Abstract
Difference in mercuric ion removal by resting and growing cells of two mercury-resistant yeast strains, identified as Yarrowia spp. (strains Idd1 and Idd2), were studied. Resting cells of strain Idd2 exhibited high maximum Hg2+ removal capacity (59 mg mercury per g dry cell weight [gdw−1]) by adsorption than those of resting cells of strain Idd1 (32 mg gdw−1). The resting cells of strain Idd2 exhibited a higher Hg2+ desorption capacity using CaCl2 (68 %) and EDTA (48 %) than strain Idd1, depicting weaker binding of Hg2+ onto strain Idd2 unlike strain Idd1. The actively growing yeast cells showed opposite Hg removal characteristics to those of the resting cells. Strain Idd1 adsorbed less Hg2+ from culture medium supplemented with Hg2+ than strain Idd2. However, the growing strain Idd1 reduced and vaporized 27 % of supplemented Hg2+ as metallic mercury (Hg0), while the growing strains Idd2 vaporized 15 % of the supplemented Hg2+. These two yeast strains are potential biotechnological tools for the eventual bioremediation of polluted aquatic systems.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-014-6279-1) contains supplementary material, which is available to authorized users.
Keywords: Mercury-resistant yeast, Resting and growing cells, Mercury bioremoval, Biosorption, Biovolatilization
Introduction
Mercury is a major pollutant and the most widely distributed toxic metal in the environment (Tchounwon et al. 2003). Natural sources are the major means through which Hg is introduced into the environment (Tomiyasu et al. 2006; 2008). However, anthropogenic activities remain an unparalleled channel through which various species of Hg polluted the biosphere (AMAP/UNEP 2013; Nimick et al. 2013; Tomiyasu et al. 2006; 2008). Hg pollution has gained global awareness because it is ubiquitous in most environments (AMAP/UNEP 2013) and because its toxicity causes several effects on human health (Guzzi and La Porta 2008). Hg often transforms into highly neurotoxic methyl-mercury (ATSDR 2008) and biomagnifies upward through the food chain in aquatic ecosystems (Kim et al. 2004).
Hg is toxic, but some microorganisms that exhibit resistance are found in systems contaminated with Hg. Various genera of bacteria and fungi have been reported to tolerate Hg (von Canstein et al. 1999 and von Canstein et al. 2002; Huang et al. 1999; Yavuz et al. 2006; Francois et al. 2012). The physicochemical and biological processes leading to the removal and/or sequestration of Hg are bio-remedial strategies proposed to alleviate systems polluted with Hg (Francois et al. 2012; Hussein et al. 2004; Velasquez and Dussan 2009). Pitfalls of non-biological approaches to Hg removal (Hussein et al. 2004) make a microbial-based technology for the detoxification of Hg in polluted systems a better remediation option. The applicability of Hg-resistant microorganisms for the determination of the fate of Hg is of great importance to environmental and public health sciences. Several reports have articulated the use of microorganisms to efficiently remove Hg from contaminated matrixes via biological reduction (von Canstein et al. 1999 and von Canstein et al. 2002), biosorption (Francois et al. 2012; Velasquez and Dussan 2009), bioaccumulation (Velasquez and Dussan 2009), and biosequestration (Francois et al. 2012). Bacterial strains that harbor merA genes are known to reduce Hg2+ to volatile Hg0 (Huang et al. 1999; von Canstein et al. 1999; 2002). Thereby, removing Hg from local environment is achieved by using mercury-resistant microorganisms.
Although yeast cells are generally known to be mediocre in terms of metal biosorption (Voleski 1994), Yarrowia species have been reported to exhibit resistance to toxic concentrations of a number of heavy metals (Bankar et al. 2009; Strouhal et al. 2003). Despite many investigations on Hg resistance and detoxification by microorganisms, little is known about Hg resistance of Yarrowia species. Based on the available information, neither Hg volatilization nor its biosorption through resting cells has been linked to Yarrowia species. The present study sought to find appropriate microbial Hg removal agents using resting and growing Hg-resistant Yarrowia yeast cells.
Materials and methods
Sampling and chemical reagents
Lagos Lagoon in Nigeria is an estuary water body more than 50 km in length and 3–13 km in width with a surface area of approximately 6400 km2. The densely populated city of Lagos spreads along the shore of more than 60 % of the lagoon, which results in the exposure of the water to multifarious contaminants. The highly polluted water empties into the Atlantic Ocean. Composite samples of water and sediment from Lagos Lagoon (06° 27′ 99.7″ N, 003° 22′ 85.1″ E) were collected and analyzed to determine their Hg content (see “Analyses of total mercury” below). HgCl2 used in this research was purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). Stock solutions (100 g l−1) of HgCl2 were prepared and sterilized at 121 °C. All of the other chemicals used were of analytical reagent grade.
