Abstract
This study characterized specific changes in the millet root zone microbiome stimulated by long-term woody-shrub intercropping at different sites in Senegal. At the two study sites, intercropping with woody shrubs and shrub residue resulted in a significant increase in millet [Pennisetum glaucum (L.) R. Br.] yield (P < 0.05) and associated patterns of increased diversity in both bacterial and fungal communities in the root zone of the crop. Across four experiments, operational taxonomic units (OTUs) belonging to Chitinophaga were consistently significantly (P < 0.001) enriched in the intercropped samples, and “Candidatus Koribacter” was consistently significantly enriched in samples where millet was grown alone. Those OTUs belonging to Chitinophaga were enriched more than 30-fold in residue-amended samples and formed a distinct subgroup from all OTUs detected in the genus. Additionally, OTUs belonging to 8 fungal genera (Aspergillus, Coniella, Epicoccum, Fusarium, Gibberella, Lasiodiplodia, Penicillium, and Phoma) were significantly (P < 0.005) enriched in all experiments at all sites in intercropped samples. The OTUs of four genera (Epicoccum, Fusarium, Gibberella, and Haematonectria) were consistently enriched at sites where millet was grown alone. Those enriched OTUs in intercropped samples showed consistently large-magnitude differences, ranging from 30- to 1,000-fold increases in abundance. Consistently enriched OTUs in intercropped samples in the genera Aspergillus, Fusarium, and Penicillium also formed phylogenetically distinct subgroups. These results suggest that the intercropping system used here can influence the recruitment of potentially beneficial microorganisms to the root zone of millet and aid subsistence farmers in producing higher-yielding crops.
INTRODUCTION
Subsistence farmers in the Sahel region of sub-Saharan Africa struggle with the challenges of drought, low organic matter in soils, and encroaching desertification (1). Resolving these issues is critical for improving crop yields in this region. Woody shrubs that occur naturally in farmers' fields might be used as part of an intercropping production system to improve crop yields. Two woody shrubs, Guiera senegalensis and Piliostigma reticulatum, are distributed throughout the Sahel region (2, 3). Currently, most farmers manage these shrubs by coppicing, collecting, and burning above-ground residue prior to planting crops. However, agricultural practices in the Sahel region can be adjusted to utilize shrub biomass as a natural source of fertilizer (4). Shrub residue can be added to the soil as a nutrient-rich organic amendment and decompose within an 8-month time period (5). Rhizosphere soils around amended shrubs show increased nutrient content (6), improved soil moisture profiles due to hydraulic lift (7), and a more diverse and complex microfauna soil food web (8). Intercropping with G. senegalensis (9) and P. reticulatum (10), with the incorporation of shrub residue, has also been shown to improve crop yields at two long-term field sites in Senegal.
Concomitantly with the physicochemical enrichments, microbial community composition and activity respond positively to shrub-based amendments. Woody-shrub intercropping drives an increase in total phospholipid fatty acid (PLFA) content for both bacterial and fungal markers, as well as increased soil enzyme activity (11). Density gradient gel electrophoresis (DGGE) profiling revealed similar patterns of increased soil microbial diversity in these intercropping systems (Sidy Diakhaté, personal communication). While such profiling methods describe some coarsely defined variations in soil microbial community structure, it is important to determine which specific organisms are associated with the phenomenon of improved crop growth in the field. Thus, the overall objective of this study was to identify and quantify specific changes in soil microbiomes associated with shrub-based enhancement of annual crop growth. Specifically, this work provides the first detailed description of the changes in the microbiome stimulated by long-term intercropping of woody shrubs with millet [Pennisetum glaucum (L.) R. Br.] in the Sahel region of Africa.
MATERIALS AND METHODS
Study sites, sample collection, and agronomic data.
Sampling was conducted at two long-term study sites near Keur Matar and Nioro, Senegal (9, 10) (see Fig. S1 in the supplemental material), which, respectively, had Dior soil (Rubic arenosol with 95% sand) and Deck-Dior soil (Haplic Ferric Lixisol) (39). The two major crops cultivated at the sites are millet and peanut (Arachis hypogaea L.). Keur Matar is in the northern region of the Peanut Basin (14′45′N, 16′51′W), with mean annual precipitation of 450 mm and mean monthly temperatures ranging from 20 to 33°C. Nioro is in the southern region of the Peanut Basin (13′45′N, 15′47′W), with a mean annual precipitation of 750 mm and mean monthly air temperatures ranging from 20.0 to 35.7°C.
At both sites, a field of approximately 0.5 ha (5,000 m2) with preexisting shrubs that had been under local farmer management for at least the last 50 years was selected. The sites had been cropped continuously with a peanut-millet rotation and then left fallow for 3 years prior to initiation of the experiment. The experimental design at both sites was a randomized complete-block design with the presence or absence of shrubs as the treatments and with four replicates each. The main plots were established in the winter (dry season) of 2003 by manually removing existing shrubs to establish no-shrub plots and transplanting shrub seedlings to achieve a standardized stand density. Specifically, stand densities of 1,500 to 1,833 Piliostigma reticulatum plants ha−1 and 888 to 1,555 Guiera senegalensis plants ha–1 were established at Nioro and Keur Matar, respectively. Shrubs were randomly but relatively evenly distributed. The following summer, millet was planted on all plots and fertilized with 68.5 kg N, 15 kg P, and 15 kg K ha−1 to allow plots to equilibrate for 1 year before initiation of the experiments. Main plot sizes were 46 m by 6 m, and subplot sizes were 10 m by 6 m. There was a 2-m gap between adjacent plots and a 3-m gap between blocks. According to the dominant farming practices in the region, all plots had a crop rotation of peanut (Arachis hypogaea var. 55-437) and millet (Pennisetum glaucum var. Souna 3) from 2004 to 2013 (except for 2008 to 2010, when no crops were planted).
For this study, the Nioro field site was sampled on 13 August 2013, and the Keur Matar site was sampled on 10 September 2013. Soil samples were taken from millet root zones of plots with and without shrubs. Samples from two independent transects were taken at each location and analyzed as four separate experiments. Five cores with 2.5-cm diameters (0- to 20-cm depths) were taken near the base of a single plant and composited into one bag. These composite samples were homogenized in the bag and then passed through a 2-mm sieve prior to being subsampled for soil chemical analysis and DNA extraction. Millet biomass was taken for each plot at the end of the growing season by cutting millet shoots at soil level and removing the panicle. Millet shoots were dried in a 65°C for 48 h and weighed to determine dry weight. At Keur Matar, a rainfall deficit caused a failure of grain production, so the weight of panicles was used to estimate millet productivity. At Nioro, yield was measured as grain weight after threshing.
Soil chemical analyses.
Soil chemical analyses were conducted at the Laboratoire des Moyens Analytiques (LAMA), a laboratory of the Institut de Recherche pour le Développement (IRD) in Dakar, Senegal. Total organic carbon was determined using a modified degtjareff method (12).
DNA extraction and Illumina MiSeq sequencing.
