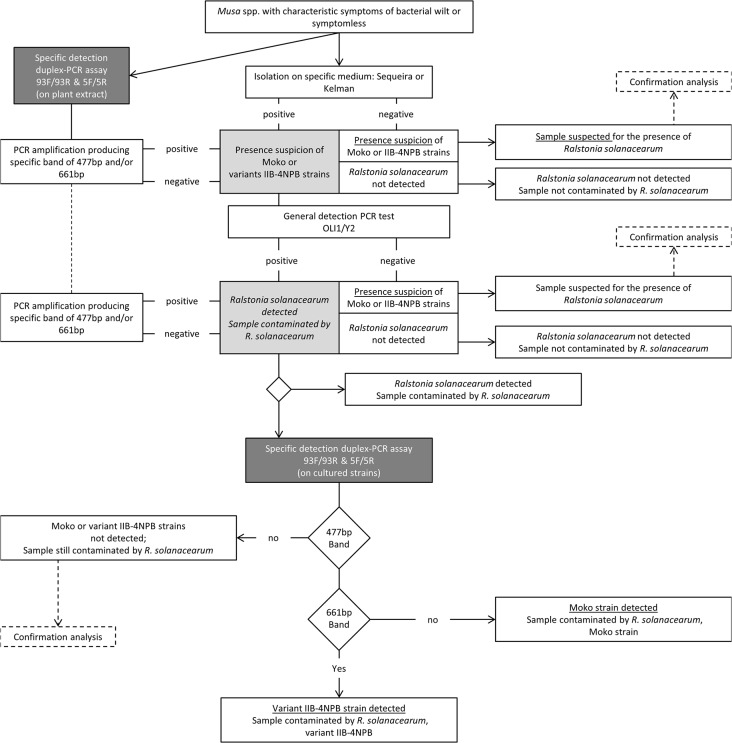
In addition, a full-length protocol assessment (Fig. 1) was conducted on a set of 15 strains (Table 5) and was repeated twice. Each targeted group was assessed for detection by following the steps of the proposed scheme (Fig. 1). A plant extract solution, composed of 2.0 g of pseudo-stems ground in 5 mL of molecular biology grade water, was used to adjust the concentration of the Rs strains to 105 CFU/mL. This spiked plant extract (50 μL) was plated on solid modified Sequeira medium for colony isolation. Typical colonies (pinkish color in the center and creamy white color on the side, smooth aspect, average elevation, and irregular border) were retrieved and characterized by PCR. We first performed PCR with the primer set Oli1/Y2 [13], then with the Mmx-PCR [19,20], and finally with the duplex PCR assay with 93F/93R and 5F/5R. PCR was conducted only on pure cultures; no DNA extraction was performed. In parallel with plating, plant extracts were also tested using the same PCR protocols.
Fig 1. Detection scheme of Ralstonia solanacearum phylotype II strains in Musa spp.: Moko and variant IIB-4NPB.
Table 5. Results of the proposed full detection protocol for detection of Ralstonia solanacearum in artificially contaminated plant extracts and identification of isolated bacteria.
| Strain | Phylotype | Sequevar | Sequeira 1 | Oli1/Y2 2 | Musa Mmx 2 | Duplex 93/5 2 , 3 | |||
|---|---|---|---|---|---|---|---|---|---|
| Plant Extracts | Pure Culture | Plant Extracts | Pure Culture | Plant Extracts | Pure Culture | ||||
| A3909 | IIA | 6 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| GMI8044 | IIA | 6 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| UW588 | IIA | 6 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| B43 | IIA | 24 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| B50 | IIA | 24 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| B91 | IIA | 24 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| Molk2 | IIB | 3 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| CFBP1183 | IIB | 3 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| UW28 | IIB | 3 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| UW163 | IIB | 4 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| UW170 | IIB | 4 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| UW179 | IIB | 4 | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| CFBP6780 | IIB | 4NPB | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| CFBP6783 | IIB | 4NPB | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
| IBSBF1503 | IIB | 4NPB | +/+ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ | ++/++ |
1 ‘+’: positive result; ‘-’: negative result; two platings were performed on the modified Sequeira semi-selective medium per sample (separated by a slash)
2 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated twice (separated by a slash)
3 Results given by the duplex-PCR 93F/R & 5F/R were in accordance with the type of strain: Moko or IIB-4NPB.
Newly described Moko disease-causing strains from Brazil
A final set of 18 DNA extracts from the newly described Brazilian Moko disease-causing strains [17] from sequevars IIA-41, IIA-53, and IIB-25, along with an extra set of 5 previously known strains of sequevars IIA-41 and IIB-25 (Table 6) associated with Solanaceae wilt but unable to cause wilt in banana plants, was assessed and repeated twice. These sets were assessed by PCR using Oli1/Y2 PCR [13], by Mmx-PCR [19,20], and by the new duplex PCR assay developed in this study.
