Abstract
Acidocin B, a bacteriocin produced by Lactobacillus acidophilus M46, was originally reported to be a linear peptide composed of 59 amino acid residues. However, its high sequence similarity to gassericin A, a circular bacteriocin from Lactobacillus gasseri LA39, suggested that acidocin B might be circular as well. Acidocin B was purified from culture supernatant by a series of hydrophobic interaction chromatographic steps. Its circular nature was ascertained by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry and tandem mass spectrometry (MS/MS) sequencing. The peptide sequence was found to consist of 58 amino acids with a molecular mass of 5,621.5 Da. The sequence of the acidocin B biosynthetic gene cluster was also determined and showed high nucleotide sequence similarity to that of gassericin A. The nuclear magnetic resonance (NMR) solution structure of acidocin B in sodium dodecyl sulfate micelles was elucidated, revealing that it is composed of four α-helices of similar length that are folded to form a compact, globular bundle with a central pore. This is a three-dimensional structure for a member of subgroup II circular bacteriocins, which are classified based on their isoelectric points of ∼7 or lower. Comparison of acidocin B with carnocyclin A, a subgroup I circular bacteriocin with four α-helices and a pI of 10, revealed differences in the overall folding. The observed variations could be attributed to inherent diversity in their physical properties, which also required the use of different solvent systems for three-dimensional structural elucidation.
INTRODUCTION
Circular bacteriocins are antimicrobial peptides that are ribosomally synthesized by bacteria and are posttranslationally modified to release a leader peptide and form a peptide bond between the N and C termini. These peptides exhibit antimicrobial activity against a broad range of Gram-positive bacteria, including Listeria spp. and Clostridium spp., which are common pathogens causing food-borne diseases (1). In addition, the circular nature of these bacteriocins imparts enhanced stability against proteolytic degradation and denaturation due to extreme temperature and pH conditions relative to linear forms (2). They thus serve as promising alternatives to traditional antimicrobial agents for food, medical, and industrial applications (3).
A number of circular bacteriocins that are composed of 58 to 70 amino acid residues have been identified to date, including enterocin AS-48 (4), gassericin A (5), circularin A (6), butyrivibriocin AR10 (7), uberolysin (8), carnocyclin A (9), lactocyclicin Q (10), garvicin ML (11), leucocyclicin Q (12), amylocyclicin (13), and aureocyclicin 4185 (14). Another peptide exhibiting N- to C-terminal cyclization is subtilosin A (15). It is, however, considered a member of the sactipeptides, which represent a class of peptides containing cross-links between cysteine sulfurs and α-carbons (16). The structure and genetics of circular bacteriocins were reviewed previously (2). More recently, a review on the biosynthesis and mode of action of these circular bacteriocins was released (1).
Circular bacteriocins are classified into two subgroups. The main difference between the two subgroups is the calculated isoelectric point (pI) of the mature peptide (2, 17). Members of subgroup I have high pI values (∼10), while those in subgroup II have low pI values (∼7 or lower). Among the circular bacteriocins identified so far, carnocyclin A and enterocin AS-48 (both subgroup I) have been structurally characterized through the elucidation of their nuclear magnetic resonance (NMR) solution structures (17, 18). A common saposin-like fold was observed in these structures and was predicted to be a conserved motif among the members of this group. The saposin-like proteins are known to interact with lipids and are composed of 4 or 5 adjacent α-helices that are folded into two leaves (19). We could not find a three-dimensional structure published for a circular bacteriocin belonging to subgroup II. It has, however, been postulated that the known members of this group, gassericin A and butyrivibriocin AR10, are composed of four α-helices that are also folded to resemble the structure of the saposins (17). Subgroup I members, having high pI values, have clusters of positively charged residues, which have been suggested to mediate binding to negatively charged bacterial membranes (20, 21). These clusters of basic residues may not be observed for subgroup II circular bacteriocins, since they contain fewer positively charged amino acid residues (1).
Acidocin B is a bacteriocin encoded by plasmid pCV461 from Lactobacillus acidophilus M46 (22, 23). It was previously reported to inhibit certain Gram-positive bacteria, such as Clostridium sporogenes C22/10, Listeria monocytogenes L2, and Brochothrix thermosphacta 39 (24). Based on an amino acid sequence deduced from DNA analysis and on amino acid composition as examined through acid hydrolysis, derivatization with phenylisothiocyanate, and quantification by reverse-phase high-performance liquid chromatography (RP-HPLC), mature acidocin B was previously predicted to be a linear peptide composed of 59 amino acids (22). However, due to its high amino acid sequence similarity (98%) with gassericin A, a circular bacteriocin from L. gasseri LA39 (5), it seems likely that acidocin B is also circular. This hypothesis is further supported by the fact that previous efforts to determine the N-terminal amino acid residue of acidocin B were unsuccessful (22). Furthermore, preliminary characterization of acidocin B revealed that it is highly stable to extreme pH and temperature conditions, properties commonly exhibited by circular peptides (24). In case the posttranslational modification of the acidocin B precursor is similar to that of gassericin A, a mature circular peptide composed of 58 amino acids would be produced, instead of 59.
In this study, we demonstrated the circular nature of acidocin B by mass spectrometry and elucidated its three-dimensional solution structure by NMR. This is a structure for a subgroup II circular bacteriocin. Acidocin B indeed consists of 58 amino acids and contains 4 α-helices that form a compact, globular bundle with a central pore. Furthermore, we report the complete biosynthetic gene cluster responsible for acidocin B production and maturation and show that it is highly similar to that of gassericin A.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. acidophilus M46, the strain that produces acidocin B, was grown in modified de Man, Rogosa, and Sharpe (MRS) medium (BD Difco, Sparks, MD) without shaking at 37°C. The modified growth medium contained twice the recommended amount of MRS powder (110 g) per liter and was further supplemented with 20 g/liter dextrose. Carnobacterium divergens LV13 (25), the indicator strain for the activity assay, was grown at 25°C in all-purpose Tween (APT) broth (BD Difco). Both strains were stored in the form of glycerol stocks at −80°C.
Plasmid isolation and sequencing.
Plasmid DNA was isolated using the GeneJET plasmid miniprep kit from Fermentas (Fermentas Canada Inc., Burlington, ON, Canada) as described by the manufacturer, except that cells were first incubated with 5 mg of lysozyme per ml for 1 h at 37°C before the cell lysis step. The acidocin B gene cluster on plasmid pCV461 was sequenced by primer walking. Sequencing reactions were performed using the ABI BigDye version 3.1 Terminator sequencing kit (Applied Biosystems, Foster City, NJ) and run on an ABI 3730 DNA analyzer (Applied Biosystems).
Bacteriocin activity assay.
