Abstract
Ticks rely exclusively on vertebrate blood for their survival. During feeding ticks inject into their hosts a sophisticated salivary potion that overcomes host hemostasis and adverse inflammatory responses. These mediators may also enhance pathogen transmission. Knowledge of the tick salivary protein repertoire may lead to vaccine targets to disrupt feeding and/or parasite transmission as well as to the discovery of novel pharmacological agents. Male saliva may also assist reproduction because males use their mouthparts to lubricate and introduce their spermatophores into the females’ genital pore. The analyses of the sialomes of male and female ticks independently allow us to understand the strategy used by each gender to feed successfully.We sequenced cDNA libraries from pools of salivary glands from adult male and female R. pulchellus feeding at different time points, using the Illumina HiSeq protocol. De novo assembly of a total of 241,229,128 paired-end reads lead to extraction of 50,460 coding sequences (CDS), 11,277 of which had more than 75% coverage to known transcripts, or represented novel sequences, and were submitted to GenBank. Additionally, we generated the proteome, from the salivary gland extracts of male and female R. pulchellus, yielding a total of 454 and 2,063 proteins respectively which were identified by one or more peptides with at least 95% confidence. The data set is presented as an annotated hyperlinked Excel spreadsheet, describing 121 putative secreted protein families. Female and male specific transcripts were identified.
Introduction
Blood-feeding arthropods depend on blood meals which are critical for their survival. During the feeding process, the host's blood vessels and tissues at the bite site are inadvertently damaged [1]. This results in the activation of the host's defense mechanisms which include hemostasis, inflammatory and immune responses, and host grooming. To counter these responses, hematophagous arthropods infuse saliva, which consists of a cocktail of anti-clotting, anti-platelet, vasodilatory, anti-inflammatory and immunomodulatory molecules, into the host. With the blood-feeding habit of arthropods having independently evolved at least 20 times [2, 3], distinct strategies, protein scaffolds and mechanisms have emerged to counter host responses. These salivary biomolecules have undergone both convergent and divergent evolution, resulting in distinct protein scaffolds exhibiting the same function and similar protein scaffolds with diverse functions. Thus large numbers of biologically active proteins with great complexity and diversity could be found in these salivary cocktails, where each arthropod may contain several hundreds to thousands of components [2, 4]. Hence, the analyses of sialomes of hematophagous arthropods shed light on numerous unique pharmacologically active components.
Some adult hematophagous arthropods are either facultative or obligatory blood-feeders. However, in certain species (e.g., mosquitoes and horse-flies), females are facultative blood-feeders while the males do not feed on blood at all [5]. This difference in feeding habit between the sexes is reflected in their salivary components. For example, in the mosquito Anopheles gambiae, anti-clotting and anti-platelet proteins are expressed exclusively in females and assist them in blood-feeding, while anti-microbials, and maltases and other enzymes that aid in digestion of plant sugar meals, are expressed in both sexes [6, 7]. On the other hand, in obligatory blood-feeders (e.g., ticks, tsetse flies, and kissing bugs) both sexes feed on blood. However, there are differences in feeding behaviors. For example, female ticks feed on a larger volume of blood as compared to males, and their body weight can increase to more than 100-fold after feeding [8-11]. This is in contrast to males, which rarely engorged to higher than two-fold their body weight. Additionally, tick saliva may have distinct roles in their reproduction. Male tick mouthparts, unlike female mouthparts, are actively engaged in reproduction; their mouthparts penetrate the female's genital pore while pushing-in the spermatophore [12]. Male ticks actively salivate on the spermatophore surface before introducing it into the female, and saliva may assist this process by lubricating it or preventing it to adhere to the tick surfaces while transiting into the female genital pore [13]. However, it is not known whether ticks salivate during their long copulation. The relationship between the salivary gland components and sexual differences in feeding or sexual behavior of the ticks is not well understood. In this study, we have shown for the first time that salivary gland extracts (SGEs) from female Rhipicephalus pulchellus (zebra tick) inhibit blood coagulation factor Xa and thrombin four fold higher than that of male ticks suggesting the possibility of male R. pulchellus using different strategies from females to obtain their blood meals. In an attempt to relate to their physiological disparity, we have unraveled the transcriptome and proteome of the salivary glands of male and female R. pulchellus separately. We have found that numerous proteins are overexpressed in a gender-specific pattern. We analyzed a single class of Kunitz-type serine protease inhibitors in detail, and further classified them into five subclasses. Quantitative PCR data suggests that male and female R. pulchellus selectively express certain subclasses of these proteins. This approach of examining the male and female sialomes of ticks independently has opened up opportunities to discover new salivary proteins and to have an initial look into various strategies deployed by each sex enabling them to feed successfully off their hosts, and determine candidate male salivary proteins that may assist reproduction.
Material and Methods
Tick rearing: Animal protocol used
The R. pulchellus tick species was the kind gift of Dr. Milan Kozánek (Institute of Zoology SAS, Bratislava) who collected it on 05/15/2007 in West Tsavo, Kenya (determined by M.Slovák). The ticks were reared under laboratory conditions [14] in the Institute of Zoology, SAS, Slovakia. Ticks used in the experiments resulted from the fourth breeding generation. Briefly, the ticks were maintained at a temperature of 24 ± 2 ⁰C in desiccators filled with concentrated KCl solution, with 85-90% relative humidity and a photoperiod of 16:8 h (L:D). White New Zealand rabbits were used as hosts for all stages and also for feeding of adult ticks for the given intervals mentioned in the experiments below. The usage of animals in these experiments was approved by the State Veterinary and Food Administration of the Slovak Republic (permit numbers 928/10-221 and 1335/12-221).
Library construction and sequencing
Salivary glands from the following time points were pooled as follows: Unfed, 1, 3 and 7 hours, 1, 2, 3, 4, 5, 6, 7 days. Six male and six female ticks per time point were used, except for 1 day where 12 ticks were used from each sex. Ticks were dissected in ice-cold sterile PBS, pH 7.2 and tissues were washed three times in the same solution before being put into RNAlater (Qiagen). The SGs in RNAlater were kept in 4 °C for a minimum of 48 hours to ensure penetration of RNAlater solution into the tissue, and kept frozen in −20 °C till ready for shipment. mRNA was isolated from the salivary glands using a Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen, San Diego, CA) according to the manufacturer's protocol. The extracted mRNA was fragmented using a Covaris E210 (Covaris, Woburn, MA). Library amplification was performed using eight cycles to minimize the risk of over-amplification. Unique barcode adapters were applied to each library. Individual libraries were quantitated by qPCR and then pooled in an equimolar ratio before sequencing on a HiSeq 2000 (Illumina) with ver. 3 flow cells and sequencing reagents. Two lanes of the HiSeq machine were used. To avoid lane bias, the two libraries were run together in both lanes, yielding a total of 102,013,516 paired ended sequences from adult females and 139,215,612 sequences from adult male ticks (100 nt long). Raw data were processed using RTA 1.12.4.2 and CASAVA 1.8.2. mRNA library construction, and sequencing was done by the NIH Intramural Sequencing Center (NISC).
Transcriptome assembly and bioinformatics
Reads were assembled with the Abyss software [15, 16] with various k values (every even number from 50 to 96). Because the Abyss software tends to miss highly expressed contigs [17], we have also run the Trinity assembler [18] on the raw data. The resulting assemblies were joined by an iterative blast and cap3 assembler [19]. Coding sequences were extracted using an automated pipeline, based on similarities to known proteins, or by obtaining coding sequences from the larger open reading frame (ORF) of the contigs containing a signal peptide. A non-redundant set of the coding and their protein sequences were mapped into a hyperlinked excel spreadsheet which is presented as supplemental file 1. Signal peptide, transmembrane domains, furin cleavage sites and mucin type glycosylation were determined with software from the Center for Biological Sequence Analysis, Denmark [20-23]. Detailed bioinformatic analysis of our pipeline can be found in our previous publication [19]. To map the raw Illumina reads to the coding sequences and determine their sex bias, raw reads from each library were blasted to the coding sequences using blastn with a word size of 25 (-W 25 switch) and allowing recovery of up to 3 matches. The 3 matches were used if they had less than two gaps and if their scores were equal to the best score. The resulting blast file was used to compile the number of reads each CDS received from each library, and also to count the number of hits at each base of the CDS, allowing for the determination of the average CDS coverage, maximum coverage and minimum coverage. These results can be statistically tested by a X2 test (using the number of reads per CDS), the results of which are reported significant if P<0.05 and no CDS had an expected value of 5 or less. Phylogenetic analyses were done with ClustalX and Mega [24, 25].
Salivary gland extracts preparation
Salivary glands were homogenized in 150 μl of PBS (pH 7.2), using a handheld homogenizer in a micro tube. The mixture was centrifuged at 15,000 rpm and the supernatant was harvested. The pellet was resuspended and the process of extraction was repeated to increase protein yield. The proteins from these two extractions were pooled and dried with either a freeze drier or a centrifugal evaporator. SGEs were stored at 4°C until further use.
Enzymatic inhibition assays
Activities of SGEs and partially purified fractions were determined with enzyme assays where they were tested for inhibition of either thrombin or FXa amidolytic activity. The enzyme activity was monitored by using a chromogenic substrate, specific to the enzyme. All assays were of a 40 μl reaction, performed in a 384-well microtiter plate at room temperature. 10 μl of enzyme (6.6 nM thrombin / 5.3 nM FXa in 50 mM Tris, 120 mM NaCl, 5 mM CaCl2 and 1% BSA) and 10 μl of protein were pre-incubated in the reaction well for 10 min. Subsequently, 20 μl of substrate (S2238, 200 μM, for thrombin; S2222, 2 mM, for FXa) was added into the mixture and the kinetics of the reaction was monitored immediately with a microplate reader (Tecan M200) at 405 nm. The substrates upon cleavage by its corresponding enzymes, release a chromogenic compound, p-nitroaniline, which can be measured at 405 nm. Percentage inhibition of the enzyme by the protein was calculated relative to the initial velocity of the control reaction without tick proteins (10 μl PBS) which was taken to be 0%. For the activities of the crude extracts, total amount of protein in SGEs were first quantitated via the Bradford assay (Bio-rad). 15 μg of proteins were used for this assay. As for the HPLC fractions, 10 μl from each fraction was used directly for the assay.
Size exclusion chromatography
Salivary gland extracts were separated by size exclusion chromatography with Superdex 75 10/300 GL (GE Healthcare). 50 mM Tris, pH 7.4, was used for equilibration of the column and salivary glands were reconstituted in 1 ml of deionized water and loaded into the column. Protein elution was monitored at a UV wavelength of 280 nm and 215 nm. The flow rate used was 0.8 ml/min and the proteins eluted were collected as 0.8 ml fractions. For both male and females, the numbers of salivary glands were standardized as 200 pairs for this experiment.
Proteomics
Salivary gland extracts were reconstituted with deionized water and 50 μg of protein of concentration 1 μg/μl was used as starting material. Reduction was performed with Tris(2-carboxyethyl)phosphine (TCEP) (5 mM) at 60 °C for 1 h, and alkylation with iodoacetamide (40 mM) at room temperature for 10 min. 1.7 μg of trypsin (Promega) was then added to the mixture and incubated at 37 °C for 16 h. Sample mixture was then desalted using Sep-Pak C18 cartridges (Waters), lyophilized and reconstituted with 50 μl of 5% acetonitrile and 0.05% formic acid for LC MS/MS analysis.
For each sample, 2 μg was injected into a reverse phase column (75 μm × 150 mm, ReproSil-Pur C18-AQ, 3 μm, 120 Å (Eksigent)). The mobile phase A used was 2% acetonitrile, 0.1% formic acid and mobile phase B was 98% acetonitrile, 0.1% formic acid. The gradient (in terms of mobile phase B percentage) for the liquid chromatography was as follows: 5% for 1 min, 5-12% for 2 min, 12-30% for 120 min, 30-90% for 1 min and lastly held at 90% for 11 min, with a flow rate of 300 nl/min. MS analysis was performed with the TripleTOF™ 5600 system (AB SCIEX). Data acquisition was performed in information dependent mode, where the mass range was set to 400-1250 m/z with an accumulation time of 250 ms per spectrum. A maximum of 20 precursor ions were subjected to MS/MS analysis, with dynamic exclusion enabled for a duration of 15 s. Proteins were identified using the ProteinPilot™ Software v. 4.5 (AB SCIEX) which uses the Paragon™ Algorithm. ID focus was set to biological modification (refer to supplemental file 2 for the full list of modifications as set by default within the program), with cysteine alkylation by iodoacetamide, digestion by trypsin, and search effort set to thorough. The database used for the search was the proteins derived from the transcriptome of R. pulchellus, containing 50,460 protein sequences. The false discover rate (FDR) analysis was also performed simultaneously with the Proteomics System Performance Evaluation Pipeline (PSPEP) feature in the software. It adopts a decoy database search strategy in which the protein database sequences were reversed and searched against. The reported FDR for both male and female proteomes for proteins with at least 95% confidence were less than 1% based on the global FDR fit. Only proteins with at least one peptide of >95% confidence are reported and hits which corresponded to the reversed sequences were removed. Peptide and protein hit summaries are available on supplemental file 2. Proteome MS/MS data is also available in supplemental file 1 (columns BG-CF), including hyperlinks to all peptides found for the particular protein (columns BN and CA).
RNA Isolation and first-strand cDNA synthesis
Salivary glands in RNAlater were thawed and weighed. Salivary glands from male (12.8 mg) and female (20.5 mg) ticks were used for RNA extraction. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Total RNA (1 μg from each sample) was used for first-strand cDNA synthesis using the Reverse Transcription & cDNA Synthesis Kit (Clontech) with the SMART Moloney Murine Leukemia Virus (MMLV) Reverse Transcriptase (Clontech), following the manufacturer's instructions.
Quantitative Real Time – Polymerase Chain Reaction (qRT-PCR)
The qRT-PCR analyses were performed using “StepOne™ Real-Time PCR Systems“ (Applied Biosystems) and analysed using the “StepOne™ Software“ (v2.1; Applied Biosystems). Each PCR reaction had a total final volume of 10 μl comprising of 5 μl of KAPA SYBR® FAST ABI Prism® 2X qPCR Master Mix (KAPA Biosystems), 25 ng of cDNA, and forward and reverse primers (125 nM each). Each reaction was set up in technical duplicates i.e. two reactions for the target gene and two for the internal control from the same cDNA. The experiment was confirmed with a biological replicate. PCR protocol used was follows: Initial denaturation and hot-start activation at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C for 3 s and annealing/extension at 63°C for 30 s. Melting curve analysis was done from 60°C to 95°C at the end of each PCR run, for detecting non-specific PCR product and/or primer-dimer co-amplification. For normalization, β-actin was used as the internal control gene. Results were expressed as relative quantitation (RQ) in reference to mRNA of the female gene for each set of genes, using the ΔΔCt method.
Availability of supporting data
Raw sequences were deposited on the Sequence Read Archive (SRA) from the NCBI under bioproject accession PRJNA170743. The individual reads received accession numbers SRR521835, SRR521944, SRR521951 and SRR521953. A total of 11,227 coding sequences and their translations were submitted to the Transcriptome Shotgun Assembly project deposited at DDBJ/EMBL/GenBank under the accessions GACK01000001-GACK01011227.