Table 6. PCR comparison for the detection of Ralstonia solanacearum new Moko disease-causing strains from Brazil (DNA) and related Solanaceae strains from same sequevar (pure culture).
| Strain | Isolation Host | Country | Phylotype | Sequevar | Oli1/Y2 1 | Musa Mmx 1 | Duplex 93F/93R & 5F/5R 1 , 2 |
|---|---|---|---|---|---|---|---|
| B105 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B54 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B64 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B66 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B73 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B74 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B75 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B95 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B96 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| B106 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| BV136 | Musa spp. | Brazil | IIA | 41 | ++/++ | —/— | ++/++ |
| Cotpin2 | Musa spp. | Brazil | IIA | 53 | ++/++ | —/— | ++/++ |
| F2 | Musa spp. | Brazil | IIA | 53 | ++/++ | —/— | ++/++ |
| F3 | Musa spp. | Brazil | IIA | 53 | ++/++ | —/— | ++/++ |
| IBSBF2572 | Musa spp. | Brazil | IIA | 53 | ++/++ | —/— | ++/++ |
| B4 | Musa spp. | Brazil | IIB | 25 | ++/++ | —/— | ++/++ |
| B7 | Musa spp. | Brazil | IIB | 25 | ++/++ | —/— | ++/++ |
| B10 | Musa spp. | Brazil | IIB | 25 | ++/++ | —/— | ++/++ |
| CFBP7032 | Solanum lycopersicum | Cameroon | IIA | 41 | ++/++ | —/— | —/— |
| 06037 | Water (irrigation) | French Guiana | IIA | 41 | ++/++ | —/— | —/— |
| CIP10 | Solanum tuberosum | Peru | IIB | 25 | ++/++ | —/— | —/— |
| IBSBF2001 | Solanum lycopersicum | Brazil | IIB | 25 | ++/++ | —/— | —/— |
| UQRS607 | Solanum tuberosum | Iran | IIB | 25 | ++/++ | —/— | —/— |
1 ‘+’: positive result; ‘-’: negative result; two PCR reactions per sample were repeated twice (separated by a slash)
2 positive results characterized strains a as part of the Moko lineage according to the only amplification of the 477bp band.
Results
Duplex PCR assay development
A genomic comparison was performed in two steps due to the high phylogenetic proximity of strains from phylotype IIB-4NPB to the Moko phylotype IIB-4. The first step compared Moko strains and variant IIB-4NPB strains to all the other strains of the Ralstonia solanacearum species complex in the Prokaryotic Genome DataBase (PkGDB) of the MicroScope platform [23], and the second step compared variant IIB-4NPB strains to all the other strains of the Ralstonia solanacearum species complex in the PkGDB of the MicroScope platform. Following extensive basic local alignment search tool (BLAST) searches against the NCBI genomic database to screen specific matches, two coding sequences were selected as markers. The first coding sequence was RALUWv1_4260003 (1,038 bp; GenBank accession KM387307), a putative KfrA protein (functional assignment based on the presence of a conserved amino acid motif, structural feature or limited homology) related to DNA binding proteins and a transcriptional modulator found in the UW181 genome and shared by both the Moko and NPB strains (92% pairwise identity). For this sequence, the primers 93F (5’ CGC TGC GCG GCC GTT TCA C 3’) and 93R (5’ CGG TCG CGG CAT GGG CTT GG 3’) were designed; they produce a 477-bp amplicon. The second coding sequence was RAL70v1_1150031 (1,140 bp; GenBank accession KM387308), a chemotaxis-related protein found in the CFBP7014 genome and only shared by NPB strains (96% pairwise identity). For this sequence, the primers 5F (5’ GCG CGC GAG GCT GGT GAT GT 3’) and 5R (5’ TGG GTT CGC AGG CGG ACA GC 3’) were designed; they produce a 661-bp amplicon.
