Abstract
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and is characterized by a heterogeneous clinical course. This variability in clinical course has spiked the search for prognostic markers able to predict patient evolution at the moment of diagnosis. Markers demonstrated to be of value are the mutation status of the immunoglobulin heavy chain variable region genes (IGHV) and lipoprotein lipase (LPL) expression. High LPL mRNA expression has been associated with short treatment free (TFS) and decreased overall survival (OS) in CLL. The LPL SNPs rs301 (T<C), rs328 (C<G) and rs13702 (T<C) have been associated with various metabolic disorders, but the association with CLL evolution is unknown. Here, in a cohort of 248 patients, we show that patients with the LPL SNP rs13702 wild-type T/T genotype had significantly shorter OS than patients with C/C and T/C genotypes (median time until CLL related death: 90 and 156 months respectively, p=0.008). The same was observed for LPL SNP rs301 (median time until CLL related death T/T: 102 and C/C, T/C: 144 months, p=0.03). Both SNPs rs301 and rs13702 were significantly associated with each other and notably, no association was found between IGHV status and presence of the SNP genotypes, indicating that these LPL SNPs are reliable prognostic markers that could add extra prognostic and predictive information to classical markers and help to improve the management of CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and mainly affects elderly people [1]. Given its highly variable disease course, prognostic markers that allow for estimating the risk of disease progression and prognosis have been identified. Interestingly, lipoprotein lipase (LPL) was among the most differentially expressed genes reported in the initial profiling studies of Rosenwald et al. and Klein et al. [2,3]. We and others have shown that LPL mRNA expression is associated with short treatment free (TFS) and decreased overall survival (OS) in CLL [4–6]. How LPL contributes to a worse outcome in CLL and which mechanisms regulate its expression in CLL cells, remains to be elucidated.
Although the genetic basis of CLL is largely unknown, strong evidence suggests that a genetic component contributes to the etiology of this disease. Approximately 10% of patients have a family history of CLL, suggesting an inherited predisposition [7,8]. Genome-wide association studies (GWAS) have shown that “common variants”, single nucleotide polymorphisms (SNPs) with a population prevalence of at least 1%, contribute to this heritable risk of CLL [9–11]. The LPL SNPs rs301 (T<C), rs328 (C<G) and rs13702 (T<C) have been associated with various metabolic disorders, such as insulin resistance and atherosclerosis [12,13], but the association with CLL disease evolution is unknown. Our aim was to evaluate whether these variants in the LPL gene are associated with the clinical course of CLL patients.
Materials and Methods
Ethics Statement
This study was approved by the Ghent University Hospital Ethics Committee and conducted according to the principles expressed in the Declaration of Helsinki. Patient samples were obtained after written informed consent.
Patients and Sample Collection
248 patients diagnosed with CLL were included in this study. The diagnosis and clinical stage for all patients were confirmed. Clinical stage was determined using the Binet staging system. Flow cytometric analysis of CD38 and zeta-chain associated protein kinase of 70 kDa (ZAP70), IGHV sequencing and cytogenetic characteristics were determined for the majority of patients as previously described [14,15]. OS was defined from the date of diagnosis to the death of the patient or the date of last follow-up before death. TFS was defined as the period between diagnosis and first CLL-specific treatment. Treatment was started when the lymphocyte count exceeded 1011 cells/L, or when patients developed massive lymphadenopathy, anemia, thrombocytopenia, splenomegaly or infections attributed to CLL related immune defects. Inclusion of patients was based on the availability of biological samples. Peripheral blood mononuclear cells (PBMCs) in fetal bovine serum (Hyclone, Thermofisher Scientific, Waltham, MA, USA) with 10% DMSO (Sigma-Aldrich, Diegem, Belgium) stored in liquid nitrogen were available from 169 patients. For the remaining 79 patients white blood cells stored at -80°C in ethanol were available.
Selection of LPL SNPs
Three SNPs, rs301, rs328 and rs13702 located in different regions of the LPL gene were selected in this study, based on the minor allele frequency (MAF) in Caucasians, availability of Taqman SNP genotyping assays and reports in literature. The location of the SNPs in the LPL gene and their MAFs in Caucasians are shown in Table 1.
Table 1. Overview of examined genetic variants in the LPL gene.
| Caucasian | ||||
|---|---|---|---|---|
| dbSNP rs number | SNP location | MAF exp | MAF obs | |
| rs301 | T<C | Intron 6 | 0.25 | 0.24 |
| rs328 | C<G | Exon 9 | 0.13 | 0.12 |
| rs13702 | T<C | 3'UTR | 0.29 | 0.30 |
Both expected (exp) and observed (obs) minor allele frequencies (MAF) are shown for Caucasians. All variants are in accordance with Hardy Weinberg law. rs indicates referenced SNP id number; T<C indicates a transition of T to C nucleotide; C<G indicates a transversion of C to G nucleotide; UTR indicates untranslated region.
Genotyping
gDNA was extracted using the wizard SV 96 genomic DNA purification system (Promega, Leiden, The Netherlands). In case of a low yield, pre-amplification of gDNA was performed with the GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Diegem, Belgium). 10 ng of gDNA was used in Taqman SNP genotyping assays (Applied Biosystems, Life Technologies, Merelbeke, Belgium) for SNPs rs301, rs328 and rs13702 on an ABI Prism 7300 Real Time PCR System (Applied Biosystems, Life Technologies).