Spot-on-lawn assays were used to determine the active fractions from the different purification steps employed. Briefly, 100 μl of an overnight culture of the indicator strain was used to inoculate 5 ml molten soft agar (0.75% [wt/vol]). The soft agar was then overlaid on an APT agar plate. Ten-microliter samples of purification fractions were spotted on the solidified soft agar. Upon drying, the plates were incubated at 25°C overnight and were examined for the appearance of zones of inhibition.
Purification of acidocin B.
A three-step activity-guided procedure was used to purify acidocin B from the supernatant of a 1-liter culture of L. acidophilus M46 (1% [vol/vol] inoculum) that was grown for 24 h at 37°C. The culture was centrifuged (10,000 × g, 10 min, 4°C), and the supernatant was applied to a column containing 80 g Amberlite XAD-16 resin (Sigma-Aldrich, St. Louis, MO). After the sample had been loaded, the resin was washed with 500 ml deionized water, 500 ml 20% isopropanol (IPA), and 750 ml 40% IPA. A 500-ml 80% IPA solution with 0.1% trifluoroacetic acid (TFA) was then used to elute acidocin B. A constant flow rate of 10 ml/min was used for all solutions. The active fraction was concentrated to 100 ml and was further purified using a Bond Elut C8 10-g, 60-ml cartridge (Agilent, Mississauga, ON, Canada). The silica was first washed with 50 ml methanol and 100 ml deionized water. After the sample had been loaded, the cartridge was consequently washed with 50 ml each of 30% ethanol, 30% acetonitrile, 20% IPA, and 100 ml 40% IPA. Elution was done using 50 ml 80% IPA with 0.1% TFA. The flow rate was maintained at 5 ml/min. The eluted fraction was concentrated to 15 ml and subjected to reverse-phase high-performance liquid chromatography (RP-HPLC) using a C4 protein column (10-μm particle size, 22 mm by 250 mm; Vydac 214TP1022). Five milliliters of sample was injected per run. Detection was done at 220 nm. A flow rate of 8 ml/min was employed with the use of solvent A (water with 0.1% TFA) and solvent B (acetonitrile with 0.1% TFA). Solvent B was initially increased from 10% to 40% over the course of 5 min, maintained at 40% for 8 min, gradually increased to 86% for 30 min, ramped up to 95% for 2 min, and held at 95% for another 2 min. Acidocin B eluted at 44 min (see Fig. S1 in the supplemental material). The combined acidocin B fractions were concentrated in vacuo to remove the organic solvent, lyophilized, and stored at −20°C.
MALDI-TOF MS.
Two-layer sample preparation (26) was performed with the use of sinapinic acid as the matrix. The data were acquired using a Perspective Biosystems Voyager Elite matrix-assisted laser desorption ionization time-of-flight mass spectrometer (MALDI-TOF MS) in positive-ion mode.
Proteolytic digestion and LC-MS/MS peptide sequencing.
A solution of acidocin B (60 mM; 100 μl) was prepared in 0.1 M ammonium bicarbonate. It was digested with trypsin (sequencing grade; Promega, Madison, WI) overnight at 37°C, with an enzyme-to-peptide molar ratio of 1:20. A 20-μl fraction of the overnight digest was treated with 1 μl of a 0.5-μg/μl solution of chymotrypsin (Roche, Indianapolis, IN), and the sample was further digested for 3 h. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to analyze the obtained fragments. A nanoAcquity column (100-Å pore size, 75 μm by 15 cm, 3-μm Atlantis dC18; Waters, MA) on a nanoAcquity ultraperformance liquid chromatograph (Waters, MA) was used to separate the fragments. A flow rate of 350 nl/min was employed with the use of a linear water-acetonitrile gradient (0.1% formic acid). MS/MS data were obtained from a quadrupole-time of flight Premier MS (Micromass, United Kingdom). The data were processed using PEAKS 5.1 software (Bioinformatics Solutions, Waterloo, ON, Canada) (27).
Circular dichroism.
Acidocin B was dissolved in 6 mM sodium dodecyl sulfate (SDS) or 6 mM N-dodecylphosphocholine (DPC) at a concentration of 0.4 mg/ml. Circular dichroism (CD) spectra of acidocin B in SDS and DPC micelles at 20°C were recorded on an Olis DSM 17 CD spectrophotometer (Olis) with the use of a quartz cell (0.2-mm path length). Five scans were run for each sample at 1-nm increments from 185 to 250 nm. Spectra of solvent reference samples were subtracted from the respective peptide samples, and the percent α-helicity was calculated as (3,000 − θ222)/39,000 (28).
NMR spectroscopy.
Acidocin B was dissolved in 350 μl of an 80 mM deuterated SDS (9:1 H2O-D2O) solution to a final concentration of ∼1 mM with a pH of ∼6. It was then transferred into a 5-mm D2O-matched Shigemi tube. 4,4-Dimethyl-4-silapentane-1-sulfonic acid was added to a final concentration of 0.01% (wt/vol) for referencing. One-dimensional 1H NMR and two-dimensional homonuclear 1H-1H total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) experiments were run at 27°C on a Varian VNMRS 700-MHz spectrometer fitted with a triple-resonance HCN cryoprobe, z-axis pulsed-field gradients, and VNMRJ 3.2 as a host control. For both experiments, a 10,000-Hz sweep width, an 80-ms mixing time, 512 complex points for the indirectly detected dimension, and 4,882 real and imaginary points for the directly detected dimension were used. The cumulative scans for each point of acquisition were 32 and 64 for TOCSY and NOESY, respectively. Suppression of the water signal was achieved by presaturation during the relaxation delay. NMRPipe (29) and NMRView (30) were used for data processing and analysis. Chemical shift assignment (see Table S1 in the supplemental material) was done manually based on a previously described procedure (31, 32).
Structure calculations.
CYANA 2.1 (33) was used to calculate the structure of acidocin B by using a combination of manually and automatically assigned nuclear Overhauser effect (NOE) cross peaks. Seven cycles were done with 10,000 steps per cycle. A total of 909 NOE cross peaks were used for the final calculation, 558 of which were short range, 296 of which were medium range, and 55 of which were long range. PyMOL (34) was used to generate the figures for the three-dimensional structure of acidocin B. Adaptive Poisson-Boltzmann software (APBS) was used for the electrostatic surface calculations (35).
Sequence alignment, homology modeling, topology prediction, and construction of a phylogenetic tree.
Clustal W (36) was used for sequence alignment. Three-dimensional structure homology modeling was done using the SWISS-MODEL server (37), wherein the sequences of gassericin A and butyrivibriocin AR10 were fitted onto the structure of acidocin B. Similar sequences were identified through BLAST (NCBI) (38) using the acidocin B precursor peptide sequence as the query and a threshold of 40% identity. The number of putative transmembrane domains was deduced using the SOSUI program (39). Phylogenetic analysis was done through the maximum likelihood method using MEGA6 software (40).
Data bank accession numbers.
The GenBank accession number for the nucleotide sequence of the acidocin B gene cluster reported in this paper is KP728900. The coordinates for the calculated structure were deposited in the Protein Data Bank (accession number 2MWR), while the chemical shift assignments were deposited in the Biological Magnetic Resonance Data Bank (accession number 25352).