DNA was extracted from 0.25 g of each soil sample using Mo-Bio PowerSoil DNA isolation kits (Mo-Bio Laboratories, Carlsbad, CA). Fifty microliters of extracted DNA was precipitated to pellet form using 3 M sodium acetate for transport to Wooster, OH. Upon arrival, DNA pellets were resuspended in 50 μl Qiagen elution buffer (Qiagen, Venlo, Netherlands) and stored in a −20°C freezer. Amplicon libraries for the 16S rRNA V4 region were generated using methods outlined by Caporaso et al. (13). Amplicon libraries for the internal transcribed spacer 1 (ITS1) region were generated by using the protocol described in the work of McGuire et al. (14). For each of these library preparation methods, we performed a single PCR experiment instead of triplicate experiments in the listed protocols. For each amplicon type, all sample libraries were pooled and purified using a Pippen Prep instrument (Sage Science, Beverly, MA). Purified library pools were quantified using the Qubit double-stranded DNA high-sensitivity assay (Life Technologies, Guilford, CT). Amplicon libraries were sequenced on an Illumina MiSeq instrument using the 250-base, paired-end kits at the Molecular and Cellular Imaging Center housed at the Ohio Agricultural Research and Development Center in Wooster, OH.
16S rRNA amplicon sequence processing.
The 16S sequence paired-end data set was demultiplexed on the MiSeq instrument itself at the time of sequencing. Each pair of reads was joined and quality filtered using the join_paired_ends.py script provided in the Quantitative Insights Into Microbial Ecology (QIIME) software suite. The open reference operational taxonomic unit (OTU) picking protocol in QIIME was used. Briefly, sequences were clustered against the 2013 Greengenes ribosomal database's 97% reference data set (http://greengenes.secondgenome.com/downloads). Sequences that did not match any entries in this reference were subsequently clustered into de novo OTUs at 97% similarity with UCLUST. Taxonomy was assigned to all OTUs using the RDP classifier (15) within QIIME and the Greengenes reference data set.
ITS1 amplicon sequence processing.
The ITS data set was demultiplexed on the MiSeq instrument itself at the time of sequencing. Not all paired-end reads overlapped due to the various lengths of the ITS1 region, so only the forward read from each sequence was used for downstream analysis. Reads were clustered into OTUs using the same open reference OTU picking protocol in QIIME as the 16S data set, using the 97% reference database of the UNITE/International Nucleotide Sequence Databases (INSD) NCBI, EMBL, and DDBJ. Taxonomy was assigned to all OTUs using the RDP classifier (15) within QIIME using the UNITE+INSD reference data set.
Statistical analyses.
Agronomic, soil, and sequence count data were subjected to analyses of variance using the general linear model and Tukey's test in R (16). A two-factor model, consisting of treatment and blocks (n = 4) for each site was used. At each site, two experiments, each with a randomized complete block design and four blocks, was conducted (see Fig. S1 in the supplemental material). Comparisons of individual OTU abundances were performed across soil treatments (shrub and crop versus sole-crop zones) for each transect at each site. OTU tables containing read counts for each OTU in each sample, taxonomy information for each OTU, and sample metadata for each sample were exported from QIIME and imported into R using phyloseq (17). In order to exclude sequences observed at very low frequencies, OTUs representing less than 0.001% of the total number of sequences from each library were removed. OTU tables were placed in a subset for each comparison and formatted for the DESeq2 package in R (18). The differential abundance of each OTU by sample type was determined using DESeq2. Sequences for all OTUs belonging to genera shared between experiments were extracted from representative sequence files in QIIME. Sequences for each genus were subsequently aligned using ClustalW (19), and trees were constructed using the neighbor-joining method with 1,000 bootstrap replicates in Geneious 6.0.3 (Biomatters). Subsets of OTUs within a genus were defined by bootstrap values over 90.
RESULTS
Effect of intercropping on millet growth and yield.
The biomass of millet plants grown in shrub and crop intercropping (residue-amended millet [MR] plots) were significantly (P < 0.05) increased compared to that of millet grown alone (plots with bare soil [MB plots]) (Table 1). This pattern of enhanced millet growth in the intercropped plots was true for the experiments conducted at both sites (Keur Matar and Nioro). Such results demonstrate the consistent response of annual crop growth to woody shrubs and annual shrub residue incorporation independently of the shrub species used. More importantly, the harvested yields of millet were also significantly (P < 0.05) increased with residue amendment at both sites.
TABLE 1.
Productivity of two cropping systems at two long-term field sites in Senegala
| Site | Cropping system | Dry biomass (103/ha) | Yield (103/ha) |
|---|---|---|---|
| Keur Matar | Shrub crop | 0.945a | 0.389a |
| Sole crop | 0.212b | 0.046b | |
| Nioro | Shrub crop | 1.491a | 0.749a |
| Sole crop | 0.833b | 0.315b |
Dry biomass and yield mean values are presented (n = 4). Values within a column and site followed by the same letter are not significantly different at a P of ≤0.05.
Soil organic C and overall microbial community size and diversity.
Soil organic C and the root zone microbiomes were increased by intercropping and by the addition of shrub residues at both long-term field sites (Table 2). As expected, intercropping increased soil organic C levels measured in the root zone of millet crops (P < 0.05 for three of four independent experimental comparisons). At Keur Matar, the increases in organic C ranged from 21% to 26%. Similarly, at Nioro, the increases ranged from 24% to 53%. Associated with these enrichments in soil carbon, an approximately 2-fold increase in the amount of microbial DNA recovered from root zone soils of the MR plots was noted. While the average amount of DNA recovered per sample was higher in the MR plots than in the MB plots in all four experiments, such differences were not individually significant but were significant overall (P < 0.10). A similar pattern was observed in the average number of bacterial sequences obtained using our high-throughput sequencing protocol. That is to say, the number of 16S sequences in the MR and MB plots varied by less than a factor of 2 in all four experiments, and, while not significant individually, the results were significant across the entire data set (P < 0.10). Similar patterns were noted for the ITS sequences, though a significant difference (P < 0.05) for the individual comparison from the Nioro B transect was noted as well.
TABLE 2.
Soil organic C, total DNA concentration, and numbers of microbial sequences for MR and MB soil samples in four independent experimentsa
| Site | Transect | Type | Total C (%) | DNA concn (ng/μl) | No. of: |
|||
|---|---|---|---|---|---|---|---|---|
| 16S sequences | 16S OTUs | ITS sequences | ITS OTUs | |||||
| Keur Matar | KM_A | MR | 0.290a | 0.186a | 247,120a | 13,971a | 91,725a | 401a |
| MB | 0.224a | 0.078a | 208,752a | 5,094b | 43,978a | 218b | ||
| KM_B | MR | 0.300a | 0.599a | 203,705a | 20,022a | 62,254a | 355a | |
| MB | 0.239a | 0.181a | 191,590a | 8,649b | 63,809a | 279a | ||
| Nioro | N_A | MR | 0.355a | 0.272a | 237,707a | 15,750a | 117,921a | 501a |
| MB | 0.286a | 0.177a | 185,360a | 9,691a | 79,330a | 305a | ||
| N_B | MR | 0.394a | 1.027a | 207,511a | 22,838a | 109,613a | 591a | |
| MB | 0.258b | 0.343a | 230,663a | 18,333a | 46,296b | 297a | ||
Values within a column and site followed by the same letter are not significantly different at a P of ≤0.05.