Isolation, culture conditions, and deposition of isolates
The sediment sample (10 g) was enriched in a sterile starch-casein broth (90 ml) supplemented with 2 mg l−1 Hg2+. The starch casein broth contained 10 g l−1 soluble starch, 2 g l−1 K2HPO4, 2 g l−1 KNO3, 2 g l−1 NaCl, 0.3 g l−1 casein, 0.05 g l−1 MgSO4·7H2O, 0.02 g l−1 CaCO3, and 0.01 g l−1 FeSO4·7H2O, pH 5.5 (approx.). After a 10-day incubation (30 °C, 150 rpm), 100 μl of the culture was spread on a dried surface of sterile Bacto YM broth (Difco, Detroit, MI, USA) solidified with 1.5 % agar (Wako, Osaka, Japan). The YM broth contained 5 g l−1 yeast extract, 3 g l−1 malt extract, 5 g l−1 peptone, and 10 g l−1 dextrose. Three days post-inoculation (30 °C), colonies were selected and stored in a water:glycerol (1:1) mixture at −80 °C. The isolated yeast strains have been deposited to Japan Collection of Microorganisms under accession no. JCM 30162 (strain Idd1) and JCM 30163 (strain Idd2).
Determination of resistance pattern
The isolated yeasts were screened to determine the minimum inhibitory concentration (MIC) of Hg, which is the concentration at which no growth of yeast was observed in the YM broth supplemented with HgCl2 (30 °C, 150 rpm, 48 h). The Hg concentrations in the YM broth used for the MIC determination were 2, 4, 8, 16, 32, and 64 mg l−1. Two strains of yeast showing resistance to 32 mg l−1 were selected and subjected to further analysis.
Molecular characterization and identification of Hg-resistant yeasts
DNA was isolated and purified using a Wizard DNA isolation kit (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions. Using universal eukaryotic primer sets (Table S1), nearly full-length 18S ribosomal RNA (rRNA) genes were amplified from the genomic DNA. Each PCR mixture contained 1 μl of DNA template, 5 μl of 10 × Taq DNA polymerase buffer (TaKaRa, Ohtsu, Japan), 4 μl of dNTP (2.5 mmol l−1 each; TaKaRa), 0.5 μl of each primer (50 μmol l−1), 0.25 μl of Ex-Taq DNA polymerase (TaKaRa), and 38.75 μl of sterile Milli-Q water in a final volume of 50 μl. The PCR protocol consisted of an initial hot incubation (95 °C, 5 min), 30 amplification cycles (denaturation at 95 °C for 45 s, annealing at 55 °C for 1 min, and extension at 72 °C for 2.5 min), and a final extension (72 °C, 7 min) using a Gradient PCR Thermal Cycler Dice (TaKaRa). The 18S rRNA gene amplicons were sequenced using the Applied Biosystems 3130xl Genetic Analyzer and a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster, CA, USA). The sequences were compared to those present in the database using BLAST (N) and aligned using the Muscle 3.7 program. Regions with significant variations were automatically removed using the Gblocks software. The phylogeny of the aligned multiple sequences were analyzed with PhyML v3.0, and a phylogenetic tree was rendered using TreeDyn. The 18S rRNA gene sequences were deposited in GenBank under accession no. AB893170 (strain Idd1) and AB893171 (strain Idd2).
Determination of mercury removal through biosorption by resting yeast cells
The resuscitated yeast strains (approx. 106 cells ml−1 as OD600 = 0.1) were starved overnight in Tris–HCl (0.1 mol l−1) and re-suspended in sterile Milli-Q water (previously supplemented with 1.0 mg l−1 Hg2+, final concentration) to prepare resting yeast cells. The yeast cell concentrations in the cell suspensions were determined as previously described (Puranik and Paknikar 1999).
To determine the Hg adsorption capacity of the yeast cells, a sufficient inoculum as prepared above (25 mgdw in 10 ml of deionized water, approx.) was introduced into an Erlenmeyer flask containing 50 ml of minimal medium (MM) supplemented with 6.0, 12.0, 24.0, 48.0, 96.0, or 192.0 mg l−1 HgCl2, and the culture was incubated (50×g; 30 °C). The MM contained 10 g l−1 glucose, 2.67 g l−1 NH4Cl, and 5.35 g l−1 Na2HPO4 in 6 ml of mineral salts solution (0.1 g of CaCl2.H2O, 10 g of MgSO4·7H2O, 0.07 g of MnSO4·7H2O, and 0.04 g of FeSO4·7H2O in 1000 ml of distilled water). At every 5 min, the quantity of Hg adsorbed by the yeast cells was determined (as described below) after centrifugation (9000×g, 20 min). The Hg adsorbed was calculated as , where Q is the specific Hg uptake (mg gdw−1), V is the volume of the Hg solution (ml), C O is the initial Hg concentration in the solution (mg l−1), C e is the final Hg concentration in the solution (mg l−1), and m is the yeast cell dry weight (gdw). The time at which the Hg-adsorption equilibrium was attained by the resting yeast cells was determined.
