Abstract
Autosomal dominant nonsyndromic hearing loss (ADNSHL/DFNA) is a highly genetically heterogeneous disorder. Hitherto only about 30 ADNSHL-causing genes have been identified and many unknown genes remain to be discovered. In this research, genome-wide linkage analysis mapped the disease locus to a 4.3 Mb region on chromosome 19q13 in SY-026, a five-generation nonconsanguineous Chinese family affected by late-onset and progressive ADNSHL. This linkage region showed partial overlap with the previously reported DFNA4. Simultaneously, probands were analyzed using exome capture followed by next generation sequencing. Encouragingly, a heterozygous missense mutation, c.505G>A (p.G169R) in exon 3 of the CEACAM16 gene (carcinoembryonic antigen-related cell adhesion molecule 16), was identified via this combined strategy. Sanger sequencing verified that the mutation co-segregated with hearing loss in the family and that it was not present in 200 unrelated control subjects with matched ancestry. This is the second report in the literature of a family with ADNSHL caused by CEACAM16 mutation. Immunofluorescence staining and Western blots also prove CEACAM16 to be a secreted protein. Furthermore, our studies in transfected HEK293T cells show that the secretion efficacy of the mutant CEACAM16 is much lower than that of the wild-type, suggesting a deleterious effect of the sequence variant.
Keywords: CEACAM16, DFNA4B, exome sequencing, linkage analysis
Introduction
Hearing loss (HL) is one of the most common sensorineural deficits in humans worldwide.1 It can be due to genetic causes, environmental factors, or a complex combination of the two. More than 50% of the profound early-onset deafness cases are caused by genetic factors in developed countries.2, 3 Genetic HL may occur as part of a multisystem disorder, or as disease restricted to the cochlea and vestibular system with no additional abnormalities. The latter is the main form accounting for about 70% of neonates failing newborn hearing screening.4 Furthermore, autosomal dominant nonsyndromic hearing loss (ADNSHL/DFNA) accounts for 20-25% of hereditary nonsyndromic sensorineural deafness cases and is most often postlingual in onset.5 Identification of the pathogenic mutations underlying ADNSHL is still difficult because of the incredible genetic heterogeneity regarding HL, and likelihood that ADNSHL is not entirely caused by monogenic mutations (also known as Mendelian disorder).6 Hitherto, only 30 distinct deafness genes have been identified while 64 DFNA loci have been mapped in the past two decades (http://hereditaryhearingloss.org). The genetic heterogeneity is a major hurdle to the efficient discovery of disease-causing genes. Traditional positional candidate approach, which was once the most promising strategy for cloning genes underlying Mendelian disorder, has amassed an impressive list of pathogenic gene discoveries for ADNSHL. Still, several challenges remain before this approach becomes firmly entrenched, such as locus heterogeneity whereby hundreds of genes assemble in the candidate region and in several cases the availability of only small families where linkage mapping would not be informative.7, 8 Recently, the application of next-generation sequencing of targeted regions has been developing dramatically and especially exome sequencing technology has already provided an new powerful tool to identify causative genes.9 In the present study, we identified efficiently a novel CEACAM16 (carcinoembryonic antigen-related cell adhesion molecule 16) mutation in a Chinese family with ADNSHL by exome sequencing in parallel with linkage analysis. In vitro, functional analyses suggest a deleterious effect of a single nucleotide variant.
Material and Methods
Ethics statement
This research involving human participants was formally approved by the Medical Ethics Committee of Xiangya Hospital, Central South University, Changsha, China. Written informed consent was obtained from all subjects (including the control individuals) or their legal guardians.
Subjects and clinical assessment
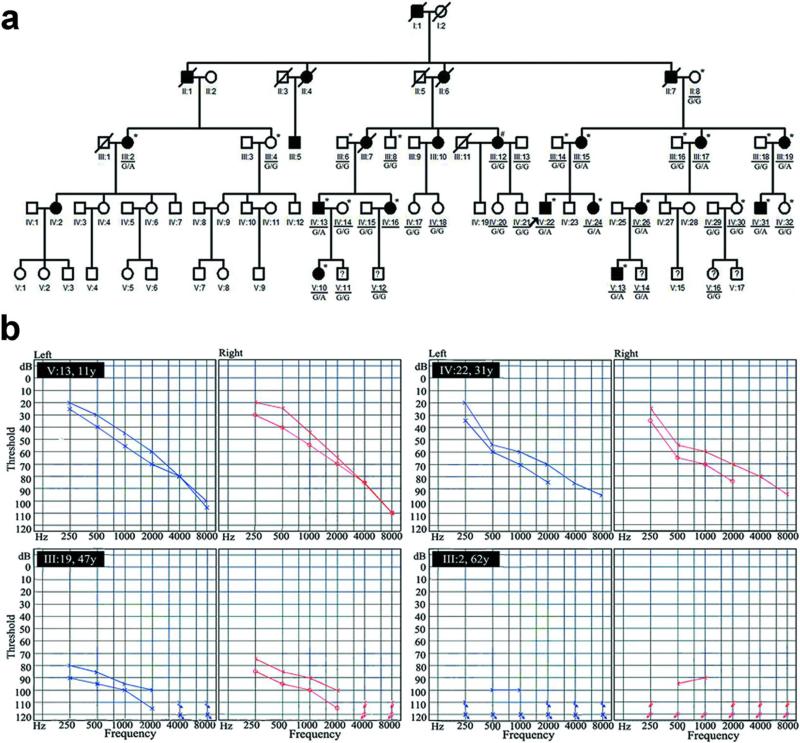
We investigated a five-generation ADNSHL family SY-026 from Hunan province of mainland China. 34 members, including 13 deaf (III:2, III:12, III:15, III:17, III:19, IV:13, IV:16, IV:22, IV:24, IV:26, IV:31, V:10 and V:13), 17 normal hearing (II:8, III:4, III:6, III:8, III:13, III:14, III:16, III:18, IV:14, IV:15, IV:17, IV:18, IV:20, IV:21, IV:29, IV:30 and IV:32), and 4 subjects younger than the onset age whose hearing status is ambiguous (V:11, V:12, V:14, V:16) , were recruited (Figure 1-a; Table 1). All participants were subjected to particular physical and otological examinations by two experienced otologists independently. Pure-tone audiometry (PTA) was performed to define hearing threshold levels (dB HL) for both air and bone conduction at frequencies of 250, 500, 1000, 2000, 4000, 6000 and 8000 Hz. Previous audiological tests were collected if available. PTA average thresholds of the air conduction, which were based on the frequencies at 500, 1000 and 