Abstract
Lymphovascular invasion (LVI) and histologic grade are clinical parameters of high prognostic value in breast cancer and indicate the level of tumor aggressiveness. Many studies have focused on the association of breast cancer subtypes with gene expression and chromosomal profiles, but considerably less genomic information is available regarding traditional prognostic factors such as histologic grade and LVI. We studied by array-CGH a group of 57 invasive ductal carcinomas of the breast to outline the DNA copy number aberration (CNA) profile linked to high histologic grades and LVI. Selected CNAs were validated using real-time quantitative PCR (qPCR). Furthermore, gene expression analysis was performed in a subset of 32 of these tumors, and findings were integrated with array-CGH data. Our findings indicated an accumulation of genomic alterations in high-grade breast tumors compared to low-grade samples. Grade III tumors showed higher number of CNAs and larger aberrations than low-grade tumors and displayed a wide range of chromosomal aberrations, which were mainly 5p, 8q, 10p, 17q12, and 19 gains, and 3p, 4, 5q proximal, 9p, 11p, 18q, and 21 losses. The presence of LVI, a well-established prognostic marker, was not significantly associated with increased genomic instability in comparison to breast tumors negative for LVI, considering the total number of chromosomal alterations. However, a slightly increase in the frequency of specific alterations could be detected in LVI-positive group, such as gains at 5p, 16p, 17q12, and 19, and losses at 8p, 11q, 18q, and 21. Three newly reported small-scale rearrangements were detected in high-risk tumors (LVI-positive grade III) harboring putative breast cancer genes (amplicons at 4q13.3 and 11p11.2, and a deletion at 12p12.3). Furthermore, gene expression analysis uncovered networks highlighting S100A8, MMP1, and MED1 as promising candidate genes involved in high-grade and LVI-positive tumors. In summary, a group of genomic regions could be associated with high-risk tumors, and expression analysis pinpointed candidate genes deserving further investigation. The data has shed some light on the molecular players involved in two highly relevant prognostic factors and may further add to the understanding of the mechanisms of breast cancer aggressiveness.
Electronic supplementary material
The online version of this article (doi:10.1007/s13277-014-2786-z) contains supplementary material, which is available to authorized users.
Keywords: Invasive ductal carcinomas of the breast, Breast cancer, DNA copy number aberrations, CNA, Lymphovascular invasion, Histologic grade III, Gene expression, S100A8, MMP1, MED1, ADAMTS3, HSD17B12
Introduction
Breast cancer is a significant cause of cancer-related death worldwide [1]. Despite significant advances in diagnosis and treatment, several considerable clinical and scientific problems remain unresolved. Invasive ductal carcinomas (IDC) of no special type (NST) represent 80 % of all invasive tumors of the breast. IDC can be associated with a poorer prognosis than certain special types of breast cancer (such as tubular and mucinous carcinomas) and shows significant biological heterogeneity. Traditional variables, such as tumor size, axillary nodal status, histologic grade, and lymphovascular invasion (LVI) status, are part of risk assessment and deliver significant prognostic information, albeit with limited predictive value [2–6].
The histologic grade [Scarff-Bloom-Richardson (SBR)] modified by Elston and Ellis (modified SBR or Nottingham system) is a widely validated prognostic factor recommended by the Union for International Cancer Control (UICC, 2009). This index grades phenotypic aggressiveness according to morphological criteria based on tubular formation, nuclear pleomorphism, and mitotic counting [7]. Although many studies have focused on the association of breast cancer subtypes with gene expression and chromosomal profiles, considerably less genomic information is available regarding traditional prognostic factors such as histologic grade and LVI [8–12].
Recent studies point to the significance of DNA copy number aberrations (CNAs) in the etiology of cancer with the number and complexity of these aberrations being indicative of overall prognosis [10, 13–15]. CNA investigation may assist the identification of regions containing oncogenes and tumor suppressor genes [16]. Grade III breast tumors frequently harbor gains at 3q and 5p, and 8q amplifications [10, 11]. More recently, a 19q12 amplification was detected, primarily associated with grade III breast tumors in estrogen-negative samples, which encompasses the CCNE1 gene, among others [17]. Grade I tumors show less complex karyotypes with recurrent gain of 16p, as found in estrogen-positive tumors [11, 18]. However, to the best of our knowledge, no study has investigated the genomic profile of CNAs related to the presence of LVI in breast carcinomas.
We performed a study outlining the CNA and gene expression patterns of invasive carcinomas of the breast to identify genomic alterations and differentially expressed genes linked to high histologic grades and LVI. We suggest several genes as potential biomarkers of breast cancer aggressiveness with the ultimate goal of improving patient care.
Materials and methods
Study approval and patient samples
This retrospective study was approved by the local Ethics and Research Committee of A.C. Camargo Cancer Center, São Paulo, Brazil (#1448/10), and informed consents were obtained from all patients. Frozen samples from 57 invasive ductal carcinomas of the breast were retrieved from the A.C. Camargo Cancer Center Biobank for DNA and RNA extractions. All selected carcinomas were tested for estrogen receptor, progesterone receptor, and HER2 status, and analyses followed the ASCO/CAP guidelines [19, 20]. Detailed clinical characteristics of the 57 breast carcinomas are given in Supplementary Table S1. These tumors were selected because of their availability as frozen samples.
DNA and RNA isolation
Genomic DNA was extracted according to the procedure of the Biobank of the institution [21]. Sample quality and quantity were assessed using NanoDrop (Thermo Scientific, Waltham, MA, USA), and molecular weight was checked by electrophoresis in agarose gels. RNA was obtained from epithelial cells from invasive ductal carcinomas samples captured by laser microdissection using the PixCell II LCM system (Arcturus Engineering, Mountain View, CA, USA). Only RNA samples with optical density of approximately 2.0 and RNA integrity number >5.0 were used for microarray experiments [22].
Comparative genome hybridization based on microarrays
We performed comparative genomic hybridization based on microarrays (array-CGH) in a commercial whole-genome 60K platform containing 60,000 oligonucleotide probes (Agilent Technologies, Santa Clara, CA, USA; design 21924). A commercially available pool of healthy human female DNA (Promega, Madison, WI, USA) was used as the reference DNA. The experimental procedure was performed as recommended by the manufacturer. Scanned images were processed using Feature Extraction Software version 10.7.3.1 (Agilent Technologies). Poor-quality hybridizations (quality control >0.2) were disregarded [23]. CNAs were identified with Nexus software 7.0 (BioDiscovery, Hawthorne, CA, USA) using the FASST2 segmentation algorithm, which is an approach based on the Hidden-Markov model that uses the log2 ratio values of the probes for CNA calling. We applied the following settings: a minimum of five consecutive affected probes (effective resolution of ∼200 kb for CNA calling), a significance threshold set at 10−8, and threshold log2 ratio Cy3/Cy5 of 0.3 and 1.4 for gain or high copy gain (named amplicons), respectively, and −0.3 and −1.1 for loss and homozygous loss, respectively. Copy number variants reported in the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation/), CNAs smaller than 50 kb, and data from the sex chromosomes were excluded. Gene annotation was performed according to the GRCh37 using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). The smallest common regions of recurrent aberrations were obtained by implementing the global frequency statistical approach of the Significance Testing for Aberrant Copy Number (STAC) method [24]. The parameters had a minimum differential threshold of 25 % and had p values of p ≤ 0.05 for LVI and p ≤ 0.01 for histologic grades. The CNA data of each breast tumor group were evaluated for number, length, and types of CNAs (gains, losses, high copy gains, and homozygous losses), and statistical analyses were performed using GraphPad Prism v5.0 (Mann-Whitney test to compare two unpaired groups and Kruskal-Wallis test to compare three unpaired groups).