Results and Discussion
Anticoagulant activity
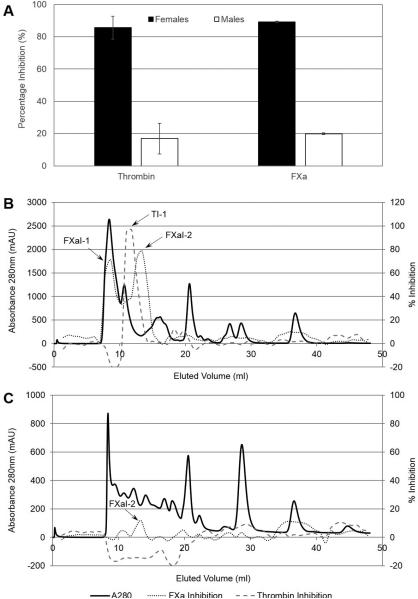
To understand the differences in blood-feeding habits between male and female ticks, we examined the anticoagulant activity of salivary gland extracts. Equivalent amounts (15 μg) of proteins from male and female SGEs were assayed for anti-FXa and anti-thrombin activities. Female SGEs inhibit both FXa and thrombin (89% and 86% inhibition, respectively), while male SGEs had lower inhibitory effects (20% and 26% inhibition, respectively) (Figure 1A). Subsequently, both SGEs (200 pairs each) were fractionated using size exclusion liquid chromatography, and each fraction was evaluated for FXa and thrombin inhibitory activities (Figure 1B and 1C). The protein elution profiles of both male and female SGEs are distinct from each other, indicating differences in saliva composition. Only some observed differences can be assigned to higher protein amounts in female SGEs (four-times) compared to males. Inhibitor elution profiles of female SGEs show two major inhibitors of FXa (FXaI-1 and FXaI-2), and one major inhibitor of thrombin (TI-1). Interestingly, in male SGEs, only a small peak of FXaI-2 was found; FXaI-1 and TI-1 were clearly absent. This corroborates poor inhibition of FXa and thrombin by male SGEs (Figure 1A), indicating distinct differences in the composition of male and female R. pulchellus SGEs, particularly in their anticoagulant protein content.
Figure 1. Anticoagulant activity of R. pulchellus salivary gland extracts.
(A) The amidolytic activities of thrombin and FXa were measured in the presence of 15 ug of female or male crude salivary gland extracts. Female extracts were able to inhibit both thrombin and FXa to higher than 80% while male extracts inhibited the enzymes lower than 20%. (B) Crude salivary gland extracts from female ticks were subjected to fractionation by size exclusion chromatography. (C) Crude salivary gland extracts from male ticks were subjected to fractionation by size exclusion chromatography. Fractions from (B) and (C) were tested for their ability to inhibit the amidolytic activity of FXa and thrombin. Female extracts inhibited FXa (FXaI-1 and FXaI-2) and thrombin (TI-1) while male extracts only inhibited FXa to a small extent (FXaI-2).
R. pulchellus sialotranscriptome
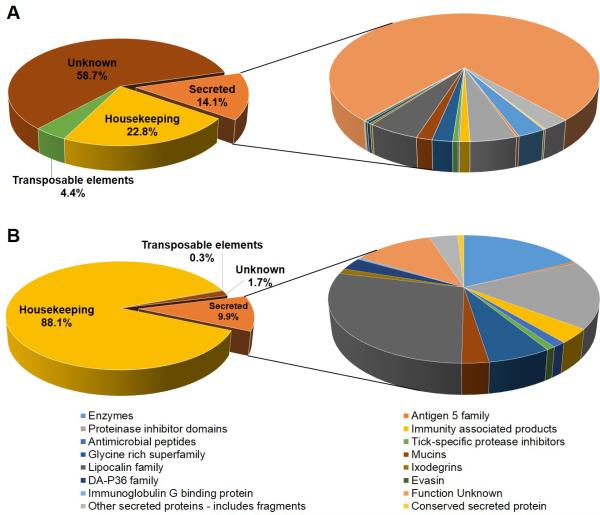
To delineate the differences in salivary composition, we generated two separate sialotranscriptomes of R. pulchellus, one for each sex. The mRNA sequences from the salivary glands of male and female ticks were determined using next generation sequencing. A total of 50,460 coding sequences (CDS) were extracted from the combined assembly of 139,215,612 and 102,013,516 paired-end sequences (100 nt long) from males and females, respectively. The CDS were classified into four main categories – housekeeping proteins (H), putative secreted proteins (S), transposable elements (TE), and proteins of unknown function (U) (Table 1 and Figure 2A). In summary, 7,134 CDS (22.8% reads) were associated with the S class, 11,499 CDS (25.6% reads) from the H class, 2,195 CDS (4.2% reads), including fragments, from the TE class. Finally, 47.4% of the reads were mapped to 29,632 putative CDS of unknown function and these CDS were not further analyzed. This class may contain secreted peptides. The proportion of S class transcripts was similar to a previously deep sequenced tick sialotranscriptome (23.7%) [19]. All CDS and their matches to several databases are available as a spreadsheet in supplemental file 1. For each CDS, the numbers of reads derived from the male and female libraries, along with a chi-squared test for the significance of the differences in their read counts are tabulated.
Table 1.
Functional classification of extracted coding sequences (CDS) from the sialotranscriptome of adult Rhipicephalus pulchellus ticks.
| Class | CDS | Associated Reads | % Total reads |
|---|---|---|---|
| Secreted | 7,134 | 16,080,727 | 22.83 |
| Housekeeping | 11,499 | 18,001,992 | 25.56 |
| Transposable elements | 2,195 | 2,989,302 | 4.24 |
| Unknown | 29,632 | 33,351,022 | 47.36 |
| Total | 50,460 | 70,423,043 | 100 |
Figure 2. Components of R. pulchellus transcriptome and proteome.
Both transcriptome (A) and proteome (B) were classified into four categories – housekeeping, secreted, transposable elements and unknown. Secretory products are shown in more details, with further classification. The charts represent proportion of CDS and proteins in the transcriptome and proteome respectively.
R. pulchellus proteome
To confirm the presence of putative proteins from the transcriptome, we used shotgun proteomics to identify proteins in the SGEs of male and female adult R. pulchellus. The transcriptome was used as a database for the proteome. In total, 2,231 proteins were identified by one or more high confidence (>95%) peptides, of which 1,777 and 169 proteins were found exclusively in females and males respectively, and 285 proteins in both sexes. Out of the 2,231 proteins, 221 (9.9%) were of the S class, 1966 (88.1%) from the H class, 7 (0.3%) from the TE class and 37 (1.7%) from the U class (Figure 2B). The discrepancies seen between the transcriptome and proteome can be due to several factors. Firstly, the proteomics work was performed on six days fed adult ticks in comparison to a mixture of unfed, and various time points of fed adults ticks (refer to methods) in the transcriptome. Thus the proteome may have a bias towards proteins that are found after six-day feeding. Secondly, the inherent constrains of proteomics limits the number of proteins that can be detected. Due to lower sensitivity in detection limits, proteins existing in lower abundance may not be identified. The nature of proteins themselves also affects detection in MS. For example, in highly glycosylated proteins (mucins, some TIL family members and Rhip 45-10), glycosylated tryptic peptides are either too large (particularly with N-glycosyl moiety) or masses do not match the peptide sequences ‘computationally generated’ from the transcriptome. In other cases (e.g. glycine-rich proteins) tryptic peptides are too large for the set limits in MS/MS due to the lower frequency and unfavorable distribution of Lys and Arg residues. Lastly, transcriptomics also contributes to the discrepancies. The number of reads may not directly represent the level of expressed proteins as other parameters such as translation efficiency and protein turnover. In addition, the assembly of reads sometimes produces fragmented transcripts resulting in more than one CDS for a single protein. Further, some peptides spanning over such fragmented regions will not be identified.
Housekeeping proteins
The 11,509 CDS associated with housekeeping function were further classified into 24 functional classes (Table 2). Protein synthesis machinery, transcription machinery and protein export machinery were among the top five classes, which were expected from an organ associated with a glandular function. The top two classes were signal transduction and hypothetical conserved proteins with unknown function [26]. It is possible that some of these conserved proteins may belong to the other four classes, once we understand their function. Several housekeeping proteins could have a role in the synthesis of small antagonists or agonists, or if secreted, they might play a direct role in saliva. Some enzymes involved in oxidative metabolism and detoxification, such as catalases, selenoproteins, peroxidases, thioredoxin peroxidases, could detoxify host oxidants associated with inflammation [27]. Cytochrome P450 enzymes could participate in the syntheses of prostaglandins known to be secreted in tick saliva [28-32]. Soluble epoxide hydrolases, if secreted, may affect host prostanoids [33, 34]. Some CDS of sphingomyelinases and deoxyribonucleases also show presence of signal peptides; such secreted enzymes could affect host immunity signaling [35] and affect host neutrophil extracellular traps (NETs) [36], respectively. Two of these deoxyribonucleases, RpIx75-684313 and RpIx75-682728, were found exclusively in females; 351 and 183 reads were found in females as compared to only two and three reads in males, respectively. Lastly, sulfotransferases may inactivate dopamine, a secretagogue found in the salivary gland of ticks [37].
Table 2.
Functional classification of extracted coding sequences (CDS) from the putative housekeeping class from the sialotranscriptome of adult Rhipicephalus pulchellus ticks.
| Class | CDS | Associated Reads | % Total reads |
|---|---|---|---|
| Unknown conserved | 2,243 | 4,083,858 | 22.6856 |
| Signal transduction | 2,092 | 2,452,172 | 13.6217 |
| Transcription machinery | 1,319 | 1,749,731 | 9.7197 |
| Protein synthesis machinery | 467 | 1,637,732 | 9.0975 |
| Protein export | 827 | 1,239,885 | 6.8875 |
| Transporters and channels | 721 | 1,068,327 | 5.9345 |
| Energy metabolism | 212 | 1,026,836 | 5.7040 |
| Extracellular matrix | 335 | 632,742 | 3.5148 |
| Proteasome machinery | 442 | 585,484 | 3.2523 |
| Lipid metabolism | 446 | 502,702 | 2.7925 |
| Protein modification | 266 | 500,861 | 2.7823 |
| Nuclear regulation | 424 | 415,586 | 2.3086 |
| Cytoskeletal proteins | 316 | 413,798 | 2.2986 |
| Carbohydrate metabolism | 304 | 403,332 | 2.2405 |
| Protein modification/Protease | 193 | 267,022 | 1.4833 |
| Transcription factor | 210 | 252,611 | 1.4032 |
| Amino acid metabolism | 140 | 223,972 | 1.2442 |
| Nucleotide Metabolism | 172 | 122,319 | 0.6795 |
| Nuclear export | 71 | 107,089 | 0.5949 |
| Oxidant metabolism/Detoxification | 72 | 100,617 | 0.5589 |
| Other immune related proteins | 77 | 74,336 | 0.4129 |
| Intermediary metabolism | 62 | 60,342 | 0.3352 |
| Detoxification | 77 | 59,983 | 0.3332 |
| Storage | 11 | 20,655 | 0.1147 |
| Total | 11,499 | 18,001,992 | 100 |
Several transcripts encoding putative housekeeping proteins were found differentially expressed in males and females. Tyrosine aminotransferase and one 4-aminobutyrate transferase were overexpressed (>50-fold) in females. This was reflected in the proteome where tyrosine aminotransferase was only found in females, and 4-aminobutyrate transferase had 18 high confidence peptides in females but only two in males. This may be explained by the increased gluconeogenesis in females as they take larger blood meals than males. An α-fucosidase (RpIx75-667813) with signal peptide, indicative of secretion, could be a lysosomal enzyme, but may be actually secreted in saliva; it is 80-fold overexpressed in females. Males, on the other hand, have increased transcription (54-58-fold) for galactosyltransferases and α-1,4-N-acetylglucosaminyltransferase indicating higher amounts of glycoproteins or glycolipids which could be associated with the lubricating function of saliva during reproduction [13]. Twenty two transcripts coding for monocarboxylate transporters were overexpressed (52-404-fold) in females, while six ABC type transporters were overexpressed (169-382-fold) in males. Prolyl-4-hydroxylases, involved in post-translational modification of proline to hydroxyproline [38], were found overexpressed (over 300-fold) in males. Males also have increased expression (100-fold) of the doublesex transcription factor [39], which is associated with male sex differentiation. Finally, males also have increased expression (270-450-fold) of dopamine β-monooxygenase, an enzyme that catalyzes oxidation of dopamine to noradrenaline. This enzyme could be associated to the detoxification of dopamine, as in the case of the sulfotransferases mentioned above, or in modifying female products during fecundation, if secreted, and indeed it has a signal peptide indicative of secretion.
Transposable elements
CDS encoding TEs are commonly found in sialotranscriptomes, and were also found in R. pulchellus. Both class I (Ty3/Gypsy, L1, Jockey, Bell, Outcast and Copia) and class II (hAT, Pogo, P, piggyback and tigger) elements [40] were identified. Several of these elements, such as tigger sequence RpTe-704693, have intact transposases. The recently discovered element sola2 [41] was also represented. A new family of TEs matching the PFAM domain (DDE_4 superfamily endonuclease) was found, and is similar to Ixodes transposons (Repbase accession: IS4EU-1_BF) [42]. Penelope-derived transcripts were also found abundantly.
Several transposons also show differential expression; RpSigp-1811, RpTe-11787 and RpTe-15723 are highly expressed in females and RpSigp-101516 and RpSigp-908814 in males. Some TE classes are abundantly transcribed, including several Gypsy elements with more than 20,000 reads mapped to their transcripts; RpTe-910969 was mapped from 65,688 reads and expressed in females 37-fold higher than in males. Most of the proteins associated with these Gypsy transcripts have frame shifts indicating these may function more as regulators of transposition than active transposition. It was recently proposed that TEs are associated with gene expression regulation [43]. The observed differential expression of TEs between sexes may contribute to gender-dependent gene expression in R. pulchellus salivary glands. Seven TEs were identified by MS from female SGEs indicative of their translation.
Putative secreted proteins
The putative secreted proteins were classified into various families following a previous review [44] and recent transcriptomes [19, 45, 46]. Accordingly, 121 different families of coding sequences were identified (Table 3). They are divided into two general groups, ubiquitous protein families and tick-specific families. The ubiquitous protein families include, enzymes such as metalloproteases, serine proteases, carboxypeptidases, dipeptidyl peptidases, lipases, pyrophosphatases and apyrases, and non-enzymatic proteins such as members of the antigen 5 protein family, and proteinase inhibitors of the Kunitz, Kazal, serpin, cystatin, thyropin and trypsin inhibitor-like (TIL) domain families. In addition, immunomodulatory proteins, such as lysozyme, defensins, MD-2-related lipid-recognition (ML) domain peptides, ixoderin, peptidoglycan recognition proteins and thioester proteins (TEP), are secreted by ticks to counter the host immune system. The tick-specific protein families include glycine-rich proteins (serve as tick cement), mucins (lubrication), evasins (bind host cytokines), DA-p36 (immunosuppressor), ixodegrins (potential anti-platelet agents) and tick-specific protease inhibitors such as carboxypeptidase inhibitors. In addition, members of lipocalin and the basic tail families were also found. Lipocalins bind to histamine, serotonin or inflammatory prostanoids. Seven additional groups of proteins, many representing multiple families, are described in Table 3 that have no known function, of which most were found uniquely in R. pulchellus.
Table 3.