2000 Hz in the better hearing ear, were used to determine the degree of HL. The list of the classification criteria was as follows: normal (< 15 dB HL), slight (16 to 25 dB HL), mild (26 to 40 dB HL), moderate (41 to 55 dB HL), moderately severe (56 to 70 dB HL), severe (71 to 90 dB HL) and profound HL (>90 dB HL).10 Two affected individuals underwent computed tomography (CT) scan of the temporal bone and vestibular testing. Vestibular function was assessed by videonystagmography (VNG) using the System 2000™ (Micromedical Technologies, Chatham, IL, USA). VNG protocol included saccade test, eye tracking test, optokinetic test, gaze test, spontaneous nystagmus test, positional test, Dix-Hallpike test, Roll test, caloric test. The detailed medical history was obtained by questionnaire to eliminate the interference of environmental factors. Genomic DNA was extracted from peripheral venous blood by standard phenol extraction protocols. Mutations in the GJB2, GJB3, EYA4 and KCNQ4 genes have been excluded in this family. After being informed, 200 Han Chinese control subjects (between the ages of 30 and 65), whose health physical examination items contained PTA and the result showed the hearing threshold was less than 15 dB HL, were also collected through health management center, Xiangya Hospital, Central South University. All control subjects had no family history of hearing loss.
Figure 1.
Pedigree of a large Chinese family (SY-026) with late-onset ADNSHL carrying the missense G169R mutation in CEACAM16 and the audiograms of four affected subjects from the family. (a) Pedigree of the family shows an autosomal-dominant inheritance pattern. The circular and square symbols represent female and male, and the black and white ones indicate affected and unaffected individuals, respectively. The subjects younger than the onset age whose hearing status is ambiguous are marked by question mark and the deceased are differentiated by a slash. Arrow shows the proband (IV:22) The bars below each symbol indicate individuals involved in this study. Twenty-two family members included in the linkage analysis are designated by asterisk on each right shoulder of the symbol. The genotypes at the c.505G>A mutation site of CEACAM16 are also presented for each enrolled individual. (Note: #The affected member of III:12 was excluded from linkage analysis because the hearing-impairment was caused by environmental factors.) (b) Audiograms of 4 affected individuals from the family. By convention, the air conduction results are displayed on audiogram using blue “×” for left ear and red “○” for right ear. And likewise, bone conduction results are displayed on audiogram using blue “greater than” angle brackets “>” for left ear and red “less than” angle brackets “<” for right ear.
Table 1.
Clinical data and genotypic characteristics of all participants from SY-026 family
| Subjects | Gender | Age at test | Age of onset | Severity1 | Tinnitus | Vertigo | Environmental factors | Alleles2 |
|---|---|---|---|---|---|---|---|---|
| III:2 | F | 62y | 25y | Profound | + | - | - | G/A |
| III:12 | F | 50y | 3y | Profound | + | - | Meningitis; aminoglycoside (+) | G/G |
| III:15 | F | 54y | 28y | Profound | + | - | - | G/A |
| III:17 | F | 49y | 22y | Profound | + | - | - | G/A |
| III:19 | F | 47y | 21y | Profound | + | - | - | G/A |
| IV:13 | M | 35y | 10y | Severe | + | - | - | G/A |
| IV:16 | F | 32y | 13y | Severe | + | - | - | G/A |
| IV:22 | M | 31y | 15y | Severe | + | - | - | G/A |
| IV:24 | F | 21y | 14y | moderately severe | - | - | - | G/A |
| IV:26 | F | 30y | 16y | Severe | + | - | - | G/A |
| IV:31 | M | 28y | 14y | moderately severe | + | - | - | G/A |
| V:10 | M | 12y | 12y | Moderate | - | - | - | G/A |
| V:13 | M | 11y | 11y | Moderate | - | - | - | G/A |
| II:8 | F | 72y | - | Normal | - | - | - | G/G |
| III:4 | F | 60y | - | Normal | - | - | - | G/G |
| III:6 | M | 61y | - | Normal | - | - | - | G/G |
| III:8 | M | 56y | - | Normal | - | - | - | G/G |
| III:13 | M | 52y | - | Normal | - | - | - | G/G |
| III:14 | M | 59y | - | Normal | - | - | - | G/G |
| III:16 | M | 53y | - | Normal | - | - | - | G/G |
| III:18 | M | 50y | - | Normal | - | - | - | G/G |
| IV:14 | F | 35y | - | Normal | - | - | - | G/G |
| IV:15 | M | 36y | - | Normal | - | - | - | G/G |
| IV:17 | F | 22y | - | Normal | - | - | - | G/G |
| IV:18 | F | 16y | - | Normal | - | - | - | G/G |
| IV:20 | F | 12y | - | Normal | - | - | - | G/G |
| IV:21 | M | 10y | - | Normal | - | - | - | G/G |
| IV:29 | M | 25y | - | Normal | - | - | - | G/G |
| IV:30 | F | 23y | - | Normal | - | - | - | G/G |
| IV:32 | F | 23y | - | Normal | - | - | - | G/G |
| V:11 | M | 9y | - | intangibility | - | - | - | G/G |
| V:12 | M | 6y | - | intangibility | intangibility | - | - | G/G |
| V:14 | M | 7y | - | intangibility | intangibility | - | - | G/A |
| V:16 | F | 3y | - | intangibility | intangibility | - | - | G/G |
Note:
According to criteria in published literature [Clark, 1981]
Alleles refer to CEACAM16 c.505, at position 45207410 (hg19)
F, female; M, male; +, present; –, not present; y, year of age.