Copy number validation by real-time quantitative PCR
Real-time quantitative PCR (real-time qPCR) was performed on three genes (ADAMTS3 F:CGTAGAAAGCCTTTGGG, R:GGTGCATGATGGAACG; HSD17B12 F:CCCTTTAAGCCATTCCG, R:GCCAATATTCAAACCGAGC, and RERGL F:CCCCACAAAGTTCCTTC, R:CTCATCTGCTCTGAAACTGG) using the SYBR Green system (Applied Biosystems, Foster City, CA, USA) on a 7500 System apparatus (Applied Biosystems, Foster City, CA, USA) with reference DNA (Promega) for copy number calibration. Values were normalized based on data from the GAPDH (12p13) and P2RX7 (12q24) genes, which were not affected by copy number changes in these groups of tumors. Duplicates were analyzed using the comparative 2-ΔΔCt cycle threshold method [25]. Values in the range of 0.8–1.2 indicate two copies, <0.6 indicates copy number loss, and >1.4 was considered a gain.
Microarray gene expression analysis
A subgroup of 32 breast carcinomas was evaluated for gene expression based on cDNA microarrays using a 244K custom platform (Agilent Technologies, Santa Clara, CA, USA). Samples were hybridized following the manufacturer’s protocol. Scanned images were processed using Feature Extraction Software version 10.7.3.1. The identification of genes differentially expressed was performed using the Agilent GeneSpring GX 12.1 software after subtracting background and filtering features flagged as not positive, not significant, or not uniform in normalization (normalization to the 65th percentile shift per array, median across all samples per transcript). To identify differentially expressed genes, we used the t test (unpaired with 5 % false-discovery ratio correction) considering a fold change of |2| with p ≤ 0.05. The genes belonging to the PAM50 list [26] were excluded from this analysis since their expression profiles predict intrinsic molecular subtypes of breast cancer and, consequently, could interfere with gene changes differentiating tumors according to histologic grades or lymphovascular invasion status. Ingenuity Pathway Analysis (IPA) software was used for the in silico analysis of the sets of differentially expressed genes. Interaction networks were obtained using a core analysis tool, which considered the Ingenuity Knowledge Base (genes + endogenous chemicals), human species, breast cancer cell lines, and all types of tissues and primary cells. The selected networks generated by the program contained at least 40 % of the set of differentially expressed genes identified in the study.
Results
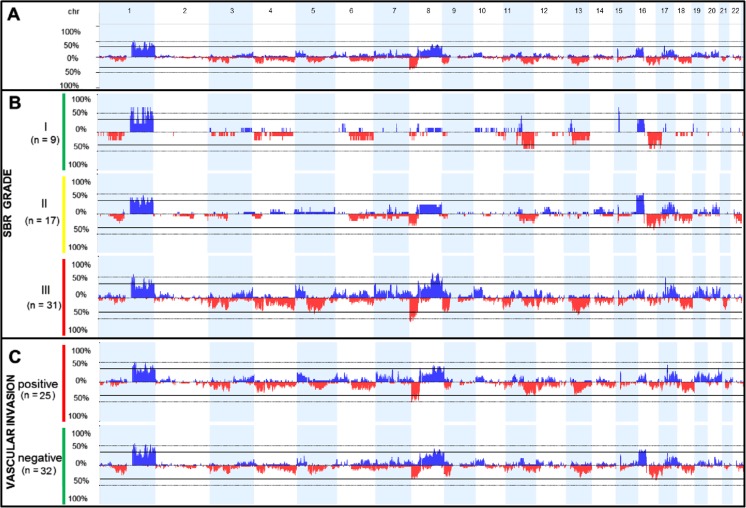
Table 1 summarizes the frequencies of the clinical categories in the group of 57 carcinomas of the breast. Of these cases, 21 % were classified as triple-negative (estrogen receptor (ER)-, progesterone receptor (PR)-, and human epidermal growth factor receptor 2 (HER2)-negative tumors) [27]. A total of 2,857 CNAs were identified in all of the breast tumors profiled (mean of ∼50 CNAs per sample). The summary of the CNA results and the full array-CGH data can be found in Supplementary Tables S2 and S3, respectively. A complex pattern of multiple chromosomal gains and losses was identified in all samples, the most frequent being gains of chromosomes 1q, 8q, 16p, and 17q and losses affecting 8p, 9p, 11q, 13q, 16q, and 18q (Fig. 1a).
Table 1.
Clinical characteristics of the group of 57 primary invasive ductal carcinomas of non-special type of the breast
| Clinical variable | Category | Number (%) |
|---|---|---|
| LVIa | Positive | 25 (43.9) |
| Negative | 32 (56.1) | |
| SBRb | I | 9 (15.8) |
| II | 17 (29.8) | |
| III | 31 (54.4) | |
| Clinical stage | I | 9 (15.8) |
| II | 21 (36.9) | |
| III | 26 (45.6) | |
| IV | 1 (1.7) | |
| Axillary lymph node | N0 | 20 (35.1) |
| N1 | 19 (33.3) | |
| N2 | 10 (17.5) | |
| N3 | 8 (14.0) | |
| Estrogen receptor | Positive | 33 (57.9) |
| Negative | 24 (42.1) | |
| ERBB2 | Positive | 5 (8.8) |
| Negative | 52 (91.2) | |
| Triple-negative | Yes | 12 (21.0) |
| No | 45 (79.0) |
aLymphovascular invasion
bSBR grade, Scarff-Bloom Richardson graduation system (histological grade)
Fig. 1.
Array-CGH genomic profile exhibiting the frequency of DNA copy number aberrations (CNAs) in the group of invasive ductal carcinomas of the breast. The x-axis represents the genomic positions of chromosomes 1p to 22q, and the y-axis denotes the percentage of gains (plotted in blue above the 0 % baseline) and losses (plotted in red below the 0 % baseline) in all selected samples at the specified genomic location (images adapted from the Nexus Copy Number 7.0 software, BioDiscovery). chr indicates the chromosome, and n is the number of samples in each group. a The genomic profile of CNAs (gains and losses) of all 57 invasive ductal carcinomas of the breast. b CNA profile of breast tumors grouped according to histologic grade (I, II, and III). c CNA profile of breast tumors grouped according to lymphovascular invasion status.
We performed a statistical comparison between subgroups of breast tumor samples according to the presence of lymph node metastasis: tumors from patients without lymph node metastasis (N0 = 20 samples) and tumors from patients harboring lymph node metastasis at diagnosis. Significant differences were not detected.
Total CNAs and their distribution and type according to histological grade
We compared different tumor subgroups according to histologic grades to assess the total number of CNAs and the CNA distribution with regard to the different types of events (gain, high copy gain, loss, homozygous loss). These results are detailed in Table 2.
Table 2.
