Abstract
The current dogma is that obesity-associated hepatic inflammation is due to increased Kupffer cell (KC) activation. However, recruited hepatic macrophages (RHMs) were recently shown to represent a sizable liver macrophage population in the context of obesity. Therefore, we assessed whether KCs and RHMs, or both, represent the major liver inflammatory cell type in obesity. We used a combination of in vivo macrophage tracking methodologies and adoptive transfer techniques in which KCs and RHMs are differentially labeled with fluorescent markers. With these approaches, the inflammatory phenotype of these distinct macrophage populations was determined under lean and obese conditions. In vivo macrophage tracking revealed an approximately sixfold higher number of RHMs in obese mice than in lean mice, whereas the number of KCs was comparable. In addition, RHMs comprised smaller size and immature, monocyte-derived cells compared with KCs. Furthermore, RHMs from obese mice were more inflamed and expressed higher levels of tumor necrosis factor-α and interleukin-6 than RHMs from lean mice. A comparison of the MCP-1/C-C chemokine receptor type 2 (CCR2) chemokine system between the two cell types showed that the ligand (MCP-1) is more highly expressed in KCs than in RHMs, whereas CCR2 expression is approximately fivefold greater in RHMs. We conclude that KCs can participate in obesity-induced inflammation by causing the recruitment of RHMs, which are distinct from KCs and are not precursors to KCs. These RHMs then enhance the severity of obesity-induced inflammation and hepatic insulin resistance.
Introduction
Insulin resistance is a major pathophysiologic defect underlying the etiology of type 2 diabetes (1). In recent years, chronic tissue inflammation has been identified as a major contributor to the decreased insulin sensitivity in obesity. This proinflammatory state is characterized by a number of changes in immune cell populations in adipose tissue, including increased proinflammatory macrophages (2,3). In addition, increased circulating levels of cytokines have been reported across several experimental rodent models as a well as in obese humans (4–7).
The liver is responsible for the metabolism, synthesis, storage, and distribution of nutrients and is a key organ with central importance in the maintenance of glucose homeostasis. Insulin stimulates storage of glucose as liver glycogen and inhibits hepatic glucose production by restricting glycogenolysis and gluconeogenesis (8). In obesity and type 2 diabetes, the liver is insulin resistant, resulting in increased gluconeogenesis and glycogenolysis leading to increased overall hepatic glucose production (9,10). Other cardinal features of obesity are nonalcoholic fatty liver disease and hepatic inflammation (11,12). Thus, obesity promotes hepatic inflammation in humans and rodents, leading to increased production of proinflammatory cytokines and acute-phase reactants. In addition, there is strong evidence that macrophages play a significant role in hepatic inflammation and insulin resistance. In support of these ideas, chemical deletion or genetic impairment of hepatic macrophages in obese mice leads to an improvement in insulin sensitivity (13–15).
Understanding this phenomenon in the liver is particularly complex, due to the heterogeneity of hepatic macrophage populations. Thus, in insulin resistance, there is an increased number of infiltrating macrophages in the liver (16,17). In addition, a large proportion of hepatic macrophages consist of resident Kupffer cells (KCs) (18). KCs are predominantly localized to the hepatic sinusoids and exhibit phagocytic activity toward blood-born materials entering the liver (18,19). Although KCs account for ∼15% of the total liver cell population (20), attempts to functionally characterize these cells have had limited success because isolating and characterizing these cells as a discrete population distinct from infiltrating macrophages have been difficult.
To elucidate the roles of recruited hepatic macrophages (RHMs) vs. KCs in the development of obesity-associated hepatic inflammation and insulin resistance, we have used a combination of in vivo macrophage tracking and adoptive transfer methodologies, in which KCs and RHMs are differentially labeled with fluorescent markers and analyzed by FACS. This allowed us to quantitate the numbers of these cells, their genomic signatures, and inflammatory phenotypes in both the lean and obese state. Our results show that obesity is associated with activation of KCs, which leads to migration of RHMs into the liver augmenting the severity of obesity-induced hepatic inflammation.
Research Design and Methods
Animal Care and Use
Male C57Bl/6 mice were fed normal chow (NC; 13.5% fat; LabDiet) or a high-fat diet (HFD; 60% fat; Research Diets, Inc.) ad libitum for 15–20 weeks from 8 weeks of age. Chemokine receptor type 2 (CCR2) knockout (KO) and wild-type (WT) littermates were provided by Taconic Inc. (Hudson, NY). Animals were housed in a specific pathogen-free facility and given free access to food and water. The University of California San Diego Animal Care and Use Committee approved all procedures.
Monocyte Preparation
Leukocyte pools from C57BL/6 male mice 12 weeks of age underwent red blood cell lysis, and monocyte subsets were enriched with the EasySep Mouse Monocyte Enrichment Kit (STEMCELL Technologies, Vancouver, BC, Canada) following the manufacturer’s instructions.
In Vitro Labeling
Isolated monocytes (5 × 106 to 10 × 106) were washed once in serum-free medium (RPMI-1640) and suspended in 2 mL Diluent Solution C (included in the PKH26 labeling kit). Two milliliters PKH26 (Sigma-Aldrich, St. Louis, MO) at 2 × 10−3 mol in Diluent C was added and mixed, and the cells were incubated for 10 min at room temperature in the dark. The staining reaction was halted by addition of an equal volume (2 mL) of medium supplemented with 10% FBS. The mixture was centrifuged, and the cells were washed once and resuspended in serum-containing medium.
In Vivo Migration
Subsequent to labeling with PKH26 dye, monocytes were counted, and ∼5 × 105 viable cells were suspended in 0.2 mL PBS and injected via the retro-orbital venous sinus of each group of mice. Five days after of injection, the nonparenchymal cells (NPCs) were immediately isolated from perfused liver and analyzed by FACS.
Fluorescence Microscopy of Mouse Liver
Immunofluorescence study of mouse liver was described in a previous study (17).
KC Labeling Model
Lean NC- and HFD-fed C57Bl/6 mice were injected via the retro-orbital venous sinus with fluorescein isothiocyanate (FITC)-dyed beads (Fluoresbrite 0.5 μm; Polysciences, Inc.) (21). Two days after the injection, the mice were irradiated with a dose of 10 Gy and were reconstituted 6 hours later with 1.5 × 106 bone marrow cells isolated from WT mice. Two weeks after reconstitution, blood cell counts were normal.