Desorption of Hg from loaded yeast cells
The desorption assay was performed as previously reported (Goyal et al. 2003): aqueous 0.1 mol l−1 NaOH, 6.0 mmol l−1 KCl, 5.0 mmol l−1 CaCl2, and 5.0 mmol l−1 EDTA were used as the desorption solutions. A cell mass previously loaded with Hg through biosorption was harvested (9000×g, 10 min), washed with physiological phosphate buffer, and treated with the desorption solution (50.0 ml) at 30 °C with shaking (100 rpm). The desorption with NaOH was allowed to continue for 20 min, whereas the other solutions were incubated for 8 h due to the disparity between the ionic strength of NaOH and that of the other solutions. The supernatant was separated (9000×g, 10 min), and its Hg content was analyzed. Sterile Hg-supplemented broth and broth without Hg inoculated with resting cells were used as the controls, and these were subjected to the same experimental conditions.
Bioaccumulation of mercury by actively growing yeast cells
The yeast inoculum (1 ml), which was prepared as described above, was inoculated into 100 ml of YM broth supplemented with Hg (0, 3.0, 6.0, 12.0, 24.0, and 48.0 mg l−1) in 500-ml Erlenmeyer flasks, and the culture was incubated at 30 °C and 50 rpm for 48 h. The growth of the yeast strains was monitored every 6 h based on the turbidity, which was determined by measuring the OD(600 nm) using a UV-visible spectrophotometer. The blank was uninoculated YM broth supplemented with the appropriate Hg concentration. The growing cell of each sample was harvested (10,000×g, 10 min), and the supernatant was separated into a new tube. The pellet was washed three times with saline solution (0.8 % NaCl), oven-dried (105 °C) overnight, and weighed. The culture, supernatant, and pellets were analyzed to determine their Hg content as explained below. The controls were the same as those described above.
Analyses of total mercury
The total mercury amounts in 100 μl of the culture medium and supernatant (obtained from the Hg removal experiments) were determined directly without any pre-treatment using a fully automated thermal vaporization mercury analysis system, i.e., Mercury/MA-3000 (Nippon Instrument Corp., Osaka, Japan). The atomic absorbance of the atomized Hg was measured at a wavelength of 253.7 nm. The instrument was calibrated with standard Hg solution (BDH, Leicestershire, England) at concentrations ranging from 0.1 to 100.0 mg l−1. The amount of volatilized Hg was determined by subtracting residual amount of Hg from initial amount in 100 μl of the culture medium.
Hg0 volatilization assay during yeast growth
The volatilization of Hg2+ to Hg0 during growth of yeast cells was confirmed qualitatively as previously described (Nakamura and Nakahara 1988) and quantitatively in terms of the depletion of Hg. For the qualitative assay, the yeast strains were cultivated in YM broth or YM agar supplemented with 40 μmol l−1 HgCl2. After 48 h of incubation (30 °C), the colonies on the YM agar plates were harvested and suspended in 100 μl of phosphate buffer (7 mmol l−1, pH 7) containing 0.5 mmol l−1 EDTA, 0.2 mmol l−1 magnesium acetate, and 5 mmol l−1 sodium thioglycolate with HgCl2 (100 μmol l−1) or without HgCl2 as the control. For the detection of Hg volatilization using the YM broth culture, 100 μl of the culture was mixed with 100 μmol l−1 HgCl2 (final concentration). The suspensions were diluted with buffer, described above, to obtain an OD(600 nm) of 0.2. The suspensions were transferred to a microplate, covered with a medical X-ray film (Fujifilm RX-U; Fujifilm Corp., Tokyo, Japan), and cultivated for 8 h at 30 °C in the dark. The foggy areas on the film, which are due to the reduction of the Ag+ emulsion of the film by gaseous Hg0, were thus interpreted as Hg volatilization. Sterile buffer with and without HgCl2 was used as the negative control, whereas Bacillus megaterium MB1, which harbors the merA gene in its chromosome and expresses Hg2+ reductase (Huang et al. 1999), was used as the positive control.