A more dramatic contrast was observed in the phylogenetic diversity of bacterial and fungal sequences obtained from MR and MB samples (Table 2). Specifically, the number of bacterial and fungal OTUs increased in the root zone soils of the intercropped plots compared to the millet-only plots. At Keur Matar, significant (P < 0.05) increases in the number of bacterial OTUs were observed in plots intercropped with G. senegalensis. DNA samples from the intercropped plots contained on average 174% and 131% more 16S OTUs for transects A and B, respectively. The same pattern was observed at Nioro, but the 63% and 25% increases were not noted to be individually significant. More strikingly, higher numbers of fungal OTUs were detected in MR samples than in MB samples in three of the four experimental transects (with increases of 84% and 27% at Keur Matar and 64% and 99% at Nioro for transects A and B, respectively).
Specific changes in soil bacterial community structure and diversity induced by woody-shrub intercropping.
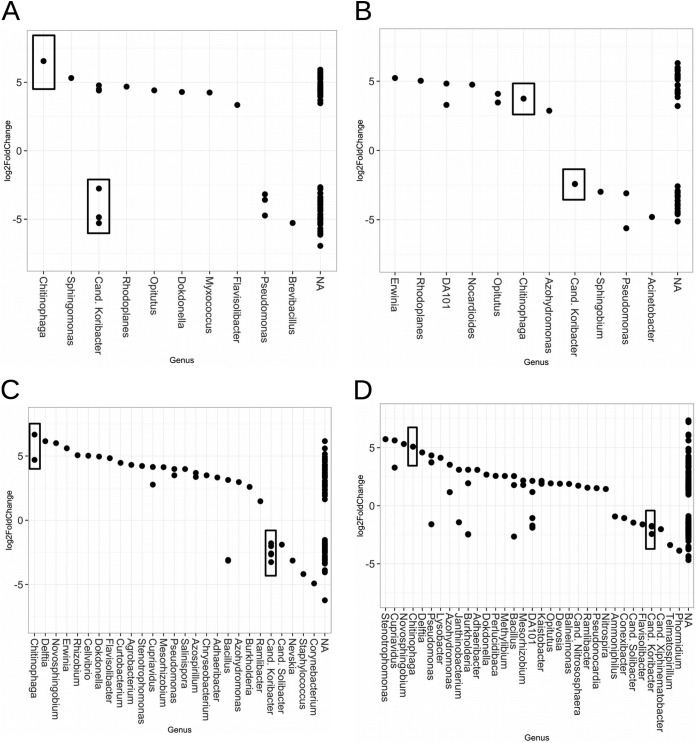
Analyses of sequence data using DESeq2 revealed small but meaningful variations in the relative abundances of just a few bacterial taxa due to crop and residue (Table 3). Between 962 and 1,272 OTUs, each representing at least 0.001% of the total number of sequences obtained, were considered in each experimental comparison. Due to these high numbers of OTUs in the analysis, an alpha of 0.001 was used to define which populations differed significantly between treatments. Comparisons of the abundance of OTUs classified to the genus level were made. Populations differing by at least 2-fold (i.e., log2 = 1) between millet samples grown in the MR and MB samples are shown in Fig. 1. The 16S markers for 8 bacterial genera at KeurMatar_A, 7 genera at KeurMatar_B, 22 genera at Nioro_A, and 25 genera at Nioro_B were enriched significantly (P < 0.001) in the root zones of millet grown in the MR compared to the MB plots. At Keur Matar, three of these genera were enriched in both experiments (i.e., Chitinophaga, Opitutus, and Rhodoplanes). At Nioro, 16S markers representing 13 bacterial genera were enriched in both experiments (i.e., Adhaeribacter, Azohydromonas, Bacillus, Burkholderia, Chitinophaga, Cupriavidus, Delftia, Dokdonella, Mesorhizobium, Novosphingobium, Pseudomonas, Ramlibacter, and Stenotrophomonas). Interestingly, all of the genera named above at Keur Matar and 60% of those named above at Nioro were enriched more than 4-fold in MR soils.
TABLE 3.
Bacteria that differed significantly by residue amendmenta
| Comparison | Exptb | No. of OTUsc | No. of generad | Genera shared between data setse |
|---|---|---|---|---|
| MR > MB | KM_A (1204) | 57 (10) | 8 (3) | Chitinophaga, Opitutus, Rhodoplanes |
| KM_B (933) | 40 (9) | 7 (3) | ||
| N_A (962) | 66 (26) | 22 (13) | Adhaeribacter, Azohydromonas, Bacillus, Burkholderia, Chitinophaga, Cupriavidus, Delftia, Dokdonella, Mesorhizobium, Novosphingobium, Pseudomonas, Ramlibacter, Stenotrophomonas | |
| N_B (977) | 156 (35) | 25 (13) | ||
| MR < MB | KM_A (1204) | 49 (7) | 3 (2) | “Candidatus Koribacter,” Pseudomonas |
| KM_B (933) | 22 (5) | 4 (2) | ||
| N_A (962) | 44 (11) | 6 (3) | Bacillus, “Candidatus Koribacter,” “Candidatus Solibacter” | |
| N_B (977) | 113 (20) | 13 (3) |
Significantly different at a P of <0.001. Abbreviations are for crop (i.e., millet [M]) and residue (i.e., residue-amended [R] or bare [B] soil).
Four independent experiments were performed at two field sites (Keur Matar [KM] or Nioro [N]) in 2013. Numbers in parentheses are the numbers of OTUs analyzed in DESeq2, which are the numbers of OTUs at a >0.001% abundance.
Numbers of bacterial OTUs enriched by more than 2-fold for a particular experiment are listed (P < 0.001). Numbers in parentheses are the numbers of OTUs belonging to the named genera.
Numbers of bacterial genera enriched by more than 2-fold for a particular experiment are listed (P < 0.001). Numbers in parentheses are the numbers of genera shared between the two data sets.
The names of only those genera whose patterns of enrichment were consistent in the two experiments at each location are listed, but those observed across all four experiments are in bold.
FIG 1.
Bacterial OTUs occurring at a >0.001% abundance which are significantly enriched in MR or MB samples. OTUs from KM_A (A), KM_B (B), N_A (C), and N_B (D). Boxes highlight OTUs from genera consistently significantly enriched in all experiments. A positive log2-fold change value indicates that the OTU is significantly enriched in MR samples, and a negative log2-fold change indicates that the OTU is significantly enriched in MB samples. Boxes highlight OTUs in genera consistently enriched across all experiments.
In contrast, the 16S markers for just 3 bacterial genera at KeurMatar_A, 4 genera at KeurMatar_B, 6 genera at Nioro_A, and 13 genera at Nioro_B were significantly (P < 0.001) enriched in the root zone of the millet grown alone. At Keur Matar, markers for two genera were enriched in both experiments (i.e., “Candidatus Koribacter” and Pseudomonas). At Nioro, three bacterial genera were enriched in both experiments (i.e., Bacillus, “Candidatus Koribacter,” and “Candidatus Solibacter”).