RNA Isolation, cDNA Synthesis and Real-Time Quantitative PCR
Total cellular RNA was extracted with the miRNeasy mini kit (Qiagen, Hilden, Germany) according to the supplier’s instructions. Contaminating DNA was removed through DNase treatment using the DNase I, Amplification Grade, kit (Life Technologies). Determination of LPL expression levels was performed as previously described by Van Bockstaele et al. [4]. PCR reactions were performed with the LightCycler 480 Probes Master mix (Roche Diagnostics, Vilvoorde, Belgium) as a reaction mix, in a final volume of 15 μl in 384-well plate (Roche Diagnostics). All reactions were done in duplicate, and each PCR run included controls and a calibration curve of 6, 2-fold dilutions of cDNA from the HL-60 cell line (ATCC). Two housekeeping genes, ACTB (primers described by [16]) and ABL1 (primers and probe described by [17]) were used to normalize LPL expression according to Pede et al. [14]. LPL mRNA expression level was measured in 192 patients.
To determine expression levels of microRNA-410 (miRNA-410), miRNA-410 specific cDNA was synthesized using the Taqman reverse transcription kit and specific stem-loop RT primers (Applied Biosystems, Life Technologies). PCR reactions were performed using an assay-on-demand hydrolysis probe based Taqman miRNA expression assay (Applied Biosystems, Life Technologies). For normalization, the two most stable small RNA controls (RNU 48, RNU 24) were used [18]. All qPCR reactions were performed in duplicate on a LightCycler 480 (Roche Diagnostics).
LPL ELISA
To determine LPL protein levels in 44 samples, patient PBMCs were lysed using a 2× concentrated lysis buffer composed of 0.25% Triton X-100 (MP Biomedicals, Solon, Ohio, USA), 50 mM KCL, 100 mM TrisHCL pH 7.4, 40% glycerol [19]. The total protein concentration was determined using the Bio-Rad Bradford protein assay (Bio-Rad Laboratories, Nazareth, Belgium). 100 μl cellular lysate was used in a human LPL enzyme-linked immunosorbent assay (ELISA) (Cusabio, Wuhan, China) following manufacturer’s instructions.
Fluorometric Lipase Activity Assay
Lipase activity of LPL was assessed by using the Confluolip fluorometric lipase activity assay (Progen, Heidelberg, Germany) according to the manufacturer’s instructions. Purified LPL from bovine milk (Sigma-Aldrich) served as a positive control.
Statistical Analysis
All statistical analyses were performed with the GraphPad statistical software (GraphPad Software, La Jolla, CA, USA). All statistical tests were two-sided and an effect was considered statistically significant at p value < 0.05. ROC curve analysis was performed to determine the LPL mRNA expression cut-off value that best distinguished between mutated and unmutated cases. Median levels of different markers were compared between two groups using Mann-Whitney non parametric tests. Associations between different clinical markers were described with Pearson χ2 statistics (with the Yates continuity correction for 2 × 2 tables) or by calculating a Spearman's rank correlation coefficient. We used the Kaplan Meier method to analyze OS and TFS. The log-rank test was used to determine significant associations between individual markers and OS or TFS.
Results
Characterization of the Patient Cohort
The clinical and biological characteristics of the 248 CLL patients included in this study are detailed in Table 2. Median age at diagnosis is 62 years (range 34–86 years) and the male:female ratio is 2:1. The median TFS is 40 months (range 0–240 months) while the median OS is 78 months (range 1.5–240 months). 39 CLL-related deaths were observed during the observation period.
Table 2. Clinical and biological characteristics among LPL rs301, rs328 and rs13702 Genotypes.
| rs301 TT | rs301 TC/CC | ||||
|---|---|---|---|---|---|
| Characteristic | n | % | n | % | p a |
| No of patients | 146 | 59.1 | 101 | 40.9 | |
| HWE c | 58.0 | 42.0 | 0.322 | ||
| Gender | |||||
| Male | 100 | 59.2 | 69 | 40.8 | 1.000 |
| Female | 46 | 59.0 | 32 | 41.0 | |
| Median age at diagnosis | 63 | 62 | 0.651 b | ||
| Age | |||||
| <60 years | 59 | 60.8 | 38 | 39.2 | 0.786 |
| ≥60 years | 85 | 58.2 | 61 | 41.8 | |
| Binet stage | |||||
| A | 106 | 57.9 | 77 | 42.1 | 0.404 |
| B/C | 31 | 66.0 | 16 | 34.0 | |
| IGHV | |||||
| Mutated | 74 | 58.7 | 52 | 41.3 | 0.972 |
| Unmutated | 48 | 60.0 | 32 | 40.0 | |