Functional classification of extracted coding sequences (CDS) from the putative secreted class from the sialotranscriptome of adult Rhipicephalus pulchellus ticks.
| Class | CDS | Associated Reads | % Total reads |
|---|---|---|---|
| Enzymes | |||
| Metalloproteases | |||
| Typical tick ZnMc_salivary_gland_MPs CDD motif | 98 | 180,020 | 1.1195 |
| Large ADAM's proteases - probably housekeeping | 5 | 4,059 | 0.0252 |
| Other Zn metalloproteases, including collagenases | 7 | 6,128 | 0.0381 |
| M13 family peptidase | 14 | 25,327 | 0.1575 |
| Dipeptidyl peptidase | 2 | 13,064 | 0.0812 |
| Serine protease | 17 | 12,921 | 0.0804 |
| Male specific salivary serine protease | 2 | 32,135 | 0.1998 |
| Suchi serine protease | 2 | 3,421 | 0.0213 |
| Zinc carboxypeptidase | 2 | 9,147 | 0.0569 |
| Serine carboxypeptidase - may be lysosomal | 19 | 43,212 | 0.2687 |
| Legumain/asparaginyl peptidase | 2 | 6,916 | 0.0430 |
| Chitinase | 2 | 2,106 | 0.0131 |
| 5'-nucleotidase/Apyrase | 20 | 28,812 | 0.1792 |
| Ribonuclease | 4 | 4,892 | 0.0304 |
| Ectonucleotide pyrophosphatase/phosphodiesterase family | 3 | 5,489 | 0.0341 |
| Lipase | 4 | 2,591 | 0.0161 |
| Inositol phosphatase | 4 | 3,734 | 0.0232 |
| Antigen 5 family | 20 | 65,252 | 0.4058 |
| Proteinase inhibitor domains | |||
| Serpins | 23 | 10,468 | 0.0651 |
| Monolaris | 81 | 125,953 | 0.7833 |
| Bilaris | 65 | 94,581 | 0.5882 |
| Trilaris | 33 | 37,042 | 0.2304 |
| Tetralaris | 6 | 32,425 | 0.2016 |
| Pentalaris | 4 | 44,321 | 0.2756 |
| Hexalaris | 1 | 368 | 0.0023 |
| Heptalaris | 2 | 4,770 | 0.0297 |
| Similar to Kunitz domain | 10 | 10,995 | 0.0684 |
| Cystatins | 20 | 73,488 | 0.4570 |
| Thyropins with two domains | 4 | 2,786 | 0.0173 |
| IGF/KAZAL/IG domain family | 2 | 2,019 | 0.0126 |
| Chymotrypsin-elastase inhibitor ixodidin - TIL domain | 33 | 636,225 | 3.9564 |
| MonoTil | 13 | 26,055 | 0.1620 |
| BiTil | 35 | 96,191 | 0.5982 |
| TriTil | 12 | 50,388 | 0.3133 |
| PolyTil | 2 | 12,311 | 0.0766 |
| Til-Like | 1 | 31 | 0.0002 |
| Immunity associated products | |||
| Lysozyme | 4 | 2,397 | 0.0149 |
| Defensins | 22 | 5,555 | 0.0345 |
| TEP proteins | 23 | 50,188 | 0.3121 |
| ML domaini | 9 | 1,533 | 0.0095 |
| Galectin | 6 | 7,046 | 0.0438 |
| Peptidoglycan recognition protein | 6 | 7,098 | 0.0441 |
| Ixoderin/Ficolin | 6 | 17,076 | 0.1062 |
| Tick specific protein families, at least one family member has known function | |||
| Antimicrobial peptides | |||
| Microplusin | 6 | 14,525 | 0.0903 |
| 5.3 kDa antimicrobial family | 4 | 317 | 0.0020 |
| Protease inhibitors | |||
| Carboxypeptidase inhibitor | 13 | 17,373 | 0.1080 |
| Basic Tail | 12 | 18,408 | 0.1145 |
| 18.3 kDa family | 14 | 12,823 | 0.0797 |
| Glycine rich superfamily | 151 | 7,640,865 | 47.5157 |
| Mucins | 118 | 1,840,933 | 11.4481 |
| Lipocalin family | |||
| Group I | 209 | 309,678 | 1.9258 |
| Group II | 20 | 34,862 | 0.2168 |
| Group III | 13 | 45,186 | 0.2810 |
| Group IV | 10 | 24,295 | 0.1511 |
| Group V | 16 | 25,277 | 0.1572 |
| Group VI | 13 | 176,753 | 1.0992 |
| Group VII | 17 | 17,945 | 0.1116 |
| Group VIII - Deorphanized Dermacentor lipocalin family | 19 | 20,816 | 0.1294 |
| New lipocalin family Rhipicephalus family XII | 5 | 7,271 | 0.0452 |
| Other putative lipocalins | 64 | 912,901 | 5.6770 |
| Ixodegrins | 21 | 45,029 | 0.2800 |
| DA-P36 family | 31 | 36,142 | 0.2248 |
| Evasin | 22 | 19,746 | 0.1228 |
| Immunoglobulin G binding protein | 12 | 1,382,686 | 8.5984 |
| Function Unknown | |||
| Found in prostriates and metastriates | |||
| Fibronectin domain containing protein family | 2 | 157 | 0.0010 |
| Hematopoietic-stem cell progenitor | 2 | 1,551 | 0.0096 |
| 8.9 kDa family | 60 | 114,278 | 0.7107 |
| 23 kDa family | 7 | 3,072 | 0.0191 |
| 24 kDa family | 9 | 3,193 | 0.0199 |
| One of each protein family - now Ixodidae | 14 | 3,505 | 0.0218 |
| Previously thought as metastriate specific | |||
| Ig domain containing secreted peptide | 4 | 1,814 | 0.0113 |
| Deorphanized Ixodidae Rhip 45-937 | 4 | 1,901 | 0.0118 |
| Insulin growth factor binding protein - also found in Ixodes | 6 | 2,065 | 0.0128 |
| Amblyomma maculatum family 40-33 | 9 | 2,599 | 0.0162 |
| Deorphanized metastriate-Rhipicephalus family XIV | 4 | 912 | 0.0057 |
| Deorphanized metastriate family 40-80 | 1 | 626 | 0.0039 |
| Somatomedin domain protein family | 4 | 599 | 0.0037 |
| Metastriate specific | |||
| 28 kDa Metastriate family | 18 | 17,340 | 0.1078 |
| 16 kDa family | 2 | 8,950 | 0.0557 |
| 8 kDa Amblyomma family | 5 | 2,135 | 0.0133 |
| 10 kDa acidic metastriate | 1 | 6,626 | 0.0412 |
| Ixostatin-like | 10 | 12,781 | 0.0795 |
| Deorphanized Dermacentor 9 kDa expansion | 5 | 1,102 | 0.0069 |
| Rhip 45-304 - deorphanized on A. maculatum sialome | 4 | 13,864 | 0.0862 |
| Rhipicephalus specific families | |||
| Rhip 45-141 | 17 | 2,401 | 0.0149 |
| Rhip 45-151 | 15 | 3,199 | 0.0199 |
| Rhip 45-236 | 2 | 22,601 | 0.1405 |
| Short protein families possibly secreted | |||
| Rhip 50-70 | 6 | 1,431 | 0.0089 |
| Rhip 50-10 | 107 | 16,133 | 0.1003 |
| Rhip 50-45 | 26 | 21,670 | 0.1348 |
| Rhip 50-18 | 79 | 10,396 | 0.0646 |
| Rhip 45-25 | 64 | 21,119 | 0.1313 |
| Rhip 45-85 | 16 | 1,445 | 0.0090 |
| Rhip 40-92 | 23 | 2,214 | 0.0138 |
| Rhip 45-2 | 623 | 179,776 | 1.1180 |
| Rhip 45-10 | 43 | 15,020 | 0.0934 |
| Rhip 45-11 | 162 | 36,474 | 0.2268 |
| Rhip 45-16 | 105 | 21,708 | 0.1350 |
| Rhip 45-22 | 56 | 13,453 | 0.0837 |
| Rhip 45-27 | 62 | 13,982 | 0.0869 |
| Rhip 45-28 | 53 | 41,876 | 0.2604 |
| Rhip 45-29 | 56 | 11,234 | 0.0699 |
| Rhip 45-37 | 43 | 2,067 | 0.0129 |
| Rhip 45-48 | 37 | 7,106 | 0.0442 |
| Rhip 45-49 | 35 | 4,196 | 0.0261 |
| Rhip 45-56 | 33 | 8,522 | 0.0530 |
| Rhip 45-61 | 30 | 52,808 | 0.3284 |
| Rhip 45-66 | 28 | 2,827 | 0.0176 |
| Rhip 45-69 | 27 | 11,930 | 0.0742 |
| Rhip 45-72 | 26 | 4,828 | 0.0300 |
| Rhip 45-73 | 26 | 2,433 | 0.0151 |
| Rhip 45-77 | 25 | 11,873 | 0.0738 |
| Rhip 45-82 | 24 | 2,064 | 0.0128 |
| Rhip 45-95 | 22 | 5,455 | 0.0339 |
| Rhip 45-130 | 18 | 2,533 | 0.0158 |
| Rhip 45-132 | 18 | 771 | 0.0048 |
| Rhip 45-133 | 18 | 1,524 | 0.0095 |
| Rhip 45-143 | 17 | 3,894 | 0.0242 |
| Rhip 45-144 | 17 | 3,781 | 0.0235 |
| Remaining peptides of large small family 40-1 | 3,340 | 619,033 | 3.8495 |
| Other secreted proteins - includes fragments | 208 | 272,152 | 1.6924 |
| Conserved secreted protein | 16 | 21,361 | 0.1328 |
| Total | 7,134 | 16,080,727 | 100 |
We have also extracted 5,265 CDS that represent putative small secreted peptides varying from 40-100 amino acid residues, grouped into 33 families according to their similarities. Except for matches to R. sanguineus peptides, no similarities are apparent to other known proteins. Although it is unclear whether they are artifacts (extracted from non-coding RNA) or real, they serve as targets for mass spectrometry experiments. Indeed a few peptides were identified by MS, such as RpSigp-496390 and RpSigp-663980, supporting existence of some of them in tick SGEs.
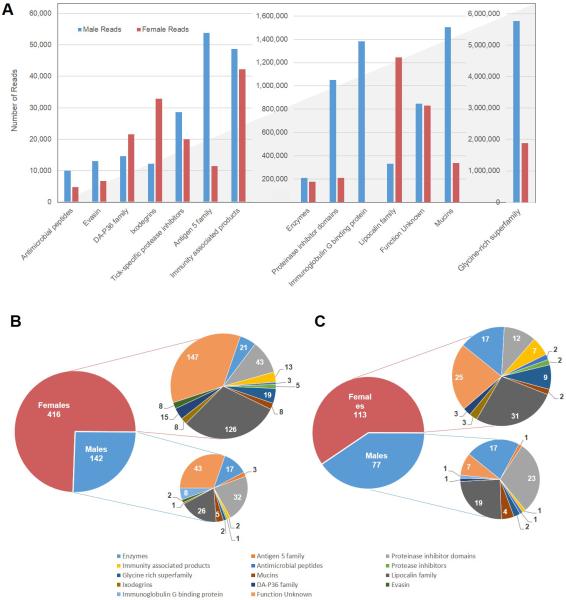
It is remarkable that 48% of S class reads map to glycine-rich proteins, which assist the tick to attach to their host (Table 2). Together with mucins (11% reads), lipocalin family (10% reads) and immunoglobulin G binding proteins (9% reads), they account for 78% of the S class reads. As glycine-rich proteins and mucins do not produce suitable tryptic peptides, as mentioned above, they were not identified in the proteome. The top classes of proteins that were identified in the proteomes were the lipocalin family (28%), proteases (12%) and the proteinase inhibitor domains (17%).
As noticed in previous sialomes of blood sucking arthropods, unique protein families were found at the genus or even at the species level [44]. This may be due to the host immune pressure which accelerates evolution of salivary gland genes. Previous families thought to be exclusively metastriate were found to have homologs in I. scapularis, albeit with a relatively low similarity, indicative of fast divergence of these salivary proteins [44].
Ubiquitous protein families
Enzymes
Proteases were conspicuously represented in tick sialomes, including metalloproteases of the reprolysin family, M13 type peptidases, serine proteases, asparaginyl proteases and dipeptidyl peptidases.
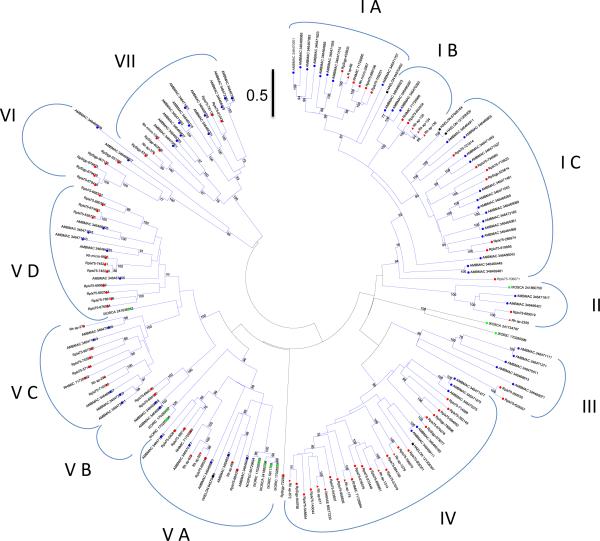
The zinc metalloproteases, ubiquitous enzymes found in metazoans, are associated with embryonic development, inflammation, angiogenesis and connective tissue remodeling, among other functions (see [47, 48]). Tick salivary metalloproteases distinctly have a Reprolysin_2 PFAM domain, typical of snake venom enzymes, as well as the ZnMc_salivary_gland motif from the Conserved Domain Database (CDD). These enzymes are important components of snake venoms where they cause hemorrhage in their prey [49]. In tick saliva, they were found to be associated with disrupting the hemostatic system of their hosts via fibrinolysis [50] and angiogenesis inhibition [51]. Although the genome of Ixodes scapularis contains only six such proteins with more than 450 amino acid residues (and some more fragments) (JMCR, unpublished), a previous deep transcriptome analysis of Amblyomma maculatum retrieved 47 near full-length sequences of this family within three superclades and nine clades, based on strong bootstrap support greater than 75% [19]. Many of these clades are due to A. maculatum specific expansions. Similarly, the sialotranscriptome of R. pulchellus allowed the extraction of 98 sequences coding for typical tick salivary metalloproteases, 51 of which are full length or near full length. Phylogenetic analysis indicates eight major clades that have more than 75% support (Figure 3), which contain at least four distinguished subclades for a total of at least 12 clades. No monospecific clade is now apparent, but expansions that are typical of tandem gene duplications are found, as on clade IV for R. pulchellus and clades IC and VIII for A. maculatum. Several R. pulchellus metalloproteases appeared distinctly sex biased. For example, RpIx75-57164, RpSigp-931308, RpIx75-721814 and RpIx75-240814 were overexpressed in females (>100-fold), while RpIx75-919895 are overexpressed in males (57-fold). In accordance with its transcript level bias among sexes, RpIx75-57164 was detected by proteomics in females with 29% coverage but was not detected in males. Proteomic analysis also revealed the product RpIx75-951936 to be highly expressed, with over 50% coverage in females and 21% coverage in males.
Figure 3. Phylogenetics of the salivary metalloproteases of ticks.
The evolutionary history was inferred using the Neighbor-Joining method [134]. The optimal tree with the sum of branch length = 75.51 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [135] and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modelled with a gamma distribution (shape parameter = 1). The analysis involved 170 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 823 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [25]. The bar at the top indicates 50% amino acid divergence. Sequences from GenBank are recognized by having the first 3 letters of the genus name, followed by the first 3 letters of the species name, followed by their accession numbers. Rhipicephalus pulchellus sequences start with RP. Other sequences were derived from a tick sialomes review [2]. Markers of the same color are used for genus, and marker shape for species differentiation.
M13 domain/neprilysin proteases are involved in the inactivation of hormone peptides. In ticks, they could function by destroying inflammatory peptidic mediators such as cytokines, anaphylatoxins or bradykinin from the hosts. While most members of this family are extracellular, some are membrane bound via a transmembrane helix. Ten R. pulchellus M13 proteases were found without this transmembrane helix indicating they are secreted, similar to A. maculatum homologs, which are 50-70% identical to the R. pulchellus sequences. RpIx75-695314 and RpIx75-901875 were identified in both male and female proteomes.
