Abstract
We report the identification, characterization, and gene cloning of a novel protein elicitor (PeBL1) secreted from Brevibacillus laterosporus strain A60. Through a purification process consisting of ion-exchange chromatography and high-performance liquid chromatography (HPLC), we isolated a protein that was identified by electrospray ionization quadrupole time of flight tandem mass spectrometry (ESI–Q-TOF–MS-MS). The 351-bp PeBL1 gene produces a 12,833-Da protein with 116 amino acids that contains a 30-residue signal peptide. The PeBL1 protein was expressed in Escherichia coli. The recombinant protein can induce a typical hypersensitive response (HR) and systemic resistance in Nicotiana benthamiana, like the endogenous protein. PeBL1-treated N. benthamiana exhibited strong resistance to the infection of tobacco mosaic virus-green fluorescent protein (TMV-GFP) and Pseudomonas syringae pv. tabaci compared to control N. benthamiana. In addition, PeBL1 triggered a cascade of events that resulted in defense responses in plants, including reactive oxygen species (ROS) production, extracellular-medium alkalization, phenolic-compound deposition, and expression of several defense-related genes. Real-time quantitative-PCR analysis indicated that the known defense-related genes PR-1, PR-5, PDF1.2, NPR1, and PAL were upregulated to varying degrees by PeBL1. This research not only provides insights into the mechanism by which beneficial bacteria activate plant systemic resistance, but also sheds new light on a novel strategy for biocontrol using strain A60.
INTRODUCTION
In nature, plants live in complicated surroundings with various beneficial microbes and potential plant pathogens. In order to prevent infection by pathogens, plants have evolved defense mechanisms leading to a basic innate immunity (1, 2). Additionally, beneficial bacteria can generate protective action that indirectly makes plants resist the infection of further pathogens through the elicitation of the plant defense system (3). This defensive capacity is systemic, for example, root treatment with beneficial bacteria could extend to above-ground plant parts, triggering resistance in the whole plant. Resistance responses triggered by nonpathogens are called induced systemic resistance (ISR), which can efficiently resist a broad spectrum of pathogens, including bacteria, fungi, viruses, nematodes, and insects (4–6). ISR is phenotypically similar to the well-studied systemic acquired resistance (SAR) motivated by an incompatible pathogen (7). Most of the ISR-inducing bacteria are plant growth-promoting bacteria (PGPB), which are related to many plant species and are generally present in a variety of environments (8). The best-studied class of PGPB are plant growth-promoting rhizobacteria (PGPR) colonizing the root surfaces and the rhizosphere (9).
ISR has been documented in many plant species, for example, Arabidopsis thaliana, tomatoes, beans, cucumbers, radishes, and tobacco (2, 6). Globally, ISR has been considered a three-step procedure that comprises perception of the elicitor, signal transduction, and defense-related gene expression, triggering resistance in the whole plant. ISR-associated signal transduction mechanisms have been demonstrated, although they are less well understood than SAR. The salicylic acid (SA)-, jasmonic acid (JA)-, and ethylene (ET)-dependent pathways are major players in the regulation of signaling networks that are involved in induced defense responses (10–12). SA is an important signaling molecule involved in the induction of systemic resistance and local defense reactions (13). Several pathogenesis-related (PR) genes, such as PR-1a, PR-1b, and PR-5, are generally used as markers of SA-dependent defenses. JA, a signaling molecule, is involved in many different aspects of plant biology, including defense and development (14). ET regulates several processes in plants and has been implicated in defense responses. Normally, ISR is mediated by a signaling pathway in which JA and ET play key roles (6, 15). However, Van Wees et al. demonstrated that activation of an SA-dependent pathway is a feature of ISR-inducing biocontrol bacteria (16). Maurhofer et al. reported that ISR induced by P. fluorescens strain CHA0 in tobacco is related to PR protein accumulation, suggesting that ISR and SAR share similar mechanisms. Thus, the defense mechanisms of ISR must be further studied (17).
The plant resistance system is a condition of enhanced defensive capacity. Plant defense responses triggered by elicitors of biotic and nonbiotic origin are part of the plant resistance and play important roles in the signal exchange between the plant and the microbe. The elicitors, derived from various organisms, including bacteria, fungi, viruses, and oomycetes, have different chemical natures and include proteins, glycoproteins, peptides, lipids, and oligosaccharides (18–20). For example, harpins are multifunctional protein elicitors produced by Gram-negative plant-pathogenic bacteria (21). The fungal elicitors Hrip1, PevD1, and MoHrip1 from Verticillium dahliae, Alternaria tenuissima, and Magnaporthe oryzae, respectively, have been applied to plants to improve their pathogen resistance (22–24). However, a few reports have focused on elicitors that were isolated from biocontrol bacteria. For example, dimethyl disulfide (DMDS), an elicitor produced by an ISR-eliciting Bacillus cereus strain, can suppress plant fungal diseases and play a crucial role in ISR by B. cereus C1L (25). Massetolide A, produced by Pseudomonas fluorescens SS101, is involved in ISR-eliciting defensive capacity in tomato against Phytophthora infestans (26). Surfactins and fengycins produced by Bacillus subtilis S499 can also act as elicitors of ISR (5). In contrast to the many research studies performed with pathogen-associated molecular patterns (PAMPs), used as models for early defense-related events, very little information is available about the perception mechanisms of ISR-specific protein elicitors (27).
In general, a defense reaction triggered by elicitors can be divided into two stages. The first stage occurs minutes after using an elicitor and includes ion fluxes across the cell membrane, extracellular-medium alkalization, and reactive oxygen species (ROS). In the plant defense reaction, ROS are considered to play an important role in the elicitor signal transduction system and also to be associated with the hypersensitive response (HR) (28) as a marker of the plant defense reaction (29, 30). ROS have been demonstrated to be sufficient for the induction of plant secondary-metabolite accumulation and are required in the plant defense reaction (27, 31, 32). The second stage occurs hours after elicitor ingestion and involves the activation of defense-related genes correlated with cytoderm reinforcement, the synthesis of phytoalexins, the accumulation of PR proteins, and induction of defense compounds, such as phenolic compounds, callose, and PAL (phenylalanine ammonia lyase) (33).
Brevibacillus laterosporus, a Gram-positive, aerobic, spore-forming bacterium previously classified as Bacillus laterosporus, can produce diverse metabolites with antifungal activity, which can control the infection of plant pathogens as biocontrol agents (34). We previously screened a novel strain, A60, that was isolated from the soil of mango plants in Changjiang, Hainan Province, China (19°15.635′N, 108°46.029′E), which was identified as B. laterosporus by phenotypic characterization and 16S rRNA sequencing (35). In addition to antimicrobial activity, strain A60 also exhibited the induction of systemic resistance in numerous types of plants, such as wheat, pepper, and Chinese cabbage. The control efficiencies against Phytophthora capsici and Peronospora parasitica in pepper and Chinese cabbage that were treated with strain A60 Aqua (5 × 109 CFU/ml) were 81.6% and 73.7%, respectively, after 10 days. In particular, the yield of Chinese cabbages after treatment with A60 Aqua (5 × 109 CFU/ml) increased by 13.2% compared to the wild type. Based on the excellent effect, we registered microbial fertilizer containing B. laterosporus strain A60 Aqua (no. 2014-2058) with the Ministry of Agriculture of China. The fertilizer has achieved large-scale production in the factory in Henan Province, with an annual output of 5,000 tons. The application area has increased to 3 million acres. In previous studies, a novel antimicrobial peptide, BL-A60, with a molecular mass of 1,602.0469 Da, was isolated and purified from strain A60 (35). However, the metabolites involved in the activation of systemic resistance by strain A60 have not been completely studied.
In this study, we report the purification and characterization of the novel protein elicitor PeBL1 from B. laterosporus strain A60. PeBL1 activated certain early plant defense responses and systemic resistance in Nicotiana benthamiana against infection by tobacco mosaic virus-green fluorescent protein (TMV-GFP) and Pseudomonas syringae pv. tabaci. Our research helps to elucidate the mechanisms of N. benthamiana systemic resistance triggered by PeBL1 and provides a novel strategy for using B. laterosporus strain A60 to control plant disease.