| CD38 | |||||
| <7% | 68 | 54.8 | 56 | 45.2 | 0.224 |
| ≥7% | 65 | 63.7 | 37 | 36.3 | |
| ZAP70 | |||||
| <20% | 75 | 60.5 | 49 | 39.5 | 1.000 |
| ≥20% | 54 | 61.4 | 34 | 38.6 | |
| LPL | |||||
| negative | 62 | 63.3 | 36 | 36.7 | 0.821 |
| positive | 57 | 60.6 | 37 | 39.4 | |
| LDT | |||||
| ≥ 1 year | 71 | 61.2 | 45 | 38.8 | 0.662 |
| < 1 year | 55 | 57.3 | 41 | 42.7 | |
| Cytogenetics | |||||
| normal | 36 | 61.0 | 23 | 39.0 | 0.590 |
| chromosomal aberrations | 87 | 55.8 | 69 | 44.2 | |
| Del13q | |||||
| absent | 68 | 63.6 | 39 | 36.4 | 0.232 |
| present | 51 | 54.3 | 43 | 45.7 | |
| Del11q | |||||
| absent | 109 | 58.3 | 78 | 41.7 | 0.606 |
| present | 11 | 50.0 | 11 | 50.0 | |
| Del17p | |||||
| absent | 106 | 55.5 | 85 | 44.5 | 0.563 |
| present | 13 | 65.0 | 7 | 35.0 | |
| rs328 CC | rs328 CG/GG | ||||
| Characteristic | n | % | n | % | P a |
| No of patients | 195 | 78.6 | 53 | 21.4 | |
| HWE c | 77.9 | 22.2 | 0.123 | ||
| Gender | |||||
| Male | 134 | 78.8 | 36 | 21.2 | 1.000 |
| Female | 61 | 78.2 | 17 | 21.8 | |
| Median age at diagnosis | 62 | 62 | 0.878 b | ||
| Age | |||||
| <60 years | 79 | 80.6 | 19 | 19.4 | 0.752 |
| ≥60 years | 114 | 78.1 | 32 | 21.9 | |
| Binet stage | |||||
| A | 144 | 78.3 | 40 | 21.7 | 0.610 |
| B/C | 39 | 83.0 | 8 | 17.0 | |
| IGHV | |||||
| Mutated | 104 | 81.9 | 23 | 18.1 | 0.421 |
| Unmutated | 61 | 76.3 | 19 | 23.8 | |
| CD38 | |||||
| <7% | 99 | 79.8 | 25 | 20.2 | 0.954 |
| ≥7% | 81 | 78.6 | 22 | 21.4 | |
| ZAP70 | |||||
| <20% | 99 | 79.2 | 26 | 20.8 | 0.867 |
| ≥20% | 68 | 77.3 | 20 | 22.7 | |
| LPL | |||||
| negative | 83 | 84.7 | 15 | 15.3 | 0.483 |
| positive | 75 | 79.8 | 19 | 20.2 | |
| LDT | |||||
| ≥ 1 year | 92 | 79.3 | 24 | 20.7 | 1.000 |
| < 1 year | 77 | 79.4 | 20 | 20.6 | |
| Cytogenetics | |||||
| Normal | 46 | 78.0 | 13 | 22.0 | 1.000 |
| Chromosomal aberrations | 123 | 78.3 | 34 | 21.7 | |
| Del13q | |||||
| absent | 85 | 78.7 | 23 | 21.3 | 1.000 |
| present | 74 | 78.7 | 20 | 21.3 | |
| Del11q | |||||
| absent | 150 | 79.8 | 38 | 20.2 | 0.144 |
| present | 14 | 63.6 | 8 | 36.4 | |
| Del17p | |||||
| Absent | 149 | 77.6 | 43 | 22.4 | 0.316 |
| Present | 18 | 90.0 | 2 | 10.0 | |
| rs13702 TT | rs13702 TC/CC | ||||
| Characteristic | n | % | n | % | p a |
| No of patients | 123 | 50.0 | 123 | 50.0 | |
| HWE c | 49.1 | 50.9 | 0.413 | ||
| Gender | |||||
| Male | 87 | 51.5 | 82 | 48.5 | 0.582 |
| Female | 36 | 46.8 | 41 | 53.2 | |
| Median age at diagnosis | 63 | 62 | 0.501 b | ||
| Age | |||||
| <60 years | 48 | 49.5 | 49 | 50.5 | 1.000 |
| ≥60 years | 73 | 50.3 | 72 | 49.7 | |
| Binet stage | |||||
| A | 87 | 47.8 | 95 | 52.2 | 0.202 |
| B/C | 28 | 59.6 | 19 | 40.4 | |
| IGHV | |||||
| Mutated | 61 | 48.8 | 64 | 51.2 | 0.369 |
| Unmutated | 45 | 56.3 | 35 | 43.8 | |
| CD38 | |||||
| <7% | 58 | 47.5 | 64 | 52.5 | 0.551 |
| ≥7% | 54 | 52.4 | 49 | 47.6 | |
| ZAP70 | |||||
| <20% | 62 | 50.0 | 62 | 50.0 | 0.663 |
| ≥20% | 47 | 54.0 | 40 | 46.0 | |
| LPL | |||||
| negative | 49 | 50.5 | 48 | 49.5 | 0.877 |
| positive | 49 | 52.7 | 44 | 47.3 | |
| LDT | |||||
| ≥ 1 year | 61 | 53.5 | 53 | 46.5 | 0.298 |
| < 1 year | 44 | 45.4 | 53 | 54.6 | |
| Cytogenetics | |||||
| Normal | 29 | 50.0 | 29 | 50.0 | 1.000 |
| Chromosomal aberrations | 77 | 49.4 | 79 | 50.6 | |
| Del13q | |||||
| absent | 55 | 51.4 | 52 | 48.6 | 0.968 |
| present | 49 | 52.7 | 44 | 47.3 | |
| Del11q | |||||
| absent | 96 | 51.6 | 90 | 48.4 | 0.126 |
| present | 7 | 31.8 | 15 | 68.2 | |
| Del17p | |||||
| absent | 93 | 48.9 | 97 | 51.1 | 0.920 |
| present | 9 | 45.0 | 11 | 55.0 | |
a Cross-tabulations of prognostic markers versus LPL SNP genotypes. p values of Pearson χ² statistics (with the Yates continuity correction for 2x2 tables)
b p value of Mann-Whitney non parametric test comparing median age at diagnosis between LPL SNP genotypes
c HWE, Hardy-Weinberg equilibrium
Three SNPs in the LPL gene listed in Table 1 were examined. LPL rs301 is located in the non-coding region of intron 6 and results in a T<C nucleotide change. LPL rs328, involving a C<G nucleotide change in exon 9, inserts a nonsense mutation leading to a change in amino acid 447 from a serine to an early stop codon. The third LPL SNP, rs13702 is located in the 3’UTR of the LPL gene. This nucleotide change disrupts a miRNA recognition element (MRE) seed site (MRESS) for the human miRNA-410 [20].