Isolation of Bone Marrow Cells for Reconstitution
Bone marrow was flushed out of the fibula and femur and washed three times with ice-cold PBS, then passed through a 25-gauge syringe to avoid clots. Red blood cell lysis was performed (eBioscience, San Diego, CA), and the cells were then counted and resuspended at a concentration of 5 × 106 cells/mL.
FACS Analysis of Liver Macrophages
For liver macrophage analysis, NPCs were prepared by two-step liver collagenase digestion and fractionation on a two-step Percoll density gradient, as previously described (22). The antibodies for surface staining were F4/80 (BM8), Ly6C (AL-21), CD11b (M1/70), and CD11c (N418; eBioscience). Numbers obtained were subsequently represented as the percentage of the highest subsets. Unstained, single-stained, and fluorescence minus one controls were used for setting compensation and gates.
RNA Isolation and Quantitative PCR
Total RNA isolation and quantitative PCR (q-PCR) were performed as described previously (23). Gene expression levels were calculated after normalization to the standard housekeeping genes RPS3 and GAPDH using the ΔΔCT method (23) and expressed as relative mRNA levels compared with the internal control. Primer information is available upon request.
Western Blot Analysis
The amount of tumor necrosis factor-α (TNF-α) in PKH26+ cells from lean and obese mouse NPCs were determined in total cellular extracts by immunoblot using the commercial antibodies given in parentheses (TNF-α and HSP90 α/β, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The cells were homogenized in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors (Roche Diagnostics, Indianapolis, IN). The protein bands were analyzed using densitometry and ImageJ image analysis software.
Hepatic Glucose Output Assay
Mice were infused through the inferior vena cava with a calcium-free HEPES-phosphate buffer, followed by Liberase TM collagenase solution (Roche Diagnostics) for another 5 min. The digested livers were excised, and cells were teased out in Earle balanced salt solution without collagenase. Hepatocyte suspension was filtered and centrifuged at 50g for 6 min, and the pellet was resuspended in Williams Medium E (Life Technologies, Inc.) fortified with glutamax, antibiotics, 10% FBS, and dexamethasone (10 nmol/L) and allowed to attach for 6 h on collagen-coated plates. After attachment, medium was switched to Williams E without serum and dexamethasone (culture medium) and incubated overnight in a 5% CO2 incubator. During this incubation, if condition medium (CM) was used for an experiment, CM was diluted 1:1 with the culture medium and incubated overnight. In control wells without CM, RPMI was used to dilute the culture medium. The next morning, cultures were washed in HEPES-phosphate–salt–bicarbonate buffer containing 0.2% BSA and incubated in the same buffer containing insulin or glucagon and 14C-pyruvate (2 mmol/L, 0.5 µCi pyruvate/incubation, PerkinElmer) as substrate. Cells were preincubated (in absence of substrate) for 30 min with insulin (10 nmol/L) or glucagon (10 ng/mL), or a combination, followed by 3 h incubation with the substrate. Tubes were vortexed and centrifuged, and the supernatants were transferred to a fresh set of tubes containing 200 mg mixed-bed ion-exchange resins, AG-501 × 8 resins (BioRad). These tubes were vortexed intermittently for 15 min and centrifuged, and the supernatants containing radiolabeled glucose were transferred to scintillation vials for counting radioactivity. Cells on the plates were dissolved in 1N NaOH for protein estimation.
Library Preparation, Sequencing, and Data Analyses
RNA was quantified using a NanoDrop 1000 (Thermo-Fisher, Waltham, MA) and assessed on a Tapestation (Agilent, Santa Clara, CA). DNA libraries were prepared according to the manufacturer’s instructions (Illumina, San Diego, CA). Each library was sequenced on the Illumina HISeq2000 platform using paired-end 100, non–strand-specific sequencing strategy. Reads were first mapped to the mouse transcriptome using the Bowtie 2 algorithm (24), counted as reads per gene, and then analyzed using the statistical algorithm DESEq (25). Significance is calculated as the q value, which is the largest false discovery rate at which a gene is deemed differentially expressed (26,27). Genes were sorted by q value and then underwent gene ontology and pathway analyses using a nonparametric variant of the Gene Set Enrichment Analysis algorithm (28). Principal component analysis was performed in R software.
Data Analysis
The values presented are expressed as the means ± SEM. The statistical significance of the differences among various treatments was determined by one-way ANOVA with the Bonferroni correction using GraphPad Prism 6.0 software (San Diego, CA). P < 0.05 was considered significant.
Results
Isolation and Characterization of Nonparenchymal Cells From Lean and Obese Mouse Livers
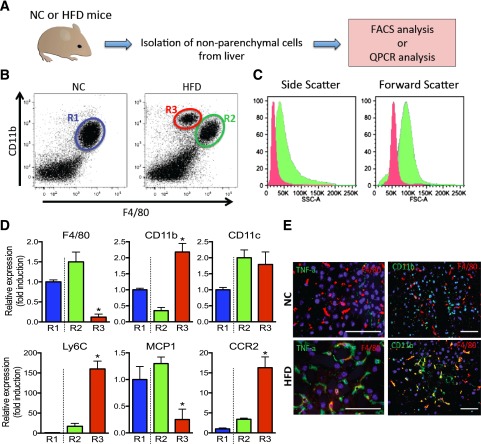
To define the characteristics of innate immune cells and KCs in the liver, we isolated NPCs from lean and obese mouse livers, followed by FACS and qPCR analyses (Fig. 1A). As shown in Fig. 1B, livers from lean mice contained F4/80+/CD11b+ cells (R1), whereas livers from obese mice exhibited two distinct populations, which are F4/80+/CD11b+ (R2) or F4/80low/CD11b+ (R3). The number of R3 cells was negligible in lean mice, with a striking expansion of this cell population in obese mice (Fig. 1B). According to forward and side-scatter analyses, cells in the R3 population were smaller and less granular than R2 cells (Fig. 1C). The gene expression levels of F4/80 and CD11b were consistent with the FACS results, showing that F4/80 levels were greater in R2 compared with R3 cells, whereas CD11b was approximately fourfold lower in R2 than in R3 cells. Interestingly, expression of Ly6C and CCR2 is markedly higher in R3 compared with R2 and R1 cells, whereas MCP-1 expression is significantly higher in R2 and R1 cells. This suggests that R3 cells represent fresh RHMs derived from circulating monocytes. By immunohistochemistry of liver sections, we assessed the colocalization of TNF-α with the macrophage marker F4/80. As seen in Fig. 1E, colocalization was low in lean NC mice, whereas both markers were highly colocalized in obese, HFD-fed mice. The same results were observed for colocalization of CD11b and F4/80.