RESULTS
Acidocin B gene cluster.
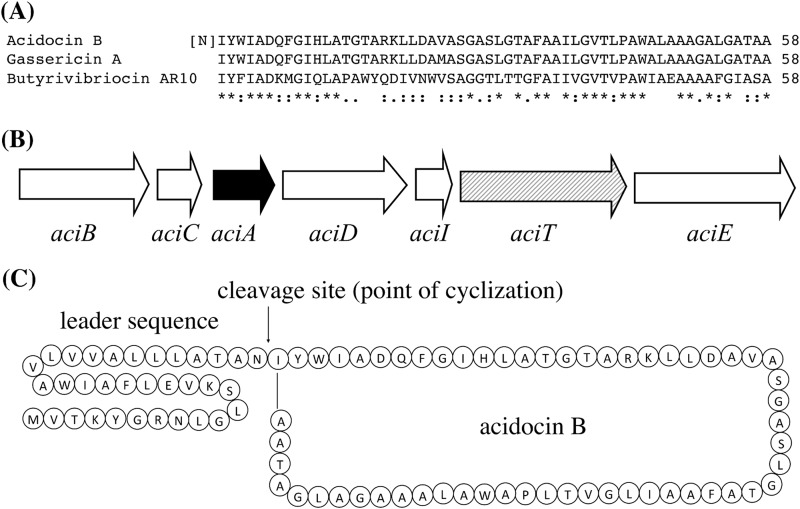
The amino acid sequence of acidocin B is highly similar to that of gassericin A, a circular bacteriocin that is encoded by a gene cluster of approximately 3.3 kb (Fig. 1A) (41). Previously, a nucleotide sequence of 2.2 kb around the structural acidocin B gene on plasmid pCV461 was determined (22). Therefore, the region surrounding the acidocin B structural gene was resequenced, and a nucleotide sequence of 3,537 bp was obtained. Analyses of the 3.5-kb nucleotide sequence revealed that the acidocin B structural gene is part of a gene cluster, aciBCADITE, that is similar to that of gassericin A (Fig. 1B). The characteristics of the proteins encoded by this gene cluster are listed in Table 1. Except for AciT, an ATP-binding protein, all other putative proteins encoded by the acidocin B gene cluster contain putative membrane-spanning domains. A BLAST search showed that a protein encoded immediately downstream of aciA, AciD, belongs to the DUF95 family of membrane proteins. The presence of genes encoding proteins belonging to DUF95 is a characteristic feature of circular bacteriocin gene clusters (42). AciD contains 162 amino acids and is larger than the putative 114-amino-acid peptide previously described to be encoded by an open reading frame (ORF) immediately downstream of aciA (22). However, the smaller size observed previously for this ORF was likely the result of a sequence error. Based on sequence homology, AciI is likely involved in immunity and AciTE may form the secretion apparatus for acidocin B (41, 43). The amino acid sequences of AciBCADITE show that these proteins are highly similar, and sometimes 100% identical, to their homologs in the gassericin A gene cluster (Table 1). The high similarities between the gene clusters of acidocin B and gassericin A and their respective encoded proteins strongly suggest that acidocin B belongs to the group of circular bacteriocins.
FIG 1.
(A) Sequence alignment of subgroup II circular bacteriocins using Clustal W (36). Conserved, conservative, and semiconservative substitutions are indicated by asterisks, colons, and semicolons, respectively. The previously proposed N-terminal amino acid for acidocin B is in brackets (22). (B) Schematic representation of the acidocin B gene cluster. The solid arrow indicates the structural bacteriocin gene aciA; the hatched arrow indicates aciT, encoding an ATP-binding protein; open arrows indicate aciB, aciC, aciD, aciI, and aciE, encoding proteins containing putative transmembrane domains. (C) Acidocin B precursor peptide. The cleavage site during maturation is indicated.
TABLE 1.
Characteristics of predicted proteins encoded by the acidocin B gene clustera
| Protein | Size (aa) | TM | Function | No. of identical amino acids/total (% identity) relative to gassericin A gene cluster homologs |
|---|---|---|---|---|
| AciB | 174 | 5 | Unknown | 173/174 (99.4) |
| AciC | 60 | 2 | Unknown | 60/60 (100) |
| AciA | 91 | 2 | Acidocin B precursor | 88/91 (96.7) |
| AciD | 162 | 4 | Unknown, DUF95 family | 160/162 (98.8) |
| AciI | 53 | 1 | Immunity | 53/53 (100) |
| AciT | 226 | 0 | ATP-binding protein | 225/226 (99.6) |
| AciE | 212 | 6 | Membrane transporter | 209/212 (98.6) |
aa, amino acids; TM, number of putative transmembrane domains.
Purification and molecular mass.
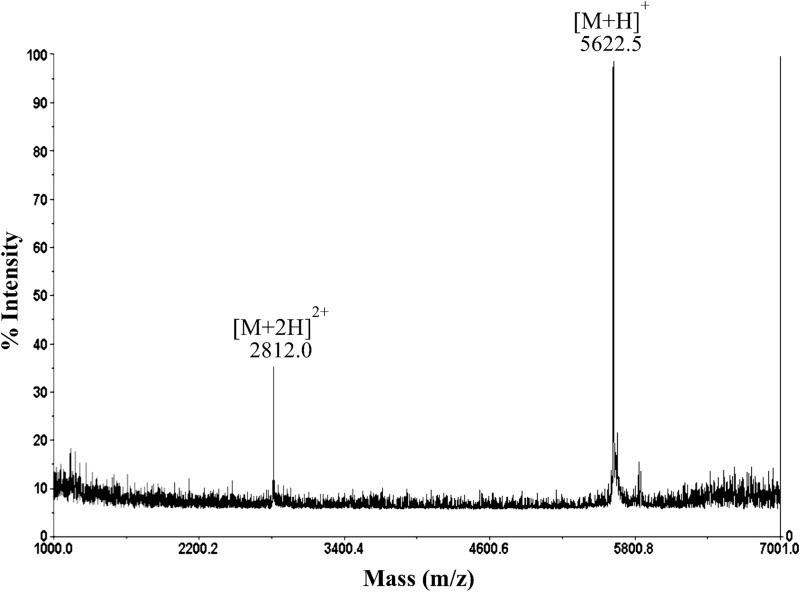
To confirm the circular nature of acidocin B and to investigate whether it consists of the same number of amino acids as gassericin A, acidocin B was isolated and characterized. Acidocin B was purified through a series of hydrophobic interaction chromatographic steps with a yield of ∼2.5 mg per liter. MALDI-TOF mass spectrometry registered a strong clean peak at 5,622.5 m/z (Fig. 2). This supported the circular nature of acidocin B, as the corresponding mass is 18 Da lower than the theoretical molecular mass of the linear peptide of 58 amino acids. This mass difference corresponds to the loss of one water molecule, which is expected during N- to C-terminal cyclization. Furthermore, the observed mass confirmed that the mature acidocin B is composed of 58 amino acids, contradicting the previously reported length of 59 amino acids (22).