Genome-wide genotyping and linkage analysis
The purification of DNA samples from 22 subjects whose phenotypes were clear (including 12 affected and 10 unaffected family members) were genotyped using commercially available HumanLinkage-24 BeadChip Kit (Illumina, San Diego, CA, USA), which contains a total of 6000 high-density single-nucleotide polymorphism (SNP) markers (Figure 1-a). The affected member III:12 and other 4 participants (V:11, V:12, V:14 and V:16) were excluded from the linkage analysis studies because of positive medical history of meningitis and exposure to ototoxic drug or less than the onset of age. Two-point LOD (logarithm of odds) scores were calculated by the Merlin programs (version 5.2) under an autosomal dominant inheritance model with a mutant allele frequency of 0.0001 and complete penetrance. The allele frequencies of markers and the recombination frequencies were assumed to be equal to each other.11, 12
Exome sequencing and bioinformatics analysis
The genomic DNA samples were randomly sheared to obtain fragments with an average length between 250 and 300 bp using adaptive focused acoustics (AFA) from Covaris. Then adapters were ligated to both ends of the fragments. Extracted DNA was then amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the custom Nimblegen SeqCap EZ Library (Roche/NimbleGen, Madison, WI, USA) for enrichment. Both non-captured and captured LM-PCR products were subjected to quantitative PCR to estimate the magnitude of enrichment. Each captured library was then loaded on a Hiseq 2000 platform (Illumina, San Diego, CA, USA). High-throughput sequencing was performed to meet the desired average sequencing depth. Raw image files were processed by Illumina basecalling Software 1.7 (Illumina, San Diego, CA, USA) for base-calling with default parameters and the sequences of each individual were generated as 90 bp pair-end reads.
SOAPaligner (soap2.21) and Burrows-Wheeler Aligner (BWA) tools were used to align clean reads to the human reference genome (hg 19 version) for single nucleotide polymorphism (SNP) and insertion/deletion (Indel), respectively.13, 14 And then, the software SOAPsnp was used to assemble the consensus sequence and call genotypes in target regions.15 We filtered candidate SNPs with the following criterion: SNP quality was ≥20, the sequencing depth was between 4 to 1000-fold, the estimated copy number was ≤2, and the distance between two SNPs was >5. Indels were detected through the alignment result with Genome Analysis Toolkit (GATK).16 Functional effects of the variants were predicted by programs of Sorting Intolerant From Tolerant (SIFT).17
Sanger sequencing
PCR was carried out using the standard protocol (Qiagen, Germantown, MD, USA). The PCR products were treated with the shrimp alkaline phosphatase and exonuclease-I to degrade deoxynucleotide triphosphates and unincorporated PCR primers. The purified amplicons were mixed with 10 picomoles of the forward and reverse PCR primers for bidirectional sequencing on an ABI-Prism 3100 DNA sequencer via dye termination chemistry (Applied Biosystems, Foster City, CA, USA). DNA sequence variants were identified using the SeqMan II program (DNA-STAR Inc., WI, USA).
Plasmid DNA construction
The pRc/CMV-hCEACAM16 was kindly provided by Tumor Immunology Laboratory, LIFE Center, University Hospital of Munich, Ludwig-Maximilians-University Munich, Germany. The hCEACAM16 was sequenced and cloned using the primers: 5’-CGATAGAATTCGCCACCATGGCGCTGACTGGGTACAG-3’ and 5’-GCATATCTAGACCCCAGGGGGGCCACCTGCA-3’. The coding sequence of CEACAM16 was inserted into pRK5-Flag at EcoR I/Xba I site to add a Flag tag to the C-terminus of CEACAM16. The mutant type G169R of CEACAM16 was generated by using QuikChange II Site-Directed Mutagenesis (GE Healthcare). All constructions were verified by Sanger sequencing.
Plasmid transfection, amplification and extraction
The two kinds of plasmid pRK5-wild type (WT) and G169R mutant (MT) CEACAM16-Flag were transformed into One Shot TOP10 competent E. coil (Life technologies) on LB culture plates with ampicillin. Then the monoclonal flora was picked up and mass cultured in LB medium with ampicillin. The amplified plasmids were isolated, purified from bacterial cultures by HiSpeed Plasmid Midi Kit (Qiagen).