Specific changes in soil fungal community structure and diversity induced by woody-shrub residues.
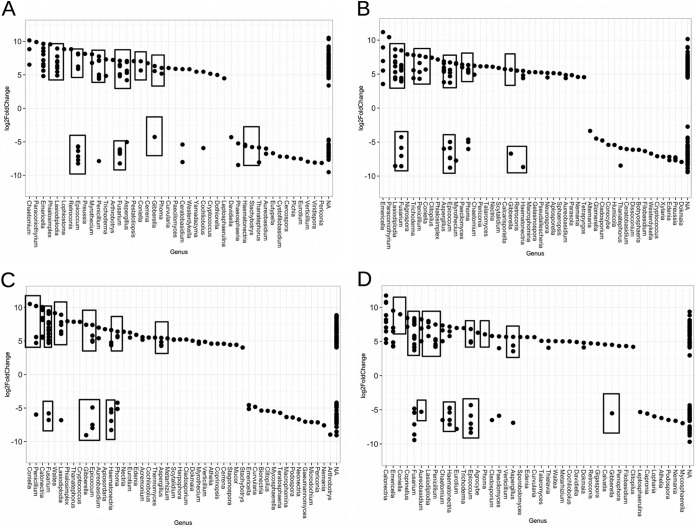
Analysis with DESeq2 was also performed with fungal microbiome sequences (Table 4). The numbers of OTUs that were present at levels that were more than 0.001% of the total sequence abundance and included in each analysis ranged between 292 and 374. Due to these lower numbers of OTUs, an alpha of 0.005 was selected. Populations differing by at least 2-fold (i.e., log2 = 1) between millet root zone samples from intercropped plots and millet-only plots are shown in Fig. 2. ITS1 markers representing 29 genera of fungi at KeurMatar_A, 34 genera at KeurMatar_B, 35 genera at Nioro_A, and 36 genera at Nioro_B were enriched significantly (P < 0.005) in MR samples compared to MB samples. At Keur Matar, 16 of these genera and, at Nioro, 19 fungal genera were enriched in both experiments (Table 4). Interestingly, the ITS1 markers for all of these genera at both sites were enriched more than 4-fold, an indication of a high degree of preference for growth in residue-amended plots. In contrast, there were markers for just 21 fungal genera at KeurMatar_A, 24 genera at KeurMatar_B, 21 genera at Nioro_A, and 17 genera at Nioro_B that were significantly (P < 0.005) enriched in MB samples. At Keur Matar and Nioro, seven genera were enriched at each site in both experiments.
TABLE 4.
Fungi that differed significantly by residue amendmenta
| Comparison | Exptb | No. of OTUsc | No. of generad | Genera shared between data setse |
|---|---|---|---|---|
| MR > MB | KM_A (292) | 136 (70) | 29 (16) | Aspergillus, Chaetomium, Coniella, Emericella, Epicoccum, Fusarium, Gibberella, Lasiodiplodia, Myrothecium, Paecilomyces, Paraconiothyrium, Penicillium, Phialosimplex, Phoma, Retroconis, Trichoderma |
| KM_B (322) | 133 (86) | 34 (16) | ||
| N_A (301) | 133 (76) | 35 (19) | Aspergillus, Aureobasidium, Calonectria, Cladosporium, Cochliobolus, Coniella, Dokmaia, Edenia, Epicoccum, Eurotium, Fusarium, Gibberella, Haemonectria, Lasiodiplodia, Metarhizium, Penicillium, Phoma, Verticillium, Waitea | |
| N_B (305) | 144 (89) | 36 (19) | ||
| MR < MB | KM_A (292) | 64 (37) | 21 (7) | Aspergillus, Ceratobasidium, Epicoccum, Fusarium, Gibberella, Haematonectria, Thanatephorus |
| KM_B (322) | 79 (37) | 24 (7) | ||
| N_A (301) | 57 (29) | 21 (7) | Epicoccum, Fusarium, Gibberella, Haemonectria, Mycosphaerella, Nemania, Podospora | |
| N_B (305) | 62 (31) | 17 (7) |
Significantly different at a P of <0.005. Abbreviations are the same as those listed in the tables above.
Four independent experiments were performed at two field sites (Keur Matar [KM] or Nioro [N]) in 2013. Numbers in parentheses are the numbers of OTUs analyzed in DESeq2, which are the numbers of OTUs at >0.001% abundance.
Numbers of fungal OTUs enriched by more than 2-fold for a particular experiment are listed (P < 0.005). Numbers in parentheses are the numbers of OTUs belonging to a named genus.
Numbers of fungal genera enriched by more than 2-fold for a particular experiment are listed (P < 0.005). Numbers in parentheses are the numbers of genera shared between the two data sets.
The names of only those genera whose patterns of enrichment were consistent in the two experiments at each location are listed, but those observed across all four experiments are in bold.
FIG 2.
Fungal OTUs occurring at a >0.001% abundance which are significantly enriched in MR or MB samples. OTUs from KM_A (A), KM_B (B), N_A (C), and N_B (D). Boxes highlight OTUs from genera consistently significantly enriched in all experiments. A positive log2-fold change value indicates that the OTU is significantly enriched in MR samples, and a negative log2-fold change indicates that the OTU is significantly enriched in MB samples. Boxes highlight OTUs in genera consistently enriched across all experiments.
Consistently enriched genera across all experiments and sites.
Because it is well established that microbiome structures can differ dramatically by site and treatment, we screened the sequence data for patterns that were consistent across multiple transects and experimental sites. Figure 1 and Table 3 show the identity and magnitude differences in abundance of bacterial OTUs which were consistently enriched in both transects at each site. We observed two genera which were responsive and common to each location and common across locations: Chitinophaga in the shrub-crop plots and “Candidatus Koribacter” in the sole-crop plots.
Among the fungal populations (Table 4) a large fraction (i.e., >10%) of the distinct OTUs were responsive to intercropping and amendment with shrub residue. When the root zone populations of millet plants grown in shrub-crop plots and in sole-crop plots were compared, eight genera were consistently enriched in the millet root zones grown in intercropped plots (i.e., Aspergillus, Coniella, Epicoccum, Fusarium, Gibberella, Lasiodiplodia, Penicillium, and Phoma) and four genera were enriched in the millet-only plots (i.e., Epicoccum, Fusarium, Gibberella, and Haemonectria) at both sites. It should be noted that the OTUs that comprise those genera that appear as significantly different in each comparison are different from one another. The consistency of responses by these noted bacterial and fungal genera indicate that they are more sensitive to the woody-shrub intercropping than to other soil and environmental variables.
Sequence variation of select taxa enriched in intercropped MR microbiomes.