FIG 2.
MALDI-TOF mass spectrum of acidocin B showing singly and doubly charged species consistent with an average molecular mass of 5,621.5 Da.
MS/MS sequencing analysis.
Acidocin B was digested with trypsin and chymotrypsin, and the resulting fragments were separated and analyzed by LC-MS/MS. A sequence coverage of 94.8% was achieved. Significant fragments obtained are presented in Table 2. The N- and C-terminal amino acid residues that are proposed to be involved in the cyclization of the peptide were observed in fragment 9. This confirmed the circular nature of acidocin B through peptide bond formation between the two terminal residues, as well as the site at which the precursor peptide is cleaved (Fig. 1C).
TABLE 2.
MS/MS sequences of fragments of acidocin B after trypsin-chymotrypsin digestion
| Fragment | Linear sequence | Observed m/z | Charge | Calculated m/z |
|---|---|---|---|---|
| 1 | --WIADQF | 779.388 | 1 | 779.365 |
| 2 | ---IADQFGIHL | 507.276 | 2 | 1,013.534 |
| 3 | ---IADQFGIHLATGT | 672.360 | 2 | 1,343.688 |
| 4 | ------------------KLLDAVASGASLGTAF | 760.929 | 2 | 1,520.825 |
| 5 | -------------------LLDAVASGASLGTAF | 696.886 | 2 | 1,392.730 |
| 6 | -----------------------------------AILGV | 472.290 | 1 | 472.306 |
| 7 | --------------------------------------GVTLPAW | 743.447 | 1 | 743.401 |
| 8 | ---------------------------------------------ALAAAGAL | 657.446 | 1 | 657.386 |
| 9 | IY---------------------------------------------AAAGALGATAA | 1,120.619 | 1 | 1,120.592 |
| Complete sequencea | IYWIADQFGIHLATGTARKLLDAVASGASLGTAFAAILGVTLPAWALAAAGALGATAA | |||
Ala-17, Arg-18, and Ala-35 were inferred from the deduced amino acid sequence from DNA sequence analysis.
Solubility and circular dichroism analysis.
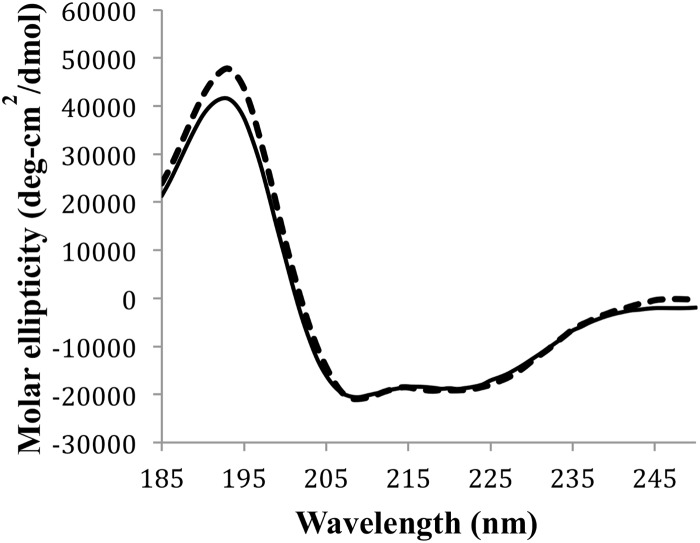
CD spectroscopy was used to screen solvent conditions suitable for further characterization of acidocin B via NMR spectroscopy. A previous report revealed that the high degree of hydrophobicity of gassericin A rendered it insoluble in aqueous solutions (44). Similarly, acidocin B could not be dissolved at a concentration required for NMR studies using the buffered aqueous solutions and slightly polar organic solvents tested; hence, the use of detergent micelles was considered. Both SDS and DPC solutions completely dissolved acidocin B. CD spectroscopic data revealed that the peptide exhibited 56% and 57% α-helicity in SDS and DPC micelles, respectively (Fig. 3). SDS was chosen for the NMR analysis of acidocin B, as it may better mimic the membranes of target bacteria (45).
FIG 3.
CD profile of acidocin B in SDS (solid line) and DPC (dashed line) micelles. The peptide exhibited similar α-helical contents in both micelles.
NMR solution structure.
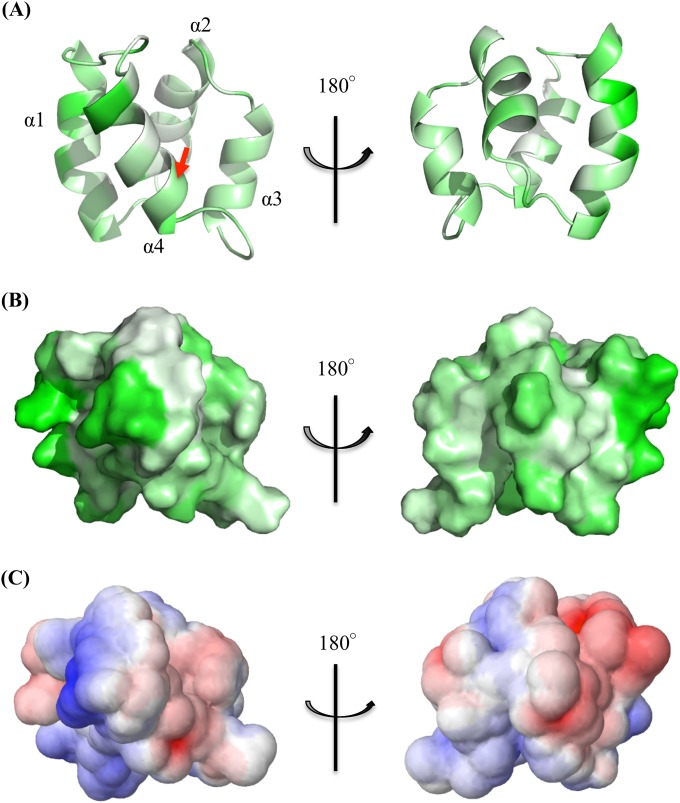
Isotopic labeling of acidocin B was attempted by growing L. acidophilus M46 in 13C-, 15N-enriched media, including Celtone complete medium (Cambridge Isotope Laboratories, Tewksbury, MA), Bioexpress-1000 (Cambridge Isotope Laboratories), and a manually prepared labeling medium previously used for another Lactobacillus strain (46). Labeling attempts were unsuccessful, as the producer organism did not grow well in the aforementioned media. However, relatively dispersed proton chemical shifts and a significant number of NOE cross peaks were obtained from the two-dimensional homonuclear TOCSY and NOESY experiments for the unlabeled peptide. These data facilitated the elucidation of the three-dimensional solution structure of acidocin B in SDS micelles. The TOCSY and NOESY spectra were overlaid and the sequential resonance assignments were determined based on the Hαi–HNi+1 cross peaks. The chemical shift assignments and NOE peak list were inputted into CYANA 2.1 (33), and a family of 20 calculated structures was obtained (see Fig. S2 in the supplemental material). These structures had no ϕ or ψ backbone angles in the disallowed region of the Ramachandran plot. The structural statistics calculated by CYANA and the data from the generated Ramachandran plot are presented in Table S2 in the supplemental material. The data indicate that acidocin B is highly structured, which is in agreement with the results of the CD spectral analysis. The calculated structure contains four right-handed α-helices. The helices are folded to form a compact globular bundle with a central pore (Fig. 4). Helix 1 is the longest helix, with 13 amino acid residues, extending from Gly15 to Ala27. The residues encompassing helices 2 and 3 are Gly31 to Val40 (10 residues) and Ala44 to Leu52 (9 residues), respectively. Helix 4 is composed of 11 residues, Ala56 to Phe8, and contains the linkage of the N and C termini. The four helices are separated by loops composed of 3 to 6 residues. When the relatively flexible loops are excluded from the root mean square deviation (RMSD) calculation, the RMSD of the backbone atoms decreases from 1.48 Å to 1.26 Å.