MATERIALS AND METHODS
Bacteria and plant cultivation.
The strain A60 was preserved at the China General Microbiological Culture Collection Center (CGMCC) (no. 5694) and maintained on Luria-Bertani (LB) medium (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter distilled water) at 37°C in the dark. N. benthamiana seeds were germinated on 1/2 Murashige-Skoog medium in a growth chamber that was maintained at 25°C with 12 h of light and 12 h of darkness. Following germination, the seedlings were transferred to an autoclaved soil mixture containing 1:3 (vol/vol) high-nutrient soil and vermiculite in 8- by 7.5- by 7.5-cm pots. One plant per pot was kept in the growth chamber at 25°C with 50% humidity and 16 h of light.
Establishment of N. benthamiana cell culture.
Tobacco (N. benthamiana) seeds were soaked in 75% ethanol for 45 s and in 10% sodium hypochlorite for 10 min, followed by three washes with sterilized water. The sterilized N. benthamiana seeds were cultivated for callus in Murashige-Skoog medium. The calluses were cut into small pieces after 15 days and suspended in liquid Murashige-Skoog medium, pH 5.0, supplemented with inositol (100 mg/ml), 0.2% KH2PO4, 3% sucrose, 2,4-dichlorophenoxyacetic acid (0.2 mg/ml), and HCl (1 mg/ml) under shaking at 130 rpm at 25°C in the dark. Subcultures were inoculated with 4 ml of 5-day-old stock suspensions (36).
Isolation and detection of crude protein.
B. laterosporus strain A60 was cultured in 3,000 ml LB medium with shaking at 180 rpm for 48 h at 37°C, and the supernatant was collected after centrifugation (4,700 × g; 15 min; 4°C). Solid ammonium sulfate was added to the supernatant to achieve 80% (wt/vol) relative saturation at 4°C overnight. The precipitate was harvested by centrifugation (12,000 × g; 20 min; 4°C), redissolved in 200 ml buffer A (25 mM MES [morpholineethanesulfonic acid]-NaOH, pH 6.2), and dialyzed against buffer A for 48 h. Before filtering the crude protein with a 0.22-μm filter (Millipore, Suzhou, China), the insoluble debris was removed from the dialysate by centrifugation (12,000 × g; 10 min; 4°C). A portion of the crude protein (50 μl) was tested for elicitor activity (HR-inducing activity).
Purification of protein.
Further purification was performed using the Äkta Explorer protein purification System (Amersham Biosciences). The crude protein was loaded onto an ion-exchange chromatography HiTrap SP FF column (GE Healthcare, Uppsala, Sweden) that was preequilibrated with elution buffer (25 mM MES-NaOH, pH 6.2). The bound proteins were eluted with a linear gradient of increasing NaCl in elution buffer at a flow rate of 2 ml/min. All fractions were collected and injected into a desalting column (GE Healthcare, Uppsala, Sweden) for elicitor activity analysis. The purified protein was monitored for elicitor activity.
The pooled active fraction after desalting was purified through high-performance liquid chromatography (HPLC) on a C18 reverse-phase column injected onto a Zorbax Eclipse XDB-C18 reverse-phase column (150 by 4.6 mm; 5 μm; Agilent) equilibrated with 5% acetonitrile (ACN), 2 mM NH4FA, 0.1% formic acid (FA), and water. The pooled active fraction was eluted with chromatography grade ACN using a linear gradient increasing from 20% (vol/vol) to 100% (vol/vol) over 30 min at a flow rate of 0.2 ml/min. All of the peaks were collected automatically by fraction collectors (Agilent). Each peak was freeze-dried, redissolved in ultrapure water (Milli-Q), and tested for elicitor activity. The fraction with elicitor activity was subjected to chromatography again to ensure its purity, and the molecular mass was determined via Tricine SDS-PAGE.
Mass spectrometry analysis and gene identification.
The protein sample was isolated on a Tricine SDS-PAGE gel and digested overnight using mass spectrometry (MS)-grade Trypsin Gold (Promega, Madison, WI, USA). The digested peptides were reacted with succinimidyl-2-morpholine acetate (SMA) for analysis by tandem MS (MS-MS). The purified peptides were sprayed into a quadrupole time of flight (Q-TOF) mass spectrometer (MicrO TOF-QII; Bruker Daltonics K.K., Tokyo, Japan) with an electrospray ionization (ESI) ion source. The MS-MS data were automatically analyzed by the Mascot search engine (Matrix Science, London, United Kingdom), using the following parameters: database, NCBInr; taxonomy, B-laterosporus; enzyme, trypsin; type of search, MS-MS ion search. The peptide and fragment mass tolerances were set at 0.1 Da. Proteins with probability-based molecular weight search (MOWSE) scores exceeding the threshold (P < 0.05) were definitely identified.
The genomic DNA was extracted from B. laterosporus strain A60 using an E.Z.N.A. bacterial DNA kit (Omega Bio-Tek, Norcross, GA, USA). A pair of gene-specific primers were designed to amplify the PeBL1 gene sequence deduced from the mass spectrometry analysis and the Mascot database search. The primer sequences were designed as follows: forward primer, 5′-ATGAAAAAAGCTGTCTCAAC-3′, and reverse primer, 5′-TTAGTAGGGAACAGTTATATT-3′. The PCR product was cloned into the pMD 18-T vector (TaKaRa, Dalian, China) and verified by DNA sequencing (Beijing Genomics Institution, Beijing, China).
Expression in E. coli and purification of recombinant protein.
The PeBL1 gene, without its predicted signal peptide, was inserted into the pET30-TEV/LIC vector (Novagen, Darmstadt, Germany). Then, the recombinant plasmid was transformed into Escherichia coli BL21(DE3) (TransGen Biotech, Beijing, China). The primers, including a fragment of the pET30-TEV/LIC vector and the 5′ and 3′ ends of the PeBL1 gene, were designed as follows: forward primer, 5′-TACTTCCAATCCAATGCCACACCAGCCAAACACTC-3′, and reverse primer, 5′-TTATCCACTTCCAATGCTATTAGTAGGGAACAGTTATATTC-3′. The DNA was isolated by electrophoresis and observed by staining with Gold View (SBS Genetech, Beijing, China) using Trans 2K DNA marker (TransGen Biotech, Beijing, China). The PCR product was cloned into the vector using ligation-independent cloning (37) and verified by DNA sequencing.
To express the recombinant PeBL1 protein, bacteria were first grown at 37°C for 4 h, and the recombinant protein was subsequently induced by the addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside (Sigma, St. Louis, MO, USA) to the medium at 16°C for 14 to 16 h. The acquired cells were disrupted two times with an ultrasonic disruptor to pool the supernatant. The purification procedure for recombinant PeBL1 was as follows: affinity chromatography with a His-Trap HP column (GE Healthcare, Waukesha, WI, USA) and a HiTrap desalting column (GE Healthcare, Waukesha, WI, USA). The purified protein was tested for elicitor activity and detected by Tricine SDS-PAGE. Protein markers (Thermo Scientific) were used to evaluate the apparent molecular masses of the purified recombinant proteins.
Characteristics of the PeBL1 protein.
The HR-inducing activity of PeBL1 was evaluated in N. benthamiana leaves. The N. benthamiana leaves were injected with samples (50 μl) or a control by using a syringe to cover areas of 1 to 2 cm2. The HR symptoms were examined after 24 h according to a previously described method (38). The amplification of the tobacco HR marker gene, HSR203, was performed by reverse transcription-PCR with the following primers: HSR203, forward, 5′-TGTACTACACTGTCTACACGC-3′, and reverse, 5′-GATAAAAGCTATGTCCCACTCC-3′), and EF-1α, forward, 5′-AGACCACCAAGTACTACTGCAC-3′, and reverse, 5′-CCACCAATCTTGTACACATCC-3′), as a positive control.
In order to ascertain the influence of pH on elicitor activity, the pH of the elicitor was adjusted to 4, 6, 8, or 11 with NaOH or HCl, and the elicitor was incubated overnight, dialyzed, and then used in the elicitor bioassay.