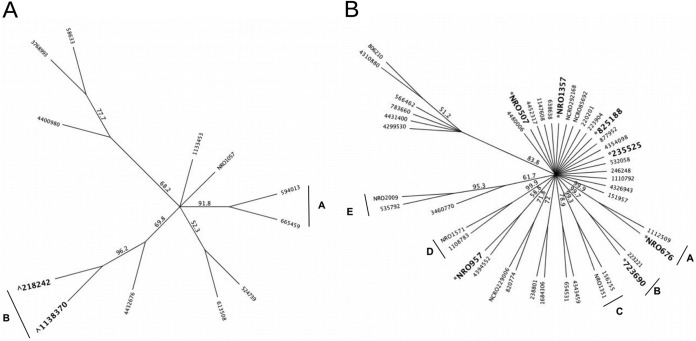
Phylogenetic analyses were performed with sequences from select taxa enriched in MR microbiomes in order to identify targets for marker-assisted recovery (20). In the 16S data set, only one genus, Chitinophaga, contained OTUs which were consistently enriched in MR samples in all four experiments. In total, 12 individual Chitinophaga OTUs were present in the >0.001%-abundance data sets (Fig. 3A). Analysis with the DESeq2 package in R software revealed that two of these Chitinophaga OTUs (17% of all types) were enriched in MR samples, both of which belong to a single cluster (Fig. 3). The boxes in Fig. 1 highlight the abundances of these OTUs, which is within a range of three to six log2-fold changes higher in MR samples than in MB samples (i.e., an 8- to 64-fold enrichment). OTU 218242 was significantly enriched in all four experiments, while OTU 1138370 was significantly enriched in one of the four experiments.
FIG 3.
Clustering of 16S OTUs within genera shown to be significantly enriched in MR or MB samples in all experiments. (A) Chitinophaga; (B) “Candidatus Koribacter.” Branches display bootstrap consensus values after 1,000 replications. Groupings classified as having a bootstrap value of >90. OTUs marked with a caret (∧) are significantly enriched (P < 0.001) in MR samples, and OTUs marked with an asterisk (*) are significantly enriched (P < 0.001) in MB samples.
In the ITS data set, eight genera (Aspergillus, Coniella, Epicoccum, Fusarium, Gibberella, Lasiodiplodia, Penicillium, and Phoma) contained OTUs which were consistently enriched in MR samples in all four experiments. There were a total of 24 Aspergillus OTUs in the >0.001%-abundance data set (Fig. 4A). DESeq2 analysis revealed 13 OTUs (54%) that were enriched in MR samples relative to MB samples. Clustering analysis revealed two groups, A and D, shown in Fig. 4A, as containing OTUs enriched only in MR samples. There were a total of three Coniella OTUs in the data set, with OTU HQ166057 shared between all sample types at all sites, and the two additional OTUs (NCRO14621 and NRO1232) that were unique to the MR samples at Keur Matar. Seventeen of 52 Epicoccum OTUs (33%) were found to be enriched in MR samples relative to MB samples after DESeq2 analysis. However, there were no distinct clusters of Epicoccum OTUs enriched in either MR or MB samples (Fig. 4B).
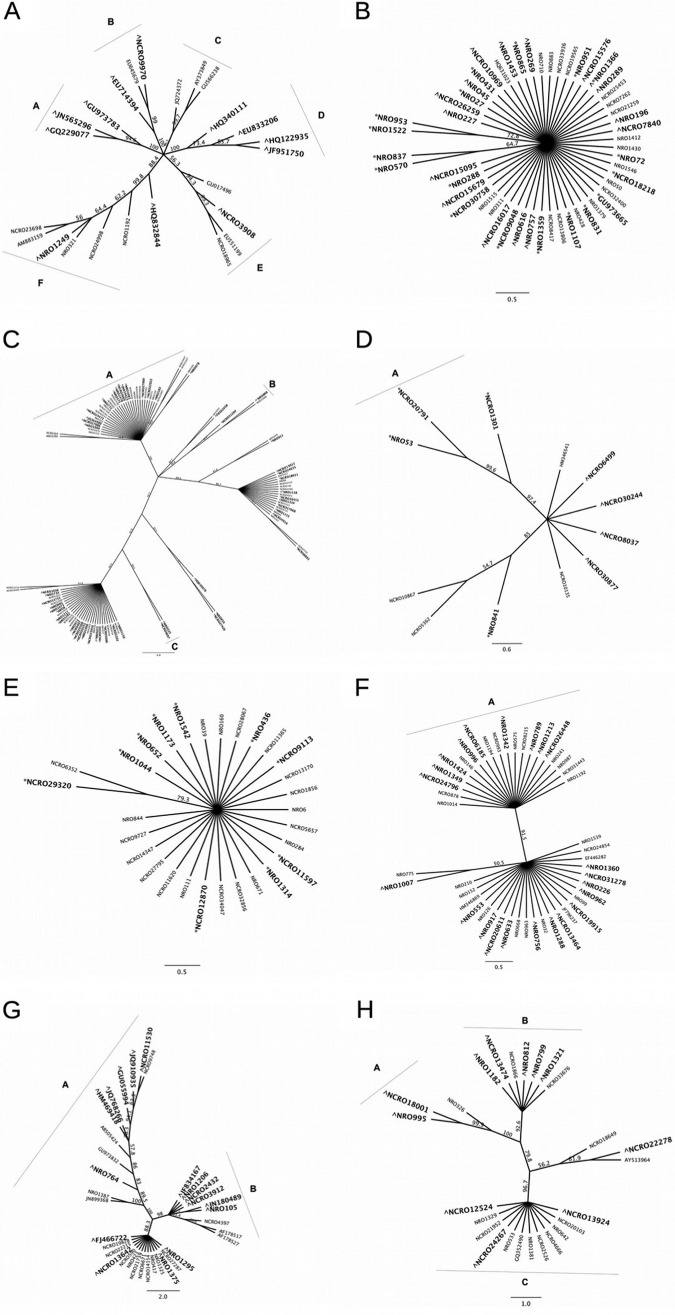
FIG 4.
Clustering of ITS OTUs within genera shown to be significantly enriched in MR or MB samples in all experiments. (A) Aspergillus; (B) Epicoccum; (C) Fusarium; (D) Gibberella; (E) Haematonectria; (F) Lasiodiplodia; (G) Penicillium; (H) Phoma. Coniella had too few OTUs to allow for clustering analysis. Branches display bootstrap consensus values after 1,000 replications. Groupings classified as having a bootstrap value of >90. OTUs marked with a caret (∧) are significantly enriched (P < 0.005) in MR samples, and OTUs marked with an asterisk (*) are significantly enriched (P < 0.005) in MB samples.
There were 103 Fusarium OTUs detected in our experiment (Fig. 4C). Analysis with DESeq2 found 38 OTUs (37%) significantly enriched in MR samples over MB samples. Phylogenetic analysis revealed two groups (A and C in Fig. 4C) that consisted of only those OTUs enriched in MR samples or not significantly differing between MR and MB samples. Five of 12 total Gibberella OTUs identified in the total data set (42%) were found to be significantly enriched in MR samples compared to MB samples (Fig. 4D). Group A contained only those OTUs enriched in MB samples (NRO 53, NCRO20791, and NCRO1301), and the Gibberella OTUs enriched in MR samples (NCRO6499, NCRO30244, NCRO8037, and NCRO30877) did not meet the standard set to be considered a distinct cluster.