Of all SNPs, MAFs were calculated and genotype frequencies were determined to be in accordance with Hardy-Weinberg equilibrium (Table 1). The observed MAFs were comparable to expected frequencies in the Caucasian population, information extracted from the HapMap database (available: http://www.ncbi.nlm.nih.gov/). There was no difference in median age at diagnosis nor in male:female ratio for patients with wild-type or SNP alleles for all three examined LPL SNPs.
LPL SNPs are Prognostic Markers in CLL
Chi-square tests showed significant associations between IGHV mutation status and its surrogate markers, CD38 status (p = 0.002), ZAP70 status (p<0.0001) and LPL status (p<0.0001) (see S1 Fig.). In this patient population no correlation was found between the examined LPL SNP genotypes and age at diagnosis, gender, Binet stage or IGHV mutation status. Furthermore no correlation was observed between CD38 or ZAP70 protein expression nor between the presence or absence of chromosomal aberrations and presence of either three SNP variants (Table 2).
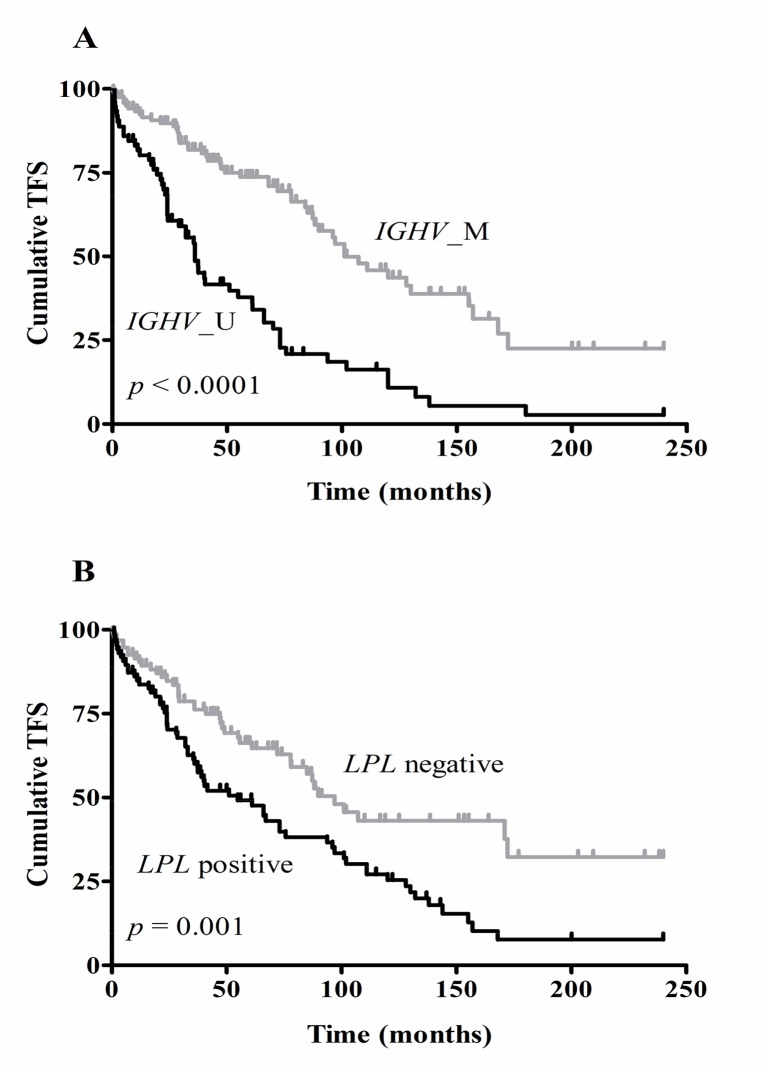
Log-rank tests showed a significant association between OS and Binet stage (p = 0.002), IGHV mutation status (p<0.0001), CD38 (p = 0.04) and ZAP70 protein expression (p = 0.006). The presence of the adverse chromosomal aberration del17p was also significantly associated with OS (p = 0.008). All prognostic markers were significantly associated with TFS, except presence or absence of del13q and del11q (see Table 3). Kaplan-Meier survival curves for TFS with regard to IGHV mutation status (1A) and LPL expression (1B) are shown in Fig. 1.