Figure 1.
Isolation and characterization of NPCs from lean and obese mouse liver. A: Schematic diagram of isolation of NPCs from NC-fed lean vs. HFD-fed obese mouse liver. B: FACS analysis of NPCs from lean (NC) and obese (HFD) mouse liver with CD11b and F4/80 gating. NC mouse liver R1 population is labeled in blue, HFD mouse liver R2 population is labeled in green, and R3 population is labeled in red. The scattergram is representative of five to six independent mice from each group. C: Representative histogram plots depict the distribution of values of side (indicating relative granularity, left panel) and forward (indicating relative size, right panel) light scatter obtained from FACS analysis of R2 and R3 population from panel B. D: The relative gene expression from R1, R2, and R3 cell populations obtained from FACS sorting is shown. Data represent mean ± SEM (n = 5–6). *P < 0.05 compared with R2 vs. R3. E: TNF-α, F4/80, and CD11b protein expression in lean and obese mouse liver analyzed by immunohistochemistry. The image is representative of six independent mice from each group. Scale bar indicates 100 μm.
Tracking and FACS Analysis of PKH26-Labeled Monocytes to Liver in Lean and Obese Mice
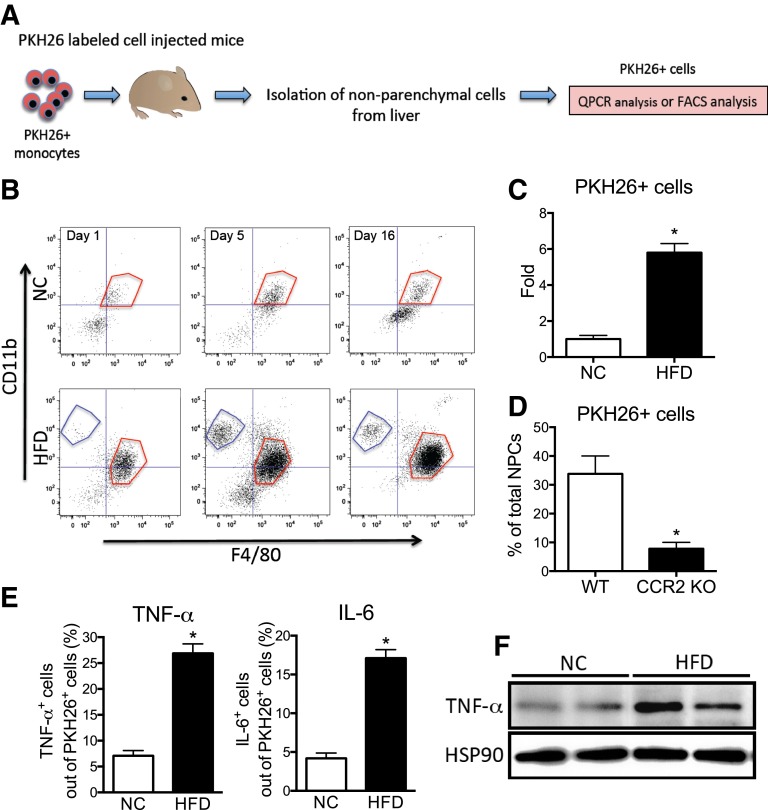
Figure 1B clearly shows two different macrophage populations in obese mouse liver. To further assess this, we performed in vivo macrophage tracking studies, as previously described (17). With this method, circulating monocytes from donor mice are labeled ex vivo with the fluorescent marker PKH26 and then injected into recipient mice. The appearance of these labeled cells as RHMs was then analyzed over time by FACS and expression analysis, as shown in Fig. 2A. After injection of PKH26+ cells, the NPCs were isolated at the indicated time and gated first for PKH26 and then plotted for CD11b and F4/80. The overall accumulation of PKH26+ cells was much greater in obese versus lean mouse liver (Fig. 2B). The RHMs in HFD mouse liver appeared as two discrete populations; F4/80low/CD11b+ (blue-gated) and F4/80high/CD11b+ (red-gated) fraction, similar to the R3 and R2 cells observed in Fig. 1B. This suggests that newly recruited monocytes initially appear in the R3 fraction and then convert into the R2 fraction as they mature into differentiated macrophages. At day 1, ∼15% of the total NPCs are fluorescently labeled in obese mice, demonstrating their origin from the injected labeled monocytes and indicating they are newly recruited RHMs: 2.6 ± 0.7% in lean and 19.6 ± 2.9% in obese mice are PKH26+ cells from the F4/80low/CD11b+ cells (R3 fraction). By 5 days after injection, there was a sixfold increase in PKH26+ cells in obese versus lean mouse livers (Fig. 2C).
Figure 2.
RHM isolation with PKH26 fluorescent labeling. A: Schematic diagram of RHM isolation and characterization with PKH26+ fluorescent-labeled monocytes. B: FACS analyses of PKH26+ cells isolated from lean or obese mouse livers 1, 5, or 16 days after injection with PKH26+-labeled monocytes. Cells were first gated out of total NPCs for PKH26 and then plotted for F4/80+ and CD11b+ fluorescence. F4/80low/CD11b+ (blue-gated) and F4/80high/CD11b+ (red-gated) fraction, similar to the R3 and R2 cells observed in Fig. 1B. The scattergram is representative of four to five independent mice from each group. C: Average number of PKH26+ cells from lean and obese mouse liver was analyzed by FACS and then plotted as fold induction ± SEM from three independent experiments (n = 4 in each group). *P < 0.05 compared with NC vs. HFD. D: FACS analysis of the average number of PKH26+ cells isolated from HFD-fed recipient mice after injection PKH26+-labeled monocytes from WT or CCR2 KO donor mice. Mean ± SEM from three independent experiments (n = 5 in each group). *P < 0.05 compared with WT vs. CCR2 KO. E: TNF-α+ (left) and interleukin-6+ (right) cells were gated out of PKH26+ cells from NPCs of lean and obese mouse liver by FACS and then plotted as the mean ± SEM from three independent experiments (n = 5 in each group). *P < 0.05 compared with NC vs. HFD. F: Western blot analysis of TNF-α protein expression in PKH26+ cells from lean and obese mouse NPCs. PKH26+ cells from lean and obese mouse NPCs were sorted by FACS and then cultured overnight before protein isolation. HSP90 was used as the loading control. The image is representative from three independent Western blots (n = 2 for each group per Western blot).