For Lasiodiplodia, a total of 46 OTUs were identified (Fig. 4F). A total of 22 Lasiodiplodia OTUs (48%) were significantly enriched in MR samples. Only one distinct subgroup was identified, but the OTUs enriched in MR samples were not concentrated in that group. In total, 34 Penicillium OTUs were identified (Fig. 4G). Analysis with DESeq2 revealed that 16 OTUs (47%) were significantly enriched in MR samples relative to MB samples. Two clusters were identified, both containing OTUs enriched in MR samples. A total of 25 Phoma OTUs were identified (Fig. 4H), and 11 of these OTUs (44%) were significantly enriched in MR samples relative to MB samples. Among these, we identified a total of three distinct groups, of which A and B contained a majority of the OTUs enriched in MR samples.
Sequence variation of select taxa enriched in sole-crop (MB) microbiomes.
In the 16S data set, only one genus was consistently enriched in MB samples, “Candidatus Koribacter.” Seven of the 44 related OTUs (16%) were found to be significantly enriched in MB samples compared to in MR samples (Fig. 3B). OTU 723690 was significantly enriched at both MB transects at Keur Matar (KM_A and KM_B) (appearing 2.5-log2-fold more than in MR samples), while NRO676 was found to be enriched at both MB transects at Nioro (N_A and N_B), with changes ranging from 1.75- to 2.5-log2-fold higher than in MR samples. The remaining OTUs marked as significant in Fig. 3 appeared in only one out of the four experiments.
In the ITS data set, Epicoccum, Fusarium, Gibberella, and Haematonectria all contained OTUs significantly enriched in MB samples compared to MR samples. A total of 19 Epicoccum (37%) (Fig. 4B), 18 Fusarium (17%) (Fig. 4C), and 4 Gibberella (33%) (Fig. 4D) OTUs were found to be significantly enriched in MB samples after analysis with DESeq2. Within these genera, only group A in the Gibberella genus was identified as a cluster of enriched MB OTUs. The genus Haematonectria contained a total of 29 OTUs (Fig. 4D). A total of 10 Haematonectria OTUs (34%) were found to be significantly enriched in MB samples compared to MR samples. However, phylogenetic analysis revealed no distinct groups in this genus.
DISCUSSION
In this study, we identified several highly significant associations between root zone-inhabiting soil microbial populations and improved millet growth mediated by woody-shrub intercropping. Specifically, bacterial OTUs in the genus Chitinophaga and fungal OTUs in the genera Aspergillus, Coniella, Lasiodiplodia, Penicillium, and Phoma increased in response to crop growth-promoting intercropping and residue amendments. We also found contrasting responses of different populations of Epicoccum, Fusarium, and Gibberella. In comparison to fungal communities, bacterial communities showed fewer significant responses, with OTUs of only Chitinophaga and “Candidatus Koribacter” consistently responding to woody-shrub intercropping. Our findings of increased diversity in soil microbial communities following application of shrub residue expand upon those of previous work done at the same sites using PLFA analysis (11). That previous study noted an increase in all PLFA types (bacterial and fungal) in shrub-crop plots compared to sole-crop plots and a higher response of fungal PLFAs than of bacterial PLFAs to residue amendment. Millet growth promotion was observed at both study sites in plots intercropped with woody shrubs. For the Keur Matar site, the results of this study are consistent with previous findings (9) in which millet (and groundnut) grown in the presence of G. senegalensis had substantially greater yields over 4 cropping seasons than sole crops. In that study, the application of an intercropping system with the addition of shrub residue took 4 cropping seasons to build the soil organic carbon and alter soil nutrient status. The elevated millet grain and biomass yield at the Nioro site with P. reticulatum were somewhat in contrast to the results of previous research on these plots (10). In the previous study, there was no significant difference between the sole-crop and shrub-crop yields in 2 of the 4 study years, with contrasting trends in the other 2 years. These divergent results at Keur Matar and Nioro may be due to the former location having a more sandy soil, allowing for a more dramatic and immediate response to shrub intercropping. Compared to Nioro, which has heavier soils and greater rainfall, Keur Matar had a less stressed environment for crop growth. Thus, it may be that at Nioro, it took longer to have a consistent crop growth response due to the generally beneficial effects of P. reticulatum on crop yields.
Phylogenetic clustering of OTU sequences revealed distinct groups of microorganisms associated with improved plant growth. These specific subgenus groups were found in the Chitinophaga (Fig. 3A), Aspergillus, Fusarium, Penicillium, and Phoma genera (Fig. 4A, C, G, and H). Additionally, we noted contrasting responses of various populations of certain genera. Previous studies have shown that some isolates of Fusarium promoted plant growth but that others acted to suppress plant growth (21). Our data suggest the potential for a similar phenomenon among millet grown in Senegal. Multiple OTUs of the Epicoccum, Fusarium, and Gibberella genera were enriched in either MR or MB samples. For the related Fusarium and Gibberella genera, we noted some clustering of OTUs enriched in MR samples, shown in Fig. 4C and D. There was also a small number of OTUs that were significantly enriched in both the MR and MB samples. These particular OTUs may show variations in niche preference related to environmental factors differing between treatments. The OTUs which were consistently enriched in MB plots may have negatively impacted millet growth as well, which would help to explain the variation in millet productivity that we observed. The techniques used in our study may miss some of the variations in the observed populations which might further distinguish subgenus clusters of OTUs. As high-throughput sequencing technology advances to allow for longer reads, more-variable components of the ITS region in particular will be accessible for differentiation of specific populations of fungi within a genus.
A pattern of both increased soil organic C and microbial diversity in plots treated with residue was observed at both field sites. Soil organic matter has been shown to have an impact on arbuscular mycorrhizal diversity in other semiarid soils (22) and on bacterial communities in polar desert soil (23). There was no detected response by the Glomeromycota in this study, but there were large increases in fungal diversity with intercropping and residue amendment. The primer set used for the ITS1 amplification and sequencing may be biased against the Glomeromycota, which would explain why we did not see a response in this group compared to other researchers (22). Van Horn et al. (23) also demonstrated that in low- to medium-saline soils, the addition of organic matter consistently shifted the bacterial community composition to a higher degree than increasing soil moisture through water addition. This result has implications for the woody-shrub intercropping system in semiarid soils, where organic matter additions may shape the microbial communities. Ng et al. (24) also found that organic matter amendment in the form of green waste significantly increased the number of PLFAs observed, particularly those coming from fungal populations. They noted that the ratio of bacteria to fungi was significantly reduced with green-waste amendment, showing a large response of the fungal community that is similar to what is shown in this study. Similarly, Cookson et al. (25) presented results showing a significant increase in total PLFAs with the application of hay, which matches the findings of Diedhiou et al. (11) and this study, indicating higher microbial diversity in residue-amended intercropped soils.