FIG 4.
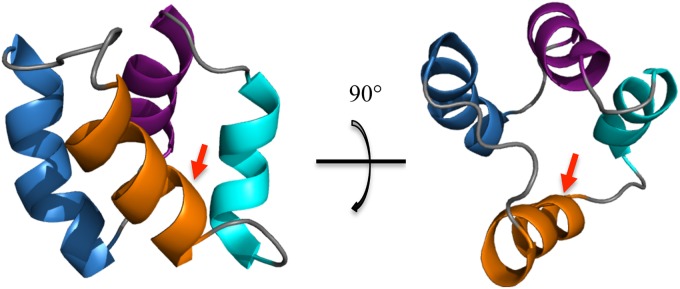
NMR solution structure of acidocin B (PDB code 2MWR): helix 1 is in blue, helix 2 is in purple, helix 3 is in cyan, and helix 4 is in orange. The arrow indicates the linkage of the N and C termini.
Helices 1, 2, and 4 of acidocin B are amphipathic, i.e., hydrophobic amino acid residues are oriented toward the core of the molecule, while the relatively hydrophilic residues are located on the surface. The amphipathic character is most evident in helix 1, as can be seen in Fig. 5A. Helices 2 and 4 are less amphipathic since they are mainly composed of hydrophobic residues. Helix 3, on the other hand, is completely hydrophobic. The hydrophobic surface map (Fig. 5B) shows that acidocin B has two distinct hydrophilic patches, but significant portions of the surface are hydrophobic. The electrostatic potential surface map of acidocin B (Fig. 5C) shows several anionic and cationic patches, while most of the exposed surface is uncharged. This was expected, since the primary sequence of acidocin B has only 2 anionic (Asp6 and Asp22) and 3 cationic (His11, Arg18, and Lys19) amino acid residues. The demonstrated surface properties could explain the difficulty encountered in dissolving the peptide in buffered aqueous solutions and slightly polar organic solvents.
FIG 5.
(A) Solution structure of acidocin B showing the amphipathicity of the helices. Hydrophobic residues are in green, while hydrophilic residues are in white. The arrow indicates the linkage of the N and C termini. (B) Hydrophobic surface map as generated from PyMOL (34). (C) Electrostatic potential surface map calculated using the APBS functionality of the PDB2PQR (version 1.8) online pipeline (35). Cationic regions are shown in red, while anionic regions are in blue.
Homology modeling and phylogenetic tree of subgroup II circular bacteriocins.
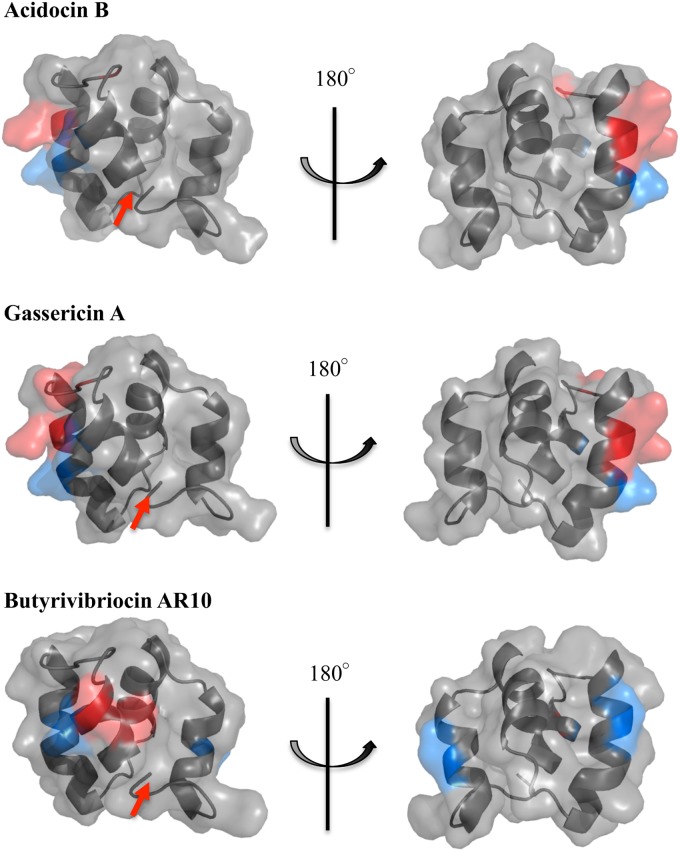
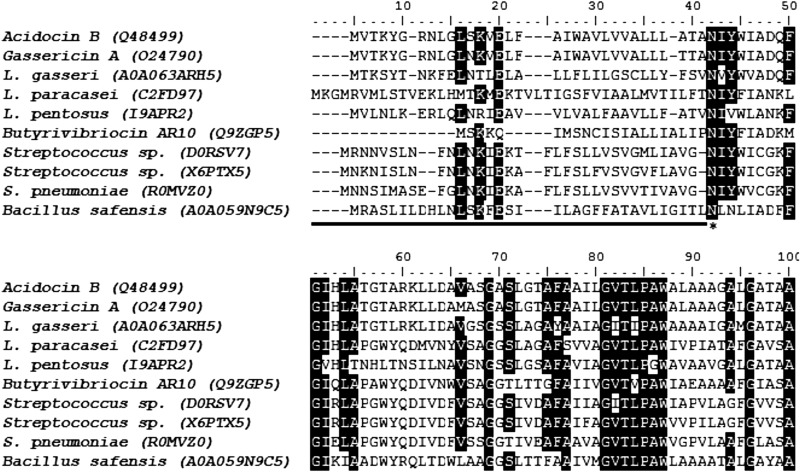
The elucidated acidocin B structure was then used for homology modeling of gassericin A and butyrivibriocin AR10, the amino acid sequences of which are 98% and 47% identical to that of acidocin B, respectively. The generated model structures revealed that these three subgroup II circular bacteriocins have similar folding and surface properties (Fig. 6; also, see Fig. S3 in the supplemental material). Other putative members of this subgroup were identified through BLAST (38) analysis. At least 7 putative subgroup II circular bacteriocins that are at least 40% identical to the acidocin B precursor peptide were identified. Alignment of their sequences identified a conserved asparaginyl cleavage site during bacteriocin maturation (Fig. 7). Considering this cleavage site, phylogenetic analysis of the mature peptides revealed that members of this subgroup could be classified further into 2 subclades (see Fig. S4 in the supplemental material).