Serine proteases are ubiquitous and commonly found in sialotranscriptomes of blood feeding ticks and insects where they may function as a fibrinolysin [52]. The sialotranscriptome of R. pulchellus reveals 19 CDS for these proteins. Remarkably, several of these appear to be male-specific, five of which have 61 to 268 times more expression in males than females. All these proteins were identified solely in males by MS experiments. These enzymes could thus be associated with the reproductive biology of ticks, which are successful practitioners of oral sex [12]. Male-specific seminal fluid serine proteases are known to occur in Drosophila [53] and the human prostate-specific antigen (PSA) is a serine protease [54, 55]. It is possible that some male salivary proteins are exerting a function homologous to seminal fluid proteins of other animals. We additionally report a legumain (RpIx75-689581) that is male specific in R. pulchellus (>600-fold more expression). This enzyme was also found only in male SGEs by MS. The only other salivary legumain (RpIx75-534217) was found to be equally expressed in both sexes, and was identified only in female glands via MS. The specific function of serine proteases and legumains in tick saliva remains unknown.
Dipeptidyl peptidases were reported as the kininase found in I. scapularis saliva [56]. These enzymes are responsible for the destruction of bradykinin, thus interfering with the host's tick rejection reactions. Two dipeptidyl peptidases were reported for R. pulchellus. The protein encoded by RpIx75-19165 was found by MS to be expressed in females, with 39% coverage, with its transcripts six times more abundant in females than in males.
Serine carboxypeptidases with unknown function in feeding or reproduction were found by MS and their transcript expression are enriched in male glands. Particularly, RpIx75-906737 and RpIx75-906740 were identified by MS with 21% coverage in male glands only. On the other hand, RpSigp-677431 and RpSigp-677433 (equivalently expressed in both sexes), were identified by MS in both sexes.
Apyrases are enzymes hydrolyzing ATP and ADP to AMP thus suppressing agonists of inflammation and platelet aggregation. Mosquito and tick apyrases belong to the 5’ nucleotidase family and lack the carboxyterminal membrane domain linking them to a phosphoglycoinositol membrane anchor [57, 58]. Twenty coding sequences were found representing this protein family. RpIx75-419942 and RpIx75-419943 were found in only female SGEs, while RpIx75-907283 and RpIx75-907282 were found only in males reflecting the transcription bias. Nucleotidases catalyzing the hydrolysis of dinucleotides (ectonucleotide hydrolase/phosphodiesterase) were found, in which some are overexpressed in male salivary glands. These enzymes may hydrolyze diadenosine polyphosphates released by platelets, attenuating their role in hemostasis and inflammation [59, 60].
Ribonuclease coding transcripts were found overexpressed in female glands; the products for two enzymes (RpIx75-670367 and RpIx75-938568) were found by MS solely in females.
Proteinase inhibitor domains
The following families of protease inhibitor domains were found abundantly expressed in the sialotranscriptome of R. pulchellus (Table 3): Kunitz, serpin, TIL (associated with serine proteinase inhibition), and cystatin and thyropin (associated with cysteine proteinase inhibition and immunosuppression).
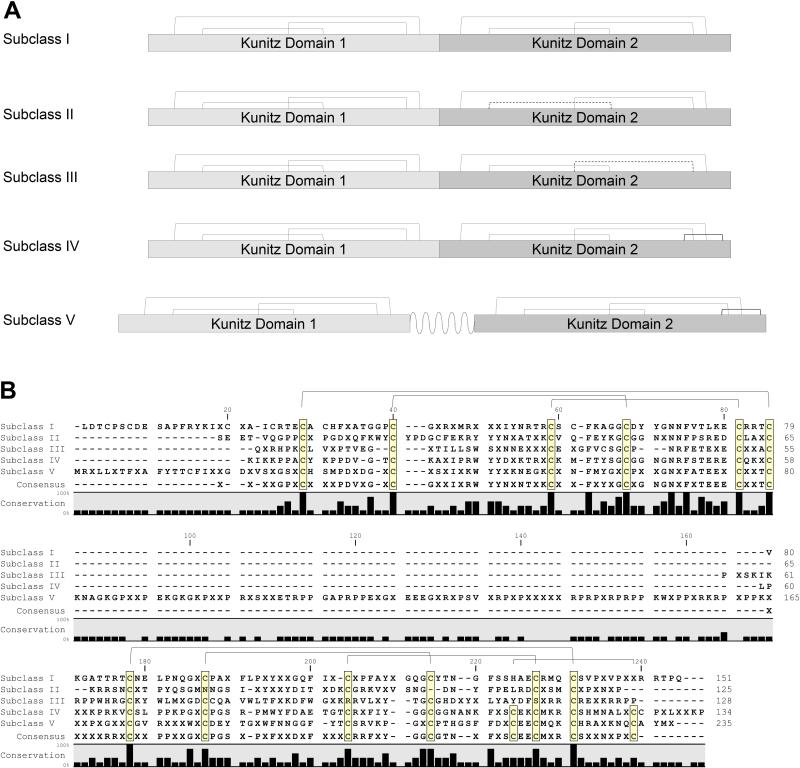
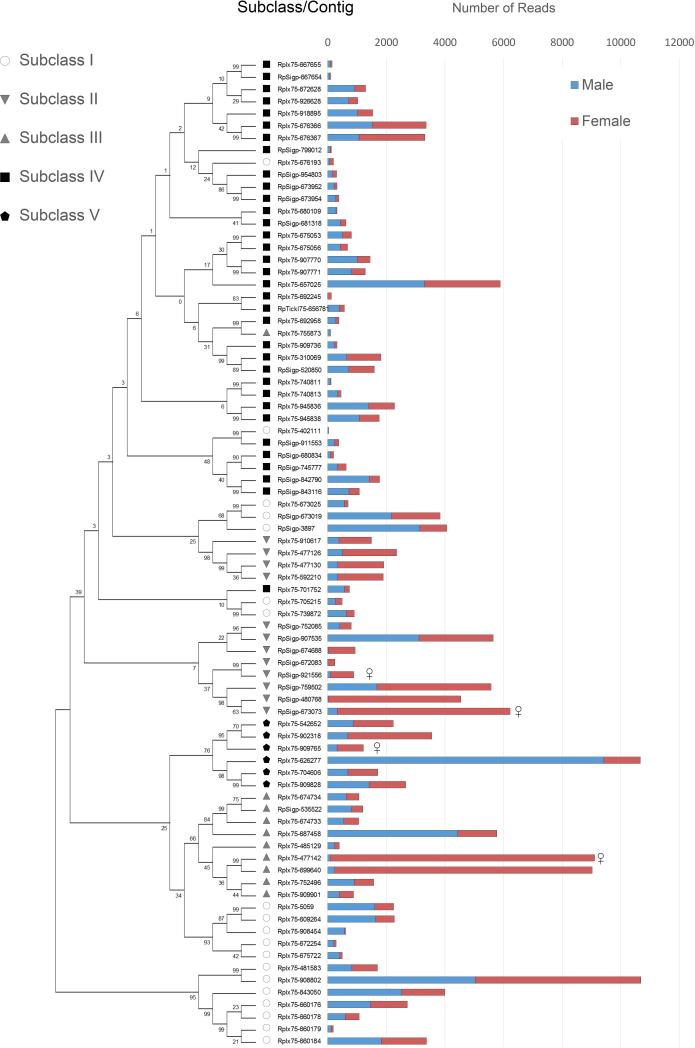
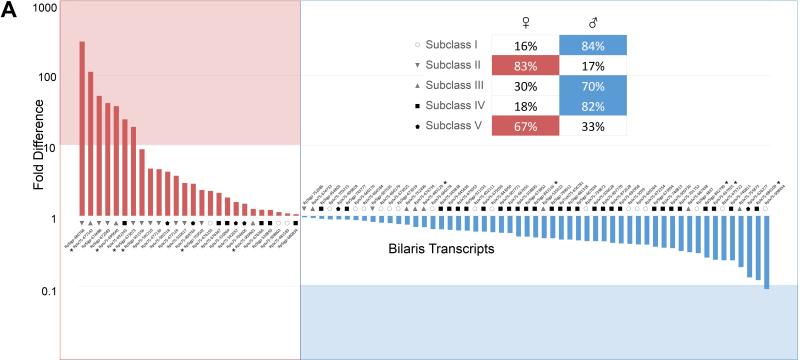
The Kunitz domain is 50-60 amino acid residues in length and their fold is conserved by three disulphide bonds. Most Kunitz domain containing proteins inhibit serine proteases, although some also block ion channels [61, 62]. In some tick saliva proteins, Kunitz domains are found in tandem; depending on the number of Kunitz domains, they are classified as monolaris, bilaris, trilaris and so on. Some are well known inhibitors of various proteases in the coagulation cascade. For example, Ixolaris, a bilaris isolated from I. scapularis, is an anticoagulant that binds both factor VIIa and factor X thus inhibiting the extrinsic pathway [63-67]. In the R. pulchellus transcriptome, 197 members containing Kunitz domains have been identified (Table 3). Six monolaris, two bilaris and one tetralaris proteins were overexpressed in females (>100-fold). In the proteome, one monolaris, five bilaris and one tetralaris proteins were identified in females, as compared to only one monolaris protein in males. Further in the text the bilaris protein family will be analyzed in detail.
Serpins are ubiquitous serine protease inhibitors and several in tick salivary glands act as clotting inhibitors or chymase inhibitor [68-70]. We have identified 23 serpins in the R. pulchellus transcriptome. Notably, several were expressed exclusively in males, such as RpIx75-772874 having 2,559 reads from males and only two from females. Not surprisingly the protein was identified only in male SGEs by MS. It is possible that this serpin plays a role in tick reproduction as does serpins in the seminal fluid of Drosophila [71, 72] and prostatic secretions of humans [73].
Cystatins are ubiquitous inhibitors of cysteinyl proteases. Some tick salivary cystatins show immunosuppressive and anti-inflammatory activities [74-81]. Several cystatin coding sequences were overexpressed in males, two of which are over 1,000-fold overexpressed (RpSigp-19398 and RpSigp-384543) and their protein products were detected by MS only in males. Cystatins were also detected in human seminal fluid [82, 83].
The TIL family in R. pulchellus contains 95 members, including peptides closely related to tick elastase inhibitors, which also have antimicrobial activity [84]. Proteins with multiple TIL domains (biTIL, triTIL and polyTIL) were also recognized. Many of these transcripts were gender biased. RpSigp-666840 was 716 times overexpressed in males and its product was detected by MS only in male glands. Several others were found by MS in either sex reflecting their expression bias.
Immunity-related
Sialotranscriptomes of hematophagous animals contain antimicrobial peptides and other proteins associated with immunity. The sialotranscriptome of R. pulchellus reveals four different lysozymes, one of which (RpIx75-909446) appears male specific and was found only in male SGEs by MS. In addition, 22 defensins were found, of which one (RpSigp-635550) was identified by MS only in males.
Full-length thioester containing proteins (TEP), involved in innate immunity [85, 86] were identified. MS identification was achieved solely from female tissues. Pathogen-recognition proteins, such as ML-domain containing proteins [87], peptidoglycan-binding proteins [88], galectins and ficolins (ixoderin) [89] were identified. Three out of six galectins were identified by MS in females only.
Tick-specific families, at least one member of which has a known function
Antimicrobial peptides
Microplusin is a histidine-rich peptide from R. microplus [90], six homologs of which were found in R. pulchellus. Two microplusins (RpIx75-675437 and RpIx75-675436) are highly female biased (190- and 257-fold) and were found by MS only in female glands. The 5.3 kDa peptide family was initially described in I. scapularis sialotranscriptomes, some members were upregulated following infection with Borrelia burgdorferi [91]. Later, one of its members was shown to be an antimicrobial [92]. R. pulchellus has four contigs matching members of this peptide family, in which one (RpIx75-680620) was highly expressed in female SGEs (>50-fold).
Tick-specific protease inhibitors
The tick salivary carboxypeptidase inhibitor is a 97 amino acid residue peptide found in R. bursa that stimulates fibrinolysis by inhibiting the thrombin-activated fibrinolysis inhibitor [93, 94]. Haemaphisalis and A. maculatum ticks also have this protein family [95]. R. pulchellus sialotranscriptome reveals 13 CDS that are 32- 88% identical to the canonical R. bursa protein, indicating the diversity of this peptide family within a single genome. One of these CDS (RpSigp-946756) was more than 80 times overexpressed in female ticks, though it was not found in the proteome. Another protein (RpSigp-946756) was found in females only.
The basic tail protein family in found in abundance in I. scapularis, one member of which was shown to inhibit clotting factor Xa [96]. In metastriates, this family is much smaller. Nine CDS were found in the R. pulchellus sialotranscriptome, one of which (RpIx75-966959) was 36-fold overexpressed in females and was found by MS only in female tissues. The 18.3 kDa family was shown by PSI-BLAST analysis to be part of the basic tail superfamily [44]. This family is abundant in both metastriate and prostriate ticks, and is well represented within the R. pulchellus sialotranscriptome. The diversity of this family is verified by their best matching metastriate tick proteins available in the NR database which varies from 33-65% identity. None of them have been functionally characterized.
Glycine-rich proteins and mucins
Ticks use a cement-like substance to attach themselves to their hosts [97]. These proteins are rich in glycine and are somewhat similar to spider silk proteins. Some of these proteins have been used as anti-tick vaccines [98-100]. The sialotranscriptome of R. pulchellus presents 151 CDS, including 26 that have over 100,000 reads mapped to them, indicating the abundance of these transcripts. Although this class of proteins constituted almost half of the total number of S class reads, only 14 were identified by MS. This low detection rate in MS can be explained by their sequence, which has fewer arginine and lysine residues. Thus, most tryptic fragments were too large for MS/MS sequence determination.
Mucins are proteins with a large number of N-acetyl-galactosamine residues linked to serine or threonine [101]. However, their primary sequence is not conserved and they have large regions of repeats with low complexity. Similar to the glycine-rich proteins, some members are highly expressed and 15 have over 10,000 reads mapped to their CDS. Four of these highly expressed CDS are male specific and may have to do with reproduction [13]. For example, RpSigp-99535 is 890-fold overexpressed in males and was found by MS only in male SGEs. Although there were 118 members found in the transcriptome, only six (four exclusively in males and two exclusively in females) were detected in the proteomes. This could be due to their high levels of glycosylation and large glycosylated peptides are not amenable for MS/MS sequencing.
Lipocalins
Although lipocalins are ubiquitous, tick salivary lipocalins have no sequence similarity to other known proteins, but are recognized by their typical barrel-like structure [102] revealed in crystal structures [103, 104]. Literally lipocalin means ‘a cup of lipid’, as their barrel structure contains normally a lipophilic interior where lipids bind and can be carried in the aqueous environment. Tick salivary lipocalins can function by scavenging biogenic amines [103-105] or lipidic mediators of inflammation [106, 107], normally using binding sites inside the barrel, but also by acting as anti-complement proteins using their side chains on the outer surface [108]. Supplemental file 1 displays 350 coding sequences that are assigned to the lipocalin class. Some of these do not have any direct match by BLAST or RPS-BLAST to known lipocalins, but they cluster at 40% similarity to members of the lipocalin family and hence are grouped as a subcategory putative lipocalins. Several lipocalins show sex biased expression, 38 and 7 are overexpressed (>100-fold) in females and males, respectively. Mass spectrometry identified 59 of the 350 predicted lipocalins.
Ixodegrins
Ixodegrins are ~100-residue polypeptides that contain an RGD tripeptide flanked by cysteines [44, 109] similar to disintegrins – platelet aggregation inhibitors found in snake venom [110, 111]. R. pulchellus sialotranscriptome contains several peptides but in most, RGD is replaced by RED sequence. Three ixodegrins were identified by MS in female glands.
DA-p36 family
DA-p36 is a 36 kDa immunosuppressive protein of isolated from the salivary glands of Dermacentor andersoni [112]. This family is widely found in metastriate ticks and 28 were identified in R. pulchellus. Fifteen were overexpressed in females (10-390-fold), while none were overexpressed in males (<3.8-fold). Seven members of this family were identified by MS – 3 exclusively in females, 1 in males, and 3 in both sexes.