In this study, fine-scale resolution of differences in microbiome structure was resolved using Illumina sequencing of ribosomal sequences. Previous studies on the effect of intercropping on the structures of microbiomes have used coarse community profiling techniques, such as terminal restriction fragment length polymorphism (T-RFLP) (26, 27), phospholipid fatty acid (PLFA) analysis (28), and denaturing gradient gel electrophoresis (DGGE) (29, 30). Such methods were able to identify changes across phyla or classes of microorganisms, but the sequence-based approach used here quantifies population responsiveness of unique members of individual genera (Fig. 1 and 2). By identifying certain microorganisms associated with a particular ecological function (such as crop growth promotion), it is possible to eventually recover those microorganisms of interest through marker-assisted selection (20). Parallel advances have been made in the analysis of functional diversity of microbial communities. Such techniques have advanced from community-level physiological profiling (CLPP) with Biolog EcoPlates (31) to the use of microarray technology in the form of GeoChip (32, 33) or metatranscriptomic analysis through the use of high-throughput sequencing (34). We used the DESeq2 R package in order to perform our analyses, following arguments in support of this methodology (35). This package was originally written for differential expression analysis of RNA sequencing data and can be very simply adapted to analyze differential abundances of OTUs between samples. We did not perform any rarification of our sequence libraries, instead preserving the original library size for our analyses. When rarification is performed, valid sequences are discarded from the analysis at random, and the use of proportional abundance discards any data about library sizes from the analysis. These two pitfalls were avoided in our analyses.
Intercropping with a shrub while using the coppicing residues as a mulch presents a workable option for small farmers in sub-Saharan Africa to improve crop growth. This study has shown that intercropping and shrub residue application can have a substantial effect on microbial communities, with individual OTUs expressing high levels of enrichment in intercropping systems (9, 10). Moving forward from these associations between individual OTUs and plant health status to the identification of microorganisms which might provide beneficial functions for improved crop growth will require the isolation and functional characterization of strains corresponding to the OTUs identified here. Previous studies have identified molecular markers associated with plant disease suppression in a disease-suppressive soil (36) using T-RFLP analysis (26). The molecular markers identified in that study were cloned, sequenced, and then used to design an isolation strategy, resulting in the recovery of novel species of Mitsuaria and Burkholderia with strong pathogen-suppressing capacities (37). This approach is generally referred to as marker-assisted selection (38) and can draw upon sequences obtained through any number of community profiling methods, including DGGE, T-RFLP, GeoChip, and high-throughput sequencing, as previously described. Subsequent work in Senegal can follow a similar course using the taxonomic information embedded in the sequenced ribosomal gene markers to design semiselective media in order to isolate organisms of interest from the amended soils. Specific primers can be designed using OTU sequences generated here, which can be used to screen culture collections isolated on semiselective media. Isolates containing these markers can then be tested in the laboratory, the greenhouse, and the field to determine their impact on crop plant growth. The OTUs in the Chitinophaga, Aspergillus, Fusarium, Penicillium, and Phoma clusters described in this work provide excellent targets for recovery and testing as novel inoculant strains. As the interest in microbials as biopesticides and biofertilizers continues to rise, new approaches to the discovery of beneficial microbes (20, 38) will help to streamline the process of identifying and introducing novel microbial inoculants. These advances in sequencing technology, analytical tools, and microbiological techniques offer new opportunities in phytobiome research and the development of novel and effective microbial inputs for agriculture.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Bright, Chelsea Delay, and Sidy Diakhaté for important contributions during fieldwork. The help of Mariama Gueye and Lamine Dieng was greatly appreciated in the lab. Maria Elena Hernandez and Jody Whittier at the Molecular and Cellular Imaging Center at the Ohio State University Ohio Agricultural Research and Development Center (http://oardc.osu.edu/mcic/) provided much assistance with the Illumina sequencing.
Spencer Debenport was supported by the Ohio State University Distinguished University Fellowship. This work was funded through National Science Foundation Partnerships for International Research and Education (PIRE) grant OISE-0968247.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04122-14.
REFERENCES
- 1.Tittonell P, Scopel E, Andrieu N, Posthumus H, Mapfumo P, Corbeels M, va Halsema GE, Lahmar R, Lugandu S, Rakotoarisoa J, Mtambanengwe F, Pound B, Chikowo R, Maudin K, Triomphe B, Mkomwa A. 2012. Agroecology-based aggradation-conservation agriculture (ABACO): targeting innovations to combat soil degradation and food insecurity in semi-arid Africa. Field Crops Res 132:168–174. doi: 10.1016/j.fcr.2011.12.011. [DOI] [Google Scholar]
- 2.Lufafa A, Diedhiou I, Samba SAN, Sene M, Khouma M, Kizito F, Dick RP, Dossa E, Moller JS. 2008. Carbon stocks and patterns in native shrub communities of Senegal's Peanut Basin. Geoderma 146:75–82. doi: 10.1016/j.geoderma.2008.05.024. [DOI] [Google Scholar]
- 3.Lufafa A, Diedhiou I, Ndiaye AS, Sene M, Kizito F, Dick RP, Noller JS. 2009. Allometric relationships and peak-season community biomass stocks of native shrubs in Senegal's Peanut Basin. J Arid Environ 73:260–266. doi: 10.1016/j.jaridenv.2008.09.020. [DOI] [Google Scholar]
- 4.Lahmar R, Bationo BA, Dan Lamso N, Guéro Y, Tittonell P. 2012. Tailoring conservation agriculture technologies to West Africa semi-arid zones: building on traditional local practices for soil restoration. Field Crops Res 132:158–167. doi: 10.1016/j.fcr.2011.09.013. [DOI] [Google Scholar]
- 5.Diack M, Sene M, Badiene AN, Diatta M, Dick RP. 2000. Decomposition of a native shrub, Piliostigma reticulatum, litter in soils of semiarid Senegal. Arid Soil Res Rehabil 14:205–218. doi: 10.1080/089030600406626. [DOI] [Google Scholar]
- 6.Dossa EL, Diedhiou S, Compton JE, Assigbetse KB, Dick RP. 2010. Spatial patterns of P fractions and chemical properties of soils in two native shrub communities in Senegal. Plant Soil 327:185–198. doi: 10.1007/s11104-009-0044-8. [DOI] [Google Scholar]
- 7.Kizito F, Sene M, Draglia MI, Lufafa A, Diedhiou I, Dossa E, Cuenca R, Selker J, Dick RP. 2007. Soil water balance of annual crop-native shrub systems in Senegal's Peanut Basin: the missing link. Agric Water Manag 90:137–148. doi: 10.1016/j.agwat.2007.02.015. [DOI] [Google Scholar]