Statistical analyses
All of the statistical tests were performed using the Prism 5 software program (GraphPad software, San Diego, CA, USA).
Definition of resting cells and growing cells of the yeasts
“Resting cells” are yeast cells that are harvested after approximately 106 cells ml−1 as OD600 0.1 growth was obtained, harvested, and washed twice; the cells are metabolically active but they are not multiplying. While “growing cells” are cells in nutrient medium that are increasing in population and consequently increasing in biomass as time of incubation changes.
Results
Sampling, isolation, Hg resistance, and identification of yeast strains
The two Hg-resistant yeast strains were isolated from lagoon sediment that had total mercury concentration of 95 mg kgdw−1. The yeast strains grew well in YM medium containing 8 % NaCl and showed resistance, as MIC, to 44.0 mg l−1 Hg (220 μM HgCl2). Based on their 18S rDNA sequences, the yeast strains were identified as species of Yarrowia because they exhibited some relatedness, as determined by their occupation of the same clade in the phylogenetic tree depicted in Fig. S1.
Hg (II) removal
Adsorption equilibrium of Hg (II) by resting yeast cells
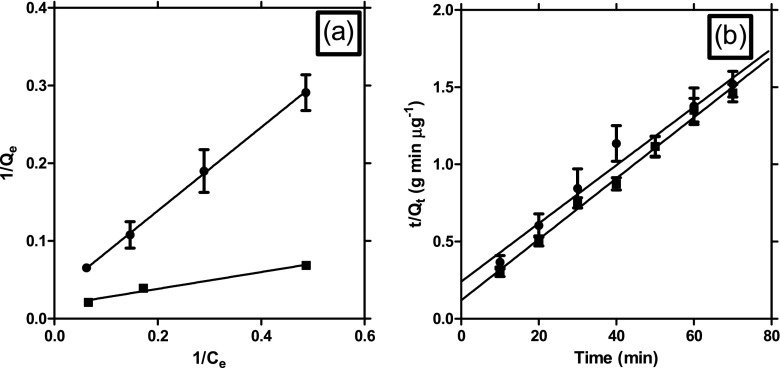
The resting cells of the two yeast strains adsorbed a major fraction of Hg2+ within the first 15-min exposure followed by an almost negligible increase after 60 min. The adsorption equilibria were attained 80 min (strain Idd1) and 100 min (strain Idd2) after inoculation. Hg adsorbed by resting yeast cells at equilibrium (Qe mg gdw−1) as a function of the concentration of Hg in the solution at equilibrium (C e) (mg l−1) was expressed by the Langmuir model equation () in a linear form as (see Fig. 1a); where Q max (maximum Hg uptake, mg gdw−1) and b (affinity between Hg2+ and the resting yeast cells) are the Langmuir constants. The Langmuir adsorption constants evaluated from the isotherms, as well as their correlation coefficients (R 2) (≥0.9) show that the Langmuir model provided a fit for the experimental data (Table 1). The Hg adsorption data indicates that strain Idd2 is characterized by a higher maximum Hg2+ (59 mg gdw−1) than strain Idd1 (32 mg gdw−1). The Langmuir affinity (saturation) constant (1/b) was marginally higher in strain Idd1 (17 mg−1) than Idd2 (6.5 mg−1). Mercury removal percentage of the resting cells is represented in Fig. 2, where more than 99 % of available Hg2+ was removed at low metal concentration (4 mg l−1). At high metal concentration (64 mg l−1), however, resting cells of Idd1 removed Hg2+ more efficiently (84 ± 2 %) than Idd2 (78 ± 4 %).
Fig. 1.
a Adsorption equilibrium by Langmuir equation and b adsorption kinetics by pseudo-second-order equation of Hg (II) on resting cells of yeast strains Idd1 (filled cycle) and Idd2 (filled square). The error bars represent the standard error of the mean from triplicate experiments
Table 1.
Hg2+ biosorption kinetics obtained from Langmuir isotherm and pseudo-second-order rate equation for resting cells of yeast strains
| Kinetic models | Parameters | Yeast strains | |
|---|---|---|---|
| Yarrowia (Idd1) | Yarrowia (Idd2) | ||
| Langmuir equation | Q max (mg gdw−1) | 32.0 | 59.0 |
| 1/b (mg l−1) | 17.0 | 6.5 | |
| R 2 | 0.91 | 0.98 | |
| P | <0.0001 | <0.0001 | |
| Pseudo second order | k 2 (mg−1 gdw min−1) | 1.5 | 3.3 |
| Q e (mg gdw−1) | 53.0 | 51.0 | |
| R 2 | 0.86 | 0.96 | |
| P | <0.0001 | <0.0001 | |
Fig. 2.