Culture and immunofluorescence of COS7 cells
The COS7 cells were dissociated with trypsin/EDTA (invitrogen) and plated onto coverslips with 2% fetal bovine serum/DMEM medium (Invitrogen) in six-well culture plates. 3 μg of plasmid was diluted into 200 μl of jetPRIME buffer and mixed with 4 μl jetRPIME reagent. The mixture was incubated at room temperature for 15 min and added to the cells (Polyplus). Cells were incubated for overnight, at 37 °C, and then washed by PBS solution gently.
COS7 cells were fixed with 4% paraformaldehyde, washed with PBS, permeabilized with 0.2% Triton/PBS, blocked with 10% goat serum in PBS for 1 h, incubated with rabbit anti-flag and phalloidin Alexa fluor-647 for 1 h, stained with Alexa Fluor 488, 568 for 30 min and then washed with PBS. At last, Gold antifade reagent was added to mount onto coverslips. The cells were captured using confocal microscope. The image data were analyzed using MetaMorph software.
HEK293T cells cultures and Western Blotting
The HEK293T cells were grown at 37 °C under 5% CO2 in DMEM which contained 10% fetal bovine serum and 100 U/ml of penicillin/streptomycin. One day before transfection, the HEK293T cells were seeded at a proximate 50% confluency in 10 cm culture plates. Cells were transfected with 6 μg plasmids for each. After 6-8 h, we replaced the cell culture medium with DMEM only with 100 U/ml of penicillin/streptomycin.
At 48 h after transfection, cells were lysed in 2×SDS sample buffer, containing 1mM phenylmethanesulfonyl fluoride and 1×protease inhibitor cocktail (Sigma). Samples were incubated for 10 min, at 98 °C, and quantified using BCA Protein Assay Kit (Thermo). Culture medium from CEACAM16-transfected cells were collected and centrifuged at 4000×rpm for 10 min, 4 °C, and filtered through 0.22 μm filters, then centrifuged at 14000×g for 10 min, 4 °C. Proteins in supernatants were precipitated by 100% trichloroacetic acid, and washed by acetone for 4 times. The same amount of protein from cell abstract and supernatant (both were 20 μg) were separated on 8% SDS-PAGE and transferred onto a PVDF membrane. The membranes were blocked in Tris-buffered saline supplemented with 5% non-fat milk for 1h at room temperature and then incubated with mouse monoclonal anti-Flag M2 antibody (1:1000 dilution, Sigma) or mouse anti-β-actin antibody (1:1000 dilution, Sigma) at room temperature for 2 h. After washing with Tris-buffered saline supplemented with 0.1% Tween 20 (Sigma), the membranes were incubated for 1h at room temperature with a horseradish peroxidase-conjugated secondary anti-mouse IgG antibody (1:10000 dilution, Sigma). Detection was performed using the ECL plus Western blotting detection system (GE Healthcare) according to manufacture's instruction.
Results
Clinical findings and the features of the family
The Chinese SY-026 family's HL was consistent with ADNSHL. The majority of the affected family members appeared to have developed a high-frequency tinnitus at the onset of HL. The affected subjects had very similar phenotypes, characterized by bilateral, postlingual, sensorineural, progressive HL (except III:12). Audiological examination of affected members suggested that HL in this pedigree was late onset and that higher frequencies were initially decreased in the first or second decade and progressed to profound HL involving all frequencies. Specially, the III:12 had normal hearing at birth but affected with sudden profound sensorineural HL at 3 years old by aminoglycoside exposure to treat meningitis. There was no any apparent evidence for vestibular dysfunction or otological associated malformations by CT and vestibular testing. The main clinical and audiological findings of affected subjects are shown in Table 1 and Figure 1-b.
Linkage analysis combined with exome sequencing
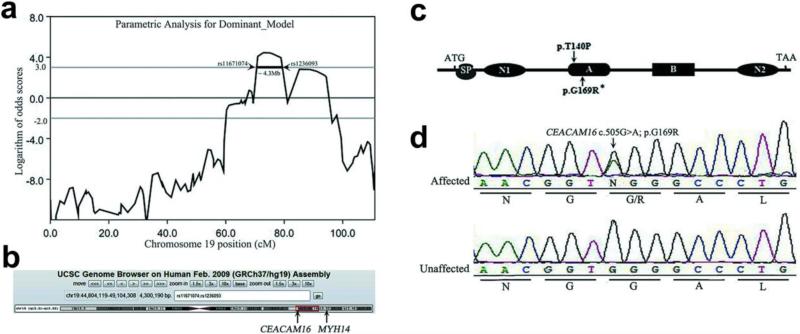
To identify the disease locus, two-point genome-wide linkage analysis was performed on the family members that were indicated by asterisks (Figure 1-a). The result identified a single locus on chromosome (chr) 19 with a maximum LOD score of 4.448 and no significant LOD scores (>3)18 were obtained for any other regions of the genome. The critical interval is flanked by genetic markers rs11671074 and rs1236093 (physical position: chr19: 44,804,119-49,104,308 and physical map: chr19q13.31-13.33), and spans approximately 4.3 Mb (Figure 2-a, b). The LOD score obtained for each genetic marker is presented in Table S1.
Figure 2.