In order to determine elicitor heat stability, four aliquots of purified protein were incubated at 4, 25, 50, and 75°C for 15 min, and subsequently, the elicitor activities of the treated proteins were tested.
Detection of hydrogen peroxide production and alkalinization of the extracellular medium.
The histological localization of hydrogen peroxide production in N. benthamiana leaves was determined as previously described (39). Briefly, PeBL1 (5 μM) or Tris-HCl (negative control) was injected into 8-week-old leaves. Subsequently, the leaves were isolated after 24 h of treatment and soaked in 3,3′-diaminobenzidine (DAB)-HCl (1 mg/ml, pH 3.8) solution. After incubation for 8 h in the dark, the treated leaves were placed in 95% ethanol at 65°C to remove chlorophyll and observed under an Olympus SZX9 stereomicroscope (Olympus America, Inc., Melville, NY, USA). ROS production in the tobacco cell suspensions was quantified by chemiluminescence using luminol (40). In brief, 250 μl of cells was incubated with 300 μl of buffer (pH 5.75) containing 10 mM HEPES, 175 mM mannitol, 0.5 mM CaCl2, 0.5 mM K2SO4 for 1 h at 26°C. Then, PeBL1 (10 μM) and luminol (0.3 mM) were mixed into the buffer and rotated in a shaker. The chemiluminescence was expressed as nanomoles of H2O2 per gram (fresh weight) of tobacco cells using a standard calibration curve and monitored with the GloMax-96 luminometer (Promega, Madison, WI, USA).
The alkalinization of extracellular medium was performed in the tobacco cell suspensions (41). The test was conducted simultaneously in three 10-ml flasks (test, negative control, and positive control), each of which contained 1 g (fresh weight) of cells per 5 ml of cell suspension. Tobacco cells were preequilibrated with an orbital shaker for approximately 45 min at 26°C until a steady pH value (5.0 to 5.2) was achieved and then treated with PeBL1 (10 μM). The pH was observed for 90 min after PeBL1 addition. Flagellin peptide Flg22 (1 μM), a bacterial PAMP, was added as a positive control. Tris-HCl buffer was added as a negative control. The changes in the pH of the suspension media were monitored using a pH meter (Sartorius Stedim, Germany).
Detection of phenolic-compound accumulation in tobacco cell culture.
For the measurement of phenolic-compound accumulation in tobacco cells, 300 μl of tobacco cell suspension was examined after incubation with 50 μl (10 μM) PeBL1 in the dark at 26°C for 108 h under epifluorescence with a Zeiss Axiovert 100 M inverted microscope equipped with a confocal laser scanner (LSM 510; Zeiss, Oberkochen, Germany). Tris-HCl buffer (pH 7.3) was used as a negative control.
Bioassay for PeBL1-induced disease resistance in N. benthamiana.
We used TMV-GFP, a recombinant virus in which the jellyfish GFP gene was extended into the coat protein (CP) open reading frame (ORF) of native TMV. The GFP was visualized by using a 100-W long-wave UV lamp (Black Ray model B 100AP; UVP, Upland, CA, USA). The recombination did not influence the infection and movements of virus in N. benthamiana (42, 43). Three leaves each from six plants were injected at one spot per leaf with 10 μM PeBL1 or with Tris-HCl buffer as a negative control. Three days later, the upper three nontreated (systemic) leaves were inoculated with TMV-GFP. The concentration of TMV-GFP solution was 0.5 g diseased leaves in 10 ml ultrapure water (Milli-Q). The signs and diameters of TMV lesions in each leaf were analyzed at 2, 4, and 6 days postinoculation (p.i.), as previously described (44, 45). For the diameters of the lesions, 10 random lesions were measured for each plant. Three replicates were performed. The inhibition of TMV lesions was calculated using the following formula: percent inhibition = {[number (size) of lesions on control leaves − number (size) of lesions on elicitor-treated leaves]/number (size) of lesions on control leaves} × 100%.
To assay whether PeBL1 can induce systemic resistance and diminish disease signs in N. benthamiana, we next included a phytopathogenic bacterium (P. syringae pv. tabaci) in our experiments. P. syringae pv. tabaci bacteria were cultivated at 28°C in King's B medium (46) for 24 h, harvested with centrifugation, and then washed three times and resuspended in sterile distilled water at an optical density at 600 nm (OD600) of 0.6 (1 × 107 CFU/ml). The method by which N. benthamiana plants were treated with PeBL1 and buffer was as described for the assay of TMV resistance. After 3 days, the upper three nontreated leaves were inoculated with P. syringae pv. tabaci by soaking for 45 s. Signs were assessed 4 days after challenge with P. syringae pv. tabaci. The leaves were detached and sterilized to remove epiphytic bacterial populations. Three samples were collected from each leaf with a sterilized hole punch and ground with a pestle in 100 μl sterile water. The sample suspensions were vortexed completely and serially diluted to 10−3. The bacteria in the dilution were inoculated on a King's B Kan25 (10 g tryptone, 10 ml glycerin, 1.5 g K2HPO4, 1.5 g MgSO4·7H2O, 25 mg kanamycin) plate and grown for 2 days, and the colonies on each plate were counted. The area of each sample was approximately 0.1963 cm2, and the number of CFU per cm2 was calculated by multiplying by the dilution factor.
Analysis of the expression of defense-related genes induced by PeBL1 using RT-qPCR.
To study the mechanisms of the defense responses induced by PeBL1 in N. benthamiana plants, N. benthamiana plants that were infiltrated with PeBL1 or buffer on three leaves were assayed for the induction of several defense-related genes. A small fragment was collected from the upper leaves at the indicated times and rapidly frozen in liquid nitrogen. The fragments were placed in RNase-free tubes and frozen at −80°C until use. Control plants were infiltrated with buffer. Total RNA was extracted with the EasyPure plant RNA kit (TransGen Biotech, Beijing, China). The cDNA was generated using the TransScript All-in-One SuperMix for qPCR kit (TransGen Biotech, Beijing, China), and the concentrations of the cDNAs were adjusted to be equal. Real-time quantitative PCR (RT-qPCR) was performed to determine the relative expression levels of several defense-related genes and conducted using TransStart Green qPCR SuperMix UDG (TransGen Biotech, Beijing, China). The specific genes were designed from the coding sequences of each gene using Beacon Designer 8.12 (Table 1). The PCR mixture was processed on an IQ-5 real-time system (Bio-Rad) under the following program: 50°C for 2 min and 94°C for 10 min, followed by 43 cycles of 94°C for 5 s and 60°C for 30 s. A melting curve was established from 65 to 95°C. Three technical replicates were amplified for each sample, including negative controls. The EF-1α (elongation factor 1α) gene was used as a reference gene for normalization. Quantification of the relative changes in gene transcript levels was performed using the comparative 2−ΔΔCT method (47). The mean deviation was calculated from the standard deviation (SD) in the ΔΔCT value using the formula 2−ΔΔCT ± SD.
TABLE 1.
Primers used for RT-qPCR of defense-related and internal control genes
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| PR-1a | 5′-CCTCGTACATTCTCATGGTCAAT-3′ | 5′-CCATTGTTACACTGAACCCTAGC-3′ |
| PR-5 | 5′-CCGAGGTAATTGTGAGACTGGAG-3′ | 5′-CCTGATTGGGTTGATTAAGTGCA-3′ |
| PDF1.2 | 5′-GGAAATGGCAAACTCCATGCG-3′ | 5′-ATCCTTCGGTCAGACAAACG-3′ |
| NPR1 | 5′-ACATCAGCGGAAGCAGTAG-3′ | 5′-GTCGGCGAAGTAGTCAAAC-3′ |
| PAL | 5′-GTTATGCTCTTAGAACGTCGCCC-3′ | 5′-CCGTGTAATGCCTTGTTTCTTGA-3′ |
| EF-1α | 5′-TGTGATGTTTTTGTTCGGTCTTTAA-3′ | 5′-TCAAAAGAAAATGCAGACAGACTCA-3′ |
Protein assay.
All protein concentrations were measured using the BCA protein assay kit (Pierce, Rockford, IL, USA).