Efficiencies of Hg removal by resting cells of yeast strains Idd1 and Idd2 in medium supplemented with various concentrations of HgCl2. The error bars represent the standard error of the mean from triplicate experiments
The pseudo-first-order rate equation, In(Q e − Q t) = InQ e − k 1 t, and the pseudo-second-order rate equation, , were represented in their linear forms. In these equations, Q t is the amount of Hg2+ adsorbed (mg gdw−1) at any given time t (min), Q e is the amount of Hg2+ adsorbed (mg gdw−1) at equilibrium, and k 1 and k 2 are the rate constants of the pseudo-first-order reaction and the pseudo-second-order reaction for biosorption, respectively. For the analysis of the applicability of the pseudo-first-order kinetic model, a plot of In (Q e − Q t) as a function of t did not yield straight lines (Fig. S1). However, a plot of t/Q t as a function of t resulted in straight lines, thereby indicating the applicability of the pseudo-second-order kinetic model (Fig. 1b). The calculated values of Q e and k 2 are shown in Table 1. The adsorption of Hg by the yeasts follows pseudo-second-order kinetics, as inferred by the high values of R 2.
Desorption of Hg from resting yeast cells
The desorption of Hg from Hg-adsorbed resting yeast cells is shown for strain Idd1 and strain Idd2 (Table 2). Solutions of CaCl2 (5.0 mM) and EDTA (5.0 mM) desorbed 68 and 48 % of the loaded Hg (II), respectively, from the resting yeast cells of strain Idd2. Hg adhered more strongly to strain Idd1, as indicated by the poorer performance of the desorbing agents (EDTA desorbs 29 % of the adsorbed Hg from the resting cell mass). In addition, the NaOH (0.1 M) solution dislodged a negligible portion (0.02 % from strain Idd1 and 0.03 % from strain Idd2).
Table 2.
Desorption of Hg2+ from yeast strains Idd1 and Idd2 using selected desorbents
| Mercury desorbed from yeast cells (%) | ||
|---|---|---|
| Desorption solution | Idd1 | Idd2 |
| EDTA | 29.0 (±1.4) | 24.0 (±6.0) |
| KCl | 3.2 (±0.53) | 11.0 (±1.0) |
| CaCl2 | 7.0 (±1.6) | 33.0 (±18.0) |
| NaOH | 2.3 (±0.2) | 1.6 (±0.085) |
Desorption conditions were EDTA (5 mmol l−1, 8 h), KCl (5 mmol l−1, 8 h), CaCl2 (5 mmol l−1, 8 h), and NaOH (0.1 mol l−1, 20 min). The Hg2+ quantities adsorbed by Idd1 and Idd2 were 55.0 (±2.4) mg gdw−1, and 49.0 (±3.2) mg gdw−1, respectively. Desorption reactions were conducted with shaking (50×g) at a temperature of 30 °C. The values indicate the mean (±SD) from triplicate experiments
Hg volatilization and Hg bioaccumulation by actively growing yeast cells
Hg2+ removal was recorded for each growing strain under the ambient pH (5.0–6.5) and temperature (30 ± 2 °C), conditions prevalent in Lagos Lagoon. The yeast strains grew without a lag phase in medium supplemented with 8.0 mg l−1 Hg2+ (and lower Hg2+ concentrations), and the characteristic growth pattern observed was similar to that obtained in medium without Hg2+ supplementation. However, at higher Hg concentrations (≥16.0 mg l−1), a lag phase (≥12 h) preceded the spontaneous growth of both strains. Adsorption and volatilization of Hg were observed during the lag phase. Hg bioaccumulation began during the exponential growth phase and increased during the stationary phase. The mass balance of Hg2+ in the batch culture system was presented in Table 3. Volatilization due to the release of Hg0 from the growing yeast cultures was observed based on the development of foggy areas on the X-ray film (Fig. S3).
Table 3.