Combinational strategy of linkage analysis and exome sequencing identifies a novel CEACAM16 mutation as causing ADNSHL. (a) SNP linkage analysis maps the disorder locus to chromosome 19 (two-point lod scores of more than 3). The critical interval is flanked by genetic markers rs11671074 and rs1236093 and spans approximately 4.3 Mb. (b) Genome bioinformatics analysis maps physical position to chr19: 44,804,119-49,104,308 and physical map to chr19q13.31-13.33 according to UCSC. (c) Schematic structure of CEACAM16 shows both mutations previously reported in American 1070 family and in the Chinese SY-026 family in this report occurred in the IgC-like domain of subtype A. (d) Identification of a novel heterozygous missense G169R mutation in the CEACAM16 gene. Arrow shows the position of the mutation.
Exome sequencing was performed on 3 affected subjects (IV:24, IV:13 and III:2) and 1 unaffected subject (III:4). The mean depth of target regions ranged from 49 to 76-fold, the coverage of target region reached up to 99% and the average read length was about 90bp. The detail information of the data production can be found in Table S2. Following alignment and quality assessment of the data, >10,000 variants (SNPs and Indels) were identified in subjects. Synonymous variants were excluded and the other were then filtered against several control data sets, including dbSNP135, the 1000 Genomes project, exome data from eight HapMap individuals previously sequenced and YH database (YanHuang Project). The remaining variants were further selected by segregating from the unaffected member (control) and the 3 affected members. Finally, a solitary heterozygous missense variant, c.505G>A in exon 3 of CEACAM16, was mapped to the linkage region on chr19q13.31-13.33 (Figure 2-b). This variant was predicted to be damaging by SIFT and co-segregated with HL in the whole family by Sanger sequencing (Figure 2-d; Figure 1-a). We further confirmed the absence of the mutation in 200 unrelated, ethnically and geographically matched controls. This missense mutation was also absent from the Human Gene Mutation Database (http://www.hgmd.org/) and has not been reported in any published literature, to our knowledge.
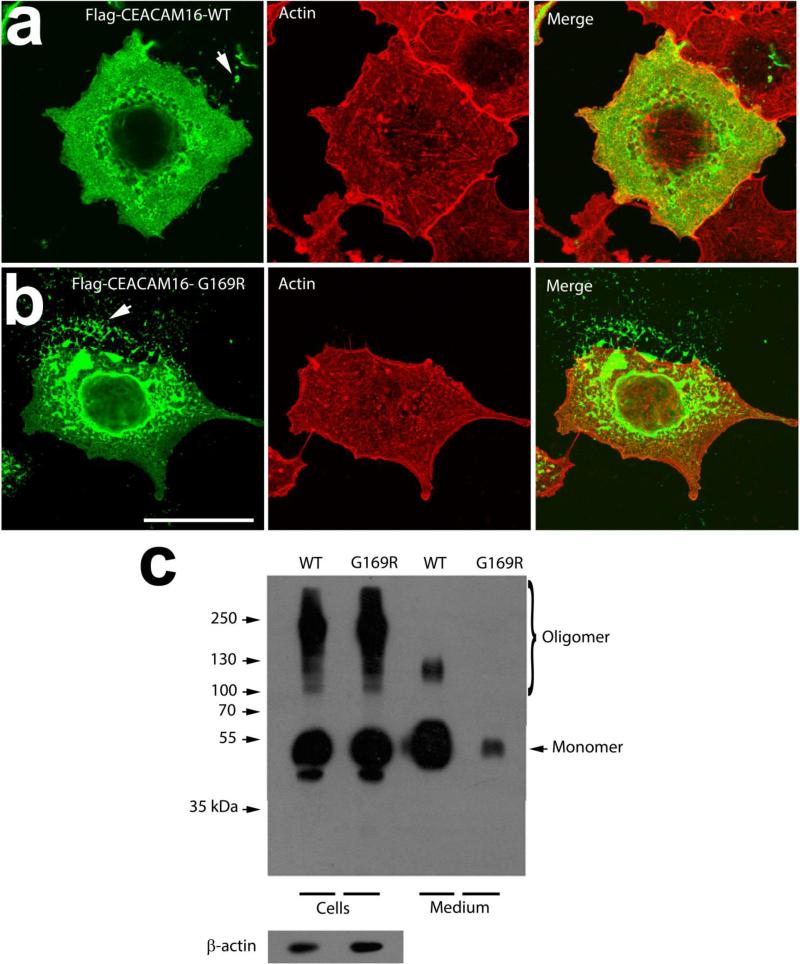
We transfected pRK5-WT and MT CEACAM16-flag plasmids into both COS7 cells and HEK293 cells. Immunocytochemical studies on transfected COS7 cells showed the expression quantity of CEACAM16 protein, both intracellular and extracellular. The cytoskeleton was counterstained for actin to observe the extracellular precipitation clearly. There was no distinct difference in the expression quantity between WT and MT CEACAM16 proteins (Figure 3-a, b). However, the Western blotting result in transfected HEK293T cells seemed to be different (Figure 3-c). The cell medium used to culture those transfected HEK293T cells were collected separately and centrifuged to remove cell and possible exosomes. The extracted proteins were detected as: a monomeric band migrating at about 50-55 kDa25, an oligomeric ladder (dimer to hexamer, ~100-300 kDa), and bands of β-actin. Although the β-actin was seen in both WT and MT CEACAM16 transfected cell pellets, either no or very little oligomeric band could be found in the culture medium of MT CEACAM16 transfected cell.