FIG 6.
Predicted structures of gassericin A and butyrivibriocin AR10 derived from homology modeling (SWISS-MODEL) (37) using the structure of acidocin B as the template. Basic residues are shown in red, and acidic residues are in blue. The arrow indicates the N- to C-terminal linkage.
FIG 7.
Multiple-sequence alignment of known and putative (indicated by source organisms) subgroup II circular bacteriocin precursors using Clustal W (36). The known circular bacteriocins are acidocin B, gassericin A, and butyrivibriocin AR10. The UniProt accession numbers are in parentheses. The region corresponding to leader peptide sequences is underlined, and the highly conserved asparaginyl cleavage site is marked by an asterisk. Conserved residues (similarity threshold of 80%) are highlighted in black.
DISCUSSION
Acidocin B was previously reported to be a linear peptide that is composed of 59 amino acid residues (22). However, MALDI-TOF mass spectrometry and MS/MS sequencing revealed that it is circular and is composed of 58 residues. Furthermore, the gene cluster responsible for its production and maturation was found to be very similar to that encoding gassericin A, a circular bacteriocin from L. gasseri LA39 (5). To date, only the three-dimensional structures of circular bacteriocins enterocin AS-48 and carnocyclin A have been published (17, 18). These structures facilitated a better understanding of the mechanism of action of circular bacteriocins. The localized positive charges on the surfaces of these bacteriocins, as revealed by the elucidated structures, have been implicated in an initial binding interaction of the bacteriocins to anionic phospholipids in the cell membrane of target organisms (9, 20). Mode of action studies further revealed that upon binding and membrane permeabilization, enterocin AS-48 creates pores that cause the discharge of ions and low-molecular-weight substances (47). On the other hand, carnocyclin A specifically transports anions through the membrane (48). Enterocin AS-48 and carnocyclin A, however, are both subgroup I circular bacteriocins. Members of this group exhibit physical properties that are very different from those of subgroup II circular bacteriocins, to which acidocin B belongs. Their characteristic high pI values (∼10) and cationic surface are not exhibited by acidocin B. These differences may indicate some variations in their mode of action. Knowing the structure of a representative member of subgroup II circular bacteriocins could shed some light on their mode of action.
In order to assess the similarities and differences between the two circular bacteriocin subgroups, the structure of acidocin B was compared with that of carnocyclin A. The two bacteriocins are of similar lengths, as acidocin B is composed of 58 residues, while carnocyclin A is comprised of 60 residues. Sequence alignment of carnocyclin A and acidocin B using Clustal W (36) revealed that the two peptides have very low amino acid sequence identity (17%). Subgroup I circular bacteriocins exhibit low sequence similarity among themselves yet were proposed to display a common overall saposin-like fold, as was demonstrated for carnocyclin A and enterocin AS-48 (17, 18). This proposal was further extended to subgroup II circular bacteriocins. Previous secondary structure predictions using Jpred3 (49) and PSIPRED (50, 51) servers suggested that subgroup II circular bacteriocins contain 4 α-helices of similar lengths (17). This was indeed observed for acidocin B, approximately at the proposed positions. Circular dichroism revealed that acidocin B has a helical content of 56% in SDS micelles, while carnocyclin A was previously reported to be approximately 36% α-helical in water and 52% α-helical in 50% trifluoroethanol, which is a structure-inducing solvent (9). The estimated α-helical content was confirmed by NMR analysis, since both acidocin B and carnocyclin A were indeed composed of a bundle of 4 helices. However, the overall fold of acidocin B did not precisely overlie that of carnocyclin A (data not shown). The observed differences could be attributed to a number of factors, such as the solvent that was used for structural elucidation. The solution structure of carnocyclin A was obtained using water as the solvent, while for acidocin B, a membrane-mimicking SDS micelle was employed. Studies have shown that certain peptides undergo conformational changes from a free state in water to a membrane-bound form in membrane mimetic solvents (52, 53, 54). The structure of carnocyclin A in water shows that helix 3 is almost perpendicular to helix 1 (17), while helix 1 and 3 of acidocin B are almost parallel. It was not possible to obtain the NMR solution structure of acidocin B in water due to solubility issues. Acidocin B was initially soluble in aqueous medium at a low concentration, since isolation and purification were done in an aqueous environment. However, the solubility of the concentrated peptide in water was not sufficient for NMR analysis, and hence comparison of its structure with carnocyclin A using the previously used solvent conditions was not possible.
The stark contrast in the physical properties of acidocin B and carnocyclin A could also explain the variations observed in their overall folding. Aside from the very low sequence similarity, one of the major differences between the two bacteriocins is the amphipathicity of their helices. All of the four helices of carnocyclin A are amphipathic, while acidocin B has only 1 amphipathic and 2 weakly amphipathic helices. The surface features of the two bacteriocins are also different. Acidocin B has a highly hydrophobic surface, which is not the case for carnocyclin A. A characteristic feature of carnocyclin A which is shared among subgroup I circular bacteriocins is the high number of basic residues and a prominent positively charged surface (17). Since acidocin B has only 3 basic residues, the presence of a highly cationic surface was not expected. Surface analysis revealed that it has both positive and negative patches but is mainly composed of uncharged, hydrophobic surfaces, which could signify that the initial recognition of acidocin B on the cell membrane could be attributed mainly to hydrophobic interaction.