Total number of DNA copy number aberrations (CNAs), their distribution in each different CNA class (gain, loss, high copy gain, homozygous loss), and genomic lengths according to histologic grades (SBR) and lymphovascular invasion status (LVI)
| Group | Total events (median) | Gains (median) | Losses (median) | High copy gains (mean) | Homozygous losses (mean) | Mean size (Mb) (median) |
|---|---|---|---|---|---|---|
| Breast carcinomas (n = 57) | 2,857 (38.0) | 1,477 (23.0) | 1,245 (16.0) | 122 (2.1) | 13 (0.2) | 70.2 (27.3) |
| SBR I (n = 9) | 290 (21.0) | 128 (12.0) | 155 (9.0) | 7 (0.8) | 0 (0) | 10.0 (27.0) |
| SBR II (n = 17) | 606 (20.0) | 315 (10.0) | 241 (9.0) | 44 (2.6) | 6 (0.3) | 16.2 (26.6) |
| SBR III (n = 31) | 1,961* (60.0) | 1,034* (33.0) | 849* (25.0) | 71 (2.3) | 7 (0.2) | 48.9* (27.3) |
| LVI positive (n = 25) | 1,289 (38.0) | 676 (25.0) | 558 (14.0) | 51 (2.0) | 4 (0.2) | 33.1 (26.6) |
| LVI negative (n = 32) | 1,568 (41.5) | 801 (22.5) | 687 (17.5) | 71 (2.2) | 9 (0.3) | 40.0 (27.6) |
*p < 0.05 (statistically significant at this level in the comparison between different SBR grades, Mann-Whitney test)
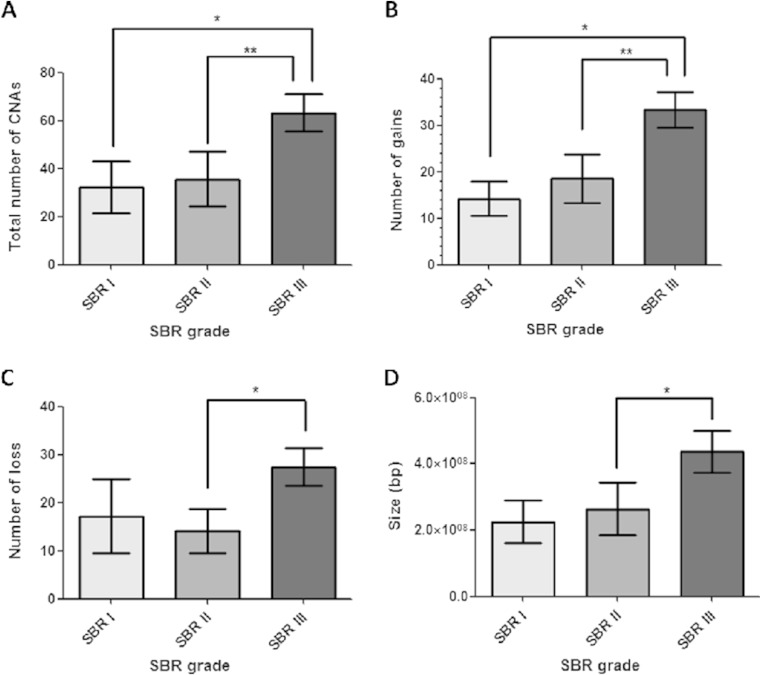
Grade III tumors showed a statistically higher total number of CNAs compared to grade I (p = 0.045) and grade II (p = 0.006) tumors (Mann-Whitney test, Fig. 2a). Additionally, grade III group had a higher number of gains (p = 0.007, Fig. 2b) and losses (p = 0.0148, Fig. 2c) compared to low-grade tumors (Mann-Whitney test), and the CNAs were larger than those detected in the grade II group (p = 0.0498, Mann-Whitney test; Fig. 2d).
Fig. 2.
Statistical analysis comparing the DNA copy number aberration (CNA) events between different breast carcinoma groups according to histologic grades. Asterisks indicate statistical significance. a Bar graph of the total number of CNAs showing significant differences between tumor grades I and II compared to grade III. b Bar graph of the number of gain events showing significant differences between tumor grades I and II compared to grade III. c Bar graph of the number of loss events showing significant differences between tumor grades II compared to grade III. d Bar graph of the genomic alteration sizes of breast tumors showing significant differences between grade II tumors compared to grade III
In a visual inspection, grade III tumors exhibited a wider range of chromosomal aberrations than grades I and II. In particular, CNAs detected preferentially or almost exclusively in this group included the following: gains affecting 5p, 8q, 10p, 17q12 (ERBB2), and 19 and losses at 3p, 4, 5q proximal, 8p, 9p, 11p, 18q, and 21 (Fig. 1b).
Using the STAC statistical analysis, we detected the smallest common regions of aberrations associated with histologic grade III. Peaks of genomic regions preferentially present in grade III tumors (minimum threshold of 25 %) were detected in several chromosomes, including peak gains at 5p15, 7p22, 8q22, 8q24, 10p12.1, 19q12, and 20p13 and peak losses at 4p13p12, 4q34q35, 9p21, 5q11q23, 12p12, and 21q21 (Supplementary Table S4).
In our samples, the manual curation of CNA data from each tumor sample revealed that the most frequent alteration identified both in grades I (33.3 %) and II (52.9 %) was a 16p gain, which was detected only in 15.2 % of the grade III samples (Fig. 1b, Supplementary Table S4). All breast tumors harboring the 16p gain were found to be positive for estrogen receptor.
Total CNAs and their distribution and type according to LVI status
We also compared the CNA profile of breast tumors according to LVI status and the results are summarized in Table 2. We did not detect statistically significant differences between LVI-positive and LVI-negative groups (p value >0.5 in all comparisons) regarding the total number of CNAs, the CNA distribution (number of gain, high copy gain, loss, homozygous loss), and genomic sizes.
Although we did not detect evidence of an increased genomic instability in LVI-positive breast tumors, some specific rearrangements could be detected at a slightly higher frequency in this group compared to the LVI-negative group, such as gains at 5p, 17q12 (ERBB2), and 19, and losses affecting 8p, 11q, 18q, and 21 (Fig. 1c).
Using the STAC analysis, we detected more frequently associated with LVI (minimum of 25 % of the samples) two deletions peaks at 11q24.1 and 11q24.3, encompassing the OR6M1 and ARHGAP32 genes, respectively, whereas the 16p gain and a 16q23 loss were detected at a higher frequency in tumors without LVI (Supplementary Table S5). In fact, the manual curation of CNA data from each tumor sample revealed that the 16p gain could be detected in about 40 % of the LVI-negative group compared to 24 % of cases presenting LVI. Considering that all these breast tumor samples carrying 16p gains were found to be positive for estrogen receptor, we performed a statistical correction (logic regression) which showed that the 16p gain is a marker independently associated with LVI-negative status.
Small-scale rearrangements: genomic amplifications and homozygous losses
We investigated the frequency of six known breast cancer amplicons [28] excluding 17q12 (ERBB2), and all of them were frequently detected in grade III tumors as genomic gains (Table 3). The 19q12 gain was detected exclusively in grade III tumors (one amplification and nine copy number gains), all of them negative for estrogen receptor. The smallest common region affected by this 19q12 gain was restricted to a ∼537 kb segment at chr19:29,777,917-30,315,215 (GRCh37) in which only six genes are mapped, including CCNE1. Regarding the LVI status, the majority of the tumors carrying the 20q13.2 gain (62.5 %) were positive for LVI.
Table 3.