Table 3. Survival data for different biological and clinical characteristics and LPL SNPs.
| Characteristic | Median OS b | p a | Median TFS b | p a |
|---|---|---|---|---|
| Binet stage | ||||
| A | UD c | 0.002 | 87.2 | < 0.0001 |
| B/C | 137.2 | 29.0 | ||
| IGHV | ||||
| Mutated | UD | < 0.0001 | 101.0 | < 0.0001 |
| Unmutated | 165.0 | 36.0 | ||
| CD38 | ||||
| <7% | UD | 0.042 | 97.0 | <0.0001 |
| ≥7% | UD | 40.4 | ||
| ZAP70 | ||||
| <20% | UD | 0.006 | 97.0 | < 0.0001 |
| ≥20% | 170.0 | 36.0 | ||
| LPL | ||||
| Negative | UD | 0.282 | 89.9 | 0.001 |
| Positive | UD | 41.6 | ||
| LDT | ||||
| ≥ 1 year | UD | 0.167 | 88.1 | 0.001 |
| < 1 year | UD | 41.6 | ||
| Cytogenetics | ||||
| Normal | UD | 0.124 | 96.0 | 0.016 |
| Chromosomal aberrations | UD | 61.0 | ||
| Del13q | ||||
| absent | UD | 0.168 | 67.0 | 0.748 |
| present | UD | 73.0 | ||
| Del11q | ||||
| absent | UD | 0.854 | 73.0 | 0.244 |
| present | UD | 36.0 | ||
| Del17p | ||||
| absent | UD | 0.008 | 75.7 | 0.016 |
| present | 133.0 | 26.7 | ||
| LPLrs301 | ||||
| TT | UD | 0.026 | 72.0 | 0.953 |
| TC/CC | UD | 93.7 | ||
| LPLrs328 | ||||
| CC | UD | 0.483 | 75.7 | 0.477 |
| CG/GG | UD | 67.0 | ||
| LPLrs13702 | ||||
| TT | UD | 0.008 | 66.0 | 0.678 |
| TC/CC | UD | 87.2 | ||
a p values of log-rank tests
b Survival curves for overall survival (OS) and treatment free survival (TFS) were estimated by the Kaplan-Meier method
c UD, undetermined, indicating that the median value was not reached
Fig 1. Kaplan Meier survival curves for TFS with regard to IGHV mutation status (A) and LPL mRNA expression (B).
IGHV gene mutation status was based on a 98% cut-off value (n = 207; M, mutated; U, unmutated). Differentiation between LPL positive and negative cases was based on the optimal cut-off value determined by ROC curve analysis (n = 192). Log-rank tests showed significantly different TFS curves for IGHV mutation status (p<0.0001) and LPL mRNA expression (p = 0.001).
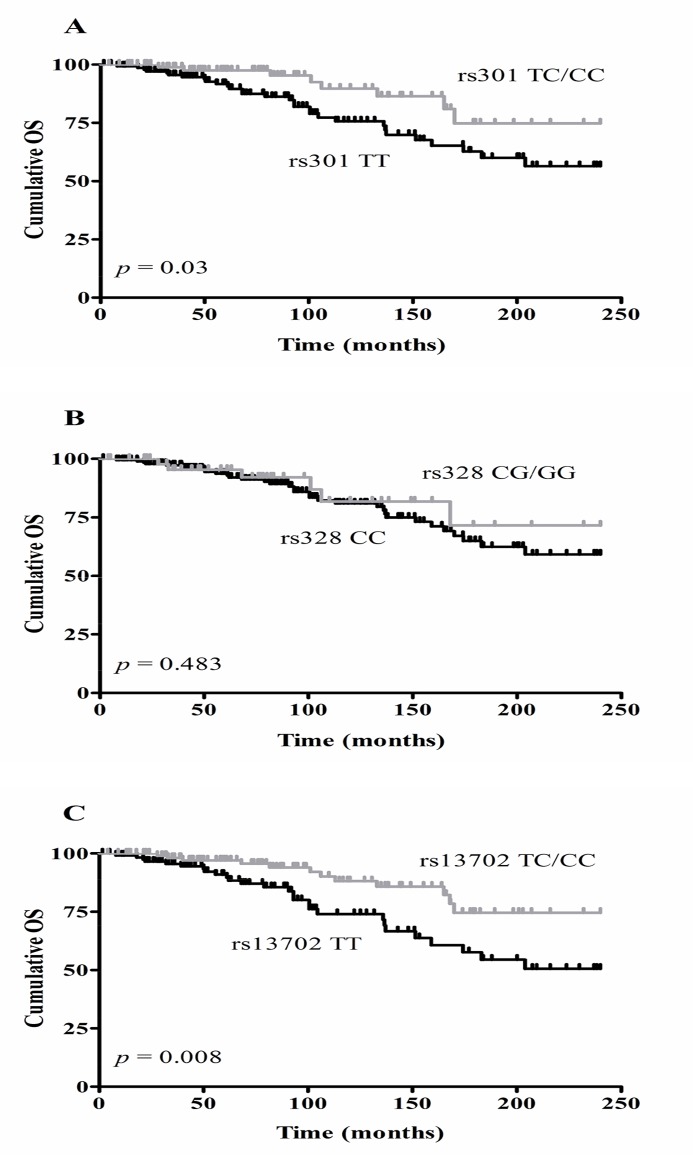
Although none of the examined SNPs correlated significantly with well-known prognostic markers in CLL, two of them, rs301 and rs13702, were significantly associated with OS (Log-rank test; p = 0.03 and p = 0.008 respectively). Indeed, patients being either heterozygous or homozygous for these SNPs showed a better outcome in terms of OS compared to patients having the wild-type genotype. This was not the case for rs328 (p = 0.483) and notably none of the SNPs affected TFS significantly. Fig. 2 shows the Kaplan Meier curves for OS.
Fig 2. Kaplan-Meier curves for OS according to LPL SNP rs301 (A), rs328 (B) and rs13702 (C) genotypes.