Statistical analysis.
All of the experiments and data presented here were from at least three repeats. The data are presented as means ± standard deviations, and significant differences between the treatments and the controls were determined by analysis of variance using SAS (SAS Institute Inc., Cary, NC, USA).
Nucleotide sequence accession number.
The PeBL1 gene sequence has been deposited in GenBank under accession no. KM668059.
RESULTS
Isolation and purification of the elicitor protein.
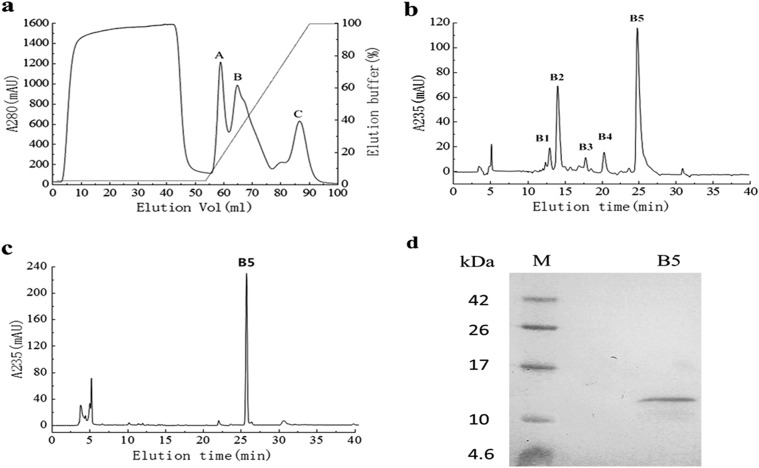
Dialyzed crude protein from B. laterosporus showed HR activity. To purify the active fractions, we performed cation-exchange chromatography on the crude protein using a HiTrap SP FF column, which produced one unadsorbed fraction and three adsorbed fractions (A, B, and C). All the fractions were injected into N. benthamiana leaves to check for elicitor activity (data not shown), and the active fraction of peak B (Fig. 1a) was further purified. Using Agilent HPLC with a Zorbax Eclipse XDB-C18 reverse-phase column, five main peaks (B1, B2, B3, B4, and B5) were obtained (Fig. 1b). Peak B5 was demonstrated to have strong activity and underwent chromatography again under the same conditions. We found that fraction B5 was a single peak (Fig. 1c) and showed a single band on Tricine SDS-PAGE, with a relative apparent molecular mass of 12 kDa (Fig. 1d). We identified the protein as PeBL1.
FIG 1.
Purification of PeBL1 from B. laterosporus. (a) The crude protein was loaded on a HiTrap SP FF 5-ml column at a flow rate of 2 ml/min. Three peaks (A, B, and C) were collected, and the target protein was included in peak B. mAU, milli-absorbance units. (b) Chromatography of peak B using a C18 reverse-phase column. The concentration of ACN in the eluted solvent was raised from 20% (vol/vol) to 100% (vol/vol) over 30 min using a linear gradient at a flow rate of 0.2 ml/min. Five main peaks (B1, B2, B3, B4, and B5) were obtained, and peak B5 showed HR activity. (c) We subjected peak B5 to chromatography again under the same conditions, and a single peak was observed. (d) Tricine SDS-PAGE analysis of peak B5 of PeBL1, showing a single band with Coomassie brilliant blue R-250 staining. Lanes: B5, PeBL1; M, protein molecular mass marker.
Characterization of the PeBL1 protein.
Infiltration of N. benthamiana leaves by the purified PeBL1 induced an apparent necrotic zone in the infiltration area at 24 to 32 h postinfiltration (Fig. 2A), while there was no necrosis on the leaves that were infiltrated with buffer. The early symptoms in the infiltrated area were clearly transparent approximately 10 to 14 h after infiltration (data not shown). PeBL1 also induced the expression of the HSR203 gene (Fig. 2B), which is regarded as the HR-specific gene (48), in N. benthamiana leaves.
FIG 2.
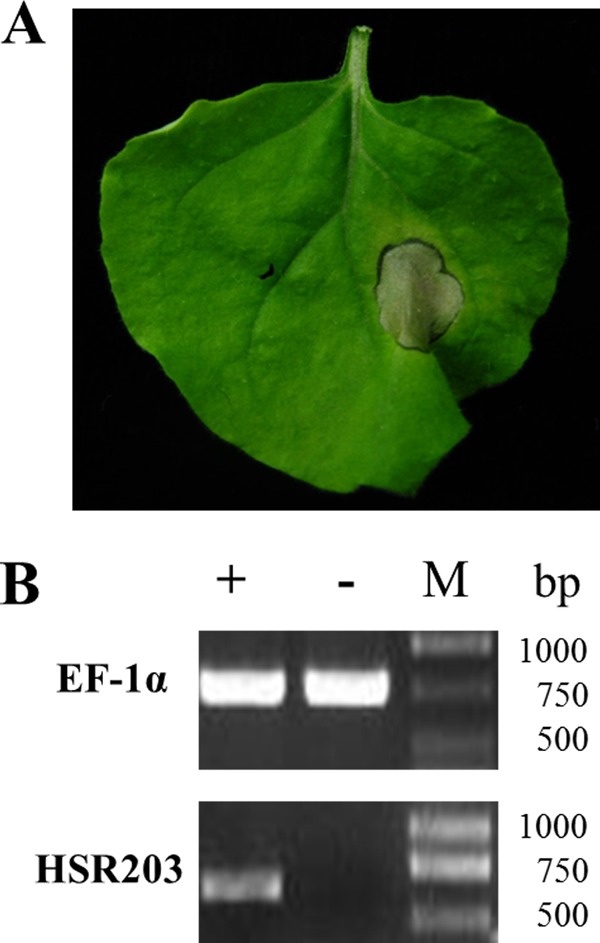
Hypersensitive response induced by PeBL1 in N. benthamiana. (A) The hypersensitive response was observed at 24 h postinjection. The right side of the leaf was treated with elicitor, and the other side was treated with Tris-HCl buffer as a negative control. (B) Total RNA prepared from N. benthamiana leaves treated with buffer (−) or PeBL1 (+) was used as a template in RT-PCR assays. The expression of the HSR203 gene was induced by PeBL1.
PeBL1 was stable at 4°C and 25°C and retained its elicitor activity. However, at 50°C and 75°C, PeBL1 was thermally denatured, with loss of HR activity (data not shown).
We also found that PeBL1 showed HR activity only within an appropriate pH range. After incubation for 12 h at 4°C in a series of solutions at different pHs (4, 6, 8, and 11), PeBL1 showed HR activity only at pH 6 or 8 (data not shown).
The Chou and Fasman Secondary-Structure Prediction (CFSSP) server analysis of the secondary structure of PeBL1 indicated that the percentages of residues of helices, sheets, and turns were 50.0%, 63.8%, and 12.9%, respectively. PeBL1 has very low identity with any known protein structure and function, and PeBL1 also lacks any conserved domains in the sequence, indicating that PeBL1 is a novel protein.
Mass spectrometry analysis and cloning of PeBL1.
The protein sample was excised from the Tricine SDS-PAGE gel for liquid chromatography (LC)-MS analysis of in-gel-digested protein to determine the amino acid sequence of PeBL1. The results of the mass spectrometry analysis were searched by Mascot. Based on the Mascot search results, we obtained the best-matching protein, which had the highest score (194) and contained two different reliable peptides: (i) 45TSNETWNLGSHIR57 (score, 67) and (ii) 88FTAVQPGNASIYVYK102 (score, 47). MOWSE scores greater than 25 were significant for these search results. The protein (GenBank accession no. ERM19151.1) was from B. laterosporus PE36. Using the sequence of strain PE36, we designed the PCR primers to clone the PeBL1 gene.
The PeBL1 gene, whose full-length was 351 bp, encoding a protein of 116 amino acids with a theoretical molecular mass of 12,833 Da, was amplified from the B. laterosporus A60 genomic DNA. The SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) was used for signal peptide analysis of the PeBL1 gene sequence, revealing that the protein contained a 30-residue signal peptide, indicating that PeBL1 is a secreted protein.