Frequency of gains affecting well-known breast cancer amplicons in high-risk breast carcinomas (histologic grade III and lymphovascular invasion positive)
| Known amplicon | Major gene | Total frequency (n affected) | Frequency among histologic grade III samples | Frequency among lymphovascular invasion positive samples |
|---|---|---|---|---|
| 8q24 | MYC | 38.6 % (22) | 81.8 % | 45.4 % |
| 8p12 | ZNF703/FGR1 | 31.6 % (18) | 72.2 % | 44.4 |
| 8p12 | FGFR1 | 29.8 % (17) | 64.7 % | 47.1 % |
| 19q12 | CCNE1 | 17.5 % (10) | 100 % | 55.5 % |
| 20q13.2 | ZNF217 | 14.0 % (8) | 62.5 % | 62.5 % |
| 11q13 | CCND1 | 14.0 % (8) | 50.0 % | 25.0 % |
In bold are indicated amplicons that were frequently detected (≥50 %) in tumors of each category
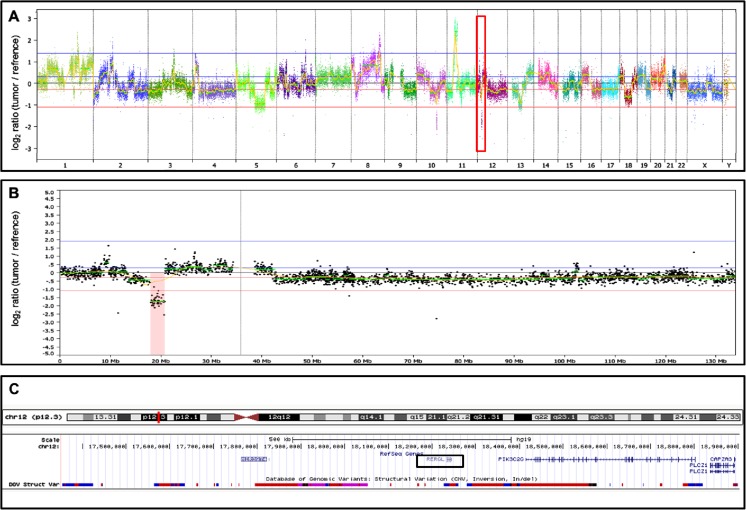
In addition to the abovementioned known breast cancer amplicons, we detected three small-scale rearrangements (< 4 Mb) not previously reported, each of them in a different high-risk tumor (grade III positive for LVI): two amplifications, one at 4q13.3 (chr4:72,209,104-73,455,050) and the other at 11p11.2 (chr11:43,830,876-46,190,463); and one homozygous deletion at 12p12.3 (chr12:17,950,780-20,811,443). Genes mapped within the two amplified regions were selected for further copy number validation (Supplementary Figure S1). Figure 3 displays the homozygous 12p12.3 deletion encompassing the RERGL and PIK3C2G genes, among others.
Fig. 3.
Homozygous deletion affecting a 1.5 Mb genomic region at 12p13.3, detected in a high-risk breast tumor (histologic grade III with lymphovascular invasion, MIC82). a Array-CGH genomic profile of the breast carcinoma -the deletion at chromosome 12 is indicated by a red box. b Array-CGH profile of chromosome 12, highlighting the ∼1.5 Mb deletion at 12p13.3 (in red). c The affected genomic segment at 12p13.3 is depicted by the UCSC Genome Browser; this region encompasses five genes, including RERGL (marked by a black box) and PIK3C2G
The 11p11.2 genomic amplification encompasses, among others, the HSD17B12 gene. In our cohort, we detected HSD17B12 gains in association with gains of the COX2 gene, in a possible 1q31/11p11 co-amplification pattern, here observed in ER-negative high-grade breast tumors.
Copy number status of previously reported breast cancer driver genes
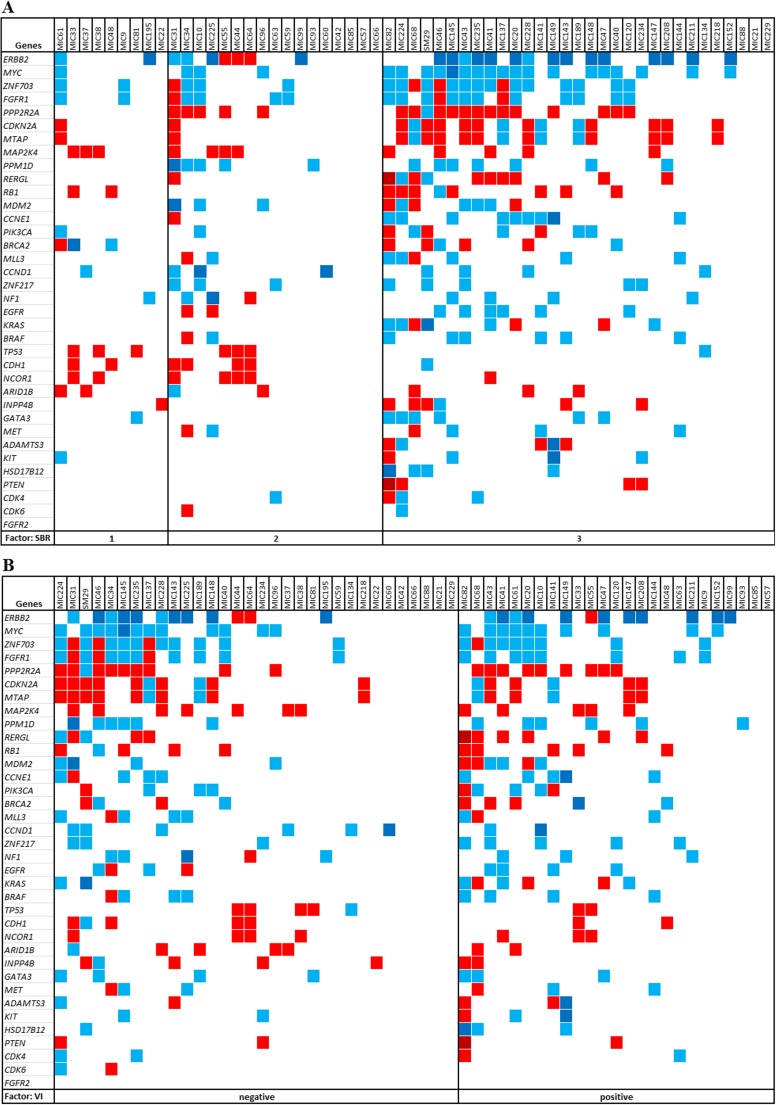
We scrutinized the copy number status of 33 driver cancer genes reported as recurrently affected by copy number changes in breast cancer [11, 17, 29–32], which includes the major genes of the aforementioned amplicons. We also examined the copy number status of three genes highlighted in the present study (ADAMTS3, HSD17B12, and RERGL). Different CNA frequencies affecting these genes were observed according to histologic grade and LVI status (Fig. 4). A panel of 16 genes exhibited copy number changes at higher frequencies in high-grade tumors compared to low-grade samples (losses affecting PPP2R2A, CDKN2A, MTAP, RERGL, RB1, PTEN, INPP4B and gains of CCNE1, ERBB2, EGFR, FGFR1, GATA3, MYC, HSD17B12, ZNF703, and ZNF217; Fig. 4a); conversely, copy number losses of TP53, CDH1, and NCOR1 were detected preferentially in low-grade breast tumors. Furthermore, ERBB2 gains and RERGL losses were more frequent in the presence of LVI (Fig. 4b).
Fig. 4.