A significant effect on OS was indicated by Log-rank tests for rs301 (n = 247; p = 0.03) and for rs13702 (n = 246; p = 0.008), but not for rs328 (n = 248; p = 0.483).
LPL SNPs and LPL expression
GWAS have reported several genetic variants in the LPL gene affecting lipid profile levels, among them the LPL SNPs analyzed in present study. For LPL rs301 a direct effect on LPL expression or activity was not reported yet. Rs328 and rs13702 however are extensively studied gain-of-function variants, known to increase LPL activity.
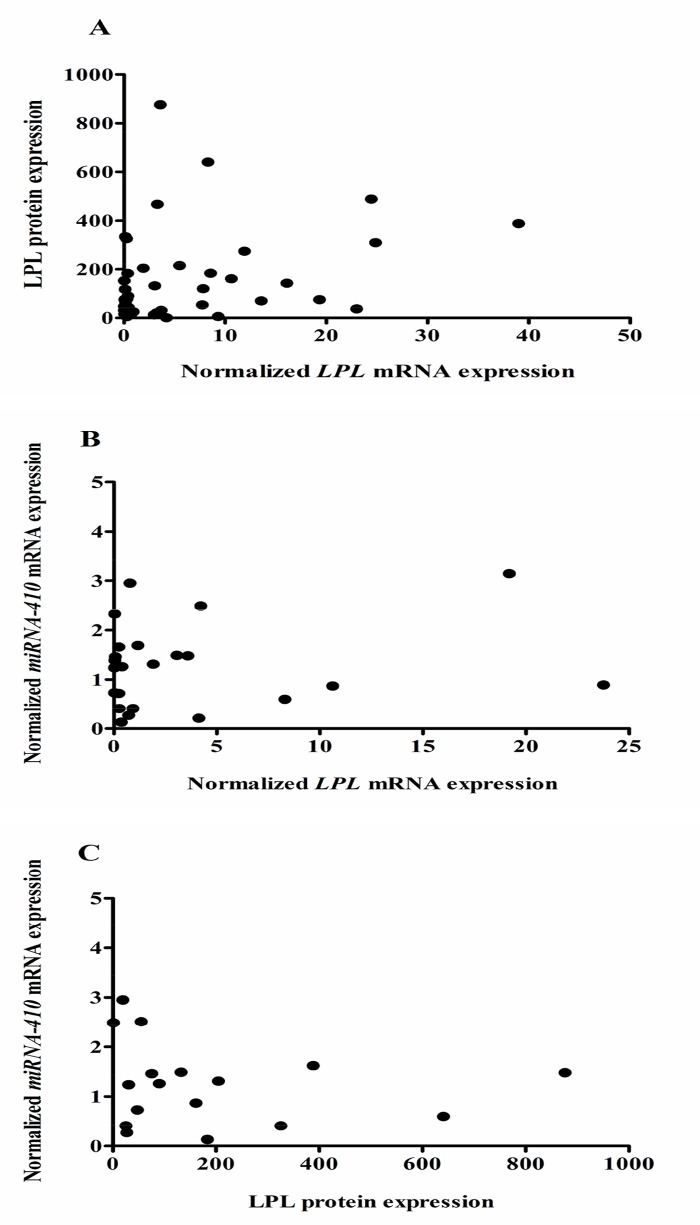
Since two of these SNPs were significantly associated with outcome of CLL patients in this cohort, we wanted to evaluate whether these SNPs affected LPL expression levels and lipase activity. For all three variants examined we couldn’t find a significant difference in LPL mRNA expression levels between patients carrying the wild-type allele or the SNP allele (rs301; p = 0.94, rs328; p = 0.64, rs13702; p = 0.67). LPL protein levels did not correlate with LPL mRNA expression levels (Spearman’s rank correlation coefficient = 0.29, Fig. 3A). Comparable to what was found on mRNA level, we couldn’t find a significant association between LPL protein levels and presence of any of the LPL SNPS (rs301; p = 0.44, rs328; p = 0.56, rs13702; p = 0.58). Finally, we also investigated whether there is a difference in lipase activity between samples of patients carrying one of the SNP alleles or not. As published before by others [21], we could not, or only at a very low level, detect lipase activity in CLL cell lysates and no further analyses were performed.
Fig 3. Correlation between LPL protein expression and LPL mRNA expression (A) and between miRNA-410 expression and LPL mRNA (B) or LPL protein (C) expression.
MiRNA-410 mRNA (n = 25) and LPL mRNA (n = 92) expression levels were determined by qPCR analysis, LPL protein levels were determined by ELISA (n = 44). No correlation was found between LPL protein and mRNA levels (Spearman’s rank correlation coefficient = 0.29) (A). No significant correlation between miRNA-410 expression and LPL mRNA (B) or protein levels (C) could be observed.
The LPL SNP rs13702 was shown to modulate lipid levels through disruption of a binding site in the 3’UTR of the LPL gene for the human miRNA-410 resulting in a gain-of-function variant [20]. To see whether this miRNA-410 affects LPL expression in CLL cells as well, we determined its expression in 25 CLL patient samples with varying LPL mRNA and protein levels. Overall the expression levels of this miRNA in the selected CLL samples were very low and no significant association with LPL mRNA or protein levels could be observed (Fig. 3B and 3C), although the latter showed a trend of inverse correlation.