Performance of the duplex PCR assay
Accuracy (analytical specificity)
Accuracy is defined by both quantitative (inclusivity) and qualitative (exclusivity) criteria, thus characterizing the method using false positive/negative analysis. A total of 111 strains were selected (Table 1) to represent the Rssc strains and closely related strains. The primer set 93F/93R demonstrated 93% inclusivity (binomial exact proportion confidence ranging from 0.86 to 0.97) for the detection of Moko disease-causing strains and variant 4NPB strains. Although some replicates produced false-negative signals, all of the target strains were detected at least once among the 3 repetitions. Exclusivity reached 100%, thus characterizing this primer set as 97% accurate (binomial exact proportion confidence ranging from 0.95 to 0.99). The primer set 5F/5R demonstrated 100% inclusivity (binomial exact proportion confidence ranging from 0.88 to 1) and 100% exclusivity (binomial exact proportion confidence ranging from 0.99 to 1), thus yielding 100% accuracy (binomial exact proportion confidence ranging from 0.99 to 1). No variation in the amplification pattern was observed with any particular group of strains. Additionally, the tested viruses associated with banana diseases did not cross-react with the duplex PCR and thus yielded no amplification signal.
All controls yielded expected results, validating the process in its entire workflow.
Detectability (analytical sensitivity) and repeatability
Detectability is evaluated as the detection limit level, whereas repeatability is evaluated as the degree of agreement among test repetitions. The primer sets were tested in simplex PCR and then in duplex PCR to assess any potential competition issues that could lead to a lower PCR yield. Strains spiked into banana pseudo-stem extract were used to identify any potential PCR inhibitors and to simulate routine laboratory conditions. The results showed comparable outputs between the two simplex PCR assays (Tables 2 and 3) and the duplex PCR assay (Table 4), and repeatability was high and stable across the entire experiment: 100% repeatability was observed until 105 CFU/mL, and the repeatability of the duplex PCR slightly dropped to 95% and 92% for 104 CFU/mL and 103 CFU/mL, respectively. The detection limit was shown to be 105 CFU/mL. The simplex PCR showed the same pattern, with a slight drop in repeatability to 80% for 103 CFU/mL when using the 93F/93R primer set. Additionally, half of the samples (n = 11) could be detected down to 103 CFU/mL with the duplex PCR assay. Finally, no primer competition was observed during the experiment, as the simplex PCR did not give a stronger confidence level for detectability or repeatability than the duplex PCR at concentrations lower than 105 CFU/mL.
All controls yielded expected results, validating the process in its entire workflow.
Selectivity
The different tested healthy cultivars did not cross-react with the duplex-PCR, and all samples did not yield any PCR amplification across three repetitions.
Full-length protocol test
The full-length protocol, consisting of both plating and PCR amplifications (Oli1/Y2, Mmx-PCR, and the 93F/93R and 5F/5R duplex PCR assay) was conducted on the historic Moko disease-causing strains and IIB-4NPB strains, both in pure culture and with spiked plant extracts. The results (Table 5) show that these two lineages were detected, both on plates and with the three PCR methods, with either pure cultures or spiked plant extracts, and for both repetitions.
Newly described Moko disease-causing strains from Brazil
The genomic DNA samples from Brazilian Moko strains showed specific amplification (Table 6) in both the Oli1/Y2 assay and the duplex 93F/93R and 5F/5R PCR assay developed in this study (characterized as part of the Moko lineage), but they were not detected by Mmx-PCR. The strains that are only pathogenic to Solanaceae, which cluster into sequevars IIA-41 and IIB-25 and are not related to the Moko lineage, were only amplified by the generic Oli1/Y2 primer set, which types Rs strains at the species level. Neither the Mmx-PCR nor the duplex 93F/93R and 5F/5R PCR resulted in amplification.
All controls yielded expected results, validating the process in its entire workflow.
Discussion
The diagnostic method presented in this study for detecting Moko disease-causing strains and variant IIB-4NPB strains was developed to fit with the requirements of officially accredited diagnostic laboratories. The proposed protocol was fully evaluated, as required by the ISO 17025 standard, by following EPPO protocols, thus ensuring the highest confidence in the method development and validation.
The focus of this study was on the development of a method for the detection of the Moko lineage, which is capable of causing wilt in Musaceae plants. This duplex PCR assay was able to detect the historical diversity of Moko strains (sequevars IIA-6, IIA-24, IIB-3, and IIB-4) and also the newly discovered Moko-related Brazilian strains that cluster into sequevars IIB-25, IIA-41, and IIA-53. The epidemiologically variant IIB-4NPB strains that cause latent infection within the vascular system of plantains were also detected in this duplex PCR assay framework. The protocol developed in this work appears suitable for research and diagnostic laboratories and showed reliable accuracy, detectability, and repeatability (Tables 1 through 6).