Copy number changes of a panel of 36 genes in all 57 breast tumor samples grouped according to SBR grades or lymphovascular invasion status. The copy number status of 33 breast cancer driver genes and 3 genes (rows) identified in the present study (ADAMTS3, HSD17B12, and RERGL) is depicted for each tumor sample (columns) by colored squares: gains and amplifications are shown in blue and dark blue, respectively; losses and homozygous losses are red and dark red, respectively. a Schematic view showing the pattern of copy number changes of the panel of 36 genes in breast tumor samples grouped according to histologic grade. b Schematic view showing the pattern of copy number changes of the panel of 36 genes in breast tumors grouped according to lymphovascular invasion status
Gene expression profiling
Table 4 displays the top differentially expressed genes of the breast tumor groups with their respective fold-change values (full differential expression analysis can be found in Supplementary Tables S6 and S7). Histological grade III tumors exhibited 12 up- and 13 downregulated genes compared to low-grade tumors (p value <0.001 and fold change of |5|). We investigated the copy number status of the detected deregulated genes. The following five genes displayed a CNA pattern concordant with the expression level in >15 % of the grade III tumors: S100A8 and MCM10 genes (upregulated) and GRP, CX3CR1, and FAM198B (downregulated). We used the network analysis tool of IPA software to reveal regulatory interconnections among these genes. The network with the highest score (26) connected 12 of the deregulated genes with 23 additional ones, showing an enrichment of the categories of Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking (Fig. 5), in which the S100A8 gene plays a central role.
Table 4.
Top differentially expressed genes (positive fold-change values indicating upregulation and negative fold-change values indicating donwregulation) identified between histologic grade III breast tumors and histologic grade I+II breast tumors (left) and between breast tumors positive or negative for lymphovascular invasion (right)
| Histologic grade IIIa | Lymphovascular invasion positiveb | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Cytoband | Fold-change | p value | Gene | Cytoband | Fold-change | p value |
| S100A8 | 1q21.3 | 22.0278 | 0.001 | C19orf33 | 19q13.2 | 4.4608 | 0.021 |
| DEFB1 | 8p23.1 | 14.2762 | 0.001 | LGALS7B | 19q13.2 | 3.5713 | 0.007 |
| ADM | 11p15.4 | 7.0605 | 0.001 | CPE | 4q32.3 | 3.0436 | 0.011 |
| MMP1 | 11q22.2 | 6.7464 | 0.001 | AGBL2 | 11p11.2 | 2.9353 | 0.028 |
| MCM10 | 10p13 | 6.4085 | 0.001 | ARSG | 17q24.2 | 2.7475 | 0.044 |
| TMSB15A | Xq22.1 | 6.2266 | 0.001 | UMOD | 16p12.3 | 2.6808 | 0.025 |
| SOX11 | 2p25.2 | 5.5269 | 0.001 | C13orf31 | 13q14.11 | 2.6631 | 0.026 |
| YBX2 | 17p13.1 | 5.3408 | 0.001 | MYCBPAP | 17q21.33 | 2.5776 | 0.032 |
| KRT81 | 12q13.13 | 5.3214 | 0.001 | CXXC4 | 4q24 | 2.5767 | 0.044 |
| E2F8 | 11p15.1 | 5.2362 | 0.001 | MED1 | 17q12 | 2.5187 | 0.033 |
| NANOS1 | 10q26.11 | 5.1092 | 0.001 | SHISA5 | 3p21.31 | 2.5084 | 0.002 |
| CDC45L | 22q11.21 | 5.0596 | 0.001 | GFRA1 | 10q25.3 | −4.4772 | 0.021 |
| GRP | 18q21.32 | −8.9074 | 0.001 | MFI2 | 3q29 | −4.0174 | 0.029 |
| CYP4X1 | 1p33 | −8.1935 | 0.001 | TBX21 | 17q21.32 | −3.8633 | 0.038 |
| SUSD3 | 9q22.31 | −8.0443 | 0.001 | KRT15 | 17q21.2 | −3.6634 | 0.016 |
| ABAT | 16p13.2 | −7.3311 | 0.001 | NFASC | 1q32.1 | −3.3951 | 0.022 |
| EVL | 14q32.2 | −6.9978 | 0.001 | FZD5 | 2q33.3 | −3.1834 | 0.008 |
| KLHDC9 | 1q23.3 | −6.9593 | 0.001 | TCF7L1 | 2p11.2 | −3.0653 | 0.047 |
| C6orf211 | 6q25.1 | −6.9305 | 0.001 | MYBPC2 | 19q13.33 | −2.8301 | 0.021 |
| NEFH | 22q12.2 | −6.3975 | 0.001 | CRABP1 | 15q25.1 | −2.7727 | 0.017 |
| CX3CR1 | 3p22.2 | −5.3859 | 0.001 | CIB2 | 15q25.1 | −2.5553 | 0.018 |
| SMOC2 | 6q27 | −5.3324 | 0.001 | DUSP3 | 17q21.31 | −2.5391 | 0.001 |
| BTG2 | 1q32.1 | −5.3206 | 0.001 | ||||
| FAM198B | 4q32.1 | −5.0859 | 0.001 | ||||
| CCDC74B | 2q21.1 | −5.0592 | 0.001 | ||||
aHistological grade III: p value <0.001 and fold change of |5|
bLymphovascular invasion positive: p value <0.05 and fold change of |2.5|
Fig. 5.
Cellular diagram of the network created by the Ingenuity Pathway Analysis based on interactions of the set of differentially expressed genes detected in histologic grade III breast tumors compared to grade I and grade II tumors. Red and green nodes represent, respectively, upregulated or downregulated genes (the intensity of the colors indicating the degree of deregulation) in grade III breast tumors in comparison to grade I and II tumors (Table 4); uncolored nodes represent genes automatically included in this network because they are biologically linked to the others based on scientific evidence. The functional categorization of this network was Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking, with the S100A8 gene playing a central role
Breast tumors with LVI exhibited 11 upregulated and 11 downregulated genes compared to negative tumors (p value <0.05 and fold change of |2.5|). The following four upregulated genes exhibited a concordant CNA pattern in tumors with LVI: UMOD (gains) and ARSG, MYCBPAP, and MED1 (gains and high-copy gains). The 22 deregulated genes were evaluated using the network analysis tool of IPA software. The more relevant network (score 12) connected seven of the differentially expressed genes with additional 28 genes, which showed an enrichment of the categories of Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function. In this network, the MED1 gene appears to play a central role (Supplementary Figure S2).
Summary of the relevant genomic regions and selected genes
Table 5 presents a summary of the relevant genomic regions and genes revealed in this work in association either with histologic grade III tumors or with LVI. Data include some of the significant recurrent CNAs, highlighting relevant gene content, as well as three newly detected small-scale rearrangements, and differentially expressed genes possibly deregulated by copy number changes.
Table 5.
Summary of the relevant genomic regions and genes revealed in this work in association either with histologic grade III or with lymphovascular invasion
| Relevant gene(s) | Chromosomal alteration | Clinical association |
|---|---|---|
| MYC | 8q24 gain | Preferentially detected in grade III samples |
| MTAP | 9p21 loss | Preferentially detected in grade III samples |
| COX6C | 8q22 gain | Preferentially detected in grade III samples |
| PMS2 | 7p22 loss | Preferentially detected in grade III samples |
| ERCC8, XRCC4, FBXL17, MEF2C | 5q11q13 loss | Preferentially detected in grade III samples |
| LMO3 | 12p12.3 loss | Preferentially detected in grade III samples |
| CCNE1, LOC284395, VSTM2B, POP4, PLEKHF1, C19orf12 | 19q12 minimum common region of gain | Exclusively detected in grade III negative for estrogen-receptor samples |
| ADAMTS3 | 4q13.3 amplification | Detected in a grade III LVI-positive sample |
| HSD17B12 | 11p11.2 amplification | Detected in a grade III LVI-positive sample |
| RERGL, PIK3C2G | 12p12.3 homozygous deletion | Detected in a grade III LVI-positive sample |
| ARHGAP32 | 11q24.3 loss | Preferentially detected in LVI-positive samples |
| S100A8, MCM10 | – | Concordant pattern of upregulated gene expression and copy number gains in grade III samples |
| GRP, CX3CR1, FAM198B | – | Concordant pattern of downregulated gene expression and copy number losses in grade III samples |
| UMOD, ARSG, MYCBPAP, MED1 | – | Concordant pattern of upregulated gene expression and copy number gains/high gains in LVI-positive samples |
| Several | 16p gain | Preferentially detected in both low-grade and LVI-negative samples (exclusively estrogen-receptor-positive tumors) |
Discussion
Increased numbers of CNAs are expected to be associated with tumor aggressiveness [12]; accordingly, we detected a particular augmentation of the number of genomic gains in histologic grade III breast tumors. Despite the fact that LVI is also associated with high-risk breast cancer, no differences in the total number of CNAs were detected in our study considering the LVI status. Taken together, our data corroborate previous studies showing an accumulation of genomic alterations in more aggressive breast tumors, although it also suggests that vascular invasion is a prognostic marker not related to increased genomic instability.