Because CCR2 is highly expressed in the R3 cells (Fig. 1D), we used labeled monocytes derived from WT and CCR2 KO donor mice and injected them into obese WT recipients (Fig. 2D). There was an ∼80% decrease in RHM recruitment with the CCR2 KO monocytes (Fig. 2D), demonstrating the importance of the MCP-1/CCR2 system for targeting monocytes to the RHM fraction (17). In the same cells, we also analyzed the amount of intracellular inflammatory cytokines by flow cytometry. Intracellular TNF-α and interleukin-6 protein levels were much higher in PKH26+ cells from obese mice compared with lean mice (Fig. 2E). In addition, we collected PKH26+ cells from the total NPC fraction sorted by FACS and cultured these cells overnight. As expected, TNF-α protein expression was significantly greater in PKH26+ cells from obese mice compared with lean mice (Fig. 2F).
Characterization of RHMs and KCs by Adoptive Transfer and Fluorescent Labeling
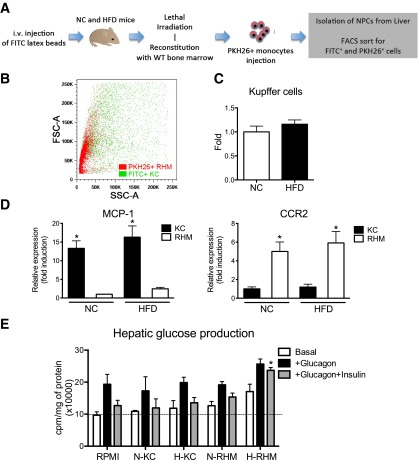
On the basis of the finding that KCs are relatively radioresistant (29), we developed a strategy to selectively label these cells with green fluorescent (FITC) beads. Before the administration of lethal irradiation, NC and HFD mice were injected with FITC beads to label all phagocytizing cells (21). The injected mice were then irradiated with 10 Gy and reconstituted with bone marrow from WT mice. Two weeks after reconstitution, mice were injected with PKH26+ monocytes. Because KCs are relatively radioresistant, with this approach, the green fluorescent cells (FITC+ cells) that survive the irradiation are KCs, whereas PKH26+ cells are RHMs (Fig. 3A). This model allowed us to separate FITC+ KCs from PKH26+ RHMs by flow cytometry. NPC fractions were prepared, and FACS analysis revealed discrete populations of PKH26+ RHMs and FITC+ KCs (Fig. 3B). Figure 3B shows that PKH26+ RHMs were significantly smaller in size, whereas FITC+ KCs were larger and more heterogeneous in size, consistent with Fig. 1C. PKH26 and FITC labeling showed there was no difference in KC numbers between lean and obese mice (Fig. 3C).
Figure 3.
Identification of KCs in mouse liver. A: Schematic diagram of the irradiation model used to distinguish KC and RHM. i.v., intravenous. B: FITC+ KCs and PKH26+ newly infiltrating macrophages from HFD mice were plotted in forward and side scatter. The scattergram is representative of five independent mice. C: Whole-liver FITC+ KC numbers from mice fed NC were compared with mice after 12 weeks of HFD feeding. Data represent mean ± SEM (n = 5–6). D: MCP-1 and CCR2 mRNA expression were measured from FACS-sorted KCs and RHMs isolated from NC and HFD mice. Data represent mean ± SEM (n = 5). *P < 0.05 compared with KC vs. RHM. E: Hepatic glucose production assay in mouse hepatocytes. Hepatocytes were cultured with conditioned media from FACS-sorted KCs and RHMs from NC (N) and HFD (H) mice. Data represent mean ± SEM (n = 5). *P < 0.05 compared with glucagon and insulin in RPMI.
FACS-sorted RHMs and KCs were analyzed for gene expression. Interestingly, CCR2, a chemokine receptor directing monocyte homing to the liver, was highly expressed in PKH26+ RHMs compared with KCs, whereas the ligand, MCP-1, was highly expressed in KCs relative to RHMs, consistent with Fig. 1D. These results suggest that KCs are the main source of MCP-1, leading to increase recruitment of RHMs through the CCR2 (Fig. 3D) (30,31).
To determine functional differences between KCs and RHMs, we measured hepatic glucose output (HGO) from primary hepatocytes treated with CM from overnight cultured KCs and RHMs. For this experiment, we cultured FACS-sorted KCs and RHMs from NC and HFD mice overnight and collected the CM to treat cultured mouse hepatocytes for 16 h. We found that glucagon stimulates HGO and that this effect was completely inhibited by insulin in RPMI (control medium), NC KC-CM, HFD KC-CM, and NC RHM-CM. In contrast, RHM-CM from HFD mice directly stimulated HGO (Fig. 3E). Most importantly, HFD RHM-CM blocked the effect of insulin to inhibit glucagon-stimulated HGO (Fig. 3E). Thus, components in RHM-CM, but not KC-CM, from HFD mice can promote HGO and attenuate insulin’s normal inhibitory effects on this aspect of hepatic metabolism.