Mercury removal by yeast strains Idd1 and Idd2 during growth in medium supplemented with 32 mg Hg2+ per liter
| Yeast strains | Mercury remaining in mediuma (%) | Mercury accumulated onto growing yeast cellsa (%) | Mercury volatilized by growing yeast cellsa (%) |
|---|---|---|---|
| Idd1 | 58.0 ± 2.9 | 16.0 ± 10.8 | 27.0 ± 13.2 |
| Idd2 | 63.0 ± 7.8 | 18.0 ± 14.0 | 18.0 ± 6.3 |
aTotal mercury is expressed as percentages of the initial Hg content in the medium upon inoculation. Values represent the mean (±standard deviation) from triplicate experiments
Discussion
Reckless waste disposal of industrial and artisanal activities are the major anthropogenic sources through which Hg contaminates the environment throughout the world (AMAP/UNEP 2013). The total Hg concentration in the sediment is higher than those reported in other climes (Tomiyasu et al. 2008) and alarmingly above the permissive level in the environment. Severe hazards to public health and estuarine ecosystems due to Hg toxicity are imminent because there is no known mitigation plan to salvage the continual pollution of the water bodies with Hg-laden effluents. The isolation of highly Hg-resistant yeast strains from the sediment in a mercury contaminated lagoon is an indication of the contamination of the estuarine water-course with mercuric ions. As a bio-indicator of mercuric pollution, the autochthonous yeast strains in the lagoon must have molecular alterations to evolve the high Hg resistance.
The equilibrium distribution of mercuric ions between the aqueous and solid phases is represented by the biosorption rate and the maximum biosorption capacity, which is commonly described by the Langmuir isotherm model (Febrianto et al. 2009). The resting cells of both strains adsorb Hg2+ from solution at an almost equal rate. The high Q max observed in Hg adsorption indicates that resting cells of strain Idd2 has a higher number of sites available for Hg binding and belongs to a single-type phenomenon with no interactions between the adsorbed Hg. The lower Langmuir saturation constant of strain Idd2 than that of strain Idd1 indicates the adsorption affinity of the resting yeast strain Idd2 for mercuric ions is higher than strain Idd1. However, certain inconsistencies in the saturation capacities (Q max) of Langmuir equation, and Hg2+ uptake at equilibrium (Q e) were observed. The rather higher Q e (53.0 mg gdw−1) than predicted Q max (32.0 mg gdw−1) recorded for strain Idd1 may be associated with the surface functional groups, and effects of several factors including number of binding sites, chemical state of the sites, and binding strength of the sites, as previously discussed by Febrianto et al. (2009).
The kinetics of Hg2+ biosorption on resting yeast cells of Yarrowia spp. was determined using the pseudo-second-order model based on solid-phase adsorption (Febrianto et al. 2009) (Fig. 1b and Table 1). The results suggest that chemisorption may be the rate-limiting step that controls the uptake of Hg2+ from aquatic systems. The rate-control may vary during the course of Hg2+ biosorption (Febrianto et al. 2009). An external surface mass transfer or a film diffusion process may control the early stages of the Hg2+ biosorption process onto resting cells of the yeast strains. This stage may be followed by a chemical reaction or constant rate stage and then by a diffusion stage in which the biosorption process slows down considerably (Djeribi and Hamdaoui 2008). It is noteworthy that good fitting model does not necessarily illustrate the nature of real adsorption between the resting cells and Hg2+ due to dynamic characteristics of all living cells unlike dead cells.
Desorption upon treatment with calcium and potassium showed a lower desorption performance of strain Idd1 than that of strain Idd2. This negates the potential reusability of strain Idd1 in the treatment of mercury-laden wastewater. From these data, the resting cells of strain Idd2 would be a better option in the treatment of wastewater laden with mercuric ions, since the adsorbed mercuric ions could be readily recovered from the resting cells. The results of Hg desorption from the resting cell masses may indicate that salt cations, particularly Ca2+, in estuarine water can readily desorb Hg from the resting cells of strain Idd2 (Table 2), and thereby spread the toxic metal to other parts of the ecosystem. However, strain Idd2 would be of great value for the treatment of industrial wastewater due to its reusability and the potential ability to recover Hg from the resting yeast cells. The poor desorption potential of strain Idd1 coupled with its higher Q max demonstrates that the organism may act as an Hg (II) sink in the limnetic zone. However, strain Idd1 would not have predominance over strain Idd2 in recovery of Hg and reusability of the yeast cells for Hg removal in a bioreactor system.
Hg2+ reduction to Hg0 and Hg0 volatilization (Table 3 and Fig. S3) were likely triggered by sulfhydryl compounds present in the biomolecules produced by the growing yeast strains in response to Hg toxicity. Larger amount of mercury was reduced and volatilized by the growing strain Idd1 than the growing strain Idd2. Recently, Rao et al. (2013) reported the role played by strong reducing agents in facilitating efficient metal removal by Yarrowia lipolytica. It has been reported that mercury is transported across the vacuolar membrane to become localized in the vacuoles of Saccharomyces cerevisiae (Diffels et al. 2006). Certain proteins (Strouhal et al. 2003) and/or interactions with sulfhydryl-containing residues have been hypothesized to be responsible for Hg bioaccumulation in microorganisms (Hansen et al. 2006; Park and Park 2007; Pulido and Parrish 2003). Concrete evidence of the actual organic biomolecules involved in Hg reduction and transportation by Yarrowia spp. remains to be obtained.