Currently, no specific diagnostic protocol related to Rssc strains that are pathogenic to Musaceae plants has been defined by the European Commission (in the European Commission Council Directive 2000/29/EC [30] on protective measures against the introduction into the community of organisms harmful to plants or plant products and against their spread within the community or in Council Directive 2006/63/EC [31]). In addition, it is known that some strains within phylotype II that cause banana and Musaceae wilt are also able to wilt Solanaceae [21] and may represent a threat to both plant families; phylotype II Musaceae-adapted strains might be carried by Solanaceae plants. Therefore, a general assay to detect Rssc strains (e.g., with primer pairs Oli1/Y2 [13] or 759/760 [32]) and a specific Brown rot detection protocol [33] for Solanaceae should be used along with a specific Rs banana wilt detection protocol to detect both types of strains.
The performance assessment of the duplex PCR assay was fully compliant with its use as a reference method for diagnostic laboratories. Moko and the epidemiologically variant IIB-4NPB strains were both specifically detected with confidence. Amplification of the 477-bp Moko band could rarely fail for the IIB-4NPB strains, positive detection status should be considered if only the specific 661-bp 4NPB band is amplified; additional confirmation might also be necessary. The samples analyzed using this duplex PCR assay may consist of a pure cell culture isolated on solid medium or of plant extract samples in molecular biology-grade water. The detection of Moko and IIB-4NPB strains may be performed using either symptomatic or latently infected plants, as the concentration within xylem vessels is usually greater than the 105 CFU/mL threshold estimated in this study [34–36]. Moreover, the new Brazilian Moko-related strains were successfully detected with this duplex PCR assay, which was even able to distinguish Brazilian Moko-related strains from non-Moko strains of sequevars IIB-25 and IIA-41 and to detect the new sequevar IIA-53. Selectivity assessment showed that no cross-amplification occurred with different banana cultivars, whether Cavendish or plantain. In the full-length protocol using a semi-selective medium and a specific PCR approach, the detection and characterization of all strains was achieved, providing a high confidence level for the integration of this protocol into official detection schemes to complete or confirm results obtained by other diagnostic technologies.
The design of Moko-specific primers first involved a genomic comparison between Moko strains and other Rssc and non-Rssc strains to obtain an optimal gene repertoire for diagnosis. The Moko lineage clusters into seven sequevars belonging to phylotype IIA and IIB: the previously reported Moko sequevars IIA-6, IIA-24, IIB-3, and IIB-4, which have been sequenced, and the newly described sequevars IIA-41, IIA-53, and IIB-25, for which sequenced genomes are not yet available. The paraphyletic nature of the Moko lineage makes the search for specific gene repertoires difficult to manage, as other non-related Moko sequevars could be genetically closer while belonging to another lineage (e.g., Brown rot IIB-1 or IIB-4NPB strains).
Unsurprisingly, given the close phylogenetic proximity between IIB-4 Moko strains and IIB-4NPB strains, no locus was found to be specific only to Moko strains; thus, IIB-4NPB strains were added to the genomic search against other Rssc and non-Rssc genomes. This similarity is consistent with the phylogenetic position of the Moko IIB-4 strains as indistinguishable from IIB-4NPB strains. We could then assume that NPB strains share almost all of the genes of the Moko lineage. Nevertheless, the IIB-4NPB strains have a specific accessory repertoire of genes, in which a specific IIB-4NPB marker was found. A total of two coding sequences were found to be suitable for primer design. The 93F/R primer set was based on the specific gene repertoire shared by both the Moko and NPB strains, while the 5F/R primer set was based solely on the specific gene repertoire of the IIB-4NPB strains. Ecologically, these two lineages are different but share common traits, such as the ability to invade plantain banana xylem vessels.