Expression and purification of recombinant protein.
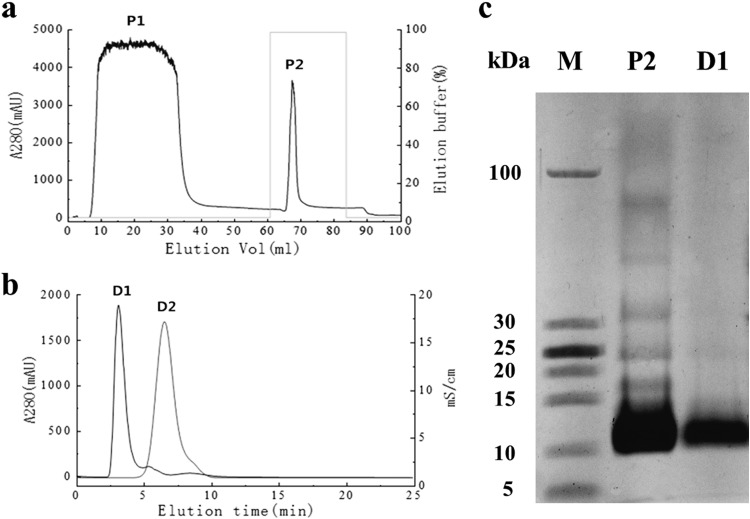
The sequence encoding residues 31 to 116, without the signal peptide, was cloned into the pET30-TEV/LIC vector downstream of a 6×His tag. After amplifying the sequence, the PCR product was cloned into the vector. Subsequently, the recombinant expression vector was transformed into E. coli BL21(DE3) cells. The prokaryotically expressed His6-PeBL1 was soluble in E. coli and was subsequently purified over a His-Trap HP column (Fig. 3a) and a HiTrap desalting column (Fig. 3b). The purified recombinant protein was characterized by a single band at 12 kDa on Tricine SDS-PAGE (Fig. 3c). The purified recombinant protein can also induce a typical HR in N. benthamiana (data not shown).
FIG 3.
Purification of recombinant PeBL1. (a) Total E. coli expressed proteins were purified with a His-Trap HP column. Peak P2, which mainly includes recombinant PeBL1, was eluted with elution buffer (25 mM Tris, 200 mM NaCl, 500 mM imidazole, pH 8.0). (b) Peak P2 was loaded on a HiTrap desalting column at a flow rate of 5 ml/min. The purified and desalted recombinant protein (peak D1) was isolated with saline ion (peak D2). (c) The purified and desalted 6×His-tagged PeBL1 protein (lane D1) showed a single band with a molecular mass of 12 kDa on Tricine SDS-PAGE. Lane M, protein molecular mass marker; lane P2, peak P2.
Induction of ROS production and alkalinization of the extracellular medium by PeBL1.
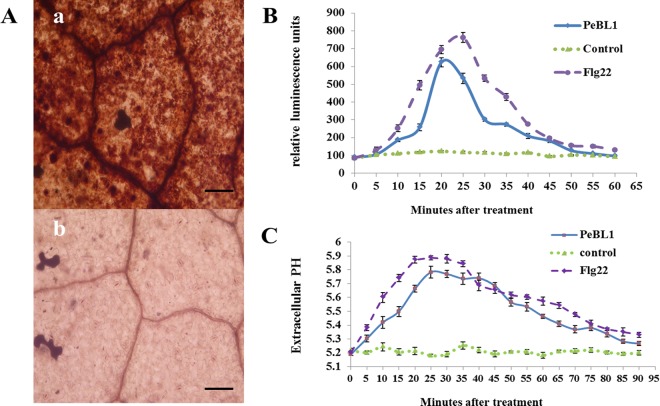
A burst in oxidative metabolism that leads to the accumulation of superoxide (O2−) and H2O2 is considered a significant early event in the plant defense system (49). The PeBL1 elicitor could induce H2O2 accumulation in N. benthamiana leaves. Hydrogen peroxide polymerized by DAB, which forms a dark red-brown precipitate, was detected, and the sites of H2O2 accumulation were obvious microscopically in the stomata and veins of N. benthamiana leaves (Fig. 4A). ROS production induced by PeBL1 was determined in the tobacco cell suspension by chemiluminescence and compared to a Tris-HCl buffer treatment negative control and an Flg22 (1 μM) treatment positive control (Fig. 4B). PeBL1 treatment caused a rapid increase in hydrogen peroxide, which reached a maximum at about 20 min, followed by a gradual decrease to a level similar to that of the negative control, just a little less than that of the well-known elicitor Flg22.
FIG 4.
ROS burst and extracellular-medium alkalinization in tobacco following PeBL1 treatment. (A) Microscopic observation of H2O2 accumulation in N. benthamiana leaves. (a) PeBL1-treated leaf; (b) buffer-treated leaf. H2O2 accumulation (as indicated by diaminobenzidine staining) appeared in the veins and stomata of elicitor-treated leaves, but not in buffer-treated leaves. Scale bars = 50 μm. (B) ROS formation in tobacco cell culture following elicitor treatment, Flg22 treatment, and buffer treatment was detected in 96-well plates by chemiluminescence. ROS formation in both the PeBL1-treated and Flg22-treated cell cultures reached maximum at approximately 20 min and declined thereafter to the level of the negative control. (C) Kinetics of extracellular-medium alkalinization induced by PeBL1 (10 μM) in a tobacco cell suspension. A distinct pH increase in the elicitor-treated cell culture was monitored for the first 5 min, and the pH reached the maximum level at 25 min. As a positive control, Flg22 induced a slightly higher and quicker maximal extracellular pH shift than PeBL1. Each data point represents three replicates. The error bars represent SD of the mean.
The alkalinization of the extracellular medium, another early key event that occurs after challenging tobacco cell suspensions with PeBL1 and which is thought to result from elicitor-induced ion fluxes caused by PeBL1, was also analyzed. The alkalinization of the tobacco culture medium following PeBL1 treatment was assessed by determining the pH of the cell medium, which significantly increased from 5.2 to 5.8 within 25 min in comparison to the negative control. Then, the pH slowly decreased to the initial pH value at 90 min. The positive control Flg22 induced a slightly higher and quicker maximal extracellular pH shift than PeBL1 (Fig. 4C).
Accumulations of phenolic compounds.
Phenolic compounds, secondary metabolism molecular components, including precursors for antifungal compounds, cytoderm reinforcement, and SA, help to control plant diseases. Under UV excitation, tobacco cells can present fluorescence attributable to the accumulation and oxidation of phenolic compounds, such as scopoletin, scopolin, and ferulic acid (50, 51). After 108 h of incubation with PeBL1, phenolic compounds accumulated in the tobacco cells and were observed in blue under fluorescence microscopy (Fig. 5b). In contrast, accumulation of phenolic compounds was not detected in control tobacco cells that were treated with Tris-HCl buffer (Fig. 5d).
FIG 5.
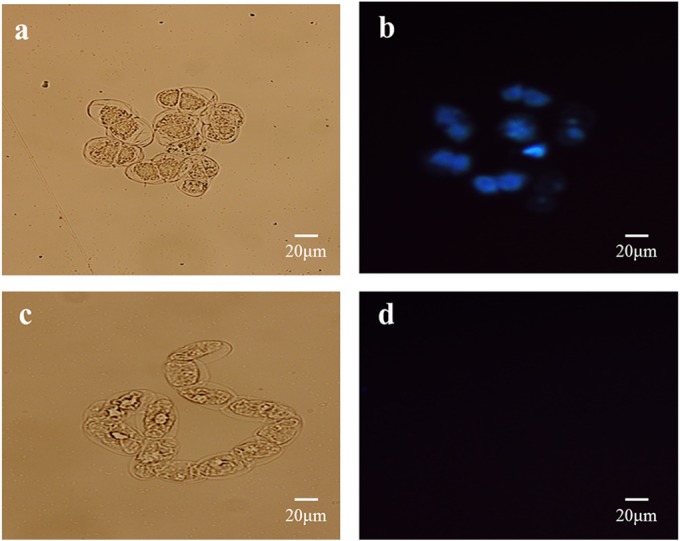
Phenolic-compound deposition in tobacco cells following PeBL1 treatment. (a and c) Tobacco cells treated with PeBL1 (a) and elicitor buffer (c) under bright-field observation. (b and d) Tobacco cells treated with PeBL1 (b) and elicitor buffer (d) under fluorescence microscopy (using a 365-nm excitation filter and a 445- to 450-nm long-wave pass filter).