Evasins
Evasins are cytokine-binding proteins isolated from the tick R. sanguineus [113, 114]. This family is found in all sialotranscriptomes of metastriate, but not in prostriate ticks. The family is extremely divergent; the best matches to R. pulchellus proteins provide only 26-56% identity at the primary sequence level. Four evasins were female specific (50-230-fold) while one evasin was male specific (54-fold).
Immunoglobulin G binding proteins
Tick immunoglobulin binding proteins help to scavenge host IgG that leaks into the hemolymph and salivary members of this family recycle host IgG from ticks back to hosts [115-118]. Male-only forms of this protein were identified in R. appendiculatus [118] and were shown to help their female counterparts feed. The sialomes of R. pulchellus reveals several transcripts with expression virtually in males; for example, the transcript coding for RpSigp-668710 has over 480,000 reads mapped to it from the male library, but only 202 reads from the female library. Other members were also overexpressed in males (>1,000-fold). RpIx75-464069, which was 1,715-fold overexpressed in male glands, was found solely in male glands by MS.
Tick-specific, unknown function
Fifty eight families of proteins are tick specific and have no known function. This group includes 34 families of small proteins (<100 residues) that have no similarities to other known proteins and could be erroneously deduced from non-coding RNA. Only one peptide match was found for members of this group.
Prostriate/metastriate families
The members of “Fibronectin-domain protein” and “Hematopoietic-stem cell progenitor” families in prostriate and metastriate ticks are quite similar, suggesting they may have housekeeping function. Alternatively, they may be of low antigenicity to their hosts either because of their low expression or lack of strong epitopes. The transcriptome confirms their low levels of expression, making up less than 0.01% reads of the S class. However, RpIx75-854185 was identified by MS in female glands.
The 8.9 kDa family was found in all tick sialomes. Sixty CDS were found in the R. pulchellus sialotranscriptome (Supplemental file 1). Some of these peptides have disintegrin domains. Both male and female specific expression were observed; RpIx75-940640, RpSigp-798933 and RpSigp-675391 were overtranscribed in females (12-26-fold) and were detected by MS only in female glands, while RpSigp-7927, RpSigp-972527, RpSigp-509737 and RpSigp-709817 were overtranscribed in male glands (4, 186, 650 and 994 -fold) and were detected by MS only in male glands.
The 23 kDa and 24 kDa protein families of ixodids were also represented in the R. pulchellus sialotranscriptome. RpIx75-677306 (24 kDa family) was found by MS in glands from both sexes.
The “one of each” family was initially thought to be only found in metatriates, where only one gene from the family was found in a tick. However, this was contradicted in the sialotranscriptome of A. maculatum, where several CDS were found. In addition, homologs were also found in I. scapularis. We here report 14 additional CDS for this diverse family, 4 of which were identified by MS in only female SGEs, in accordance with their female-biased transcription (39-172 times overexpressed).
Previously thought to be metastriate-specific families:
Seven protein families that were identified previously as metastriate specific were found to have homologs in the I. scapularis proteome (Table 3). Two members of “Insulin growth factor binding proteins” (IGFBP) family were found in A. variegatum, a short form with only the insulin growth factor-binding protein homologs (IB) SMART domain, and a long form with additional Kazal and immunoglobulin (IG) domains [119]. The sialomes of R. pulchellus reveals six proteins of the short variety, which matches not only metastriate, but also I. scapularis proteins (Supplemental file 1).
A deduced protein from R. appendiculatus ESTs [44] was annotated as orphan as it had no similarities to other known proteins, but it matches R. pulchellus proteins having a weak Somatomedin SMART domain.
Metastriate specific families
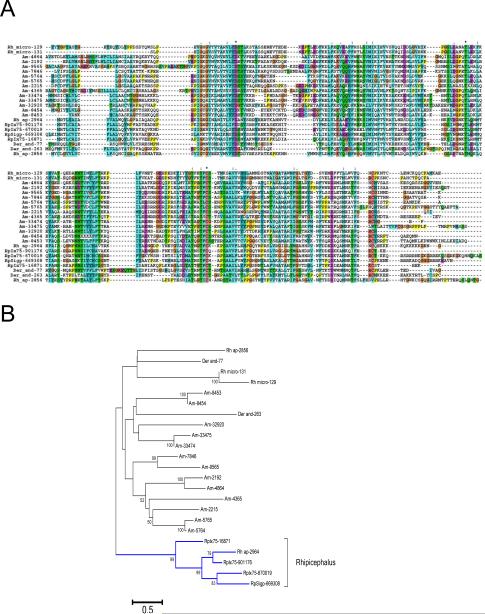
There are seven additional protein families which are metastriate-specific. Five were previously described, while two, the 9 kDa protein family and the Rhip 45-304 family, were deorphanized with the R. pulchellus sialotranscriptome. The female-biased Rhip 45-304 family (~200-250 residues) was so named because the members cluster at 45% similarity within cluster 304 of the database protein clusterization algorithm. An alignment of this family of proteins with sequences from other ticks revealed three identities and 36 similarities (Figure 4A). Phylogenetic analysis indicates a robust clade with R. appendiculatus and R. pulchellus sequences, but no other robust clade of mixed species (Figure 4B). RpIx75-16871 protein was identified by MS only in female glands.
Figure 4. Novel metastriate tick salivary family.
(A) Clustal alignment. The symbols above the alignment indicates for * identity of residues, “:” similarity and “.” indicates less similarity. (B) Phylogenetic evolutionary history was inferred using the Neighbor-Joining method [134].The bootstrap consensus tree inferred from 10000. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates). All ambiguous positions were removed for each sequence pair. There were a total of 360 positions in the final dataset. The bar at the bottom indicates 50% amino acid divergence. Other conditions as in figure 1. The sequences were obtained from previous publications [2, 19]. The R. pulchellus sequences start with Rp and can be obtained from supplemental file 1.
Rhipicephalus specific families
Three protein families from R. pulchellus have similarities exclusively to R. appendiculatus proteins and may represent genus-specific proteins. RpIx75-569044 and RpIx75-697446 from the 45-236 family were identified by MS in male SGEs.
Gender differences in transcript and protein expression
As indicated in the above sections, many transcripts were found differentially expressed in male or female salivary glands, in many cases remarkably so, with hundreds to thousands fold more reads in one sex. Figure 5A shows an overall number of reads from each class of secretory proteins. It is evident that some classes were more highly expressed in one sex. It is interesting to note that the total number of upregulated transcripts (10-fold or more) in females was almost twice that of males (1148 vs. 612) (Table 4 and 5, and supplemental file 1). This may be attributable to the difference in feeding habits between male and female ticks, as previously mentioned. Females secrete more proteins that allow a larger blood meal. This increased intake of blood necessitates metabolic difference to process it.
Figure 5. Differential expression of secretory proteins.
(A) The number of reads from male and female R. pulchellus belonging to the secretory class in increasing order. (B) Transcripts which are expressed more than 10 times in either sex, together with their classification. (C) Proteins which were found only in one sex in the proteome.
Table 4.
Differential expression of genes in male and female salivary glands in the transcriptome
| Class | Females* | % Total | Males* | % Total |
|---|---|---|---|---|
| Secreted | 416 | 34.87 | 142 | 22.87 |
| Cytoskeletal | 14 | 1.17 | 0 | 0.00 |
| Extracellular matrix/cell adhesion | 12 | 1.01 | 4 | 0.64 |
| Metabolism, amino acid | 5 | 0.42 | 6 | 0.97 |
| Metabolism, carbohydrate | 7 | 0.59 | 12 | 1.93 |
| Metabolism, energy | 3 | 0.25 | 2 | 0.32 |
| Metabolism, lipid | 12 | 1.01 | 7 | 1.13 |
| Metabolism, nucleotide | 6 | 0.50 | 0 | 0.00 |
| Oxidant metabolism/detoxification | 2 | 0.17 | 11 | 1.77 |
| Proteasome machinery | 1 | 0.08 | 1 | 0.16 |
| Protein export machinery | 1 | 0.08 | 1 | 0.16 |
| Protein modification machinery | 11 | 0.92 | 7 | 1.13 |
| Signal transduction | 15 | 1.26 | 8 | 1.29 |
| Signal transduction, apoptosis | 1 | 0.08 | 0 | 0.00 |
| Storage | 1 | 0.08 | 0 | 0.00 |
| Transcription factor | 3 | 0.25 | 4 | 0.64 |
| Transcription machinery | 3 | 0.25 | 4 | 0.64 |
| Transporters/storage | 67 | 5.62 | 16 | 2.58 |
| Transposable element | 43 | 3.60 | 18 | 2.90 |
| Unknown | 521 | 43.67 | 354 | 57.00 |
| Unknown, conserved | 49 | 4.11 | 24 | 3.86 |
| Total | 1193 | 100 | 621 | 100 |
More than 10 times overexpressed
Table 5.
Differential expression of genes that encode secretory proteins in male and female salivary glands in the transcriptome
| Females | Males | |
|---|---|---|
| Enzymes | ||
| Metalloproteases | 18 | 5 |
| Serine protease | 0 | 6 |
| Male specific salivary serine protease | 0 | 2 |
| Legumain/asparaginyl peptidase | 0 | 1 |
| 5'-nucleotidase/Apyrase | 0 | 2 |
| Ribonuclease | 2 | 0 |
| Ectonucleotide pyrophosphatase/phosphodiesterase family | 0 | 1 |
| Lipase | 1 | 0 |
| Antigen 5 family | 0 | 3 |
| Proteinase inhibitor domains | ||
| Serpins | 1 | 4 |
| Monolaris | 17 | 3 |
| Bilaris | 7 | 1 |
| Trilaris | 1 | 0 |
| Tetralaris | 1 | 0 |
| Similar to Kunitz domain | 4 | 1 |
| Cystatins | 1 | 11 |
| Chymotrypsin-elastase inhibitor ixodidin - TIL domain | 9 | 11 |
| MonoTil | 0 | 1 |
| BiTil | 2 | 0 |
| Immunity associated products | ||
| Lysozyme | 1 | 1 |
| Defensins | 8 | 1 |
| TEP proteins | 3 | 0 |
| Ixoderin/Ficolin | 1 | 0 |
| Tick specific protein families, at least one family member has known function | ||
| Antimicrobial peptides | ||
| Microplusin | 2 | 0 |
| 5.3 kDa antimicrobial family | 1 | 0 |
| Protease inhibitors | ||
| Carboxypeptidase inhibitor | 1 | 0 |
| Basic Tail | 2 | 1 |
| 18.3 kDa family | 2 | 0 |
| Glycine rich superfamily | 19 | 2 |
| Mucins | 8 | 5 |
| Lipocalin family | ||
| Group I | 62 | 15 |
| Group II | 7 | 1 |
| Group III | 9 | 0 |
| Group IV | 7 | 0 |
| Group V | 5 | 2 |
| Group VI | 4 | 1 |
| Group VII | 3 | 0 |
| Group VIII - Deorphanized Dermacentor lipocalin family | 9 | 1 |
| New lipocalin family Rhipicephalus family XII | 0 | 0 |
| Other putative lipocalins | 20 | 6 |
| Ixodegrins | 8 | 1 |
| DA-P36 family | 15 | 0 |
| Evasin | 8 | 2 |
| Immunoglobulin G binding protein | 0 | 8 |
| Function Unknown | ||
| Found in prostriates and metastriates | ||
| 8.9 kDa family | 22 | 6 |
| 24 kDa family | 3 | 0 |
| One of each protein family - now Ixodidae | 11 | 0 |
| Previously thought as metastriate specific | ||
| Insulin growth factor binding protein - also found in Ixodes | 3 | 0 |
| Amblyomma maculatum family 40-33 | 2 | 0 |
| Deorphanized metastriate-Rhipicephalus family XIV | 0 | 1 |
| Somatomedin domain protein family | 2 | 0 |
| Metastriate specific | ||
| 28 kDa Metastriate family | 3 | 0 |
| 8 kDa Amblyomma family | 1 | 0 |
| Ixostatin-like | 2 | 1 |
| Deorphanized Dermacentor 9 kDa expansion | 2 | 0 |
| Rhip 45-304 - deorphanized on A. maculatum sialome | 4 | 0 |
| Short protein families possibly secreted | 64 | 25 |
| Other secreted proteins - includes fragments | 25 | 10 |
| Conserved secreted protein | 3 | 0 |
| Total | 416 | 142 |
The majority of the transcripts overexpressed in females (45%) and males (58%) were of the U class, but many have signal peptides indicative of secretion and could be novel peptides. The S class transcripts that were upregulated in females (32%, 371 CDS) belong to metalloproteases, lipocalins, glycine-rich proteins and other typical tick specific families, such as evasins and 8.9 kDa family members (Table 5). In contrast, the S class transcripts that were upregulated in males (21%, 133 contigs) include metalloproteases, serine proteases, an asparaginyl peptidase and an ectonucleotide pyrophosphatase along with eight IgG binding proteins [115-117, 120], and protease inhibitors of the serpin, cystatin, Kunitz or TIL families (Table 4 and 5, and Figure 5B). As mentioned above, IgG binding proteins may help their female counterparts feed, while four serpins and 11 cystatins (females have only one of each in their enriched group) may be involved in reproduction as previously proposed [13] (Table 5). The H class transcripts that are upregulated in females (19%) included transporters and enzymes involved in gluconeogenesis (4-aminobutyrate aminotransferase and tyrosine transaminase). DNAses may actually be secreted and prevent NET formation [36]. Similarly, female lipases may be secreted [121]. On the contrary, the H class transcripts upregulated in males (17%) included sulfotransferases, sulfatases and multi-drug transporters for detoxification, and amino acid, ABC, and monoamine transporters. They also code for enzymes targeting aromatic amino acids, such as dopamine β-monooxygenase and aromatic-L-amino-acid/L-histidine decarboxylase, which may be associated with hormone or pheromone metabolism [122]. Lipases (with >400-fold more reads in males) and long-chain acyl-CoA synthetase indicate exceptional lipid metabolism in male salivary glands. The TE class transcripts that are upregulated in males (3%) include transcription factor doublesex, which is associated with sex-specific gene expression [123, 124].
The proteome data, although not geared for detailed quantitative measurements, similarly indicated many differentially expressed proteins, mainly when we consider those proteins found in only one of the two sexes. Of 2,243 proteins identified with at least one high-confidence peptide match, 286 were detected in both sexes, while 174 and 1,783 were identified only in male or female tissues, respectively (Table 6 and 7, Figure 5C). Although the number of proteins identified in females was about 10 times that of males, most of them are accounted for by housekeeping proteins (1,466 in females vs. 74 males). Among these proteins are notably those responsible for the transcription, synthesis, modification and export of proteins. This strongly corroborates with the transcriptome data where there were almost twice as many transcripts that were overexpressed in females as compared to males. In addition, a significant number of signal transduction proteins were found in females. A total of 226 secretory proteins were identified in the proteome – 31 found in both sexes, 77 exclusively in males, and 118 exclusively in females. The lipocalin family was found abundantly in both genders, which corresponds to the transcriptome where this family had the third most abundant reads. The signal intensity of the MS-identified proteins presented no correlation with the transcript abundance (not shown). This may result from the continuous infusion of this class of proteins since the organ was taken from feeding ticks.
Table 6.