Conclusions
The research presented herein shows that the 93F/R and 5F/R duplex PCR assay offers reliable detection of phylotype II strains of the Rssc that can be retrieved from banana plants, from other Musaceae, or from ornamentals. The test relies on two fully validated primer sets (as required by the ISO 17025 standard) and can be implemented in accredited laboratories or research laboratories. This duplex PCR assay showed high accuracy (inclusivity and exclusivity), with correct detection of target strains without any cross-reaction within the Rssc, high sensitivity for detecting low-concentration samples, and high repeatability, even when banana pseudo-stems were added to the sample. We recommend performing a total of two PCR repetitions for a given sample, with the results interpreted as follows: two positive results imply specific amplification and two negative results imply no specific amplification. In the case of one positive and one negative result, we suggest that the PCR should be performed again; if the results again give one positive and one negative result, it will imply (as for two positive results) specific amplification. Avoiding the introduction and spread of quarantined organisms is of utmost concern, and diagnostic methods should be implemented as early as possible in the plant breeding process (in vitro plantlet production) to ensure a safe phytosanitary framework. This assay was designed to detect strains within the Rssc phylotype II that are able to cause wilt or latent infections in bananas or plantains. This method can also be easily used as an epidemiological surveillance protocol (Fig. 1) for phylotype II Rs strains on both symptomatic and asymptomatic Musaceae plants and on ornamentals. This assay could complement EC policies on protective measures to be deployed to prevent the introduction and spread of organisms that are harmful to plants or plant products.
Acknowledgments
We thank Jean-Jacques Chéron (CIRAD UMR PVBMT) for his laboratory support with regard to the Ralstonia solanacearum strain collection.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was funded by the French Agency for Food, Environmental and Occupational Health & Safety (ANSES) - Plant Health Laboratory (LSV) - Tropical Pests and Diseases unit. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012; 13: 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dale JL. Banana bunchy top: an economically important tropical plant virus disease. Adv Virus Res. 1987; 33: 301–325. [DOI] [PubMed] [Google Scholar]
- 3. Tripathi L, Mwangi M, Abele S, Aritua V, Tushemereirwe WK, et al. Xanthomonas Wilt: A Threat to Banana Production in East and Central Africa. Plant Dis. 2009; 93: 440–451. [DOI] [PubMed] [Google Scholar]
- 4. Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012; 13: 614–629. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sequeira L. Bacterial wilt: the missing element in international banana improvement programs In: Prior P, Allen C, Elphinstone J, editors. Bacterial Wilt Disease: Molecular and Ecological Aspects. Paris, France: INRA Editions; 1998. pp. 6–14. [Google Scholar]
- 6. Fegan M, Prior P. How complex is the "Ralstonia solanacearum species complex" In: Allen C, Prior P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum species complex. St Paul, MN: APS Press; 2005. pp. 449–461. [Google Scholar]
- 7. Villa JE, Tsuchiya K, Horita M, Natural M, Opina N, et al. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and hrpB gene sequences. J Gen Plant Pathol. 2005; 71: 39–46. [Google Scholar]
- 8. Taghavi M, Hayward C, Sly LI, Fegan M. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996; 46: 10–15. [DOI] [PubMed] [Google Scholar]
- 9. Vaneechoutte M, Kampfer P, De Baere T, Falsen E, Verschraegen G. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int J Syst Evol Microbiol. 2004; 54: 317–327. [DOI] [PubMed] [Google Scholar]
- 10. Safni I, Cleenwerck I, De Vos P, Fegan M, Sly L, et al. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int J Syst Evol Microbiol. 2014; 64: 3087–3103. 10.1099/ijs.0.066712-0 [DOI] [PubMed] [Google Scholar]
- 11. Fegan M, Prior P. Diverse members of the Ralstonia solanacearum species complex cause bacterial wilts of banana. Australas Plant Pathol. 2006; 35: 93–101. [Google Scholar]
- 12. Raymundo AK, Orlina ME, Lavina WA, Opina NL. Comparative genome plasticity of tomato and banana strains of Ralstonia solanacearum in the Philippines In: Allen C, Prior P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum species complex. St Paul, MN: American Phytopathological Society (APS Press); 2005. pp. 387–393. [Google Scholar]