PeBL1-induced disease resistance in N. benthamiana.
The numbers and diameters of TMV lesions in systemic leaves of PeBL1-treated plants were obviously smaller than in control plants (Table 2). The greatest reductions in both the numbers and the diameters of TMV lesions were approximately 43% at 4 days p.i. (Fig. 6A).
TABLE 2.
PeBL1 induces significant resistance against TMV in tobaccoa
| Time (days) after TMV inoculation | No. of lesions |
Inhibition (%)b | Diameter of lesions (mm) |
Inhibition (%)b | ||
|---|---|---|---|---|---|---|
| Control | PeBL1 | Control | PeBL1 | |||
| 2 | 14.33 ± 4.15 | 9.61 ± 3.22 | 32.94 A | 1.22 ± 0.12 | 0.86 ± 0.13 | 29.51 B |
| 4 | 37.17 ± 9.22 | 21.22 ± 5.57 | 42.91 A | 2.66 ± 0.14 | 1.51 ± 0.15 | 43.23 A |
| 6 | 58.39 ± 16.56 | 44.28 ± 10.74 | 24.17 A | 4.74 ± 0.24 | 3.97 ± 0.15 | 16.24 C |
The data are representative of three replicates, with nine plants for each replicate. The values are means ± standard deviations.
Inhibition of either the number or the diameter of lesions (i.e., reduction of the PeBL1 value relative to the control value). Values followed by the same letter in the same column are not statistically different at a 5% significance level.
FIG 6.
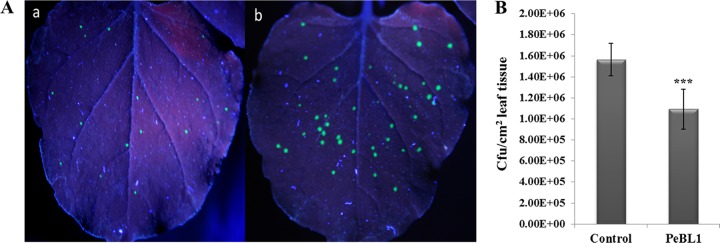
Systemic resistance induced by PeBL1 against infection by TMV-GFP and P. syringae pv. tabaci in N. benthamiana. (A) The numbers and diameters of TMV-GFP lesions (green fluorescent spots) in systemic leaves of protein-treated plants were significantly smaller than in control plants. The GFP images were taken under UV illumination. (a) The upper PeBL1-treated leaves were imaged at 4 days p.i. (b) The upper buffer-treated leaves were imaged at 4 days p.i. (B) Resistance against P. syringae pv. tabaci. The number of CFU per cm2 of buffer-treated N. benthamiana was compared with the number of CFU per cm2 of PeBL1-treated N. benthamiana. The latter significantly reduced infection by P. syringae pv. tabaci, with an inhibition ratio of 30% at 4 days p.i. The results are mean values from three independent experiments ± SD. The statistical analyses were performed using Student's t test. The asterisks indicate a significant difference between the treated and control samples (***, P < 0.001).
We also found that PeBL1 can enhance systemic resistance against P. syringae pv. tabaci in N. benthamiana. In the PeBL1-pretreated plants, the occurrence of disease signs was delayed and the bacterial population had decreased by 30% at 4 days p.i. (Fig. 6B).
Expression of defense-related genes induced by PeBL1 in N. benthamiana.
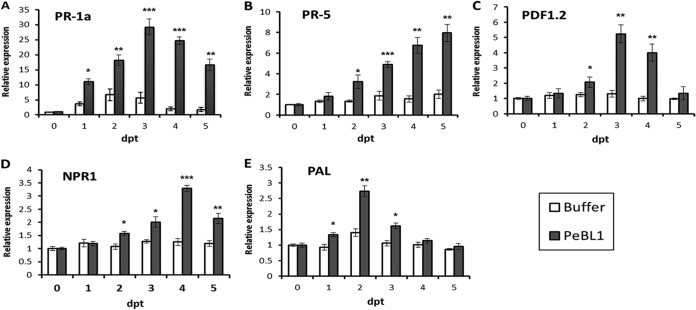
To further study the mechanism of PeBL1-induced plant systemic resistance, we assayed the expression levels of PR proteins of the PR-1a and PR-5 (encoding thaumatin-like proteins) gene families, which are markers of the SA-dependent defense pathway. The expression levels of the two genes were both significantly upregulated within 5 days in PeBL1-pretreated N. benthamiana in relation to untreated plants. The maximum level of the PR-1a gene increased by 30-fold at 4 days posttreatment (p.t.) (Fig. 7A). The expression of the PR-5 gene continuously increased over 5 days and achieved a maximum increase of 8-fold at 5 days p.t. (Fig. 7B). The expression levels of PR-1a and PR5 were also upregulated during the majority of growth stages for buffer-infiltrated plants, but apparently at a lower intensity than was observed for the PeBL1-treated plants (Fig. 7A and B). To analyze the JA/ET-dependent pathways, we examined the expression of the plant defensin 1.2 (PDF1.2) gene (52). The expression level of PDF1.2 was not influenced by the negative control but was 5-fold greater in PeBL1-treated N. benthamiana (Fig. 7C). The nonexpressor of pathogenesis related 1 (NPR1) gene is important for activating PR gene expression and is regarded as a key regulator of the cross talk between SA- and JA-dependent defense pathways (53). Measurement of transcript accumulation for the NPR1 gene indicated a slight increase in expression between 2 and 5 days p.t. compared to untreated plants, while the expression level did not increase in buffer-infiltrated controls during the same time period (Fig. 7D). The transcription level of the defense-related enzyme PAL was also determined. The expression level of PAL increased 3-fold at 2 days p.t. and then began to decrease, while PAL was upregulated by only 1.5-fold at 2 days p.t. in the buffer-infiltrated controls (Fig. 7E).
FIG 7.
Expression levels of N. benthamiana defense-related genes relative to the control. N. benthamiana leaves were infiltrated with 10 μM PeBL1 or buffer. At the indicated times, a small fragment was taken from the upper leaves, and the amounts of mRNAs of five defense-related genes were measured by RT-qPCR. Shown are the relative expression levels of the PR-1a (A), PR-5 (B), PDF1.2 (C), NPR1 (D), and PAL (E) genes. The samples were normalized against EF-1α. The expression levels are represented as the fold change in relation to the untreated control. The results are mean values from three independent experiments ± SD. The statistical analyses were performed using Student's t test. The asterisks indicate significant differences between the PeBL1-treated samples and buffer-infiltrated controls at the same point (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
The B. laterosporus strain A60 has a well-known ability to act as a biocontrol agent, to induce systemic resistance in plants, and to produce various antagonistic factors, such as a parasporal crystal, an extracellular protease, a lipopeptide antibiotic, and a pseudopeptide (35, 54–56). However, the molecular mechanisms involved in the activation of systemic resistance by strain A60 have not yet been completely elucidated. In this study, we report a novel secreted protein elicitor of 12.833 kDa (PeBL1) from the culture supernatant of B. laterosporus that elicits systemic resistance in N. benthamiana. The PeBL1 protein contains a secretory signal sequence and 116 amino acid residues. The amino acid sequence of PeBL1 does not exhibit homology or identity to other reported protein elicitors, indicating that PeBL1 is a novel elicitor. We also successfully cloned the PeBL1 gene and purified the prokaryotically expressed recombinant protein. Both the recombinant and native PeBL1 proteins can induce HR in N. benthamiana leaves. A concentration of 2.5 μM PeBL1 protein was sufficient to induce HR within 24 h, and the induced HR did not increase the size of the infiltrated zone, even when high concentrations of the protein were applied, suggesting that HR induced by PeBL1 was typical. Furthermore, the HSR203 gene, which is considered an HR-specific gene (48), was expressed in PeBL1-infiltrated N. benthamiana leaves. In general, HR is part of the plant innate immunity and induces a signaling cascade that activates plant defense responses, leading to systemic resistance (57).