Figure 3.
Effects of G169R mutation on CEACAM16 trafficking and secretion. (a, b) Confocal images of COS7 cells expressing Flag-tagged wild type (WT, a) and G169R mutant (b) CEACAM16 (green), counterstained for actin (red). Arrows in a and b show secreted proteins. (c) Western blot of protein extracts of HEK293T cells expressing wild type (WT) and G169R mutant Flag-CEACAM16 and of culture media secreted protein precipitates (upper panel). Total protein expression of WT and G169R CEACAM16 is very similar (Cells); total β-actin loads from both preparations are comparable (lower panel). However, the secreted (Medium) amount of mutant CEACAM16 is much lower than the WT CEACAM16. Scale bar: 10 μm.
Discussion
In this study, we described the clinical features and genetic pattern of a 5-generation Chinese family (SY-026) with clinical traits of ADNSHL. We identified a candidate region mapping to chr19q13.31-13.33 that partially overlaps with the previously reported DFNA4 locus.19 Prior to this research, the DFNA4 locus was originally mapped to chromosome 19q13 in an American family (1070) in 1995.19 Subsequently, the second family from Germany (Leipzig 9) and then several other families around the world were identified whose mutations could be mapped to DFNA4 locus.20-24 Because the MYH14 gene, encoding a non-muscle myosin heavy-chain, was the first gene known to correspond to DFNA4, we sequenced it in the probands of family SY-026 and no pathogenic mutation was revealed. Then, we performed exome sequencing analysis on 4 family members and successfully identified a c.505G>A (p.G169R) in exon 3 of the CEACAM16 gene. The p.G169 is conserved in some species, including elephant and a number of primates (such as Pan troglodytes, Gorilla gorilla, Pongo abelii, Nomascus leucogenys, and Callithrix jacchus).36 This mutation co-segregated with the phenotype and was absent in 200 controls. At the time of the study, Zheng et al. reported their identification of a heterozygous missense CEACAM16 mutation in the family 1070 (i.e. originally reported by Chen et al. in 1995) by positional candidate gene approach.19, 25 The clinical features of that family were fluctuating, progressive, sensorineural HL that began in the second decade and led to severe-to-profound HL by the age of 40 years old. Likewise, the phenotypes in our family was very similar to those of the family 1070, except III:12 whose HL was definitely caused by meningitis and aminoglycoside exposure. By contrast, HL was non-fluctuating and several individuals manifested an earlier onset age (the youngest was 10 years old) in our family. Moreover, the concomitant symptom of bilateral continuous high-frequency tinnitus was first reported in most of cases with DFNA4, to our knowledge.
The CEACAM16 gene belongs to the CEACAMs family, a subgroup of the immunoglobulin (Ig) superfamily. CEACAMs comprise a cluster of both conserved and independently evolved proteins that are involved in various physiological and pathophysiological processes, including cell differentiation and proliferative, tissue architecture, immune responses, angiogenesis, tumorigenesis and metastasis, and bacteria mucosal colonization.26-33 Mouse CEACAM16 was initially cloned and detected only in the cerebellum with a low level expression in 2005.34 Mouse CEACAM16 and its human counterpart are well conserved. Analysis by bioinformatics shows CEACAM16 molecules consists of an Ig variable (IgV)-like N domain at its NH2-terminus (N1), followed by Ig constant (IgC)-like domains of subtypes A (A) and B (B) and another IgV-like N domain at its COOH-terminal (N2) (Figure 2-c). The protein encoded by CEACAM16 is a secreted glycoprotein with a signal peptide at its NH2-terminal and lacks a glycosylphosphatidylinositol anchor or transmembrane domain present in other CEACAM members.25, 34, 35 EST database analysis showed CEACAM16 expression in inner/organ of Corti. The causative gene of the previous American DFNA4B family was first focused on the CEACAM16 gene in 2011, but its function is still unknown.25 TM is an extracellular connective tissue that covers the mechanically-sensitive hair bundles of the exquisite sensory receptor cells in the organ of Corti and plays a pivotal role in transforming sound to mechanical stimulation. In vivo and in vitro studies of CEACAM16 indicated that CEACAM16 protein was highly expressed at the tip of cochlear outer hair cells (OHC) and interacts with α-tectorin, a tectorial membrane (TM) protein coded by the TECTA gene.25, 36 Previous studies have identified mutations in TECTA can lead to DFNA12 or DFNB21.37-39 In 2014, a recent study showed that TECTB (coded by the TECTB gene) levels were reduced, a clearly defined striated-sheet matrix did not develop, and Hensen's stripe, a prominent feature in the basal two-thirds of the TM, was absent in CEACAM16-null mutant mice. Furthermore, CEACAM16 was confirmed to interact with TECTB.40 Therefore, CEACAM16 protein, as another mammal-specific TM component, may play a key role in the maintenance of proper hearing function, just like α-tectorin. We had observed that the surface membrane of transfected COS7 cells was intact using confocal microscope. Figure 3-a, b shows that the borders of cells are defined by the red fluorescence dye (the actin cytoskeleton), and a part of the Flag-tagged wild type (a) and G169R mutant (b) CEACAM16 (green) seems to be secreted from the cells, without background staining. This suggests that the CEACAM16 protein may be a secreted protein, which is consistent with the Western blotting result (Figure 3c). Furthermore, our studies in transfected HEK293T cells show that the secretion efficacy of MT CEACAM16 is much lower than that of the WT, and