- 13. Seal SE, Jackson LA, Young JP, Daniels MJ. Differentiation of Pseudomonas solanacearum, Pseudomonas syzygii, Pseudomonas pickettii and the Blood Disease Bacterium by partial 16S rRNA sequencing: construction of oligonucleotide primers for sensitive detection by polymerase chain reaction. J Gen Microbiol. 1993; 139: 1587–1594. [DOI] [PubMed] [Google Scholar]
- 14. Thwaites R, Eden-Green SJ, Black R. Diseases caused by bacteria In: Jones DR, editor. Diseases of Banana, Abac and Enset. Wallingford, United Kingdom: CABI Publishing; 2000. pp. 213–239. [Google Scholar]
- 15. Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, et al. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl Environ Microbiol. 2007; 73: 6790–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buddenhagen IW, Elsasser TA. An insect-spread bacterial wilt epiphytotic of bluggoe banana. Nature. 1962; 194: 164–165. [Google Scholar]
- 17.Albuquerque GM, Santos LA, Felix KC, Rollemberg CL, Silva AM, et al. Moko disease-causing strains of Ralstonia solanacearum from Brazil extend known diversity in paraphyletic phylotype II. Phytopathology. 2014. [DOI] [PubMed]
- 18. Cellier G, Remenant B, Chiroleu F, Lefeuvre P, Prior P. Phylogeny and population structure of brown rot- and Moko disease-causing strains of Ralstonia solanacearum phylotype II. Appl Environ Microbiol. 2012; 78: 2367–2375. 10.1128/AEM.06123-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prior P, Fegan M. Diversity and molecular detection of Ralstonia solanacearum race 2 strains by multiplex PCR In: Allen C, Prior P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum species complex. St Paul, MN: APS Press; 2005. pp. 405–414. [Google Scholar]
- 20. Das RN, Sly LI, Fegan M. Molecular diversity of Moko disease causing strains of Ralstonia solanacearum; 2006. 17–20 July; York, United Kingdom: Central Science Laboratory; pp. 75. [Google Scholar]
- 21. Cellier G, Prior P. Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology. 2010; 100: 1250–1261. 10.1094/PHYTO-02-10-0059 [DOI] [PubMed] [Google Scholar]
- 22. Fegan M, Taghavi M, Sly LI, Hayward AC. Phylogeny, diversity and molecular diagnostics of Ralstonia solanacearum In: Prior P, Allen C, Elphinstone J, editors. Bacterial Wilt Disease: Molecular and Ecological Aspects. Paris, France: INRA; Editions; 1998. pp. 19–33. [Google Scholar]
- 23. Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford). 2009; 2009: bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007; 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- 25. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012; 40: e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.General requirements for the competence of testing and calibration laboratories. In: AFNOR, editor. EN ISO/CEI 17025. La Plaine Saint-Denis. France. 2005; pp. 28.
- 27. EPPO. PM 7/76 Use of EPPO diagnostic protocols. EPPO Bul. 2006; 40: 350–352. [Google Scholar]
- 28. EPPO. PM 7/98 Specific requirements for laboratories preparing accreditation for a plant pest diagnostic activity. EPPO Bul. 2010; 40: 5–22. [Google Scholar]
- 29.R Development Core Team. R: A Language and Environment for Statistical Computing. 2011.
- 30.European Commision (2000) Council Directive 2000/29/EC concerning protective measures against the introduction into the Community of organisms harmful to plants or plant products and against their spread within the Community. In: Communities E, editor. Official Journal L169. 2000; pp. 112.
- 31.Council Directive 2006/63/EC amending Annexes II to VII to Council Directive 98/57/EC on the control of Ralstonia solanacearum (Smith) Yabuuchi et al. In: Communities E, editor. Official Journal L206/36. 2006; pp. 71.
- 32. Opina N, Tavner F, Hollway G, Wang JF, Li TH, et al. A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formerly Pseudomonas solanacearum). Asia Pac J Mol Biol Biotechnol. 1997; 5: 12. [Google Scholar]
- 33.Li X, Nie J, Hammill DL, Smith D, Xu H, et al. A comprehensive comparison of assays for detection and identification of Ralstonia solanacearum race 3 biovar 2. J Appl Microbiol. 2014. [DOI] [PubMed]
- 34. Grimault V, Anais G, Prior P. Distribution of Pseudomonas solanacearum in the stem tissues of tomato plants with different levels of resistance to bacterial wilt. Plant Pathol. 1994; 43: 663–668. [Google Scholar]
- 35. Lenarcic R, Morisset D, Pirc M, Llop P, Ravnikar M, et al. Loop-mediated isothermal amplification of specific endoglucanase gene sequence for detection of the bacterial wilt pathogen Ralstonia solanacearum . PLoS ONE. 2014; 9: e96027 10.1371/journal.pone.0096027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swanson JK, Yao J, Tans-Kersten J, Allen C. Behavior of Ralstonia solanacearum Race 3 Biovar 2 During Latent and Active Infection of Geranium. Phytopathology. 2005; 95: 136–143. 10.1094/PHYTO-95-0136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.