R. pulchellus salivary gland proteins identified in the proteome
| All proteins* | Found in both sexes | Found in male only | Found in female only | |||||
|---|---|---|---|---|---|---|---|---|
| Class | Number | % of total | Number | % of total | Number | % of total | Number | % of total |
| Secreted | 221 | 9.91 | 31 | 10.88 | 77 | 45.56 | 113 | 6.36 |
| Cytoskeletal | 126 | 5.65 | 9 | 3.16 | 3 | 1.78 | 114 | 6.42 |
| Extracellular matrix/cell adhesion | 46 | 2.06 | 3 | 1.05 | 3 | 1.78 | 40 | 2.25 |
| Immunity | 12 | 0.54 | 3 | 1.05 | 0 | 0.00 | 9 | 0.51 |
| Metabolism, amino acid | 62 | 2.78 | 21 | 7.37 | 2 | 1.18 | 39 | 2.19 |
| Metabolism, carbohydrate | 89 | 3.99 | 24 | 8.42 | 6 | 3.55 | 59 | 3.32 |
| Metabolism, energy | 63 | 2.82 | 30 | 10.53 | 5 | 2.96 | 28 | 1.58 |
| Metabolism, intermediate | 29 | 1.30 | 5 | 1.75 | 0 | 0.00 | 24 | 1.35 |
| Metabolism, lipid | 81 | 3.63 | 15 | 5.26 | 5 | 2.96 | 61 | 3.43 |
| Metabolism, nucleotide | 45 | 2.02 | 12 | 4.21 | 7 | 4.14 | 26 | 1.46 |
| Nuclear export | 18 | 0.81 | 1 | 0.35 | 0 | 0.00 | 17 | 0.96 |
| Nuclear regulation | 89 | 3.99 | 1 | 0.35 | 2 | 1.18 | 86 | 4.84 |
| Oxidant metabolism/detoxification | 39 | 1.75 | 14 | 4.91 | 2 | 1.18 | 23 | 1.29 |
| Proteasome machinery | 74 | 3.32 | 2 | 0.70 | 0 | 0.00 | 72 | 4.05 |
| Protein export machinery | 134 | 6.01 | 10 | 3.51 | 2 | 1.18 | 122 | 6.87 |
| Protein modification machinery | 197 | 8.83 | 29 | 10.18 | 13 | 7.69 | 155 | 8.72 |
| Protein synthesis machinery | 131 | 5.87 | 16 | 5.61 | 1 | 0.59 | 114 | 6.42 |
| Signal transduction | 243 | 10.89 | 16 | 5.61 | 5 | 2.96 | 222 | 12.49 |
| Signal transduction, apoptosis | 42 | 1.88 | 2 | 0.70 | 3 | 1.78 | 37 | 2.08 |
| Storage | 5 | 0.22 | 3 | 1.05 | 0 | 0.00 | 2 | 0.11 |
| Transcription factor | 20 | 0.90 | 1 | 0.35 | 0 | 0.00 | 19 | 1.07 |
| Transcription machinery | 167 | 7.49 | 8 | 2.81 | 9 | 5.33 | 150 | 8.44 |
| Transporters/storage | 57 | 2.55 | 4 | 1.40 | 6 | 3.55 | 47 | 2.64 |
| Unknown, conserved | 197 | 8.83 | 22 | 7.72 | 9 | 5.33 | 166 | 9.34 |
| Transposable element | 7 | 0.31 | 0 | 0.00 | 0 | 0.00 | 7 | 0.39 |
| Unknown | 37 | 1.66 | 3 | 1.05 | 9 | 5.33 | 25 | 1.41 |
| Total | 2231 | 100 | 285 | 100 | 169 | 100 | 1777 | 100 |
Identified by at least 1 peptide of 95% confidence
Table 7.
Functional classification of secretory R. pulchellus salivary gland proteins identified in the proteome
| All proteins* | Found in both sexes | Found in males only | Found in females only | |||||
|---|---|---|---|---|---|---|---|---|
| Class | Number | % of total | Number | % of total | Number | % of total | Number | % of total |
| Enzymes | ||||||||
| Metalloproteases | 6 | 2.71 | 1 | 3.23 | 1 | 1.30 | 4 | 3.54 |
| M13 family peptidase | 2 | 0.90 | 2 | 6.45 | 0 | 0.00 | 0 | 0.00 |
| Dipeptidyl peptidase | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Serine protease | 3 | 1.36 | 0 | 0.00 | 2 | 2.60 | 1 | 0.88 |
| Male specific salivary serine protease | 2 | 0.90 | 0 | 0.00 | 2 | 2.60 | 0 | 0.00 |
| Zinc carboxypeptidase | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Serine carboxypeptidase - may be lysosomal | 10 | 4.52 | 0 | 0.00 | 9 | 11.69 | 1 | 0.88 |
| Legumain/asparaginyl peptidase | 2 | 0.90 | 0 | 0.00 | 1 | 1.30 | 1 | 0.88 |
| 5'-nucleotidase/Apyrase | 8 | 3.62 | 2 | 6.45 | 2 | 2.60 | 4 | 3.54 |
| Ribonuclease | 2 | 0.90 | 0 | 0.00 | 0 | 0.00 | 2 | 1.77 |
| Inositol phosphatase | 2 | 0.90 | 0 | 0.00 | 0 | 0.00 | 2 | 1.77 |
| Antigen 5 family | 1 | 0.45 | 0 | 0.00 | 1 | 1.30 | 0 | 0.00 |
| Proteinase inhibitor domains | ||||||||
| Serpins | 7 | 3.17 | 1 | 3.23 | 5 | 6.49 | 1 | 0.88 |
| Monolaris | 2 | 0.90 | 0 | 0.00 | 1 | 1.30 | 1 | 0.88 |
| Bilaris | 5 | 2.26 | 0 | 0.00 | 0 | 0.00 | 5 | 4.42 |
| Tetralaris | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Similar to Kunitz domain | 1 | 0.45 | 0 | 0.00 | 1 | 1.30 | 0 | 0.00 |
| Cystatins | 2 | 0.90 | 0 | 0.00 | 2 | 2.60 | 0 | 0.00 |
| Chymotrypsin-elastase inhibitor ixodidin - TIL domain | 1 | 0.45 | 0 | 0.00 | 1 | 1.30 | 0 | 0.00 |
| BiTil | 11 | 4.98 | 2 | 6.45 | 5 | 6.49 | 4 | 3.54 |
| TriTil | 8 | 3.62 | 0 | 0.00 | 8 | 10.39 | 0 | 0.00 |
| Immunity associated products | ||||||||
| Defensins | 1 | 0.45 | 0 | 0.00 | 1 | 1.30 | 0 | 0.00 |
| TEP proteins | 4 | 1.81 | 0 | 0.00 | 0 | 0.00 | 4 | 3.54 |
| Galectin | 3 | 1.36 | 0 | 0.00 | 0 | 0.00 | 3 | 2.65 |
| Tick specific protein families, at least one family member has known function | ||||||||
| Antimicrobial peptides | ||||||||
| Microplusin | 3 | 1.36 | 0 | 0.00 | 1 | 1.30 | 2 | 1.77 |
| Protease inhibitors | ||||||||
| Carboxypeptidase inhibitor | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Basic Tail | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Glycine rich superfamily | 14 | 6.33 | 3 | 9.68 | 2 | 2.60 | 9 | 7.96 |
| Mucins | 6 | 2.71 | 0 | 0.00 | 4 | 5.19 | 2 | 1.77 |
| Lipocalin family | ||||||||
| Group I | 34 | 15. 38 | 7 | 22. 58 | 13 | 16. 88 | 14 | 12. 39 |
| Group II | 3 | 1.36 | 1 | 3.23 | 0 | 0.00 | 2 | 1.77 |
| Group III | 4 | 1.81 | 0 | 0.00 | 0 | 0.00 | 4 | 3.54 |
| Group IV | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Group V | 3 | 1.36 | 0 | 0.00 | 0 | 0.00 | 3 | 2.65 |
| Group VI | 2 | 0.90 | 1 | 3.23 | 0 | 0.00 | 1 | 0.88 |
| Group VIII - Deorphanized Dermacentor lipocalin family | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| New lipocalin family | ||||||||
| Rhipicephalus family XII | 2 | 0.90 | 0 | 0.00 | 2 | 2.60 | 0 | 0.00 |
| Other putative lipocalins | 13 | 5.88 | 4 | 12.90 | 4 | 5.19 | 5 | 4.42 |
| Ixodegrins | 3 | 1.36 | 0 | 0.00 | 0 | 0.00 | 3 | 2.65 |
| DA-P36 family | 7 | 3.17 | 3 | 9.68 | 1 | 1.30 | 3 | 2.65 |
| Immunoglobulin G binding protein | 1 | 0.45 | 0 | 0.00 | 1 | 1.30 | 0 | 0.00 |
| Function Unknown | ||||||||
| Found in prostriates and metastriates | ||||||||
| Hematopoietic-stem cell progenitor | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| 8.9 kDa family | 7 | 3.17 | 0 | 0.00 | 4 | 5.19 | 3 | 2.65 |
| 24 kDa family | 1 | 0.45 | 1 | 3.23 | 0 | 0.00 | 0 | 0.00 |
| One of each protein family - now Ixodidae | 4 | 1.81 | 0 | 0.00 | 0 | 0.00 | 4 | 3.54 |
| Metastriate specific | ||||||||
| 28 kDa Metastriate family | 4 | 1.81 | 1 | 3.23 | 1 | 1.30 | 2 | 1.77 |
| 10 kDa acidic metastriate | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Ixostatin-like | 2 | 0.90 | 2 | 6.45 | 0 | 0.00 | 0 | 0.00 |
| Rhip 45-304 - deorphanized on A. maculatum sialome | 1 | 0.45 | 0 | 0.00 | 0 | 0.00 | 1 | 0.88 |
| Rhipicephalus specific families | ||||||||
| Rhip 45-236 | 2 | 0.90 | 0 | 0.00 | 2 | 2.60 | 0 | 0.00 |
| Short protein families possibly secreted | 2 | 0.90 | 0 | 0.00 | 0 | 0.00 | 2 | 1.77 |
| Other secreted proteins - includes fragments | 9 | 4.07 | 0 | 0.00 | 0 | 0.00 | 9 | 7.96 |
| Conserved secreted protein | 2 | 0.90 | 0 | 0.00 | 0 | 0.00 | 2 | 1.77 |
| Total | 221 | 100 | 31 | 100 | 77 | 100 | 113 | 100 |
Do male salivary products display seminal-fluid properties?
Male tick saliva assists reproduction following the observation that both soft and hard ticks salivate copiously on the surface of their spermatophores before pushing them with their mouthparts into the female's genital pore [13]. Male saliva was postulated to avoid stickiness of the spermatophore when it is being transferred from the male genitalia into the females’. It is possible that in addition to this lubricating or anti-stickiness property of saliva, male salivary molecules may play a role traditionally played by arthropod male accessory glands to increase the female's fecundity [125]. Male ticks remain with their mouthparts inside the female's genital pore for several hours after successful spermatophore transfer, perhaps as a mechanism of guarding the females from other males and thus avoid sperm competition [12], but whether they transfer additional fluids into the females is unknown. Histological studies of tick salivary glands have described male-specific alveolus and cells [126-128] that were postulated to assist tick reproduction [13]. Using a differential display approach, differential male salivary gene expression was observed when males were feeding together with and without female conspecific ticks [129, 130]. Another previously reported salivary gland transcriptome of male and female R. haemaphysaloides ticks have detected, by suppression subtractive hybridization protocols, 17 male-specific transcripts that include a serine protease, cytochrome P450, an IgG binding protein and metalloproteases [131], in concordance with our findings. We here propose that at least some male-specific salivary gland proteins play a role in tick reproduction. Among these candidates are serpins, cystatins and serine proteases which are ubiquitously found in seminal fluids from Drosophila to humans, and also metalloproteases and legumains mentioned above. The differentially male expressed cytochrome P450 may be involved in the production of prostaglandins or other eicosanoids. Male housekeeping enzymes linked to lipid metabolism may relate to additional secreted lipidic products. Male-specific mucins may also fall under this class of seminal-fluid like proteins and the male upregulated galactosyl-transferase may assist the post-translational modifications abundant in mucins. While hypothesis-driven research states their hypothesis in the introduction of their work, discovery driven research states its hypothesis in their discussion. This work allows hypothesizing the several transcripts mentioned above to be involved in tick reproduction. RNAi experiments can now be designed and the hypothesis tested.
Gender-dependent expression of Bilaris proteins
While male differentially-expressed transcripts can be hypothesized to play a role in reproduction, it is also possible that they result from males expressing a different anti-hemostatic cocktail than females, due to their different feeding requirements and/or to diversify their antigenic cocktail.
As shown in Figure 1, the saliva of male ticks contained less FXa and thrombin inhibitors. Thus, in an attempt to search for male-specific anticoagulants, we further analyzed the bilaris proteins (with two tandem Kunitz domains) in the transcriptome, as this class of proteins has some well-characterized anticoagulants [132, 133].
We have identified 81 coding sequences for bilaris proteins in the R. pulchellus transcriptome. By systematic analysis, we classified them into five subclasses based on their number of cysteine residues, as well as their length (Figure 6A). In a typical Kunitz domain, the fold is conserved by three conserved disulphide bonds: C1-C6, C2-C4 and C3-C5. In the first subclass, both Kunitz domains have all three disulphide bonds conserved. In all other subclasses, the first Kunitz domain contains all three conserved disulphide bridges. However, marked differences are seen in the second Kunitz domain. In the second subclass, cysteine residues C2 and C4 are substituted, resulting in the loss of one disulphide bridge in the second Kunitz domain. This is seen in ixolaris [63]. Similarly, proteins in subclass III has cysteine residues C3 and C5 substituted, thus it also has only two disulphide bridges in the second Kunitz domain. In subclass IV and V, an additional pair of cysteine residues, is present at the C-terminal of the second Kunitz domain. However, subclass V has a long proline-rich inter-domain segment (73 - 85 residues). It is interesting to note that even within each subclass, similarity between the individual proteins, not taking into account isoforms, are low; the majority of the proteins within a subclass had less than 40% identity to each other. Proteins within each subclass were aligned and the consensus sequence for each subclass was generated and aligned with other subclasses (Figure 6B). This alignment further highlights the characteristic differences across the five subclasses and the low conservation of sequences in the inter-cysteine regions within as well as out of each subclass. The variations in the inter-cysteine regions suggest diverse functions for these proteins despite similar structures. They might target different proteases and their complexes, and inhibit them through different mechanisms.
Figure 6. Five subclasses of bilaris proteins.
(A) Schematic representation of the five subclasses of bilaris proteins. Subclass I has two full tandem Kunitz domains, each three conserved disulphide bonds, represented by the solid lines. A typical Kunitz domain disulphide bond pairing is as follows: C1-C6, C2-C4 and C3-C5. Subclass II and III has two missing cysteine residues C2, C4 and C3, C5, respectively, in the second Kunitz domain, resulting in a loss of a disulphide bond. Missing disulphide bonds are represented by the bold dashed line. Subclass IV and V has two extra cysteine residues in the second Kunitz domain, one located between the typical C4 and C5 cysteine residues, and the other after the C6 cysteine residue. This extra disulphide bond is represented by a bold solid line. However, subclass V contains a long inter-domain, proline-rich segment (~ 80 residues and represented by the bold dotted line. (B) Alignment of consensus sequences of the five bilaris subclasses showing conserved disulphide bridges.
All bilaris proteins were aligned using ClustalW and a phylogenetic tree was constructed (Figure 7). Proteins from the same subclass clustered together even though they had low sequence homology. Figure 7 also shows the abundance of reads from the transcriptome for each contig. In order to have a better comparison of the difference in number of reads of each contig from each sex, the normalized number of female reads was divided by the normalized number of male reads. Figure 8A illustrates the number of fold differences of each contig. There were altogether eight contigs in which one sex had more than 10 times the number of reads than the other; seven were higher in females while one was higher in males. It was interesting to see that the majority of subclass II bilaris proteins (10 of 12) were more highly expressed in females, while the majority of subclass I and IV bilaris proteins (16 of 19 and 28 of 34 respectively) were higher in males. This further suggests that male and female ticks employ different sets of proteins to target the host hemostatic system.
Figure 7. Phylogenetics and associated number of reads of bilaris CDS.
81 CDS belonging to the bilaris class were aligned with ClustalW and a phylogenetic tree was drawn using the Neighbor-Joining method in MEGA5 [25]. The subclass of each CDS is indicated by the symbol on the left of the accession number. The number of reads associated with each CDS is indicated by the bar chart, where red represents reads from the female library, and blue from the male library. Proteins that were identified in the female proteome are marked with a ♀ beside the bar chart.