only CEACAM16 monomers could be detected. The decreased secretion and the defection of polymerization suggest the effect of the G169R mutation of CEACAM16. In previous studies, the second IgV domains were shown to mediate cell-cell adhesion and enable CEACAM16 to possibly link α-tectorin to the tip of stereocilia of OHC as a fifth member of non-collagenous proteins specific to the acellular extracellular matrix.25, 36 The p.G169R in CEACAM16 is found in this deafness family. The Glycine is small, with 2 carbons and 1 nitrogen. It is neutral, and plays a hinge-like role in protein structures. In the protein sequence, G169 is located in the middle, and probably part of a loop. It is part of a predicted IgC2 type 1 domain in CEACAM16, which is mainly a beta sheet domain where glycines typically play a hinge-like role in the loops between the sheets. In hugely sharp contrast, Arginine has 5 carbons and 3 nitrogens, it is positively charged, and plays a role in protein folding by creating hydrogen bonds with other amino acids in the peptide chain.41 So apart from a natural disturbance of having to physically fit this large amino acid in place of the glycine, this substitute amino acid will communicate with others around it, potentially causing folds that would certainly disrupt the precise beta sheet arrangement that is typical of Ig–like type 1 domains. Interestingly, lgC2 has been shown to be associated with CEACAM1-mediated bacterial internalization.42 A study suggested that the absence of the IgC2(A) domain could not affect the CEACAM16 protein to form the oligomer,36 but we suggest the G169R mutation at the IgC2(A) domain may change the spatial structure and thereby decrease the stability and polymerizing ability of the protein, and possibly speed up its degradation. This alteration in turn may lead to a significant impact on the cochlear amplification mechanism and subsequently cause hearing impairment. However, future in vivo research on cochleae in mice with mutated CEACM16 cochlea is indicated.
In summary, we have reported the clinical characteristics of a Chinese family with ADNSHL and the high-efficiency identification of a novel missense mutation in the CEACAM16 gene. Our study demonstrates that CEACAM16 is a secreted protein and that the G169R mutation in this gene may lead to a defect in the CEACAM16 protein, which can results in disruption of the tectorial membrane amplification mechanism.
Supplementary Material
Acknowledgment
We gratefully thank all the subjects in this study for their collaboration. We appreciate professor Wolfgang Zimmermann and his lab (Tumor Immunology Laboratory, LIFE Center, University Hospital of Munich, Ludwig-Maximilians-University Munich, Germany) for providing the pRc/CMV-hCEACAM16 plasmid. We thank Dr. Denise Yan and Dr. Robert Cunningham Gerring of the University of Miami, Miller School of Medicine for comments on the manuscript. This study was partly supported by National Basic Research Program of China (also called 973 Program) (2014CB541702); by National Nature Science Foundation of China (81170923, 81470705 and 81300833); by Special Scientific Research Fund for Public Welfare Industry of the Ministry of Health in China (201302001); by Natural Science Foundation of Hunan Province, China (14JJ7009 and 13JJ4023), and by the National Institutes of Health grants R01DC005575 and R01DC012115 to XZL.
Footnotes
Disclosure statement: The authors declare no conflict of interest.
References
- 1.Kalatzis V, Petit C. The fundamental and medical impacts of recent progress in research on hereditary hearing loss. Human molecular genetics. 1998;7:1589–1597. doi: 10.1093/hmg/7.10.1589. [DOI] [PubMed] [Google Scholar]
- 2.Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. American journal of medical genetics. 1993;46:486–491. doi: 10.1002/ajmg.1320460504. [DOI] [PubMed] [Google Scholar]
- 3.Reardon W. Genetics of deafness: clinical aspects. British journal of hospital medicine. 1992;47:507–511. [PubMed] [Google Scholar]
- 4.Van Camp G, Willems PJ, Smith RJ. Nonsyndromic hearing impairment: unparalleled heterogeneity. American journal of human genetics. 1997;60:758–764. [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen MB. Non-syndromic autosomal-dominant deafness. Clinical genetics. 2002;62:1–13. doi: 10.1034/j.1399-0004.2002.620101.x. [DOI] [PubMed] [Google Scholar]
- 6.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutation research. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature genetics. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. International journal of biological sciences. 2007;3:420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews. Genetics. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 10.Clark JG. Uses and abuses of hearing loss classification. Asha. 1981;23:493–500. [PubMed] [Google Scholar]
- 11.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nature genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 12.Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, et al. SNP detection for massively parallel whole-genome resequencing. Genome research. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton NE. Sequential tests for the detection of linkage. American journal of human genetics. 1955;7:277–318. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen AH, Ni L, Fukushima K, Marietta J, O'Neill M, Coucke P, et al. Linkage of a gene for dominant non-syndromic deafness to chromosome 19. Human molecular genetics. 1995;4:1073–1076. doi: 10.1093/hmg/4.6.1073. [DOI] [PubMed] [Google Scholar]