- 8.Diakhaté S, Villenave C, Diallo NH, Ba AO, Djigal D, Masse D, Sembène PM, Chapuis-Lardy L. 2013. The influence of a shrub-based intercropping system on the soil nematofauna when growing millet in Senegal. Eur J Soil Biol 57:37–41. doi: 10.1016/j.ejsobi.2013.04.003. [DOI] [Google Scholar]
- 9.Dossa EL, Diedhiou I, Khouma M, Sene M, Lufafa A, Kizito F, Samba SAN, Badiene AN, Diedhiou S, Dick RP. 2012. Crop productivity and nutrient dynamics in a shrub (Guiera senegalensis)-based farming system of the Sahel. Agron J 104:1255–1264. doi: 10.2134/agronj2011.0399. [DOI] [Google Scholar]
- 10.Dossa EL, Diedhiou I, Khouma M, Sene M, Badiene AN, Ndiaye SA, Assigbetse KB, Sall A, Lufafa A, Kizito F, Dick RP, Saxena J. 2013. Crop productivity and nutrient dynamics in a shrub-based farming system in the Sahel. Agron J 105:1237–1246. doi: 10.2134/agronj2012.0432. [DOI] [Google Scholar]
- 11.Diedhiou S, Dossa EL, Badiane AN, Diedhiou I, Sene M, Dick RP. 2009. Decomposition and spatial microbial heterogeneity associated with native shrubs in soils of agroecosystems in semi-arid Senegal. Pedobiologia 52:273–286. doi: 10.1016/j.pedobi.2008.11.002. [DOI] [Google Scholar]
- 12.Walkley A, Black IA. 1934. An examination of the degetjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci 37:29–38. doi: 10.1097/00010694-193401000-00003. [DOI] [Google Scholar]
- 13.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, Gedallovich SM, Discenza J, Rangamannar R, Koshner JA, Massmann AL, Orazi G, Essene A, Leff JW, Fierer N. 2013. Digging the New York City skyline: soil fungal communities in green roofs and city pars. PLoS One 8(3):e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulan-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Core Team R. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 17.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. BioRxiv http://dx.doi.org/10.1101/002832. [DOI] [PMC free article] [PubMed]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Park JK, Lee SW, Han S, Kim JC, Kim YC, McSpadden Gardener B. 2013. Marker-assisted selection of novel bacteria contributing to soilborne disease suppression, p 637–642. In de Bruijn FJ. (ed), Molecular microbial ecology of the rhizosphere. Wiley, Hoboken, NJ. [Google Scholar]
- 21.Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim YO, Redman RS. 2008. Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 22.Del Mar Alguacil M, Díaz-Pereira E, Caravaca F, Fernández DA, Roldán A. 2009. Increased diversity of arbuscular mycorrhizal fungi in a long-term field experiment via application of organic amendments to a semiarid degraded soil. Appl Environ Microbiol 75:4254–4263. doi: 10.1128/AEM.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Horn DJ, Okie JG, Buelow HN, Gooseff MN, Barrett JE, Takacs-Vesbach CD. 2014. Soil microbial responses to increased moisture and organic resources along a salinity gradient in a polar desert. Appl Environ Microbiol 80:3034–3043. doi: 10.1128/AEM.03414-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng EL, Patti AF, Rose MT, Schefe CR, Kilkinson K, Cavagnaro TR. 2014. Functional stoichiometry of soil microbial communities after amendment with stabilized organic matter. Soil Biol Biochem 76:170–178. doi: 10.1016/j.soilbio.2014.05.016. [DOI] [Google Scholar]
- 25.Cookson WR, Abaye DA, Marschner P, Murphy DV, Stockdale EA, Goulding KWT. 2005. The contribution of soil organic matter fractions to carbon and nitrogen mineralization and microbial community size and structure. Soil Biol Biochem 37:1726–1737. doi: 10.1016/j.soilbio.2005.02.007. [DOI] [Google Scholar]
- 26.Benitez MS, Baysal F, Rotenberg D, Kleinhenz MD, Cardina J, Stinner D, Miller SA, McSpadden Gardener BB. 2007. Multiple statistical approaches of community fingerprint data reveal bacterial populations associated with general disease suppression arising from the application of different organic field management strategies. Soil Biol Biochem 39:2289–2301. doi: 10.1016/j.soilbio.2007.03.028. [DOI] [Google Scholar]
- 27.Sun YM, Zhang NN, Wang ET, Yuan HL, Yang JS, Chen WX. 2009. Influence of intercropping and intercropping plus rhizobial inoculation on microbial activity and community composition in rhizosphere of alfalfa (Medicago sativa L.) and Siberian wild rye (Elymus sibiricus L.). FEMS Microb Ecol 70:218–226. doi: 10.1111/j.1574-6941.2009.00752.x. [DOI] [PubMed] [Google Scholar]
- 28.Lacombe S, Bradley RL, Hamel C, Beaulieu C. 2009. Do tree-based intercropping systems increase the diversity and stability of soil microbial communities? Agric Ecosyst Environ 131:25–31. doi: 10.1016/j.agee.2008.08.010. [DOI] [Google Scholar]
- 29.Song YN, Zhang FS, Marschner P, Fan FL, Gao HM, Gao XG, Sun JH, Li L. 2007. Effect of intercropping on crop yield and chemical and microbiological properties in rhizosphere of wheat (Triticum aestivum L.), maize (Zea mays L.), and faba bean (Vica faba L.). Biol Fertil Soils 43:565–574. doi: 10.1007/s00374-006-0139-9. [DOI] [Google Scholar]
- 30.Zhou X, Yu G, Wu F. 2011. Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Eur J Soil Biol 47:279–287. doi: 10.1016/j.ejsobi.2011.07.001. [DOI] [Google Scholar]
- 31.Liu F, Wu J, Ying GG, Luo Z, Feng H. 2012. Changes in functional diversity of soil microbial community with addition of antibiotics sulfamethoxazole and chlortetracycline. Appl Microbiol Biotechnol 95:1615–1623. doi: 10.1007/s00253-011-3831-0. [DOI] [PubMed] [Google Scholar]
- 32.Bai S, Li J, He Z, Van Nostrand JD, Tian Y, Lin G, Zhou J, Zheng T. 2013. GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Appl Microbiol Biotechnol 97:7035–7048. doi: 10.1007/s00253-012-4496-z. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Zhao X, Liang Y, Li G, Zhou J. 2013. Microbial functional genes reveal selection of microbial community by PAHs in polluted soils. Environ Chem Lett 11:11–17. doi: 10.1007/s10311-012-0370-6. [DOI] [Google Scholar]
- 34.Tveit A, Urich TT, Svenning MM. 2014. Metatranscriptomic analysis of arctic peat soil microbiota. Appl Environ Microbiol 80:5761–5772. doi: 10.1128/AEM.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarifying microbiome data is inadmissible. PLoS Comput Biol 10(4):e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baysal F, Benitez MS, Kleinhenz MD, Miller SA, McSpadden Gardener BB. 2008. Field management effects on damping-off and early season vigor of crops in a transitional organic cropping system. Phytopathology 98:562–570. doi: 10.1094/PHYTO-98-5-0562. [DOI] [PubMed] [Google Scholar]
- 37.Benitez MS, McSpadden Gardener BB. 2009. Linking sequence to function in soil: sequence-directed isolation of novel bacteria contributing to soilborne disease suppression. Appl Environ Microbiol 75:914–924. doi: 10.1128/AEM.01296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JK, Lee SH, Lee JH, Han S, Kang H, Kim JC, Kim YC, McSpadden Gardener B. 2013. Sampling and selection factors that enhance the diversity of microbial collections: application to biopesticide development. Plant Pathol J 29:144–153. doi: 10.5423/PPJ.SI.01.2013.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.FAO. 2006. The state of food insecurity in the world. FAO, Rome, Italy. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.