Discussion
In the present study we examined three SNPs in the LPL gene, rs301, rs328 and rs13702, in a cohort of 248 CLL patients and found that rs301 and rs13702 affected OS significantly, whereas no association with OS could be observed for rs328.
Based on these findings, we also investigated whether the LPL SNP genotypes correlated with other well known prognostic markers in this CLL patient population. Notably, no association with the IGHV mutation status, the gold standard in CLL prognostication, could be observed. This indicates that the SNPs rs301 and rs13702, showing a significant correlation with OS, are no surrogate markers for the IGHV mutation status, but could function as independent prognostic markers in CLL. In line with these results none of the SNPs showed a correlation with other prognostic markers such as ZAP70, CD38 or FISH. This independency was highlighted by Malek as one of the reasons positively affecting the applicability of a CLL biomarker [22]. However, in a multivariate analysis including age at diagnosis, IGHV, ZAP70, LPL, CD38, del11q, del17p and the three LPL SNPs, only age, ZAP70 and del17p were significant and independent predictors of OS (age; p = 0.003, ZAP70; p = 0.009, del17p; p = 0.02). We could not show that the LPL SNPs have additional prognostic value, which is probably due to a lack of power in our study.
Other characteristics of a perfect CLL biomarker that are fulfilled for the SNPs we describe, include the use of a widely available, reliable and valid measuring technique, the fact that genotyping can be performed in peripheral blood samples, the stability of the SNP status over time and the fact that these SNP markers don’t require the use of arbitrary cut-off values for marker positivity, unlike most currently available biomarkers. Moreover, in case no RNA or protein is available, this DNA based marker can still provide prognostic information. This is in contrast with the labor-intensive procedure required for IGHV mutation status analysis or the ZAP70 analysis which is extremely difficult to standardize.
A major shortcoming of these SNP prognostic markers is the fact that these LPL SNPs, as far as we know now, do not represent a biological mechanism directly involved in CLL cell biology. For LPL mRNA or protein however such a relationship with an intrinsic feature of the leukemic cells was not identified either.
The LPL SNPs rs301 and 13702 affected OS significantly, but did not correlate with TFS in this CLL patient cohort, indicating that disease progression is not affected. We speculate that CLL patients carrying these SNPs are in an overall better shape and are less vulnerable to infections or respond better to therapy, explaining the observed association with better disease outcome. These genotypes might not affect disease outcome or therapy responsiveness due to an intrinsic effect on CLL cell biology, but could affect CLL comorbidity. Since CLL is mainly a disease of the elderly, patients are often compromised by co-existent pathological conditions or deterioration of their overall health. In a study investigating the effect of CLL on the quality of life, 71.5% of 1482 patients that were included had at least one co-morbid health problem, with hypertension and increased cholesterol being the most frequent comorbidities [23]. Both conditions were demonstrated to be affected by the presence of the LPL SNPs under study [12,13]. Indeed these SNPs were found to be among the list of LPL variants positively affecting ‘healthy’ HDL cholesterol (HDL-C) and triglyceride (TG) levels, being important risk factors for these and other pathological conditions [12,13]. The mechanism by which rs301 affects HDL-C and TG levels is unknown yet, for rs328 and rs13702 however possible explanations for the observed phenotypes were reported. The LPL SNP rs13702 was shown to modulate lipid levels through disruption of a MRESS in the 3’UTR of the LPL gene for the human miRNA-410. Richardson et al. found that while in the majority of the people binding of miRNA-410 to the LPL mRNA reduced synthesis of LPL, carriers of the genetic variant rs13702 showed no miRNA activity, presumably higher LPL levels and as a consequence lower TG levels and higher HDL-C levels [20]. Also for rs328 a mechanism has been described by which this variant contributes to favorable lipid profiles and reduces the risk for common disease. This nucleotide polymorphism in exon 9 of the LPL gene, introduces a premature stop codon resulting in a truncated protein with a higher activity giving a comparable phenotype (lower TG, higher HDL-C) as observed for rs13702 carriers [24].
Despite the observed effect of rs13702 and rs328 on lipase activity reported by others, we couldn’t find an association between presence of any of these SNPs and LPL mRNA or protein levels, suggesting that LPL expression is controlled by other not yet defined mechanisms in CLL cells. We also tried to compare the lipase activity between CLL samples with different genotypes for the SNPs under study, but overall lipase activity was very low to undetectable, which is in accordance with previous findings [21]. However, the biological role of LPL in CLL remains to be elucidated, indicating that a not yet defined mechanism could be responsible for the observed results as well.
The LPL SNPs that we evaluated in this study display important features that other currently available markers often lack, making them reliable prognostic markers that could help to improve the management of CLL. In this respect, it would be interesting to evaluate the clinical utility, stability and robustness of these genetic prognostic indicators, alone or in combination with each other and other clinical markers in large patient cohorts [25].
Supporting Information
CD38 (n = 218) and ZAP70 (n = 167) protein expression were determined by flow cytometry, LPL mRNA expression was determined by qPCR analysis (n = 92). IGHV gene mutation status was based on a 98% cut-off value (n = 207). Mann-Whitney non parametric tests showed statistically significant differences between the median CD38 (p = 0.002), ZAP70 (p<0.0001) and LPL (p<0.0001) expression levels in IGHV mutated (M) and unmutated (U) CLL cases.