Transcriptome Analysis of RHMs and KCs
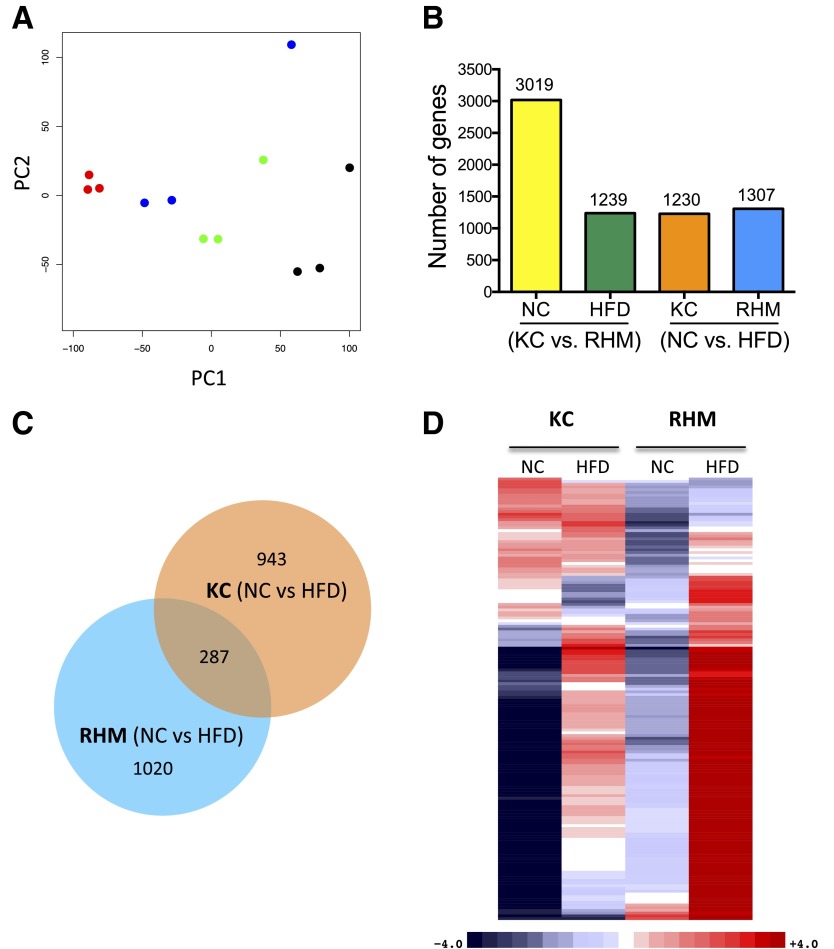
RNA sequencing was used to determine the transcriptome of FACS-sorted RHMs and KCs from mice fed NC or the HFD (three biological replicates per group). Principal component analysis of the expressed genes showed that replicate samples clustered together (Fig. 4A). Bioinformatics analyses revealed marked differences in the transcriptome of KCs and RHMs (Fig. 4B), with 3,019 differentially expressed genes (q < 0.05) between KCs and RHMS isolated from mice fed NC and 1,239 differentially expressed genes from mice fed the HFD. We then investigated the gene expression changes induced by the HFD within each cell type. We determined that 1,230 genes were differentially expressed between NC and the HFD within KCs and that 1,307 genes were differentially expressed in RHMs. Venn diagram analyses revealed that these two cell types responded quite differently to the HFD (Fig. 4C). Thus, the HFD induced different gene sets in these two distinct cell types (KCs and RHMs), with only 287 (23.3%) differentially expressed genes in common.
Figure 4.
RNA-seq analysis of KCs and RHMs from NC- and HFD-fed mice. A: Principal component analysis of expressed genes in KCs and RHMs from NC- or HFD-fed mice. Samples representing three biological replicates cluster together (KC-NC, red; KC-HFD, blue; RHM-NC, green; RHM-HFD, black). B: Bar chart shows the number of differentially expressed genes between cell types (KC vs. RHM) and diets (NC vs. HFD). C: Venn diagram shows common and distinct differentially expressed genes induced by the HFD within cell types. D: Heat map shows differentially expressed inflammatory response genes between KCs and RHMs from mice fed NC and the HFD in a four-way comparison. Gene counts were adjusted for cell number as KC increase 1.17-fold on HFD and RHMs increase 5.8-fold. Upregulated genes are colored red, downregulated genes are colored blue, and darker colors reflect higher fold changes. RNA-seq studies used three biological replicates per each group.
We performed Gene Ontology (GO) analysis of differentially expressed genes within these four comparisons and identified enriched GO terms to gain insights into the biological processes (BP) within these cells types (Supplementary Table 1). We identified 656 BPs that were significantly different between these cell types (Bonferroni-adjusted P < 0.01). The immune system was a recurrent theme in these enriched BP terms; in particular, “inflammatory response,” (GO:0006954) was significantly different (Bonferroni P = 2.58E-36) when comparing the diet effects within cell types (Supplementary Table 1). This enriched BP term “inflammatory response” contained 162 genes that were differentially expressed (q < 0.05) and are displayed in a heat map reflecting the fold change between groups (Fig. 4D). We adjusted these gene counts to reflect the number of KCs (×1.17) and RHMs (×5.7) present in the liver in HFD-fed mice compared with NC-fed mice. To identify potential surface markers to distinguish KCs from RHMs, we identified the differentially expressed cell surface genes (extracellular region, GO:0044421). Table 1 reports the most discriminating genes based on high expression and fold changes between the two cell types on both diets.
Table 1.
Cell surface markers to distinguish between KCs and RHMs*
| Symbol | Description | NC | HFD | ||||
|---|---|---|---|---|---|---|---|
| KC | RHM | f(KC/RHM) | KC | RHM | f(KC/RHM) | ||
| KC marker† | |||||||
| F8 | Coagulation factor VIII | 20,355 | 4,238 | 5 | 12,074 | 1,198 | 10 |
| Kit | Kit oncogene | 11,300 | 2,186 | 5 | 5,430 | 492 | 11 |
| Wnt2 | Wingless-related MMTV integration site 2 | 4,836 | 818 | 6 | 2,580 | 233 | 11 |
| Prelp | Proline arginine-rich end leucine-rich repeat | 3,485 | 699 | 5 | 2,383 | 234 | 10 |
| Masp1 | Mannan-binding lectin serine peptidase 1 | 2,396 | 450 | 5 | 1,915 | 182 | 11 |
| Vwf | Von Willebrand factor homolog | 2,320 | 416 | 6 | 787 | 121 | 7 |
| Wnt9b | Wingless-type MMTV integration site 9B | 1,149 | 156 | 7 | 288 | 21 | 14 |
| Selp | Selectin, platelet | 973 | 178 | 5 | 1,006 | 87 | 12 |
| Cdh13 | Cadherin 13 | 929 | 114 | 8 | 982 | 64 | 15 |
| Dkk3 | Dickkopf homolog 3 (Xenopus laevis) | 911 | 156 | 6 | 447 | 49 | 9 |
| Tnxb | Tenascin XB | 828 | 154 | 5 | 577 | 41 | 14 |
| Col3a1 | Collagen, type III, α 1 | 752 | 100 | 8 | 711 | 46 | 15 |
| Fbln2 | Fibulin 2 | 355 | 61 | 6 | 397 | 32 | 12 |
| Pdgfd | Platelet-derived growth factor, D polypeptide | 343 | 66 | 5 | 231 | 23 | 10 |
| Col1a2 | Collagen, type I, α 2 | 381 | 59 | 6 | 379 | 29 | 13 |
| Col1a1 | Collagen, type I, α 1 | 357 | 50 | 7 | 423 | 20 | 21 |
| RHM marker† | |||||||
| C1qa | Complement component 1, q subcomponent, α polypeptide | 15,635 | 80,760 | 5 | 39,139 | 107,329 | 3 |
| C1qc | Complement component 1, q subcomponent, C chain | 15,459 | 74,157 | 5 | 34,129 | 98,710 | 3 |
| Apoc1 | Apolipoprotein C-I | 2,510 | 12,325 | 5 | 2,734 | 10,236 | 4 |
| C4b | Complement component 4B (Chido blood group) | 2,361 | 7,773 | 3 | 2,798 | 8,656 | 3 |
| Gbp2b | Guanylate binding protein 2b | 1,156 | 5,569 | 5 | 3,824 | 13,625 | 4 |
| C6 | Complement component 6 | 927 | 8,118 | 9 | 1,142 | 6,303 | 6 |
| Kif23 | Kinesin family member 23 | 912 | 2,633 | 3 | 1,257 | 3,436 | 3 |
| Cpq | Carboxypeptidase Q | 661 | 2,884 | 4 | 1,571 | 4,243 | 3 |
| C4a | Complement component 4A (Rodgers blood group) | 221 | 869 | 4 | 224 | 780 | 3 |
| Kcp | Kielin/chordin-like protein | 75 | 471 | 6 | 63 | 231 | 4 |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 36 | 220 | 6 | 72 | 228 | 3 |
*Differentially expressed genes between KC and RHMs that were classified within the GO cellular component term “extracellular region part” are listed.