We examined several defense-related early events involved in plant-elicitor interactions. One of the early events during the HR process is the production of ROS in an oxidative burst. Numerous reports have demonstrated that ROS play a key role in the whole plant defense system and often appear in host or nonhost plants after treatment with an elicitor (28). ROS regulate multiple cellular functions in plants, including alterations of redox status, that directly affect specific plant transcription factors and regulate antimicrobial activity (58). In this study, we detected H2O2 in N. benthamiana leaves via histological imaging and ROS production in tobacco suspension cells induced by PeBL1. In comparison with a known elicitor, Flg22, used as a positive control, PeBL1-treated and Flg22-treated tobacco suspension cells showed similar ROS production patterns, indicating that PeBL1 performs similarly to this well-known elicitor. ROS act as signaling molecules and interact with various molecules, including calcium ions, NO (nitric oxide), lipids, and mitogen-activated protein (MAP) kinase, which are general regulatory elements (59, 60). These signaling molecules may be involved in physiological phenomena and can regulate several processes in interconnected branch pathways and defense signaling pathways (61, 62). However, the involvement of these signaling molecules in PeBL1-regulated signal transduction is still unclear, and further study is required. As observed for other elicitors, PeBL1 induces the extracellular-medium alkalinization of tobacco cell suspensions, indicating that it might be involved in the restoration of the ion influx and pH gradient between the cytosol and the apoplast. All of the data indicate that PeBL1 is a real elicitor and the key player for inducing defense responses in N. benthamiana.
To clarify the downstream signaling pathways of defense responses induced by PeBL1 in N. benthamiana, we investigated the behavior of the PR-1a, PR-5, PDF1.2, and NPR1 genes using RT-qPCR. Considering that the plant defense responses can also be elicited by the damage associated with infiltration, we analyzed the buffer-infiltrated plants as negative controls. We found that the relative expression levels of these defense-related genes were differentially upregulated after infiltration with PeBL1 and higher than those caused by damage at certain stages (Fig. 7). The coordinated expression of SA-responsive PR genes, the JA/ET-responsive PDF1.2 gene, and the signaling regulatory gene NPR1 may indicate that PeBL1 elicits systemic resistance using a complex signaling network, which most likely includes SA- and JA/ET-dependent pathways. However, the exact signaling pathway induced by PeBL1 in N. benthamiana requires further elucidation.
The phenylpropanoid-biosynthetic pathway is known to be involved in the plant defense system because the antimicrobial compounds produced by the pathway, such as lignin, phytoalexin, and phenolic compounds, act as plant barriers to pathogens (50). In particular, phenolic compounds play key roles in interactions between plants and soil microorganisms and are involved in preformed plant defenses (63). The first enzyme in the phenylpropanoid-biosynthetic pathway is PAL, which provides precursors for the formation of antimicrobial compounds (64). In this study, we found (using RT-qPCR) that the PAL gene was upregulated and (using fluorescence microscopy) that phenolic compounds were produced after PeBL1 treatment, indicating that PeBL1 can induce the accumulation of antimicrobial compounds involved in defense responses.
To determine if the activation of the defense system in N. benthamiana can impart resistance to phytopathogens, experiments were performed by inoculating N. benthamiana with a TMV-GFP recombinant virus and virulent P. syringae pv. tabaci. The numbers and diameters of TMV lesions in systemic leaves of PeBL1-treated plants were obviously smaller than in control plants (Table 2). PeBL1 may potentially transfer TMV resistance via the accumulation of antiviral compounds and creation of a physical barrier, which suppress virus replication or movement, respectively, in N. benthamiana. In addition, the disease signs of the wildfire pathogen P. syringae pv. tabaci were alleviated and the bacterial population was significantly decreased after PeBL1 treatment, potentially because the pretreatment of N. benthamiana with PeBL1 influences the perception of and invasion by the pathogen. In conclusion, the defense systems activated by PeBL1 in N. benthamiana are effective against a broad spectrum of pathogens, including viruses and pathogenic bacteria.
In summary, we have reported the purification, characterization, and gene cloning of a secreted protein elicitor, PeBL1, from B. laterosporus strain A60. Both the recombinant and native PeBL1 proteins can induce a typical HR and activate defense-related early events and antimicrobial compound production in N. benthamiana, indicating that PeBL1 is an excellent candidate as a plant defense activation agent to challenge infections with pathogens. PeBL1 could be effectively utilized in biopesticides and transgenic crops to reduce the application of chemicals to food plants, with benefits to human health. Furthermore, we have provided a foundation for expanding the understanding of the signaling transduction mechanism of plant defense responses triggered by PeBL1. Our researches not only provide insights into molecular mechanisms and biological functions of PeBL1 in the activation of plant systemic resistance, but also strongly support the potential applications of B. laterosporus strain A60 in biocontrol and sustainable agriculture.
ACKNOWLEDGMENTS
This work was supported by the development program (863 Program) project (2011AA10A205) and international cooperation project (2014DFG32270) national technology research.
REFERENCES
- 1.Díez-Navajas AM, Wiedemann-Merdinoglu S, Greif C, Merdinoglu D. 2008. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 98:776–780. doi: 10.1094/PHYTO-98-7-0776. [DOI] [PubMed] [Google Scholar]
- 2.Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. 2009. Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 3.Haas D, Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 4.Kloepper JW, Ryu CM, Zhang SA. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 5.Ongena M, Adam A, Jourdan E, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P. 2007. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol 9:1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Loon LC, Bakker P, Pieterse CMJ. 1998. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 7.Durrant WE, Dong X. 2004. Systemic acquired resistance. Annu Rev Phytopathol 42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 8.Bashan Y, Holguin G. 1998. Proposal for the division of plant growth promoting rhizobacteria into two classifications: biocontrol-PGPB (plant growth-promoting bacteria) and PGPB. Soil Biol Biochem 30:1225–1228. doi: 10.1016/S0038-0717(97)00187-9. [DOI] [Google Scholar]
- 9.Kloepper JW, Rodriguez-Kabana R, Zehnder GW, Murphy J, Sikora E, Fernandez C. 1999. Plant root-bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Aust Plant Pathol 28:27–33. doi: 10.1071/AP99004. [DOI] [Google Scholar]
- 10.Pieterse CMJ, Van Loon LC. 1999. Salicylic acid-independent plant defense pathways. Trends Plant Sci 4:52–58. [DOI] [PubMed] [Google Scholar]
- 11.Thomma BPHJ, Penninckx IAMA, Cammue BPA, Broekaert WF. 2001. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13:63–68. doi: 10.1016/S0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 12.Glazebrook J. 2001. Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4:301–308. doi: 10.1016/S1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- 13.Durner J, Shah J, Klessig DF. 1997. Salicylic acid and disease resistance in plants. Trends Plant Sci 2:266–274. doi: 10.1016/S1360-1385(97)86349-2. [DOI] [Google Scholar]
- 14.Reymond P, Farmer EE. 1998. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1:404–411. doi: 10.1016/S1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 15.Pieterse CMJ, Van Pelt JA, Verhagen BWM, Ton J, van Wees ACM, Léon-Kloosterziel KM, van Loon LC. 2003. Induced systemic resistance by plant growth promoting rhizobacteria. Symbiosis 35:39–54. [Google Scholar]
- 16.Van Wees SC, Pieterse CMJ, Trijssenaar A, Van't Westende YAM, Hartog F, Van Loon LC. 1997. Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant Microbe Interact 10:716–724. doi: 10.1094/MPMI.1997.10.6.716. [DOI] [PubMed] [Google Scholar]
- 17.Maurhofer M, Hase C, Meuwly P, Métraux J-P, Défago G. 1994. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology 84:139–146. doi: 10.1094/Phyto-84-139. [DOI] [Google Scholar]