Aside from acidocin B, there are two other known circular bacteriocins that belong to subgroup II, namely, gassericin A and butyrivibriocin AR10. Gassericin A is produced by L. gasseri LA39, while butyrivibriocin AR10 is produced by Butyrivibrio fibrisolvens AR10. The bacteriocin reutericin 6 from L. reuteri LA6 was previously thought to be another subgroup II circular bacteriocin but was later found to be identical to gassericin A (55). Acidocin B shares 98% sequence identity to gassericin A, differing only in residue 24, where acidocin B has valine and gassericin A has methionine (Fig. 1A). A previous study established the insolubility of gassericin A in aqueous solution (44), similar to what was encountered with acidocin B. In the same study, CD data showed that gassericin A is α-helical in 60% isopropanol. The amino acid sequence of butyrivibriocin AR10, on the other hand, is 47% identical to that of acidocin B (Fig. 1A). Given the sequence similarity among the known subgroup II circular bacteriocins, homology modeling using the SWISS-MODEL server (37) was done. As expected, gassericin A and acidocin B have similar electrostatic and hydrophobic surface properties (Fig. 6; also, see Fig. S3 in the supplemental material). The hydrophobicity of the surface of the butyrivibriocin AR10 model is also similar to that of acidocin B. It also has minimal charges on the surface, but unlike gassericin A and acidocin B, where the charged residues are located in helices 1 and 4, butyrivibriocin AR10 has an additional acidic residue in helix 3.
Although only three subgroup II circular bacteriocins have been characterized, putative genes encoding other members of this subgroup are more widespread. BLAST (38) analysis identified at least 7 putative subgroup II circular bacteriocins that show at least 40% identity to the acidocin B precursor peptide. These putative bacteriocin-like peptides are encoded by species within the genera Lactobacillus, Streptococcus, and Bacillus. Sequence alignment of these putative bacteriocins revealed that the asparaginyl cleavage site identified for gassericin A, acidocin B, and butyrivibriocin AR10 is totally conserved among putative subgroup II circular bacteriocins (Fig. 7). This suggests that an endopeptidase that specifically cleaves C-terminally to this highly conserved asparagine residue may be responsible for the release of the leader peptide and may indicate that the members of this subgroup share similar biosynthetic machinery. The endopeptidase may have an S1 subsite highly specific for Asn, an S1′ subsite specific for residues with a branched aliphatic side chain (Ile/Val/Leu), and an S2′ subsite preferring those with bulky side chains (Tyr/Val/Asn). Subgroup II bacteriocins have leader sequences ranging from 22 to 42 residues, which are generally longer than those of subgroup I circular bacteriocins. Based on this putative cleavage site, the known circular bacteriocins and the predicted mature bacteriocin-like peptides are shown in the phylogenetic tree in Fig. S4 in the supplemental material, together with their pI values. Phylogenetic analysis of these known and putative bacteriocins revealed that members of this subgroup could be classified further into 2 subclades. The first subclade consists of bacteriocins found solely in members of the genus Lactobacillus and contains acidocin B and gassericin A. On the other hand, the second subclade is a more diverse group, with members coming from the genera Bacillus, Lactobacillus, Butyrivibrio, and Streptococcus, and contains butyrivibriocin AR10.
In conclusion, this study confirms the circular nature of acidocin B and describes the three-dimensional solution structure of this bacteriocin in the membrane-mimicking SDS micelle solvent system. This is a structure of a circular bacteriocin belonging to subgroup II. The elucidated structure revealed that acidocin B is composed of four helices that are folded to form a compact, globular bundle having a central pore. Surface analysis showed that hydrophobic and uncharged residues dominate the surface of acidocin B, which could potentially signify that initial binding to the cell membrane of target organisms is mediated by hydrophobic interaction. Elucidating the solution structure of acidocin B is a prerequisite to acquiring a deeper understanding of its properties and better molecular insights on how it operates in nature. Future work to determine the mode of action of subgroup II circular bacteriocins, like acidocin B, may ultimately lead to their increased utilization in food preservation, probiotics, and other applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jos M. B. M. van der Vossen (TNO Earth, Environmental and Life Sciences, Zeist, The Netherlands) for providing us the L. acidophilus M46 strain, Randy Whittal and Jing Zheng for their assistance in the MS/MS analysis, and Leah A. Martin-Visscher for her help with NMR data analysis.
Financial support was provided by the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chair in Bioorganic and Medicinal Chemistry.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04265-14.
REFERENCES
- 1.Gabrielsen C, Brede DA, Nes IF, Diep DB. 2014. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol 80:6854–6862. doi: 10.1128/AEM.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol 19:411–418. doi: 10.1016/j.tim.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Gálvez A, Abriouel H, Lopez RL, Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol 120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Samyn B, Martinez-Bueno M, Devreese B, Maqueda M, Gálvez A, Valdivia E, Coyette J, Van Beeumen J. 1994. The cyclic structure of the enterococcal peptide antibiotic AS-48. FEBS Lett 352:87–90. doi: 10.1016/0014-5793(94)00925-2. [DOI] [PubMed] [Google Scholar]
- 5.Kawai Y, Saito T, Kitazawa H, Itoh T. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci Biotechnol Biochem 62:2438–2440. doi: 10.1271/bbb.62.2438. [DOI] [PubMed] [Google Scholar]
- 6.Kemperman R, Kuipers A, Karsens H, Nauta A, Kuipers O, Kok J. 2003. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl Environ Microbiol 69:1589–1597. doi: 10.1128/AEM.69.3.1589-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalmokoff ML, Cyr TD, Hefford MA, Whitford MF, Teather RM. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can J Microbiol 49:763–773. doi: 10.1139/w03-101. [DOI] [PubMed] [Google Scholar]
- 8.Wirawan RE, Swanson KM, Kleffmann T, Jack RW, Tagg JR. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630. doi: 10.1099/mic.0.2006/005967-0. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl Environ Microbiol 74:4756–4763. doi: 10.1128/AEM.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawa N, Zendo T, Kiyofuji J, Fujita K, Himeno K, Nakayama J, Sonomoto K. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl Environ Microbiol 75:1552–1558. doi: 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrero J, Brede DA, Skaugen M, Diep DB, Herranz C, Nes IF, Cintas LM, Hernández PE. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl Environ Microbiol 77:369–373. doi: 10.1128/AEM.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda Y, Ono H, Kitagawa H, Ito H, Mu F, Sawa N, Zendo T, Sonomoto K. 2011. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl Environ Microbiol 77:8164–8170. doi: 10.1128/AEM.06348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholz R, Vater J, Budiharjo A, Wang Z, He Y, Dietel K, Schwecke T, Herfort S, Lasch P, Borriss R. 2014. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J Bacteriol 196:1842–1852. doi: 10.1128/JB.01474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potter A, Ceotto H, Coelho ML, Guimarães AJ, Bastos MDCDF. 2014. The gene cluster of aureocyclicin 4185: the first cyclic bacteriocin of Staphylococcus aureus. Microbiology 160:917–928. doi: 10.1099/mic.0.075689-0. [DOI] [PubMed] [Google Scholar]