Figure 8. Expression difference of bilaris proteins between male and female ticks.
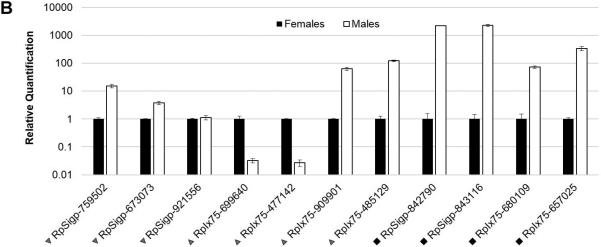
(A) Fold difference in the transcriptome is expressed over number of male reads. Seven transcripts were highly expressed in females while one was highly expressed in males. These sex biased transcripts which were more than 10 fold expressed in either sex are highlighted in grey. The subclass of the bilaris proteins are indicated beside the transcript name. It is noted that most of subclass I, III and IV were expressed by males, and subclass II and V in females, as tabulated in the inset. Transcripts marked with an asterisk (*) were used for RT-PCR studies. (B) RT-PCR was performed on selected bilaris proteins from subclass II, III and IV. Comparison of expression is in reference to females. Two independent biological samples were used to obtain the standard deviation.
As data from the transcriptome only gives us a rough estimate on the level of expression of these proteins, we performed quantitative real time – polymerase chain reaction (qRT-PCR) experiments. This allowed us to compare the relative abundance of mRNA of each gene in males and females, and thus the relative level of expression. As we were interested in subclass II, III and IV, four genes were chosen from each subclass (full sequences of chosen genes are listed in the Appendix). However, we were unable to successfully amplify one gene from subclass II. qRT-PCR was performed using β-actin as an internal control and the level of expression of males are shown in reference to the female genes (Figure 8B). The proteins from subclass IV were indeed highly expressed in males, with two of them expressed more than 2000 times higher in males than females. This corresponds to the elevated number of male reads we see in the transcriptome for proteins belonging to subclass IV, and serves to validate the overall transcriptional differences observed between genders.
Bilaris proteins are potentially important in anticoagulant function and may target various parts of the coagulation cascade. There appears to be distinct pattern of expression of this group of proteins. The functional studies of these proteins may help us delineate additional aspects of differential feeding habits between male and female ticks.
Conclusions
This work represents the first deep-sequencing transcriptome of ticks aimed at distinguishing salivary gender-specific transcripts, and the first sialotranscriptome of R. pulchellus. As a product of discovery-driven research, this work deposited 11,227 publicly available CDS to GenBank that will serve as a discovery platform for future identification of pharmacologically interesting molecules and other studies. These coding sequences served the basis for a preliminary proteomics study where 460 and 2,069 proteins were identified from male and female salivary glands, respectively. The identification of male specific products is hypothesized, for at least some of them, to play a role in tick reproduction taking into account the involvement of tick mouthparts in spermatophore transfer [13]. Candidate “seminal fluid-like” salivary proteins identified can be the substrate for RNAi experiments to attempt disruption of tick reproduction. On the other hand, a gender specialization in the anti-hemostatic salivary cocktail is postulated, and a detailed study of the bilaris family of serine protease inhibitors was performed, followed by qPCR experiments that served to validate the overall transcriptional differences observed between genders.
Supplementary Material
Significance.
This annotated Rhipicephalus pulchellus database represents a mining field for future experiments involving the resolution of time-dependent transcript expression in this tick species, as well as to define novel vaccine targets and discover novel pharmaceuticals. Gender specific proteins may represent different repertoires of pharmacological reagents to assist feeding by each sex, and in males may represent proteins that assist reproduction similarly to seminal proteins in other animals.
Highlights.
We report the first deep-sequencing transcriptome of ticks aimed at distinguishing salivary gender-specific transcripts.
Over 11,000 coding sequences have been publicly deposited, serving as a discovery platform.
These proteins may be functionally analogous to seminal proteins of both vertebrates and invertebrates.
A detailed study of a dual Kunitz family of peptides is presented.
Acknowledgments
J.M.C.R. and I.F. were supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH; USA). MS was supported by grant VEGA (Vedecká grantová agentúra) 2/0060/12. AWLT and RMK were supported by grant MOE2010-T2-2-023 from the Singapore Ministry of Education.
Because J.M.C.R. is a government employee and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- CDS
Coding sequence
- CDD
Conserved Domain Database
- H
Housekeeping proteins
- PSA
Human prostate-specific antigen
- IGFBP
Insulin growth factor binding protein
- ML
MD-2-related lipid-recognition
- NET
Neutrophil extracellular trap
- U
Protein of unknown function
- S
Putative secreted proteins
- SGE
Salivary gland extract
- TEP
Thioester proteins
- TE
Transposable elements
- TIL
Trypsin inhibitor-like
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nava S, Guglielmone AA, Mangold AJ. An overview of systematics and evolution of ticks. Front Biosci. 2009;14:2857–77. doi: 10.2741/3418. [DOI] [PubMed] [Google Scholar]
- 2.Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Frontiers in Biosciences. 2009;14:2051–88. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mans BJ, Francischetti IMB. Sialomic perspectives on the evolution of blood-feeding behavior in Arthropods: Future therapeutics by natural design. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and Hemostasis: From Bench to Bed-side. Springer; 2010. [Google Scholar]
- 4.Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Lehane MJ. The biology of blood-sucking in insects. 2 ed. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 6.Calvo E, Pham VM, Lombardo F, Arca B, Ribeiro JM. The sialotranscriptome of adult male Anopheles gambiae mosquitoes. Insect Biochem Mol Biol. 2006;36:570–5. doi: 10.1016/j.ibmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, et al. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Li Y, Liu Z, Yang J, Yin H. The life cycle of Hyalomma rufipes (Acari: Ixodidae) under laboratory conditions. Experimental & applied acarology. 2012;56:85–92. doi: 10.1007/s10493-011-9490-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Yu Z, Guo L, Li L, Meng H, Wang D, et al. Life cycle of Haemaphysalis doenitzi (Acari: Ixodidae) under laboratory conditions and its phylogeny based on mitochondrial 16S rDNA. Experimental & applied acarology. 2012;56:143–50. doi: 10.1007/s10493-011-9507-8. [DOI] [PubMed] [Google Scholar]
- 10.Ma M, Guan G, Chen Z, Liu Z, Liu A, Gou H, et al. The life cycle of Haemaphysalis qinghaiensis (Acari: Ixodidae) ticks under laboratory conditions. Experimental & applied acarology. 2013;59:493–500. doi: 10.1007/s10493-012-9617-y. [DOI] [PubMed] [Google Scholar]
- 11.Zheng H, Yu Z, Chen Z, Zhou L, Zheng B, Ma H, et al. Development and biological characteristics of Haemaphysalis longicornis (Acari: Ixodidae) under field conditions. Experimental & applied acarology. 2011;53:377–88. doi: 10.1007/s10493-010-9415-3. [DOI] [PubMed] [Google Scholar]
- 12.Kiszewski AE, Matuschka FR, Spielman A. Mating strategies and spermiogenesis in ixodid ticks. Annual review of entomology. 2001;46:167–82. doi: 10.1146/annurev.ento.46.1.167. [DOI] [PubMed] [Google Scholar]
- 13.Feldman-Muhsam B, Borut S, Saliternik-Givant S. Salivary secretion of the male tick during copulation. J Insect Physiol. 1970;16:1945–9. doi: 10.1016/0022-1910(70)90239-8. [DOI] [PubMed] [Google Scholar]
- 14.Slovak M, Labuda M, Marley SE. Mass laboratory rearing of Dermacentor reticulatus ticks (Acarina, Ixodidae). Biologia (Bratisl) 2002;57:261–6. [Google Scholar]
- 15.Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, et al. De novo transcriptome assembly with ABySS. Bioinformatics (Oxford, England) 2009;25:2872–7. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 16.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19:1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics. 2011;12(Suppl 14):S2. doi: 10.1186/1471-2105-12-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the Gulf Coast tick, Amblyomma maculatum. PLoS ONE. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein engineering. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 21.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–12. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 23.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galperin MY, Koonin EV. 'Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic acids research. 2004;32:5452–63. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nature reviews Drug discovery. 2007;6:662–80. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson RG, O'Hagan JE, Shotz M, Binnington KC, Hegarty MP. Prostaglandin in the saliva of the cattle tick Boophilus microplus. Aust J Exp Biol Med Sci. 1976;54:475–86. doi: 10.1038/icb.1976.48. [DOI] [PubMed] [Google Scholar]
- 29.Higgs GA, Vane JR, Hart RJ, Porter C, Wilson RG. Prostaglandins in the saliva of the cattle tick, Boophilus microplus(Canestrini) (Acarina, Ixodidae). Bull Ent Res. 1976;66:665–70. [Google Scholar]
- 30.Ribeiro JM, Makoul GT, Robinson DR. Ixodes dammini: evidence for salivary prostacyclin secretion. The Journal of parasitology. 1988;74:1068–9. [PubMed] [Google Scholar]
- 31.Ribeiro JMC, Evans PM, MacSwain JL, Sauer J. Amblyomma americanum: Characterization of salivary prostaglandins E2 and F2alpha by RP-HPLC/bioassay and gas chromatography-mass spectrometry. Exp Parasitol. 1992;74:112–6. doi: 10.1016/0014-4894(92)90145-z. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira CJ, Sa-Nunes A, Francischetti IM, Carregaro V, Anatriello E, Silva JS, et al. Deconstructing Tick Saliva: Non protein molecules with potent immunomodullatory properties. The Journal of biological chemistry. 2011;286:10960–9. doi: 10.1074/jbc.M110.205047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marino JP., Jr Soluble epoxide hydrolase, a target with multiple opportunities for cardiovascular drug discovery. Curr Top Med Chem. 2009;9:452–63. doi: 10.2174/156802609788340805. [DOI] [PubMed] [Google Scholar]
- 34.Spector AA, Fang X, Snyder GD, Weintraub NL. Epoxyeicosatrienoic acids (EETs): metabolism and biochemical function. Prog Lipid Res. 2004;43:55–90. doi: 10.1016/s0163-7827(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 35.Utermohlen O, Herz J, Schramm M, Kronke M. Fusogenicity of membranes: the impact of acid sphingomyelinase on innate immune responses. Immunobiology. 2008;213:307–14. doi: 10.1016/j.imbio.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Wartha F, Beiter K, Normark S, Henriques-Normark B. Neutrophil extracellular traps: casting the NET over pathogenesis. Curr Opin Microbiol. 2007;10:52–6. doi: 10.1016/j.mib.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Pichu S, Yalcin EB, Ribeiro JM, King RS, Mather TN. Molecular characterization of novel sulfotransferases from the tick, Ixodes scapularis. BMC Biochem. 2011;12:32. doi: 10.1186/1471-2091-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–98. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 39.Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes & development. 1988;2:477–89. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- 40.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nature reviews. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 41.Bao W, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Molecular biology and evolution. 2009;26:983–93. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nature reviews. 2008;9:411–2. doi: 10.1038/nrg2165-c1. author reply 4. [DOI] [PubMed] [Google Scholar]
- 43.Kelley DR, Rinn JL. Transposable elements reveal a stem cell specific class of long noncoding RNAs. Genome biology. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–88. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anatriello E, Ribeiro JM, de Miranda-Santos IK, Brandao LG, Anderson JM, Valenzuela JG, et al. An insight into the sialotranscriptome of the brown dog tick, Rhipicephalus sanguineus. BMC genomics. 2010;11:450. doi: 10.1186/1471-2164-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francischetti IM, Anderson JM, Manoukis N, Pham VM, Ribeiro JM. An insight into the sialotranscriptome and proteome of the coarse bontlegged tick, Hyalomma marginatum rufipes. Journal of proteomics. 2011;74:2892–908. doi: 10.1016/j.jprot.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. Journal of proteome research. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- 48.Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131:275–8. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hati R, Mitra P, Sarker S, Bhattacharyya KK. Snake venom hemorrhagins. Crit Rev Toxicol. 1999;29:1–19. doi: 10.1080/10408449991349168. [DOI] [PubMed] [Google Scholar]
- 50.Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochemical and biophysical research communications. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francischetti IM, Mather TN, Ribeiro JM. Tick saliva is a potent inhibitor of endothelial cell proliferation and angiogenesis. Thrombosis and haemostasis. 2005;94:167–74. doi: 10.1267/THRO05010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, et al. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–90. doi: 10.1074/mcp.M700497-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.LaFlamme BA, Ram KR, Wolfner MF. The Drosophila melanogaster seminal fluid protease “seminase” regulates proteolytic and post-mating reproductive processes. PLoS genetics. 2012;8:e1002435. doi: 10.1371/journal.pgen.1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bilhartz DL, Tindall DJ, Oesterling JE. Prostate-specific antigen and prostatic acid phosphatase: biomolecular and physiologic characteristics. Urology. 1991;38:95–102. doi: 10.1016/s0090-4295(05)80066-4. [DOI] [PubMed] [Google Scholar]
- 55.Veveris-Lowe TL, Kruger SJ, Walsh T, Gardiner RA, Clements JA. Seminal fluid characterization for male fertility and prostate cancer: kallikrein-related serine proteases and whole proteome approaches. Seminars in thrombosis and hemostasis. 2007;33:87–99. doi: 10.1055/s-2006-958467. [DOI] [PubMed] [Google Scholar]
- 56.Ribeiro JM, Mather TN. Ixodes scapularis: salivary kininase activity is a metallo dipeptidyl carboxypeptidase. Experimental parasitology. 1998;89:213–21. doi: 10.1006/expr.1998.4296. [DOI] [PubMed] [Google Scholar]
- 57.Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5'-nucleotidase family. Proc Natl Acad Sci U S A. 1995;92:694–8. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stutzer C, Mans BJ, Gaspar AR, Neitz AW, Maritz-Olivier C. Ornithodoros savignyi: soft tick apyrase belongs to the 5'-nucleotidase family. Experimental parasitology. 2009;122:318–27. doi: 10.1016/j.exppara.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Schluter H, Tepel M, Zidek W. Vascular actions of diadenosine phosphates. Journal of autonomic pharmacology. 1996;16:357–62. doi: 10.1111/j.1474-8673.1996.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 60.Ogilvie A, Blasius R, Schulze-Lohoff E, Sterzel RB. Adenine dinucleotides: a novel class of signalling molecules. Journal of autonomic pharmacology. 1996;16:325–8. doi: 10.1111/j.1474-8673.1996.tb00045.x. [DOI] [PubMed] [Google Scholar]