†The top 16 KC marker genes listed are expressed at least fivefold higher in KC vs. RHM from NC- or HFD-fed mice. The top 11 RHM markers are all expressed at least threefold higher in RHMs vs. KCs. All markers are expressed at 200 counts per gene or more. RNA-seq studies used three biological replicates per each group.
Discussion
These studies show that in the context of obesity, liver macrophages are made up of heterogeneous, discrete populations. The first is the resident hepatic macrophage, the KC, which shows high levels of inflammatory markers after being activated and appears to be a major source of liver-derived MCP-1 after HFD. In addition, upon HFD feeding, there is a marked increase in the accumulation of fresh RHMs, and these cells are distinct from KCs. The RHMs initially appear in the liver as monocytic cells, expressing high levels of LY6c and CCR2 (R3 in Fig. 1) (16,17); over time, these cells take on the characteristics of differentiated macrophages, which we term RHMs. Obesity not only increases the numbers of RHMs but also enhances their proinflammatory potential, as seen by increased expression of a variety of inflammatory pathway genes (Fig. 4D). These cells express high levels of CCR2 and low levels of MCP-1, whereas KCs express high levels of MCP-1 and low levels of CCR2. This leads to the view that in HFD mice, obesity-resident KCs secrete increased amounts of MCP-1. This serves as a migration signal to recruit monocytic cells into the liver, which eventually mature into differentiated RHMs.
Our results are consistent with previous reports (16,32,33), which show that monocyte-derived infiltrating liver macrophages, which we term RHMs, are smaller, contain fine granules in the cytoplasm, and are F4/80dmCD11b+CCR2+ (16), CD11b+Ly6ChiLy6G− (32), or CD45+CD11c−F4/80lowCD11bhigh cells (33), whereas KCs appear as larger cells with multiple phagocytic granules and are F4/80highCD11blow. A complicating factor in comparing results from different published reports relates to the technical difficulties in isolating and distinguishing KCs from RHMs. Different isolation methods, including cell adherence, density-gradient centrifugation, centrifugal elutriation, and cell sorting, have been used to enrich or purify KCs and the RHM fraction (16,31–33). Moreover, distinction of KCs from RHMs has been based on the expression of F4/80, CD11b (16,32,33), and/or CD68 (34). However, these markers are not expressed exclusively on KCs or RHMs but are expressed by both and on other myeloid cells, such as dendritic cells and polymononuclear cells (34).
To overcome these difficulties in clearly defining KCs from RHMs, our methods use fluorescent markers to differentially label RHMs and KCs. To identify RHMs, we use a macrophage tracking technique in which donor blood monocytes are labeled ex vivo with PKH26 and then injected into recipient mice (17). We found the appearance of these fluorescently labeled monocytes as monocytic cells and subsequently as differentiated macrophages in the liver. KCs are labeled by injecting FITC-dyed beads, which are taken up by all phagocytic cell types in the liver (21). That the latex beads might also be taken up by liver sinusoidal endothelial cells (LSECs) is unlikely, because LSECs possess a maximum capacity of uptake for particles sized at ∼0.1 μm and have very low capacity for the uptake of particles ∼0.5 μm in diameter (35). In contrast, the capacity of KCs to ingest the larger particles is not decreased, even when the particle diameter increases much more (36,37), possibly up to the size of cells. To be certain of this, we assessed LSEC markers (Pecam-1, VAP-1, Stabillin-2, and CD209a) (38) in our RNA-seq database from FITC-labeled KCs and PKH26+ RHMs. Overall, LSEC marker gene expression counts were low and not different between FITC-labeled KCs versus PKH26+ RHMs (data not shown). Therefore, contamination by LSEC in FITC-labeled KCs is negligible.
After lethal irradiation, the radioresistant KCs survive and are marked by the FITC fluorescent beads. Using these approaches, we were able to monitor the numbers of RHMs and KCs in the lean and obese state and to assess their function after cell sorting. Interestingly, KC numbers were similar in the lean compared with the obese state, whereas RHM content increased approximately sixfold in obesity. In addition, a large set of inflammatory pathway genes was induced in the RHMs from HFD mice, whereas inflammatory pathway induction in KCs was only modest. Given this approximately sixfold increase in RHM numbers in HFD mice combined with the increased RHM inflammatory gene induction per cell, we suggest that RHMs are the major contributors to the hepatic inflammatory state in obesity. In addition, we provide evidence that factors secreted from HFD RHMs can cause hepatic insulin resistance, because CM derived from these cells directly stimulated HGO in hepatocytes and blocked the normal effect of insulin to inhibit hepatocyte HGO. In contrast, CM derived from KCs did not exhibit this effect.
Steatosis is the characteristic hepatic manifestation in insulin-resistant states (39,40). This steatotic state, termed nonalcoholic fatty liver disease, can progress to nonalcoholic steatohepatitis, which imparts a high risk for development of fibrosis and cirrhosis (41,42). Immune cell accumulation and activation in the livers from obese rodents and humans are clearly the underlying cause of obesity-induced hepatic inflammation but, most likely, also play a role in the development of steatosis (43,44). Thus, inflammatory cytokines, such as TNF-α, can stimulate hepatocyte lipogenesis, ceramide production, and adipocyte lipolysis, all of which can contribute to hepatic steatosis (45–49). Furthermore, depletion of hepatic phagocytic cells by clodronate administration has a marked effect to ameliorate hepatic insulin resistance and steatosis (50,51). Thus, RHMs and KCs are important contributors to the underlying hepatic inflammatory state in obesity and may also be pathophysiologically important in the development of steatosis.