- 15.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J Biochem 98:585–603. [DOI] [PubMed] [Google Scholar]
- 16.Murphy K, O'Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. 8 July 2011. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS One 6:e20852. doi: 10.1371/journal.pone.0020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Visscher LA, Gong X, Duszyk M, Vederas JC. 2009. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J Biol Chem 284:28674–28681. doi: 10.1074/jbc.M109.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González C, Langdon GM, Bruix M, Gálvez A, Valdivia E, Maqueda M, Rico M. 2000. Bacteriocin AS-48, a microbial cyclic polypeptide structurally and functionally related to mammalian NK-lysin. Proc Natl Acad Sci U S A 97:11221–11226. doi: 10.1073/pnas.210301097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruhn H. 2005. A short guided tour through functional and structural features of saposin-like proteins. Biochem J 389:249–257. doi: 10.1042/BJ20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maqueda M, Gálvez A, Bueno MM, Sanchez-Barrena MJ, González C, Albert A, Rico M, Valdivia E. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci 5:399–416. doi: 10.2174/1389203043379567. [DOI] [PubMed] [Google Scholar]
- 21.Miteva M, Andersson M, Karshikoff A, Otting G. 1999. Molecular electroporation: a unifying concept for the description of membrane pore formation by antibacterial peptides, exemplified with NK-lysin. FEBS Lett 462:155–158. doi: 10.1016/S0014-5793(99)01520-3. [DOI] [PubMed] [Google Scholar]
- 22.Leer RJ, van der Vossen JMBM, van Giezen M, van Noort JM, Pouwels PH. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629–1635. doi: 10.1099/13500872-141-7-1629. [DOI] [PubMed] [Google Scholar]
- 23.van der Vossen JMBM, van Herwijnen MHM, Leer RJ, ten Brink B, Pouwels PH, Huis int Veld JHJ. 1994. Production of acidocin B, a bacteriocin of Lactobacillus acidophilus M46 is a plasmid-encoded trait: plasmid curing, genetic marking by in vivo plasmid integration, and gene transfer. FEMS Microbiol Lett 116:333–340. doi: 10.1111/j.1574-6968.1994.tb06724.x. [DOI] [Google Scholar]
- 24.ten Brink B, Minekus M, van der Vossen JMBM, Leer RJ, Huis in't Veld JH. 1994. Antimicrobial activity of lactobacilli: preliminary characterization and optimization of production of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus M46. J Appl Bacteriol 77:140–148. doi: 10.1111/j.1365-2672.1994.tb03057.x. [DOI] [PubMed] [Google Scholar]
- 25.Worobo RW, Van Belkum MJ, Sailer M, Roy KL, Vederas JC, Stiles ME. 1995. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol 177:3143–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Y, Whittal RM, Li L. 1999. Two-layer sample preparation: a method for MALDI-MS analysis of complex peptide and protein mixtures. Anal Chem 71:1087–1091. doi: 10.1021/ac980684h. [DOI] [PubMed] [Google Scholar]
- 27.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. 2003. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom 17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 28.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. 2000. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BA. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 31.Wider G, Macura S, Kumar A, Ernst RR, Wüthrich K. 1984. Homonuclear two-dimensional 1H NMR of proteins. Experimental procedures. J Magn Reson 56:207–234. [Google Scholar]
- 32.Wüthrich K. 1986. NMR of proteins and nucleic acids, p 1–292. John Wiley & Sons, New York, NY. [Google Scholar]
- 33.Guntert P, Mumenthaler C, Wuthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 35.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin M, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito Y, Kawai Y, Arakawa K, Honme Y, Sasaki T, Saito T. 2009. Conjugative plasmid from Lactobacillus gasseri LA39 that carries genes for production of and immunity to the circular bacteriocin gassericin A. Appl Environ Microbiol 75:6340–6351. doi: 10.1128/AEM.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2010. Cloning and characterization of the gene cluster involved in the production of the circular bacteriocin carnocyclin A. Probiotics Antimicrob Proteins 2:218–225. doi: 10.1007/s12602-010-9056-1. [DOI] [PubMed] [Google Scholar]
- 43.Kawai Y, Kusnadi J, Kemperman R, Kok J, Ito Y, Endo M, Arakawa K, Uchida H, Nishimura J, Kitazawa H, Saito T. 2009. DNA sequencing and homologous expression of a small peptide conferring immunity to gassericin A, a circular bacteriocin produced by Lactobacillus gasseri LA39. Appl Environ Microbiol 75:1324–1330. doi: 10.1128/AEM.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai Y, Ishii Y, Arakawa K, Uemura K, Saitoh B, Nishimura J, Kitazawa H, Yamazaki Y, Tateno Y, Itoh T, Saito T. 2004. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl Environ Microbiol 70:2906–2911. doi: 10.1128/AEM.70.5.2906-2911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strandberg E, Ulrich AS. 2004. NMR methods for studying membrane-active antimicrobial peptides. Concepts Magn Reson 23A:89–120. doi: 10.1002/cmr.a.20024. [DOI] [Google Scholar]
- 46.Wutzke KD, Oetjens I. 2005. 13C- and 15N-incorporation of doubly stable isotope labelled Lactobacillus johnsonii in humans. Eur J Clin Nutr 59:1167–1172. doi: 10.1038/sj.ejcn.1602227. [DOI] [PubMed] [Google Scholar]
- 47.Gálvez A, Maqueda M, Martínez-Bueno M, Valdivia E. 1991. Permeation of bacterial cells, permeation of cytoplasmic and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J Bacteriol 173:886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong X, Martin-Visscher LA, Nahirney D, Vederas JC, Duszyk M. 2009. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim Biophys Acta 1788:1797–1803. doi: 10.1016/j.bbamem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 51.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 52.Bandyopadhyay S, Junjie RL, Lim B, Sanjeev R, Xin WY, Yee CK, Hui Melodies SM, Yow N, Sivaraman J, Chatterjee C. 2014. Solution structures and model membrane interactions of Ctriporin, an anti-methicillin-resistant Staphylococcus aureus peptide from scorpion venom. Biopolymers 101:1143–1153. doi: 10.1002/bip.22519. [DOI] [PubMed] [Google Scholar]
- 53.Bandyopadhyay S, Ng BY, Chong C, Lim MZ, Gill SK, Lee KH, Sivaraman J, Chatterjee C. 2014. Micelle bound structure and DNA interaction of brevinin-2-related peptide, an antimicrobial peptide derived from frog skin. J Pept Sci 20:811–821. doi: 10.1002/psc.2673. [DOI] [PubMed] [Google Scholar]
- 54.Barbosa SC, Cilli EM, Dias LG, Fuzo CA, Degrève L, Stabeli RG, Itri R, Ciancaglini P, Barber JD, Barton GJ. 2014. Interaction of cyclic and linear labaditin peptides with anionic and zwitterionic micelles. J Colloid Interface Sci 438:39–46. doi: 10.1016/j.jcis.2014.09.059. [DOI] [PubMed] [Google Scholar]
- 55.Arakawa K, Kawai Y, Ito Y, Nakamura K, Chujo T, Nishimura J, Kitazawa H, Saito T. 2010. HPLC purification and reevaluation of chemical identity of two circular bacteriocins, gassericin A and reutericin 6. Lett Appl Microbiol 50:406–411. doi: 10.1111/j.1472-765X.2010.02810.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.