- 20.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). American journal of human genetics. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirghomizadeh F, Bardtke B, Devoto M, Pfister M, Oeken J, Konig E, et al. Second family with hearing impairment linked to 19q13 and refined DFNA4 localisation. European journal of human genetics : EJHG. 2002;10:95–99. doi: 10.1038/sj.ejhg.5200769. [DOI] [PubMed] [Google Scholar]
- 22.Pusch CM, Meyer B, Kupka S, Smith RJ, Lalwani AK, Zenner HP, et al. Refinement of the DFNA4 locus to a 1.44 Mb region in 19q13.33. J Mol Med (Berl) 2004;82:398–402. doi: 10.1007/s00109-004-0538-z. [DOI] [PubMed] [Google Scholar]
- 23.Yang T, Pfister M, Blin N, Zenner HP, Pusch CM, Smith RJ. Genetic heterogeneity of deafness phenotypes linked to DFNA4. American journal of medical genetics. Part A. 2005;139:9–12. doi: 10.1002/ajmg.a.30989. [DOI] [PubMed] [Google Scholar]
- 24.Zong L, Lu C, Zhao Y, Li Q, Han D, Yang W, et al. Clue to a new deafness gene: a large Chinese nonsyndromic hearing loss family linked to DFNA4. Journal of genetics and genomics = Yi chuan xue bao. 2012;39:653–657. doi: 10.1016/j.jgg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng J, Miller KK, Yang T, Hildebrand MS, Shearer AE, DeLuca AP, et al. Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4). Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4218–4223. doi: 10.1073/pnas.1005842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benchimol S, Fuks A, Jothy S, Beauchemin N, Shirota K, Stanners CP. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57:327–334. doi: 10.1016/0092-8674(89)90970-7. [DOI] [PubMed] [Google Scholar]
- 27.Boulton IC, Gray-Owen SD. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nature immunology. 2002;3:229–236. doi: 10.1038/ni769. [DOI] [PubMed] [Google Scholar]
- 28.Eidelman FJ, Fuks A, DeMarte L, Taheri M, Stanners CP. Human carcinoembryonic antigen, an intercellular adhesion molecule, blocks fusion and differentiation of rat myoblasts. The Journal of cell biology. 1993;123:467–475. doi: 10.1083/jcb.123.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ergun S, Kilik N, Ziegeler G, Hansen A, Nollau P, Gotze J, et al. CEA-related cell adhesion molecule 1: a potent angiogenic factor and a major effector of vascular endothelial growth factor. Molecular cell. 2000;5:311–320. doi: 10.1016/s1097-2765(00)80426-8. [DOI] [PubMed] [Google Scholar]
- 30.Muenzner P, Bachmann V, Zimmermann W, Hentschel J, Hauck CR. Human-restricted bacterial pathogens block shedding of epithelial cells by stimulating integrin activation. Science. 2010;329:1197–1201. doi: 10.1126/science.1190892. [DOI] [PubMed] [Google Scholar]
- 31.Sarantis H, Gray-Owen SD. Defining the roles of human carcinoembryonic antigen-related cellular adhesion molecules during neutrophil responses to Neisseria gonorrhoeae. Infection and immunity. 2012;80:345–358. doi: 10.1128/IAI.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitter T, Agerer F, Peterson L, Munzner P, Hauck CR. Granulocyte CEACAM3 is a phagocytic receptor of the innate immune system that mediates recognition and elimination of human-specific pathogens. The Journal of experimental medicine. 2004;199:35–46. doi: 10.1084/jem.20030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmer R, Thomas P. Mutations in the carcinoembryonic antigen gene in colorectal cancer patients: implications on liver metastasis. Cancer research. 2001;61:2822–2826. [PubMed] [Google Scholar]
- 34.Zebhauser R, Kammerer R, Eisenried A, McLellan A, Moore T, Zimmermann W. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Current opinion in cell biology. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kammerer R, Ruttiger L, Riesenberg R, Schauble C, Krupar R, Kamp A, et al. Loss of mammal-specific tectorial membrane component carcinoembryonic antigen cell adhesion molecule 16 (CEACAM16) leads to hearing impairment at low and high frequencies. The Journal of biological chemistry. 2012;287:21584–21598. doi: 10.1074/jbc.M111.320481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balciuniene J, Dahl N, Borg E, Samuelsson E, Koisti MJ, Pettersson U, et al. Evidence for digenic inheritance of nonsyndromic hereditary hearing loss in a Swedish family. American journal of human genetics. 1998;63:786–793. doi: 10.1086/302012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer NC, Alasti F, Nishimura CJ, Imanirad P, Kahrizi K, Riazalhosseini Y, et al. Identification of three novel TECTA mutations in Iranian families with autosomal recessive nonsyndromic hearing impairment at the DFNB21 locus. American journal of medical genetics. Part A. 2007;143A:1623–1629. doi: 10.1002/ajmg.a.31718. [DOI] [PubMed] [Google Scholar]
- 39.Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, et al. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Human molecular genetics. 1999;8:409–412. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- 40.Cheatham MA, Goodyear RJ, Homma K, Legan PK, Korchagina J, Naskar S, et al. Loss of the tectorial membrane protein CEACAM16 enhances spontaneous, stimulus-frequency, and transiently evoked otoacoustic emissions. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:10325–10338. doi: 10.1523/JNEUROSCI.1256-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borders CL, Jr., Broadwater JA, Bekeny PA, Salmon JE, Lee AS, Eldridge AM, et al. A structural role for arginine in proteins: multiple hydrogen bonds to backbone carbonyl oxygens. Protein science : a publication of the Protein Society. 1994;3:541–548. doi: 10.1002/pro.5560030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voges M, Bachmann V, Naujoks J, Kopp K, Hauck CR. Extracellular IgC2 constant domains of CEACAMs mediate PI3K sensitivity during uptake of pathogens. PloS one. 2012;7:e39908. doi: 10.1371/journal.pone.0039908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.