CNA pattern according to histological grade
Grade III tumors exhibited a wide range of genomic aberrations previously reported for this category [11, 12]; the previously reported 19q12 amplification [17] was detected in only one grade III tumor, although gains affecting 19q12 were identified in additional nine grade III samples, which allowed us to narrow the critical region to a ∼537 kb segment. It is noteworthy that all grade III tumors carrying the 19q12 gain were found to be negative for estrogen receptor, a finding that corroborates the link with poor outcome [11, 28].
Some regions statistically associated with grade III tumors in our study harbor genes either frequently amplified in aggressive breast cancer such as the oncogenes MYC (8q24) and CCNE1 (19q12) or already reported as deleted in breast tumors such as MTAP (9p21) [30]. The 8q22 gain is reportedly related to poor prognosis and chemoresistance possibly due to the activation of MTDH [33], although the 8q22 segment here detected did not harbor this gene and included the cancer gene COX6C, which is involved in oxidative metabolism, upregulated in prostate tumors, and fusioned to HMGIC gene in uterine leiomyomas [34, 35]. Conversely, some of the detected regions in the present work are not strongly related to breast cancer; recurrent losses affecting 7p22 (segment encompassing, among others, the PMS2 gene) and 5q11-q13 (ERCC8 and XRCC4) could suggest the involvement of DNA repair pathways in cancer progression. The 5q loss also includes FBXL17, a gene reported to be a biomarker for breast cancer resistance [36], and MEF2C, which was previously identified in association with breast cancer invasion [37]. The loss at 12p12.3 includes LMO3, a key gene implicated in the onset and progression of several cancers [38].
Consistent with the literature [12, 18, 39], the 16p gain was detected here preferentially in low-grade tumors (33.3 % of grade I samples and 52.9 % of grade II samples), and all of them were estrogen-positive samples.
CNA pattern according to LVI status
Although the LVI-positive tumors were not associated with an increased genomic instability compared to LVI-negative tumors, they showed a few genomic alterations detected at a slightly higher frequency than the negative group, such as gains at 5p, 17q12 and 19, and losses at 8p, 11q, 18q, and 21. The most interesting alteration was an 11q24.3 loss harboring ARHGAP32, a gene earlier reported to be involved in fusion events in breast cancer cell lines and to participate in autophagy processes [40–43]. Additionally, the 16p gain was also preferentially associated with the absence of LVI (40 % of LVI-negative tumors), which reinforces this CNA as a marker of good prognosis [12, 18, 39].
Copy number status of known driver genes and new genes proposed for breast cancer
Known breast cancer amplicons were detected as gains either exclusively (19q12) or predominantly in grade III tumors [15, 44]. The 20q13.2 amplicon, previously associated with poor outcome [30, 44–47], was related in our study both to high-grade and LVI-positive tumors. We identified three new focal rearrangements in grade III/LVI-positive breast tumors: 4q13 and 11q11 amplifications (harboring the ADAMTS3 and the HSD17B12 genes, respectively) and a homozygous deletion at 12p12.3 (containing, among others, the RERGL gene). These highlighted regions because of the small size and the amplitude of the copy number changes indicate potential driver genes for breast cancer. The ADAMTS3 gene belongs to the ADAMTS metalloproteinase family that has been implicated in tumor progression [44–46]. Overexpression of HSD17B12 was recently associated with COX2 (1q31) overexpression in breast carcinomas [47], and in our cohort, HSD17B12 gains were detected in association with COX2 gains, which may indicate a new 1q31/11p11 co-amplification pattern in ER-negative high-grade breast tumors. However, functional studies are necessary to validate the biological relevance of the emphasized genes.
Additionally, we assembled a panel of genes exhibiting differences in the frequency of copy number changes between high-grade and low-grade breast tumors groups (see Table 5), including two of those genes here reported (RERGL and HSD17B12). Finally, the combination of frequencies of copy number changes of two genes was detected in LVI-positive breast cancer group: gains of ERBB2 and losses of RERGL.
Gene expression profiling
Concerning the gene expression pattern of grade III breast tumors, the S100A8 gene, the top upregulated gene in grade III compared to low-grade tumors, appeared to be a central gene in the main regulatory network. This gene can act as a potent amplifier of inflammation in autoimmunity as well as of cancer development and metastasis [48–50]. S100A8 overexpression in high-grade breast tumors in comparison to low-grade tumors has been previously reported [51] and associated with poor prognosis, poor tumor differentiation, LVI, and node metastasis when co-expressed with S100A9 [52]. Another promising candidate gene for breast cancer aggressiveness is MMP1, which belongs to a matrix metalloproteinase family associated with cell growth, metastasis, and the progression of different neoplasias, including breast cancer [53–56]. A recent paper by our group disclosed another gene of the same family, MMP2, as a key player in the progression of ductal carcinomas of the breast [57].
Some of the genes with the highest differential expression (fold change >|5| for histological grade, fold change >|3| for LVI) exhibited direct correlations between copy number changes and expression levels, such as gains affecting S100A8 and MCM10, which suggests that expression levels could be driven by CNAs [17, 58, 59]. We also detected a downregulation pattern for RERG, a gene from the family of the abovementioned deleted RERGL gene.
Regarding the LVI tumors, the most upregulated gene in the LVI-positive group compared to the negative group was C19orf33 which is also known as IMUP (immortalization upregulated protein), a gene previously shown to be overexpressed in lung, colon, and ovarian carcinoma cell lines [60, 61]. Another remarkable upregulated gene was LGALS7B, a basal cell marker which enhances metastasis to the lungs and bones in breast cancer [62]. Additionally, the upregulated MED1 gene, earlier reported in association with metastasis and therapy resistance in breast tumors [63], was uncovered as a relevant gene in a network enriched for genes in the categories of Cellular Growth and Proliferation and Hematological System Development and Function. A concordant pattern of association between CNAs and the expression profile was detected in our study for some genes already implicated in invasion and metastasis, such as gains and amplifications of MYCBPAP and MED1 [63, 64]. Furthermore, downregulated genes in tumors exhibiting LVI included MFI2, a gene associated with breast cancer metastasis when downregulated [65].