KCs are the largest population of resident macrophages in the liver. They resemble other resident tissue macrophages in nonlymphoid organs; however, their location within hepatic sinusoids enables intimate contact with the circulation, facilitating their activity to clear bacterial products and interact with blood-derived molecules (52,53). Because the morphology (Fig. 1) and gene expression patterns (Table 1) of KCs and RHMs are strikingly different, this raises the question about the origin of these two distinct cell types. Obviously, RHMs are derived from circulating monocytes, which, in turn, are derived from bone marrow progenitor cells. The origin of KCs, and how KC populations are maintained over time, is more controversial. One view is that KCs have a slow rate of turnover, are not self-renewing, and are replenished from bone marrow–derived monocytes (54,55). This view is supported by bone marrow radiation chimera experiments and chemical depletion studies, which find that circulating monocytes can replenish depleted KC populations (50,54,56). However, other transplantation studies have not found evidence for bone marrow repopulation of hepatic KCs, suggesting these cells are derived from another source (29,57). This would be consistent with reports for other tissue-resident macrophages, such as microglia, showing an origin from non–bone marrow–derived cells (58,59). With this formulation, KCs might arise during early development from a liver progenitor cell type. In this event, KCs would have self-renewing properties, and their population would not be sustained from circulating monocytes. Because our studies showed that the number of KCs was unchanged during HFD, whereas the number of RHMs was increased several fold, and because the gene expression patterns under both NC and HFD conditions are so different between KCs and RHMs, our studies would certainly be consistent with the concept of an intrahepatic, non–bone marrow–derived origin of KCs with an extrahepatic, bone marrow origin for RHMs.
Obesity is associated with a large increase in RHMs, which are a major cause of the inflammatory state in the livers of HFD/obese mice, whereas KCs provide a trigger for this massive RHM infiltration. Factor(s) secreted from HFD mouse RHMs appear to contribute to hepatic insulin resistance, whereas KC-derived factors do not. Moreover, we provide the transcriptome analysis of KCs and RHMs. This analysis shows that KCs widely differ in their transcriptome compared with RHMs (Fig. 4). We also identified and verified the cell surface markers that distinguish KCs from RHMs (Table 1). This latter finding could provide the opportunity for a direct isolation strategy for KCs using specific surface markers.
Supplementary Material
Article Information
Acknowledgments. The authors thank Angela Tyler at the Department of Medicine, University of California, San Diego (UCSD), for editorial assistance, Neal Sekiya at the VA San Diego Hospital for assistance with FACS analysis, and the UCSD Histology Core Laboratory for technical help with processing liver specimens.
Funding. This study was funded in part by the Marshall Plan Scholarship to A.R.P., Austrian Science Fund (FWF) (FWF Doktoratskolleg DK-MCD W1226 and the FWF project P24143) to J.G.B.-S., and by grants to J.M.O. (DK-033651, DK-074868, DK-063491, DK-09062) and D.Y.O. (P30-DK-063491). The UCSD Microscope Resource for Microscopy Analysis was funded by UCSD Neuroscience Microscopy Shared Facility Grant P30-NS-047101.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. H.M. performed RHM isolation from mouse liver, FACS analysis, gene expression measurements, and immunohistochemistry. R.M. performed most of the KC and RHM isolation experiments. J.H. designed the KC isolation protocol and performed KC isolation. O.O., N.F., and N.H. prepared samples for RNA sequencing and carried out RNA sequencing data analysis. E.W. assisted with KC and RHM isolation, FACS analysis, and gene expression measurements. G.B. performed HGO assay. A.R.P., T.J.C., and H.C. assisted with KC/RHM isolation and gene expression measurement. J.G.B.-S. and R.M.E. contributed to discussion. J.M.O. and D.Y.O. analyzed and interpreted data, supervised the project, and cowrote the manuscript. D.Y.O. designed most of the studies and performed macrophage tracking experiments and FACS analysis. D.Y.O. and J.M.O. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-1238/-/DC1.
H.M. is currently affiliated with the Department of Endocrinology and Diabetes Mellitus School of Medicine, Fukuoka University, Fukuoka, Japan.
References
- 1.Olefsky JM, Courtney CH. Type 2 diabetes mellitus: etiology, pathogenesis, and natural history. In Endocrinology. 5th ed. DeGroot LJ, Jameson JL, Eds. Philadelphia, W.B. Saunders and Company, 2005, p. 1093–1117 [Google Scholar]
- 2.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab 2012;15:635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860–867 [DOI] [PubMed] [Google Scholar]
- 4.Curat CA, Wegner V, Sengenès C, et al. Macrophages in human visceral adipose tissue: increased accumulation in obesity and a source of resistin and visfatin. Diabetologia 2006;49:744–747 [DOI] [PubMed] [Google Scholar]
- 5.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 2004;25:4–7 [DOI] [PubMed] [Google Scholar]
- 6.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gribble FM. Metabolism: a higher power for insulin. Nature 2005;434:965–966 [DOI] [PubMed] [Google Scholar]
- 9.Magnusson I, Rothman DL, Katz LD, Shulman RG, Shulman GI. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest 1992;90:1323–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olefsky JM, Garvey WT, Henry RR, Brillon D, Matthaei S, Freidenberg GR. Cellular mechanisms of insulin resistance in non-insulin-dependent (type II) diabetes. Am J Med 1988;85:86–105 [DOI] [PubMed] [Google Scholar]
- 11.Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 2014;510:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006;116:1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai D, Yuan M, Frantz DF, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med 2005;11:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, Metlakunta A, Dedousis N, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010;59:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia L, Vianna CR, Fukuda M, et al. Hepatocyte Toll-like receptor 4 regulates obesity-induced inflammation and insulin resistance. Nat Commun 2014;5:3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obstfeld AE, Sugaru E, Thearle M, et al. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes 2010;59:916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh DY, Morinaga H, Talukdar S, Bae EJ, Olefsky JM. Increased macrophage migration into adipose tissue in obese mice. Diabetes 2012;61:346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheth K, Bankey P. The liver as an immune organ. Curr Opin Crit Care 2001;7:99–104 [DOI] [PubMed] [Google Scholar]