Based on the results described above, characteristics of mercury removal by the resting and growing cells of yeast strains Yarrowia sp. Idd1 and Idd2 isolated in this study were observed. The resting cells of yeast strain Idd2 would be applicable to Hg removal and recovery from mercury-laden water as a more appropriate and reusable bio-adsorbent. In contrast, the growing cells of yeast strain Idd1 would be applicable as an appropriate Hg2+ bio-reduction and volatilization agent. Therefore, the most appropriate Hg removal method should be selected according to the removal characteristics of resting cells and growing cells of the yeast strains. However, further studies are needed to understand detailed physiological Hg removal mechanisms of the resting and growing cells of yeast strains, by analyzing biomolecules involved in the Hg reduction and adsorption.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 443 kb)
Acknowledgments
We thank Dr. Mei-Fang Chien and Toshiki Kurosawa for their useful discussion and assistance during the experiment. GOO and GE appreciate the Japan Society for the Promotion of Science (JSPS) for the awarding of Postdoctoral Research Fellowship FY2012-2013. This work was funded by Grants-in-Aid (No. 24-02373) for Scientific Research from the JSPS.
Contributor Information
Ganiyu Oladunjoye Oyetibo, Email: goyetibo@unilag.edu.ng, Email: goyetibo@mail.tohoku-gakuin.ac.jp.
Shakirat Titilayo Ishola, Email: ishola_titilayo@yahoo.com.
Wakako Ikeda-Ohtsubo, Email: wohtsubo@mail.tohoku-gakuin.ac.jp.
Keisuke Miyauchi, Email: kmiya@mail.tohoku-gakuin.ac.jp.
Matthew Olusoji Ilori, Email: milori@unilag.edu.ng.
Ginro Endo, Email: gendo@mail.tohoku-gakuin.ac.jp.
References
- AMAP/UNEP (2013) Global releases of mercury to aquatic environments. In: Technical background report for global mercury assessment. Pp 263
- ATSDR (2008) Public health statement: toxic substances and health. Agency for toxic substances and disease registry, Atlanta, GA: Division of toxicology and environmental medicine September bulletin
- Bankar AV, Kumar AR, Zinjarde SS. Removal of chromium (VI) ions from aqueous solution by adsorption onto two marine isolates of Yarrowia lipolytica. J Hazard Mater. 2009;170:487–494. doi: 10.1016/j.jhazmat.2009.04.070. [DOI] [PubMed] [Google Scholar]
- Diffels JF, Seret ML, Goffeau A, Baret PV. Heavy metal transporters in Hemiascomycete yeasts. Biochimie. 2006;88:1639–1649. doi: 10.1016/j.biochi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Djeribi R, Hamdaoui O. Sorption of copper (II) from aqueous solutions by cedar sawdust and crushed brick. Desalination. 2008;225:95–112. doi: 10.1016/j.desal.2007.04.091. [DOI] [Google Scholar]
- Febrianto J, Kosasih AN, Sunarso J, Ju Y-H, Indraswati N, Ismadji S. Equilibrium and kinetic studies in adsorption of heavy metals using biosorbent: a summary of recent studies. J Hazard Mater. 2009;162:616–645. doi: 10.1016/j.jhazmat.2008.06.042. [DOI] [PubMed] [Google Scholar]
- Francois F, Lombard C, Guigner J-M, Soreau P, Brian-Jaisson F, Martino G, Vandervennet M, Garcia D, Molinier A-L, Pignol D, Peduzzi J, Zirah S, Rebuffat S. Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl Environ Microbiol. 2012;78:1097–1106. doi: 10.1128/AEM.06522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N, Jain SC, Banerjee UC. Comparative studies on the adsorption of heavy metals. Adv Environ Res. 2003;7:311–319. doi: 10.1016/S1093-0191(02)00004-7. [DOI] [Google Scholar]
- Guzzi G, La Porta CA. Molecular mechanisms triggered by mercury. Toxicology. 2008;244:1–12. doi: 10.1016/j.tox.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Zhang H, Jones DP. Differential oxidation of thioredoxin-1, thioredoxin-2, and glutathione by metal ions. Free Radic Biol Med. 2006;40:138–145. doi: 10.1016/j.freeradbiomed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Huang CC, Narita M, Yamagata T, Itoh Y, Endo G. Structure analysis of a class II transposon encoding the mercury resistance of the Gram positive bacterium, Bacillus megaterium MB1, a strain isolated from Minamata Bay, Japan. Gene. 1999;234:361–369. doi: 10.1016/S0378-1119(99)00184-5. [DOI] [PubMed] [Google Scholar]