(TIF)
Acknowledgments
We thank the patients for sample donation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Research Foundation - Flanders (FWO) to AR and JP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ghia P, Ferreri AM, Caligaris-Cappio F. Chronic lymphocytic leukemia. Crit Rev Oncol Hematol. 2007;64: 234–46. [DOI] [PubMed] [Google Scholar]
- 2. Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194: 1639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194: 1625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Bockstaele F, Pede V, Janssens A, Callewaert F, Offner F, Verhasselt B, et al. Lipoprotein lipase mRNA expression in whole blood is a prognostic marker in B cell chronic lymphocytic leukemia. Clin Chem. 2007;53: 204–12. [DOI] [PubMed] [Google Scholar]
- 5. van't Veer MB, Brooijmans AM, Langerak AW, Verhaaf B, Goudswaard CS, Graveland WJ, et al. The predictive value of lipoprotein lipase for survival in chronic lymphocytic leukemia. Haematologica. 2006;91: 56–63. [PubMed] [Google Scholar]
- 6. Heintel D, Kienle D, Shehata M, Krober A, Kroemer E, Schwarzinger I, et al. High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia. 2005;19: 1216–23. [DOI] [PubMed] [Google Scholar]
- 7. Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009;94: 647–53. 10.3324/haematol.2008.003632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104: 1850–4. [DOI] [PubMed] [Google Scholar]
- 9. Dicker F, Rauhut S, Kohlmann A, Kern W, Schoch C, Haferlach T, et al. Re: Prognostic significance of a short sequence insertion in the MCL-1 promoter in chronic lymphocytic leukemia. J Natl Cancer Inst. 2005;97: 1092–3; author reply 3–5. [DOI] [PubMed] [Google Scholar]
- 10. Dong HJ, Fang C, Fan L, Zhu DX, Wang DM, Zhu HY, et al. MDM2 promoter SNP309 is associated with an increased susceptibility to chronic lymphocytic leukemia and correlates with MDM2 mRNA expression in Chinese patients with CLL. Int J Cancer. 2012;130: 2054–61. 10.1002/ijc.26222 [DOI] [PubMed] [Google Scholar]
- 11. Nuckel H, Frey UH, Bau M, Sellmann L, Stanelle J, Durig J, et al. Association of a novel regulatory polymorphism (-938C>A) in the BCL2 gene promoter with disease progression and survival in chronic lymphocytic leukemia. Blood. 2007;109: 290–7. [DOI] [PubMed] [Google Scholar]
- 12. Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60: 1329–39. 10.2337/db10-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deo RC, Reich D, Tandon A, Akylbekova E, Patterson N, Waliszewska A, et al. Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: the Jackson Heart Study. PLoS Genetics. 2009;5: e1000342 10.1371/journal.pgen.1000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pede V, Rombout A, Vermeire J, Naessens E, Mestdagh P, Robberecht N, et al. CLL cells respond to B-Cell receptor stimulation with a microRNA/mRNA signature associated with MYC activation and cell cycle progression. PLoS One. 2013;8: e60275 10.1371/journal.pone.0060275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113: 5237–45. 10.1182/blood-2008-11-189407 [DOI] [PubMed] [Google Scholar]
- 16. Cheung VG, Nayak RR, Wang IX, Elwyn S, Cousins SM, Morley M, et al. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biology. 2010;8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using 'real-time' quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)—a Europe against cancer program. Leukemia. 2003;17: 2474–86. [DOI] [PubMed] [Google Scholar]
- 18. Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biology. 2009;10: R64 10.1186/gb-2009-10-6-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, et al. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7: e50859 10.1371/journal.pone.0050859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richardson K, Nettleton JA, Rotllan N, Tanaka T, Smith CE, Lai CQ, et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am J Hum Genet. 2013;92: 5–14. 10.1016/j.ajhg.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mansouri M, Sevov M, Fahlgren E, Tobin G, Jondal M, Osorio L, et al. Lipoprotein lipase is differentially expressed in prognostic subsets of chronic lymphocytic leukemia but displays invariably low catalytical activity. Leuk Res. 2010;34: 301–6. 10.1016/j.leukres.2009.07.032 [DOI] [PubMed] [Google Scholar]
- 22. Malek S. Molecular biomarkers in chronic lymphocytic leukemia. Adv Exp Med Biol. 2013;792: 193–214. 10.1007/978-1-4614-8051-8_9 [DOI] [PubMed] [Google Scholar]
- 23. Shanafelt TD, Bowen D, Venkat C, Slager SL, Zent CS, Kay NE, et al. Quality of life in chronic lymphocytic leukemia: an international survey of 1482 patients. Br J Haematol. 2007;139: 255–64. [DOI] [PubMed] [Google Scholar]
- 24. Ranganathan G, Unal R, Pokrovskaya ID, Tripathi P, Rotter JI, Goodarzi MO, et al. The lipoprotein lipase (LPL) S447X gain of function variant involves increased mRNA translation. Atherosclerosis. 2012;221: 143–7. 10.1016/j.atherosclerosis.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pflug N, Bahlo J, Shanafelt TD, Eichhorst BF, Bergmann MA, Elter T, et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood. 2014;124: 49–62. 10.1182/blood-2014-02-556399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD38 (n = 218) and ZAP70 (n = 167) protein expression were determined by flow cytometry, LPL mRNA expression was determined by qPCR analysis (n = 92). IGHV gene mutation status was based on a 98% cut-off value (n = 207). Mann-Whitney non parametric tests showed statistically significant differences between the median CD38 (p = 0.002), ZAP70 (p<0.0001) and LPL (p<0.0001) expression levels in IGHV mutated (M) and unmutated (U) CLL cases.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.