- 19.Papackova Z, Palenickova E, Dankova H, et al. Kupffer cells ameliorate hepatic insulin resistance induced by high-fat diet rich in monounsaturated fatty acids: the evidence for the involvement of alternatively activated macrophages. Nutr Metab (Lond) 2012;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wisse E. Kupffer cell reactions in rat liver under various conditions as observed in the electron microscope. J Ultrastruct Res 1974;46:499–520 [DOI] [PubMed] [Google Scholar]
- 21.Tacke F, Alvarez D, Kaplan TJ, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest 2007;117:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nnalue NA, Shnyra A, Hultenby K, Lindberg AA. Salmonella choleraesuis and Salmonella typhimurium associated with liver cells after intravenous inoculation of rats are localized mainly in Kupffer cells and multiply intracellularly. Infect Immun 1992;60:2758–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh DY, Talukdar S, Bae EJ, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010;142:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 1995;57:289–300 [Google Scholar]
- 27.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B 2002;64:479–498 [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein I, Cornejo JC, Polakos NK, et al. Kupffer cell heterogeneity: functional properties of bone marrow derived and sessile hepatic macrophages. Blood 2007;110:4077–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandrekar P, Ambade A, Lim A, Szabo G, Catalano D. An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology 2011;54:2185–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura K, Yang L, van Rooijen N, Ohnishi H, Seki E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol Gastrointest Liver Physiol 2012;302:G1310–G1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng ZB, Liu Y, Liu C, et al. Immature myeloid cells induced by a high-fat diet contribute to liver inflammation. Hepatology 2009;50:1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Movita D, Kreefft K, Biesta P, et al. Kupffer cells express a unique combination of phenotypic and functional characteristics compared with splenic and peritoneal macrophages. J Leukoc Biol 2012;92:723–733 [DOI] [PubMed] [Google Scholar]
- 34.Lloyd CM, Phillips AR, Cooper GJ, Dunbar PR. Three-colour fluorescence immunohistochemistry reveals the diversity of cells staining for macrophage markers in murine spleen and liver. J Immunol Methods 2008;334:70–81 [DOI] [PubMed] [Google Scholar]
- 35.Kamimoto M, Rung-Ruangkijkrai T, Iwanaga T. Uptake ability of hepatic sinusoidal endothelial cells and enhancement by lipopolysaccharide. Biomed Res 2005;26:99–107 [DOI] [PubMed] [Google Scholar]
- 36.Ogawara K, Yoshida M, Higaki K, et al. Hepatic uptake of polystyrene microspheres in rats: effect of particle size on intrahepatic distribution. J Control Release 1999;59:15–22 [DOI] [PubMed] [Google Scholar]
- 37.Rung-ruangkijkrai T, Fujikura D, Kitamura H, Saito M, Iwanaga T. The expression of src-suppressed C kinase substrate (SSeCKS) and uptake of exogenous particles in endothelial and reticular cells. Arch Histol Cytol 2004;67:135–147 [DOI] [PubMed] [Google Scholar]
- 38.DeLeve LD. Vascular liver disease and the liver sinusoidal endothelial cell. In Vascular Liver Disease: Mechanisms and Management. DeLeve LD, Garcia-Tsao G, Eds. Berlin, Germany, Springer Science+Business Media, 2011, p. 25–40
- 39.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med 2002;346:1221–1231 [DOI] [PubMed] [Google Scholar]
- 40.Sanyal AJ; American Gastroenterological Association . AGA technical review on nonalcoholic fatty liver disease. Gastroenterology 2002;123:1705–1725 [DOI] [PubMed] [Google Scholar]
- 41.Clark JM, Brancati FL, Diehl AM. Nonalcoholic fatty liver disease. Gastroenterology 2002;122:1649–1657 [DOI] [PubMed] [Google Scholar]
- 42.Marchesini G, Brizi M, Bianchi G, et al. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes 2001;50:1844–1850 [DOI] [PubMed] [Google Scholar]
- 43.Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology 2014;147:577–, e1. [DOI] [PubMed] [Google Scholar]
- 44.Meli R, Mattace Raso G, Calignano A. Role of innate immune response in non-alcoholic Fatty liver disease: metabolic complications and therapeutic tools. Front Immunol 2014;5:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bikman BT, Summers SA. Ceramides as modulators of cellular and whole-body metabolism. J Clin Invest 2011;121:4222–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dressler KA, Mathias S, Kolesnick RN. Tumor necrosis factor-alpha activates the sphingomyelin signal transduction pathway in a cell-free system. Science 1992;255:1715–1718 [DOI] [PubMed] [Google Scholar]
- 47.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest 1995;95:2409–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 1993;259:87–91 [DOI] [PubMed] [Google Scholar]
- 49.Wiegmann K, Schütze S, Machleidt T, Witte D, Krönke M. Functional dichotomy of neutral and acidic sphingomyelinases in tumor necrosis factor signaling. Cell 1994;78:1005–1015 [DOI] [PubMed] [Google Scholar]
- 50.Clementi AH, Gaudy AM, van Rooijen N, Pierce RH, Mooney RA. Loss of Kupffer cells in diet-induced obesity is associated with increased hepatic steatosis, STAT3 signaling, and further decreases in insulin signaling. Biochim Biophys Acta 2009;1792:1062–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stienstra R, Saudale F, Duval C, et al. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology 2010;51:511–522 [DOI] [PubMed] [Google Scholar]
- 52.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol 2009;27:147–163 [DOI] [PubMed] [Google Scholar]
- 53.Parker GA, Picut CA. Liver immunobiology. Toxicol Pathol 2005;33:52–62 [DOI] [PubMed] [Google Scholar]
- 54.Diesselhoff-den Dulk MM, Crofton RW, van Furth R. Origin and kinetics of Kupffer cells during an acute inflammatory response. Immunology 1979;37:7–14 [PMC free article] [PubMed] [Google Scholar]
- 55.van Furth R. Monocyte origin of Kupffer cells. Blood Cells 1980;6:87–92 [PubMed] [Google Scholar]
- 56.Parwaresch MR, Wacker HH. Origin and kinetics of resident tissue macrophages. Parabiosis studies with radiolabelled leucocytes. Cell Tissue Kinet 1984;17:25–39 [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto T, Naito M, Moriyama H, et al. Repopulation of murine Kupffer cells after intravenous administration of liposome-encapsulated dichloromethylene diphosphonate. Am J Pathol 1996;149:1271–1286 [PMC free article] [PubMed] [Google Scholar]
- 58.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014;14:392–404 [DOI] [PubMed] [Google Scholar]
- 59.Naito M, Umeda S, Yamamoto T, et al. Development, differentiation, and phenotypic heterogeneity of murine tissue macrophages. J Leukoc Biol 1996;59:133–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.