- Hussein H, Ibrahim SF, Kandeel K, Moawad H. Biosorption of heavy metals from waste water using Pseudomonas sp. Electron J Biotechnol. 2004;7:38–46. doi: 10.2225/vol7-issue1-fulltext-2. [DOI] [Google Scholar]
- Kim E-H, Mason RP, Porter ET, Soulen HL. The effect of resuspension on the fate of total mercury and methyl mercury in a shallow estuarine ecosystem: a mesocosm study. Mar Chem. 2004;86:121–137. doi: 10.1016/j.marchem.2003.12.004. [DOI] [Google Scholar]
- Nakamura K, Nakahara H. Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl Environ Microbiol. 1988;54:2871–2873. doi: 10.1128/aem.54.11.2871-2873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimick DA, Caldwell RR, Skaar DR, Selch TM. Fate of geothermal mercury from Yellowstone National Park in the Madison and Missouri Rivers, USA. Sci Total Environ. 2013;443:40–54. doi: 10.1016/j.scitotenv.2012.10.080. [DOI] [PubMed] [Google Scholar]
- Park EJ, Park K. Induction of reactive oxygen species and apoptosis in BEAS-2B cells by mercuric chloride. Toxicol in Vitro. 2007;21:789–794. doi: 10.1016/j.tiv.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Pulido MD, Parrish AR. Metal-induced apoptosis: mechanisms. Mutat Res/FMMM. 2003;533:227–241. doi: 10.1016/j.mrfmmm.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Puranik PR, Paknikar KM. Biosorption of lead, cadmium, and zinc by Citrobacter strain MCM B-181: characterisation studies. Biotechnol Prog. 1999;15:228–237. doi: 10.1021/bp990002r. [DOI] [PubMed] [Google Scholar]
- Rao A, Bankar A, Kumar AR, Gosavi S, Zinjarde S. Removal of hexavalent chromium ions by Yarrowia lipolytica cells modified with phyto-inspired Fe0/Fe3O4 nanoparticles. J Contam Hydrol. 2013;146:63–73. doi: 10.1016/j.jconhyd.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Strouhal M, Kizek R, Vacek J, Trnkova L, Nemec M. Electrochemical study of heavy metals and metallothionein in yeast Yarrowia lipolytical. Biogeosciences. 2003;60:29–36. doi: 10.1016/s1567-5394(03)00043-4. [DOI] [PubMed] [Google Scholar]
- Tchounwon PB, Ayensu WK, Ninashvili N, Sutton Environmental exposure to mercury and its toxicopathologic implications for public health. Environ Toxicol. 2003;18:149–175. doi: 10.1002/tox.10116. [DOI] [PubMed] [Google Scholar]
- Tomiyasu T, Matsuyama A, Eguchi T, Fuchigami Y, Oki K, Horvat M, Rajar R, Akagi H. Spatial variations of mercury in sediment of Minamata Bay, Japan. Sci Total Environ. 2006;368:283–290. doi: 10.1016/j.scitotenv.2005.09.090. [DOI] [PubMed] [Google Scholar]
- Tomiyasu T, Matsuyama A, Eguchi T, Marumoto K, Oki K, Akagi H. Speciation of mercury in water at the bottom of Minamata Bay, Japan. Mar Chem. 2008;112:102–106. doi: 10.1016/j.marchem.2008.07.003. [DOI] [Google Scholar]
- Velasquez L, Dussan J. Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J Hazard Mater. 2009;167:713–716. doi: 10.1016/j.jhazmat.2009.01.044. [DOI] [PubMed] [Google Scholar]
- Voleski B. Advances in biosorption of metals: selection of biomass types. FEMS Microbiol Rev. 1994;14:291–302. doi: 10.1111/j.1574-6976.1994.tb00102.x. [DOI] [PubMed] [Google Scholar]
- von Canstein H, Li Y, Timmis KN, Deckwer W-D, Wagner-Döbler I. Removal of mercury from chlor-alkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl Environ Microbiol. 1999;65:5279–5284. doi: 10.1128/aem.65.12.5279-5284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Canstein H, Kelly S, Le Y, Wagner-Döbler I. Species diversity improves the efficiency of mercury-reducing biofilms under changing environmental conditions. Appl Environ Microbiol. 2002;68:2829–2837. doi: 10.1128/AEM.68.6.2829-2837.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz H, Denizli A, Gungunes H, Safarikova M, Safarik I. Biosorption of mercury on magnetically modified yeast cells. Sep Purif Technol. 2006;52:253–260. doi: 10.1016/j.seppur.2006.05.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 443 kb)