Conclusion
Therefore, we could delineate a group of genomic alterations and relevant genes (see Table 5) that seems to be associated with high-grade breast cancer as well as with LVI. We recognize that histologic grade and LVI are easily evaluated in routine histological analysis, but CNA screening upon the diagnosis of low-grade breast tumors could assist in the identification of potentially more aggressive tumors requiring further investigation. Our study has shed some light on the molecular players involved in two highly relevant prognostic factors and may enable a better understanding of the mechanisms of breast cancer aggressiveness.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Real-time qPCR validation of two small-scale rearrangements not previously reported in the literature. The graphs depict the copy number average of three replicates of each investigated gene in the breast tumor sample carrying the copy number alteration detected by array-CGH. (A) Real-time qPCR data showing a high number of copies of the HSD17B12 gene, confirming the 11p11.2 amplification detected in the MIC82 breast tumor sample. (B) Real-time qPCR data showing a high number of copies of the ADAMTS3, confirming the 4q13.3 amplification detected in the MIC149 breast tumor sample. (PPTX 53 kb)
Cellular diagram of the network created by the Ingenuity Pathway Analysis based on interactions of the set of differentially expressed genes detected in breast tumors positive for lymphovascular invasion compared to tumors without invasion. Red and green nodes represent up-regulated or down-regulated genes, respectively, in breast tumors positive for lymphovascular invasion in comparison to negative tumors (Table 4) (the intensity of the colors indicating the degree of deregulation); uncolored nodes represent genes automatically included in this network because they are biologically linked to the others based on scientific evidence. The functional categorization of this network revealed Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function; in this network, the MED1 gene appeared to be a key gene. (GIF 1765 kb)
(XLSX 928 kb)
(DOCX 35 kb)
(DOCX 22 kb)
(DOCX 17 kb)
(DOCX 14 kb)
(DOCX 36 kb)
(DOCX 21 kb)
Acknowledgments
This work was supported by the National Institute of Science and Technology in Oncogenomics (2008/57887-9), and FAPESP (2012/12139-5, 2012/21932-6, 2011/24007-9).
Conflicts of interest
None.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Kothari MS, Rusby JE, Agusti AA, MacNeill FA. Sentinel lymph node biopsy after previous axillary surgery: a review. Eur J Surg Oncol. 2012;38:8–15. doi: 10.1016/j.ejso.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 3.McGuire WL. Prognostic factors for recurrence and survival in human breast cancer. Breast Cancer Res Treat. 1987;10:5–9. doi: 10.1007/BF01806129. [DOI] [PubMed] [Google Scholar]
- 4.Foster RS. The biologic and clinical significance of lymphatic metastases in breast cancer. Surg Oncol Clin N Am. 1996;5:79–104. [PubMed] [Google Scholar]
- 5.Schoppmann SF, Bayer G, Aumayr K, Taucher S, Geleff S, Rudas M, et al. Group ABaCCS: prognostic value of lymphangiogenesis and lymphovascular invasion in invasive breast cancer. Ann Surg. 2004;240:306–12. doi: 10.1097/01.sla.0000133355.48672.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee AH, Pinder SE, Macmillan RD, Mitchell M, Ellis IO, Elston CW, et al. Prognostic value of lymphovascular invasion in women with lymph node negative invasive breast carcinoma. Eur J Cancer. 2006;42:357–62. doi: 10.1016/j.ejca.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer: I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–10. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 8.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Walker LC, Harris GC, Wells JE, Robinson BA, Morris CM. Association of chromosome band 8q22 copy number gain with high grade invasive breast carcinomas by assessment of core needle biopsies. Genes Chromosom Cancer. 2008;47:405–17. doi: 10.1002/gcc.20545. [DOI] [PubMed] [Google Scholar]
- 11.Natrajan R, Lambros MB, Rodríguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchió C, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–22. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 12.Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosom Cancer. 2006;45:1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 13.Bowcock AM. Invited review DNA copy number changes as diagnostic tools for lung cancer. Thorax. 2014;69:495–6. doi: 10.1136/thoraxjnl-2013-204681. [DOI] [PubMed] [Google Scholar]
- 14.Hirose Y, Sasaki H, Abe M, Hattori N, Adachi K, Nishiyama Y, et al. Subgrouping of gliomas on the basis of genetic profiles. Brain Tumor Pathol. 2013;30:203–8. doi: 10.1007/s10014-013-0148-y. [DOI] [PubMed] [Google Scholar]
- 15.Ueno T, Emi M, Sato H, Ito N, Muta M, Kuroi K, et al. Genome-wide copy number analysis in primary breast cancer. Expert Opin Ther Targets. 2012;16(Suppl 1):S31–5. doi: 10.1517/14728222.2011.636739. [DOI] [PubMed] [Google Scholar]
- 16.Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28:550–9. doi: 10.1016/j.tig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Natrajan R, Mackay A, Wilkerson PM, Lambros MB, Wetterskog D, Arnedos M, et al. Functional characterization of the 19q12 amplicon in grade iii breast cancers. Breast Cancer Res. 2012;14:R53. doi: 10.1186/bcr3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol. 2010;4:255–66. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. Oncology ASoC, Pathologists CoA: American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 20.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos AH, Silva AA, Mota LD, Olivieri ER, Prescinoti VC, Patrão D, et al. The value of a tumor bank in the development of cancer research in Brazil: 13 years of experience at the a c Camargo hospital. Biopreserv Biobank. 2012;10:168–73. doi: 10.1089/bio.2011.0032. [DOI] [PubMed] [Google Scholar]
- 22.Mueller O, Lightfoot S, Schroeder A (2004) RNA integrity number (RIN)—standardization of RNA quality control. BMC Mol Biol. 2006;317:3. [DOI] [PMC free article] [PubMed]
- 23.Dokidis A, Delenstarr G (2005) Use of Agilent Feature Extraction Software (v8.1)—QC report to evaluate microarray performance. Agilent Technologies, Santa Clara.