- 61.Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, et al. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Current protein & peptide science. 2003;4:231–51. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 62.Castaneda O, Harvey AL. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon. 2009;54:1119–24. doi: 10.1016/j.toxicon.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 63.Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–12. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- 64.Monteiro RQ, Rezaie AR, Ribeiro JM, Francischetti IM. Ixolaris: a factor Xa heparin-binding exosite inhibitor. The Biochemical journal. 2005;387:871–7. doi: 10.1042/BJ20041738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nazareth RA, Tomaz LS, Ortiz-Costa S, Atella GC, Ribeiro JM, Francischetti IM, et al. Antithrombotic properties of Ixolaris, a potent inhibitor of the extrinsic pathway of the coagulation cascade. Thrombosis and haemostasis. 2006;96:7–13. doi: 10.1160/TH06-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monteiro RQ, Rezaie AR, Bae JS, Calvo E, Andersen JF, Francischetti IM. Ixolaris binding to factor X reveals a precursor state of factor Xa heparin-binding exosite. Protein Sci. 2008;17:146–53. doi: 10.1110/ps.073016308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carneiro-Lobo TC, Konig S, Machado DE, Nasciutti LE, Forni MF, Francischetti IM, et al. Ixolaris, a tissue factor inhibitor, blocks primary tumor growth and angiogenesis in a glioblastoma model. J Thromb Haemost. 2009;7:1855–64. doi: 10.1111/j.1538-7836.2009.03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez-Valle M, Vance M, Moolhuijzen PM, Tao X, Lew-Tabor AE. Differential recognition by tick-resistant cattle of the recombinantly expressed Rhipicephalus microplus serine protease inhibitor-3 (RMS-3). Ticks and tick-borne diseases. 2012;3:159–69. doi: 10.1016/j.ttbdis.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Chmelar J, Oliveira CJ, Rezacova P, Francischetti IM, Kovarova Z, Pejler G, et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–44. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chalaire KC, Kim TK, Garcia-Rodriguez H, Mulenga A. Amblyomma americanum (L.) (Acari: Ixodidae) tick salivary gland serine protease inhibitor (serpin) 6 is secreted into tick saliva during tick feeding. J Exp Biol. 2011;214:665–73. doi: 10.1242/jeb.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, Shirras AD, et al. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a procathepsin and a soluble gamma-glutamyl transpeptidase. Proteome science. 2006;4:9. doi: 10.1186/1477-5956-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolfner MF, Harada HA, Bertram MJ, Stelick TJ, Kraus KW, Kalb JM, et al. New genes for male accessory gland proteins in Drosophila melanogaster. Insect biochemistry and molecular biology. 1997;27:825–34. doi: 10.1016/s0965-1748(97)00056-8. [DOI] [PubMed] [Google Scholar]
- 73.Seregni E, Botti C, Ballabio G, Bombardieri E. Biochemical characteristics and recent biological knowledge on prostate-specific antigen. Tumori. 1996;82:72–7. doi: 10.1177/030089169608200116. [DOI] [PubMed] [Google Scholar]
- 74.Horka H, Staudt V, Klein M, Taube C, Reuter S, Dehzad N, et al. The tick salivary protein sialostatin L inhibits the Th9-derived production of the asthma-promoting cytokine IL-9 and is effective in the prevention of experimental asthma. J Immunol. 2012;188:2669–76. doi: 10.4049/jimmunol.1100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwarz A, Valdes JJ, Kotsyfakis M. The role of cystatins in tick physiology and blood feeding. Ticks and tick-borne diseases. 2012;3:117–27. doi: 10.1016/j.ttbdis.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kotsyfakis M, Horka H, Salat J, Andersen JF. The crystal structures of two salivary cystatins from the tick Ixodes scapularis and the effect of these inhibitors on the establishment of Borrelia burgdorferi infection in a murine model. Molecular microbiology. 2010;77:456–70. doi: 10.1111/j.1365-2958.2010.07220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salat J, Paesen GC, Rezacova P, Kotsyfakis M, Kovarova Z, Sanda M, et al. Crystal structure and functional characterization of an immunomodulatory salivary cystatin from the soft tick Ornithodoros moubata. The Biochemical journal. 2010;429:103–12. doi: 10.1042/BJ20100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaji K, Tsuji N, Miyoshi T, Islam MK, Hatta T, Alim MA, et al. A salivary cystatin, HlSC-1, from the ixodid tick Haemaphysalis longicornis play roles in the blood-feeding processes. Parasitology research. 2009;106:61–8. doi: 10.1007/s00436-009-1626-3. [DOI] [PubMed] [Google Scholar]
- 79.Kotsyfakis M, Anderson JM, Andersen JF, Calvo E, Francischetti IM, Mather TN, et al. Cutting edge: Immunity against a “silent” salivary antigen of the Lyme vector Ixodes scapularis impairs its ability to feed. J Immunol. 2008;181:5209–12. doi: 10.4049/jimmunol.181.8.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kotsyfakis M, Karim S, Andersen JF, Mather TN, Ribeiro JM. Selective cysteine protease inhibition contributes to blood-feeding success of the tick Ixodes scapularis. The Journal of biological chemistry. 2007;282:29256–63. doi: 10.1074/jbc.M703143200. [DOI] [PubMed] [Google Scholar]
- 81.Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J Biol Chem. 2006;281:26298–307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 82.Moura AA, Souza CE, Stanley BA, Chapman DA, Killian GJ. Proteomics of cauda epididymal fluid from mature Holstein bulls. Journal of proteomics. 2010;73:2006–20. doi: 10.1016/j.jprot.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Yadav VK, Chhikara N, Gill K, Dey S, Singh S, Yadav S. Three low molecular weight cysteine proteinase inhibitors of human seminal fluid: purification and enzyme kinetic properties. Biochimie. 2013;95:1552–9. doi: 10.1016/j.biochi.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 84.Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2005;27:667–74. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 85.Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Molecular immunology. 2004;40:903–8. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 86.Kopacek P, Hajdusek O, Buresova V, Daffre S. Tick innate immunity. Advances in experimental medicine and biology. 2010;708:137–62. [PubMed] [Google Scholar]
- 87.Horackova J, Rudenko N, Golovchenko M, Havlikova S, Grubhoffer L. IrML - a gene encoding a new member of the ML protein family from the hard tick, Ixodes ricinus. J Vector Ecol. 2010;35:410–8. doi: 10.1111/j.1948-7134.2010.00100.x. [DOI] [PubMed] [Google Scholar]
- 88.Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, et al. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:32721–7. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 89.Rego RO, Hajdusek O, Kovar V, Kopacek P, Grubhoffer L, Hypsa V. Molecular cloning and comparative analysis of fibrinogen-related proteins from the soft tick Ornithodoros moubata and the hard tick Ixodes ricinus. Insect biochemistry and molecular biology. 2005;35:991–1004. doi: 10.1016/j.ibmb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 90.Fogaca AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Developmental and comparative immunology. 2004;28:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 91.Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–29. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 92.Pichu S, Ribeiro JM, Mather TN. Purification and characterization of a novel salivary antimicrobial peptide from the tick, Ixodes scapularis. Biochemical and biophysical research communications. 2009;390:511–5. doi: 10.1016/j.bbrc.2009.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arolas JL, Lorenzo J, Rovira A, Castella J, Aviles FX, Sommerhoff CP. A carboxypeptidase inhibitor from the tick Rhipicephalus bursa: isolation, cDNA cloning, recombinant expression, and characterization. The Journal of biological chemistry. 2005;280:3441–8. doi: 10.1074/jbc.M411086200. [DOI] [PubMed] [Google Scholar]
- 94.Arolas JL, Popowicz GM, Lorenzo J, Sommerhoff CP, Huber R, Aviles FX, et al. The three-dimensional structures of tick carboxypeptidase inhibitor in complex with A/B carboxypeptidases reveal a novel double-headed binding mode. Journal of molecular biology. 2005;350:489–98. doi: 10.1016/j.jmb.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 95.Gong H, Zhou J, Liao M, Hatta T, Harnnoi T, Umemiya R, et al. Characterization of a carboxypeptidase inhibitor from the tick Haemaphysalis longicornis. J Insect Physiol. 2007;53:1079–87. doi: 10.1016/j.jinsphys.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 96.Narasimhan S, Koski RA, Beaulieu B, Anderson JF, Ramamoorthi N, Kantor F, et al. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect molecular biology. 2002;11:641–50. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- 97.Sauer JR, McSwain JL, Bowman AS, Essenberg RC. Tick salivary gland physiology. Annual review of entomology. 1995;40:245–67. doi: 10.1146/annurev.en.40.010195.001333. [DOI] [PubMed] [Google Scholar]
- 98.Bishop R, Lambson B, Wells C, Pandit P, Osaso J, Nkonge C, et al. A cement protein of the tick Rhipicephalus appendiculatus, located in the secretory e cell granules of the type III salivary gland acini, induces strong antibody responses in cattle. International journal for parasitology. 2002;32:833–42. doi: 10.1016/s0020-7519(02)00027-9. [DOI] [PubMed] [Google Scholar]
- 99.Maruyama SR, Anatriello E, Anderson JM, Ribeiro JM, Brandao LG, Valenzuela JG, et al. The expression of genes coding for distinct types of glycine-rich proteins varies according to the biology of three metastriate ticks, Rhipicephalus (Boophilus) microplus, Rhipicephalus sanguineus and Amblyomma cajennense. BMC genomics. 2010;11:363. doi: 10.1186/1471-2164-11-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou J, Gong H, Zhou Y, Xuan X, Fujisaki K. Identification of a glycine-rich protein from the tick Rhipicephalus haemaphysaloides and evaluation of its vaccine potential against tick feeding. Parasitology research. 2006;100:77–84. doi: 10.1007/s00436-006-0243-7. [DOI] [PubMed] [Google Scholar]
- 101.Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2009;26:325–34. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 103.Mans BJ, Ribeiro JM, Andersen JF. Structure, function, and evolution of biogenic amine-binding proteins in soft ticks. The Journal of biological chemistry. 2008;283:18721–33. doi: 10.1074/jbc.M800188200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI. Tick histamine-binding proteins: isolation, cloning, and three- dimensional structure. Molecular cell. 1999;3:661–71. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- 105.Paesen GC, Adams PL, Nuttall PA, Stuart DL. Tick histamine-binding proteins: lipocalins with a second binding cavity. Biochim Biophys Acta. 2000;1482:92–101. doi: 10.1016/s0167-4838(00)00168-0. [DOI] [PubMed] [Google Scholar]
- 106.Mans BJ, Ribeiro JM. Function, mechanism and evolution of the moubatin-clade of soft tick lipocalins. Insect biochemistry and molecular biology. 2008;38:841–52. doi: 10.1016/j.ibmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mans BJ, Ribeiro JM. A novel clade of cysteinyl leukotriene scavengers in soft ticks. Insect biochemistry and molecular biology. 2008;38:862–70. doi: 10.1016/j.ibmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, et al. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174:2084–91. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 109.Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, et al. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae). Insect biochemistry and molecular biology. 2005;35:1142–61. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calvete JJ, Moreno-Murciano MP, Theakston RD, Kisiel DG, Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. The Biochemical journal. 2003;372:725–34. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang TF, Niewriarowski S. Disintegrins: The naturally-occurring antagonists of platelet fibrinogen receptors. J Toxicol Toxin Rev. 1994;13:253–73. [PubMed] [Google Scholar]
- 112.Bergman DK, Palmer MJ, Caimano MJ, Radolf JD, Wikel SK. Isolation and molecular cloning of a secreted immunosuppressant protein from Dermacentor andersoni salivary gland. J Parasitol. 2000;86:516–25. doi: 10.1645/0022-3395(2000)086[0516:IAMCOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 113.Deruaz M, Frauenschuh A, Alessandri AL, Dias JM, Coelho FM, Russo RC, et al. Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. The Journal of experimental medicine. 2008;205:2019–31. doi: 10.1084/jem.20072689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frauenschuh A, Power CA, Deruaz M, Ferreira BR, Silva JS, Teixeira MM, et al. Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. The Journal of biological chemistry. 2007;282:27250–8. doi: 10.1074/jbc.M704706200. [DOI] [PubMed] [Google Scholar]
- 115.Wang H, Nuttall PA. Immunoglobulin-binding proteins in ticks: new target for vaccine development against a blood-feeding parasite. Cell Mol Life Sci. 1999;56:286–95. doi: 10.1007/s000180050430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang H, Nuttall PA. Immunoglobulin G binding proteins in male Rhipicephalus appendiculatus ticks. Parasite Immunol. 1995;17:517–24. doi: 10.1111/j.1365-3024.1995.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 117.Wang H, Nuttall PA. Immunoglobulin-G binding proteins in the ixodid ticks, Rhipicephalus appendiculatus, Amblyomma variegatum and Ixodes hexagonus. Parasitology. 1995;111:161–5. doi: 10.1017/s0031182000064908. [DOI] [PubMed] [Google Scholar]
- 118.Wang H, Paesen GC, Nuttall PA, Barbour AG. Male ticks help their mates to feed. Nature. 1998;391:753–4. doi: 10.1038/35773. [DOI] [PubMed] [Google Scholar]
- 119.Ribeiro JM, Anderson JM, Manoukis NC, Meng Z, Francischetti IM. A further insight into the sialome of the tropical bont tick, Amblyomma variegatum. BMC genomics. 2011;12:136. doi: 10.1186/1471-2164-12-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang H, Nuttall PA. Excretion of host immunoglobulin in tick saliva and detection of IgG-binding proteins in tick haemolymph and salivary glands. Parasitology. 1994;109(Pt 4):525–30. doi: 10.1017/s0031182000080781. [DOI] [PubMed] [Google Scholar]
- 121.Bowman AS, Gengler CL, Surdick MR, Zhu K, Essenberg RC, Sauer JR, et al. A novel phospholipase A2 activity in saliva of the lone star tick, Amblyomma americanum (L.). Experimental parasitology. 1997;87:121–32. doi: 10.1006/expr.1997.4201. [DOI] [PubMed] [Google Scholar]
- 122.Sonenshine DE. Pheromones and other semiochemicals of the acari. Annual review of entomology. 1985;30:1–28. doi: 10.1146/annurev.en.30.010185.000245. [DOI] [PubMed] [Google Scholar]
- 123.Shukla JN, Nagaraju J. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. Journal of genetics. 2010;89:341–56. doi: 10.1007/s12041-010-0046-6. [DOI] [PubMed] [Google Scholar]
- 124.Portman DS. Genetic control of sex differences in C. elegans neurobiology and behavior. Advances in genetics. 2007;59:1–37. doi: 10.1016/S0065-2660(07)59001-2. [DOI] [PubMed] [Google Scholar]
- 125.Kaufman WR. Assuring paternity in a promiscuous world: are there lessons for ticks among the insects ? Parasitology. 2004;129:S145–S60. doi: 10.1017/s0031182004004846. [DOI] [PubMed] [Google Scholar]
- 126.Chinery WA. Studies on the various glands of the tick Haemaphysalis spinigera Neumann 1897. 3. The salivary glands. Acta tropica. 1965;22:321–49. [PubMed] [Google Scholar]
- 127.Till WM. A contribution to the anatomy and histology of the brown ear tick Rhipicephalus appendiculatus Neumann. Mem ent Sot S Afr. 1961;6:3–122. [Google Scholar]
- 128.Furquim KC, Bechara GH, Camargo Mathias MI. Morpho-histochemical characterization of salivary gland cells of males of the tick Rhipicephalus sanguineus (Acari: Ixodidae) at different feeding stages: description of new cell types. Experimental & applied acarology. 2010;50:59–70. doi: 10.1007/s10493-009-9282-y. [DOI] [PubMed] [Google Scholar]
- 129.Anyomi FM, Bior AD, Essenberg RC, Sauer JR. Gene expression in male tick salivary glands is affected by feeding in the presence of females. Archives of insect biochemistry and physiology. 2006;63:159–68. doi: 10.1002/arch.20151. [DOI] [PubMed] [Google Scholar]
- 130.Bior AD, Essenberg RC, Sauer JR. Comparison of differentially expressed genes in the salivary glands of male ticks, Amblyomma americanum and Dermacentor andersoni. Insect Biochem Mol Biol. 2002;32:645–55. doi: 10.1016/s0965-1748(01)00143-6. [DOI] [PubMed] [Google Scholar]
- 131.Xiang FY, Zhou YZ, Zhou JL. Identification of differentially expressed genes in the salivary gland of Rhipicephalus haemaphysaloides by the suppression subtractive hybridization approach. J Integr Agr. 2012;11:1528–36. [Google Scholar]
- 132.van de Locht A, Stubbs MT, Bode W, Friedrich T, Bollschweiler C, Hoffken W, et al. The ornithodorin-thrombin crystal structure, a key to the TAP enigma? The EMBO journal. 1996;15:6011–7. [PMC free article] [PubMed] [Google Scholar]
- 133.Soares TS, Watanabe RM, Tanaka-Azevedo AM, Torquato RJ, Lu S, Figueiredo AC, et al. Expression and functional characterization of boophilin, a thrombin inhibitor from Rhipicephalus (Boophilus) microplus midgut. Veterinary parasitology. 2012;187:521–8. doi: 10.1016/j.vetpar.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 134.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 135.Zuckerkand E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.