- 18.Ellis JG, Rafiqi M, Gan P, Chakrabarti A, Dodds PN. 2009. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr Opin Plant Biol 12:399–405. doi: 10.1016/j.pbi.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 19.De Wit PJ, Mehrabi R, Van den Burg HA, Stergiopoulos I. 2009. Fungal effector proteins: past, present and future. Mol Plant Pathol 10:735–747. doi: 10.1111/j.1364-3703.2009.00591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nürnberger T. 1999. Signal perception in plant pathogen defense. Cell Mol Life Sci 55:167–182. doi: 10.1007/s000180050283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Z, Laby RJ, Zumoff CH, Bauer DW, He SY, Collmer A, Beer SV. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 22.Kulye M, Liu H, Zhang Y, Zeng H, Yang X, Qiu D. 2012. Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defense-related genes and systemic acquired resistance in tobacco. Plant Cell Environ 35:2104–2120. doi: 10.1111/j.1365-3040.2012.02539.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Yang X, Zeng H, Liu H, Zhou T, Tan B, Qiu D. 2012. The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl Microbiol Biotechnol 93:191–201. doi: 10.1007/s00253-011-3405-1. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Zeng H, Qiu D, Guo L, Yang X, Zhou T, Zhao J. 2012. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS One 7:e37654. doi: 10.1371/journal.pone.0037654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Tsay J, Chang S, Yang H, Wu W, Chen C. 2012. Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag Sci 68:1306–1310. doi: 10.1002/ps.3301. [DOI] [PubMed] [Google Scholar]
- 26.Tran H, Ficke A, Asiimwe T, Höfte M, Raaijmakers JM. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol 175:731–742. doi: 10.1111/j.1469-8137.2007.02138.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Davis LC, Verpoorte R. 2005. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 29.Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. 2004. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 30.Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 31.Asai S, Ohta K, Yoshioka H. 2008. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20:1390–1406. doi: 10.1105/tpc.107.055855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDowell JM, Dangl JL. 2000. Signal transduction in the plant immune response. Trends Biochem Sci 25:79–82. doi: 10.1016/S0968-0004(99)01532-7. [DOI] [PubMed] [Google Scholar]
- 33.Blumwald E, Aharon GS, Lam BC-H. 1998. Early signal transduction pathways in plant-pathogen interactions. Trends Plant Sci 3:342–346. doi: 10.1016/S1360-1385(98)01289-8. [DOI] [Google Scholar]
- 34.Sunita C, Eunice JA, Steve W. 2010. Biological control of Fusarium oxysporum f. sp. lycopersici on tomato by Brevibacillus brevis. J Phytopathol 158:470–478. doi: 10.1111/j.1439-0434.2009.01635.x. [DOI] [Google Scholar]
- 35.Zhao J, Guo L, Zeng H, Yang X, Yuan J, Shi H, Xiong Y, Chen M, Han L, Qiu D. 2012. Purification and characterization of a novel antimicrobial peptide from Brevibacillus laterosporus strain A60. Peptides 33:206–211. doi: 10.1016/j.peptides.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Lippmann R, Kaspar S, Rutten T, Melzer M, Kumlehn J, Matros A, Mock HP. 2009. Protein and metabolite analysis reveals permanent induction of stress defense and cell regeneration processes in a tobacco cell suspension culture. Int J Mol Sci 10:3012–3032. doi: 10.3390/ijms10073012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Silva I, Heath MC. 1997. Purification and characterization of two novel hypersensitive response-inducing specific elicitors produced by the cowpea rust fungus. J Biol Chem 272:3924–3927. doi: 10.1074/jbc.272.7.3924. [DOI] [PubMed] [Google Scholar]
- 39.Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB. 1997. Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- 40.Pugin A, Frachisse JM, Tavernier E, Bligny R, Gout E, Douce R, Guern J. 1997. Early events induced by the elicitor cryptogein in tobacco cells: involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate pathway. Plant Cell 9:2077–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brault M, Amiar Z, Pennarun A-M, Monestiez M, Zhang Z, Cornel D, Dellis O, Knight H, Bouteau F, Rona J-P. 2004. Plasma membrane depolarization induced by abscisic acid in Arabidopsis suspension cells involves reduction of proton pumping in addition to anion channel activation, which are both Ca2+ dependent. Plant Physiol 135:231–243. doi: 10.1104/pp.104.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivprasad S1, Pogue GP, Lewandowski DJ, Hidalgo J, Donson J, Grill LK, Dawson WO. 1999. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255:312–323. doi: 10.1006/viro.1998.9579. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 44.Mao J, Liu Q, Yang X, Long C, Zhao M, Zeng H, Liu H, Qiu D. 2010. Purification and expression of a protein elicitor from Alternaria tenuissima and elicitor-mediated defense responses in tobacco. Ann Appl Biol 156:411–420. doi: 10.1111/j.1744-7348.2010.00398.x. [DOI] [Google Scholar]
- 45.Zhang Y, Yang X, Liu Q, Qiu D, Zhang Y, Zeng H. 2010. Purification of novel protein elicitor from Botrytis cinerea that induces disease resistance and drought tolerance in plants. Microbiol Res 165:142–151. doi: 10.1016/j.micres.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 46.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real time quantitative PCR and 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Govrin EM, Rachmilevitch S, Sagar Tiwari B, Solomon M, Levine A. 2006. An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology 96:299–307. doi: 10.1094/PHYTO-96-0299. [DOI] [PubMed] [Google Scholar]
- 49.Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson RL, Hammerschmidt R. 1992. Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30:369–386. [Google Scholar]
- 51.Chaerle L, Lenk S, Hagenbeek D, Buschmann C, Van Der Straeten D. 2007. Multicolor fluorescence imaging for early detection of the hypersensitive reaction to tobacco mosaic virus. J Plant Physiol 164:253–262. doi: 10.1016/j.jplph.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Guo X, Stotz HU. 2007. Defense against Sclerotinia sclerotiorum in Arabidopsisis dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Microbe Interact 20:1384–1395. doi: 10.1094/MPMI-20-11-1384. [DOI] [PubMed] [Google Scholar]
- 53.Pieterse CMJ, Van Loon LC. 2004. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7:456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Smirnova TA, Minenkova IB, Orlova MV, Lecadet MM, Azizbekyan RR. 1996. The crystal-forming strains of Bacillus laterosporus. Res Microbiol 147:343–350. doi: 10.1016/0923-2508(96)84709-7. [DOI] [PubMed] [Google Scholar]
- 55.Huang X, Tian B, Niu Q, Yang J, Zhang L, Zhang K. 2005. An extracellular protease from Brevibacillus laterosporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes. Res Microbiol 156:719–727. doi: 10.1016/j.resmic.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Desjardine K, Pereira A, Wright H, Matainaho T, Kelly M, Andersen RJ. 2007. Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: structure elucidation and synthesis. J Nat Prod 70:1850–1853. doi: 10.1021/np070209r. [DOI] [PubMed] [Google Scholar]
- 57.Hammond-Kosack KE, Parker JE. 2003. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 2:177–193. doi: 10.1016/S0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 58.Bowler C, Fluhr R. 2000. The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci 5:241–246. doi: 10.1016/S1360-1385(00)01628-9. [DOI] [PubMed] [Google Scholar]
- 59.Srivastava N, Gonugunta VK, Puli MR, Raghavendra AS. 2009. Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativum. Planta 229:757–765. doi: 10.1007/s00425-008-0855-5. [DOI] [PubMed] [Google Scholar]
- 60.Bóka K, Orbán N. 2007. New aspect of H2O2 signaling. Plant Signal Behav 2:498–500. doi: 10.4161/psb.2.6.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, Wendehenne D, Pugin A. 2006. Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact 19:711–724. doi: 10.1094/MPMI-19-0711. [DOI] [PubMed] [Google Scholar]
- 62.Bouché N, Yellin A, Snedden WA, Fromm H. 2005. Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56:435–466. doi: 10.1146/annurev.arplant.56.032604.144224. [DOI] [PubMed] [Google Scholar]
- 63.De Werra P, Huser A, Tabacchi R, Keel C, Maurhofer M. 2011. Plant- and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl Environ Microbiol 77:2807–2812. doi: 10.1128/AEM.01760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang LJ. 2002. The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3:371–390. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]