- 24.Diskin SJ, Eck T, Greshock J, Mosse YP, Naylor T, Stoeckert CJ, et al. STAC: a method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006;16:1149–58. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–92. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 28.Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, et al. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spellman P, Gray J. A new treasure in the breast cancer gene hunt. Nat Med. 2011;17:422–3. doi: 10.1038/nm0411-422. [DOI] [PubMed] [Google Scholar]
- 33.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurose K, Mine N, Doi D, Ota Y, Yoneyama K, Konishi H, et al. Novel gene fusion of COX6C at 8q22-23 to HMGIC at 12q15 in a uterine leiomyoma. Genes Chromosom Cancer. 2000;27:303–7. doi: 10.1002/(SICI)1098-2264(200003)27:3<303::AID-GCC11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Wang FL, Wang Y, Wong WK, Liu Y, Addivinola FJ, Liang P, et al. Two differentially expressed genes in normal human prostate tissue and in carcinoma. Cancer Res. 1996;56:3634–7. [PubMed] [Google Scholar]
- 36.Xiao GG, Zhou BS, Somlo G, Portnow J, Juhasz A, Un F, et al. Identification of F-box/LLR-repeated protein 17 as potential useful biomarker for breast cancer therapy. Cancer Genomics Proteomics. 2008;5:151–60. [PubMed] [Google Scholar]
- 37.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–86. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 38.Matthews JM, Lester K, Joseph S, Curtis DJ. LIM-domain-only proteins in cancer. Nat Rev Cancer. 2013;13:111–22. doi: 10.1038/nrc3418. [DOI] [PubMed] [Google Scholar]
- 39.Jönsson G, Staaf J, Vallon-Christersson J, Ringnér M, Holm K, Hegardt C, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulte I, Batty EM, Pole JC, Blood KA, Mo S, Cooke SL, et al. Structural analysis of the genome of breast cancer cell line ZR-75-30 identifies twelve expressed fusion genes. BMC Genomics. 2012;13:719. doi: 10.1186/1471-2164-13-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–84. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okabe T, Nakamura T, Nishimura YN, Kohu K, Ohwada S, Morishita Y, et al. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-d-aspartate receptor signaling. J Biol Chem. 2003;278:9920–7. doi: 10.1074/jbc.M208872200. [DOI] [PubMed] [Google Scholar]
- 44.Fernandes RJ, Hirohata S, Engle JM, Colige A, Cohn DH, Eyre DR, et al. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J Biol Chem. 2001;276:31502–9. doi: 10.1074/jbc.M103466200. [DOI] [PubMed] [Google Scholar]
- 45.Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10:2429–40. doi: 10.1158/1078-0432.CCR-0398-3. [DOI] [PubMed] [Google Scholar]
- 46.Molokwu CN, Adeniji OO, Chandrasekharan S, Hamdy FC, Buttle DJ. Androgen regulates ADAMTS15 gene expression in prostate cancer cells. Cancer Investig. 2010;28:698–710. doi: 10.3109/07357907.2010.489538. [DOI] [PubMed] [Google Scholar]
- 47.Visus C, Ito D, Dhir R, Szczepanski MJ, Chang YJ, Latimer JJ, et al. Identification of hydroxysteroid (17β) dehydrogenase type 12 (HSD17b12) as a CD8+ T-cell-defined human tumor antigen of human carcinomas. Cancer Immunol Immunother. 2011;60:919–29. doi: 10.1007/s00262-011-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srikrishna G. S100a8 and S100a9: new insights into their roles in malignancy. J Innate Immunol. 2012;4:31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berthier S, Nguyen MV, Baillet A, Hograindleur MA, Paclet MH, Polack B, et al. Molecular interface of S100a8 with cytochrome b558 and NADPH oxidase activation. PLoS One. 2012;7:e40277. doi: 10.1371/journal.pone.0040277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanamandra K, Alexeyev O, Zamotin V, Srivastava V, Shchukarev A, Brorsson AC, et al. Amyloid formation by the pro-inflammatory S100a8/A9 proteins in the ageing prostate. PLoS One. 2009;4:e5562. doi: 10.1371/journal.pone.0005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKiernan E, McDermott EW, Evoy D, Crown J, Duffy MJ. The role of S100 genes in breast cancer progression. Tumour Biol. 2011;32:441–50. doi: 10.1007/s13277-010-0137-2. [DOI] [PubMed] [Google Scholar]
- 52.Arai K, Takano S, Teratani T, Ito Y, Yamada T, Nozawa R. S100a8 and S100a9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr Cancer Drug Targets. 2008;8:243–52. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- 53.Pulukuri SM, Rao JS. Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int J Oncol. 2008;32:757–65. [PMC free article] [PubMed] [Google Scholar]
- 54.Schütz A, Schneidenbach D, Aust G, Tannapfel A, Steinert M, Wittekind C. Differential expression and activity status of MMP-1, MMP-2 and MMP-9 in tumor and stromal cells of squamous cell carcinomas of the lung. Tumour Biol. 2002;23:179–84. doi: 10.1159/000064034. [DOI] [PubMed] [Google Scholar]
- 55.Slattery ML, John E, Torres-Mejia G, Stern M, Lundgreen A, Hines L, et al. Matrix metalloproteinase genes are associated with breast cancer risk and survival: the breast cancer health disparities study. PLoS One. 2013;8:e63165. doi: 10.1371/journal.pone.0063165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fanjul-Fernández M, Folgueras AR, Fueyo A, Balbín M, Suárez MF, Fernández-García MS, et al. Matrix metalloproteinase Mmp-1a is dispensable for normal growth and fertility in mice and promotes lung cancer progression by modulating inflammatory responses. J Biol Chem. 2013;288:14647–56. doi: 10.1074/jbc.M112.439893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carraro DM, Elias EV, Andrade VP. Ductal carcinoma in situ of the breast: morphological and molecular features implicated in progression. Biosci Rep. 2013;34(1):e00090. doi: 10.1042/BSR20130077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rachidi SM, Qin T, Sun S, Zheng WJ, Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PLoS One. 2013;8:e57911. doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batra R, Harder N, Gogolin S, Diessl N, Soons Z, Jäger-Schmidt C, et al. Time-lapse imaging of neuroblastoma cells to determine cell fate upon gene knockdown. PLoS One. 2012;7:e50988. doi: 10.1371/journal.pone.0050988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JK, Ryll R, Ishizuka Y, Kato S. Identification of cDNAs encoding two novel nuclear proteins, IMUP-1 and IMUP-2, upregulated in SV40-immortalized human fibroblasts. Gene. 2000;257:327–34. doi: 10.1016/S0378-1119(00)00414-5. [DOI] [PubMed] [Google Scholar]
- 61.Ryoo ZY, Jung BK, Lee SR, Kim MO, Kim SH, Kim HJ, et al. Neoplastic transformation and tumorigenesis associated with overexpression of IMUP-1 and IMUP-2 genes in cultured NIH/3T3 mouse fibroblasts. Biochem Biophys Res Commun. 2006;349:995–1002. doi: 10.1016/j.bbrc.2006.08.137. [DOI] [PubMed] [Google Scholar]
- 62.Demers M, Rose AA, Grosset AA, Biron-Pain K, Gaboury L, Siegel PM, et al. Overexpression of galectin-7, a myoepithelial cell marker, enhances spontaneous metastasis of breast cancer cells. Am J Pathol. 2010;176:3023–31. doi: 10.2353/ajpath.2010.090876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moelans CB, van der Groep P, Hoefnagel LD, van de Vijver MJ, Wesseling P, Wesseling J, et al. Genomic evolution from primary breast carcinoma to distant metastasis: few copy number changes of breast cancer related genes. Cancer Lett. 2013;344(1):138–146. doi: 10.1016/j.canlet.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 64.Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, et al. The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic. 2009;10:982–93. doi: 10.1111/j.1600-0854.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bemmo A, Dias C, Rose AA, Russo C, Siegel P, Majewski J. Exon-level transcriptome profiling in murine breast cancer reveals splicing changes specific to tumors with different metastatic abilities. PLoS One. 2010;5:e11981. doi: 10.1371/journal.pone.0011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time qPCR validation of two small-scale rearrangements not previously reported in the literature. The graphs depict the copy number average of three replicates of each investigated gene in the breast tumor sample carrying the copy number alteration detected by array-CGH. (A) Real-time qPCR data showing a high number of copies of the HSD17B12 gene, confirming the 11p11.2 amplification detected in the MIC82 breast tumor sample. (B) Real-time qPCR data showing a high number of copies of the ADAMTS3, confirming the 4q13.3 amplification detected in the MIC149 breast tumor sample. (PPTX 53 kb)
Cellular diagram of the network created by the Ingenuity Pathway Analysis based on interactions of the set of differentially expressed genes detected in breast tumors positive for lymphovascular invasion compared to tumors without invasion. Red and green nodes represent up-regulated or down-regulated genes, respectively, in breast tumors positive for lymphovascular invasion in comparison to negative tumors (Table 4) (the intensity of the colors indicating the degree of deregulation); uncolored nodes represent genes automatically included in this network because they are biologically linked to the others based on scientific evidence. The functional categorization of this network revealed Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function; in this network, the MED1 gene appeared to be a key gene. (GIF 1765 kb)
(XLSX 928 kb)
(DOCX 35 kb)
(DOCX 22 kb)
(DOCX 17 kb)
(DOCX 14 kb)
(DOCX 36 kb)
(DOCX 21 kb)