Abstract
The hyperthermophilic archaeon Ferroglobus placidus can utilize a wide variety of electron donors, including hydrocarbons and aromatic compounds, with Fe(III) serving as an electron acceptor. In Fe(III)-reducing bacteria that have been studied to date, this process is mediated by c-type cytochromes and type IV pili. However, there currently is little information available about how this process is accomplished in archaea. In silico analysis of the F. placidus genome revealed the presence of 30 genes coding for putative c-type cytochrome proteins (more than any other archaeon that has been sequenced to date), five of which contained 10 or more heme-binding motifs. When cell extracts were analyzed by SDS-PAGE followed by heme staining, multiple bands corresponding to c-type cytochromes were detected. Different protein expression patterns were observed in F. placidus cells grown on soluble and insoluble iron forms. In order to explore this result further, transcriptomic studies were performed. Eight genes corresponding to multiheme c-type cytochromes were upregulated when F. placidus was grown with insoluble Fe(III) oxide compared to soluble Fe(III) citrate as an electron acceptor. Numerous archaella (archaeal flagella) also were observed on Fe(III)-grown cells, and genes coding for two type IV pilin-like domain proteins were differentially expressed in Fe(III) oxide-grown cells. This study provides insight into the mechanisms for dissimilatory Fe(III) respiration by hyperthermophilic archaea.
INTRODUCTION
Dissimilatory Fe(III) reduction is a common form of anaerobic respiration among a diversity of Archaea and Bacteria (1). It has an important impact on the biogeochemistry of anaerobic soils and sediments and is involved in the cycling of such nutrients as carbon, nitrogen, and sulfur in the biosphere (2). Much of the research on this type of metabolism has been conducted on mesophilic organisms; however, dissimilatory iron reduction is also an important terminal electron-accepting process in many geothermally heated ecosystems (3–5). Conditions in these hot environments are similar to those found on early Earth when Fe(III) was continuously being formed by photochemical oxidation of Fe(II) in archaean seas and from hydrothermal vent fluids (6–8). Therefore, any information regarding mechanisms involved in dissimilatory Fe(III) reduction by hyperthermophilic archaea isolated from these environments not only will shed light on present-day biogeochemical cycling patterns but also may provide information about what may have been occurring when life first appeared on the planet.
Although Fe(III) reduction is significant from both an ecological and evolutionary point of view, the mechanisms behind this form of respiration still are largely unknown. The most prevalent form of Fe(III) in subsurface environments is insoluble Fe(III) oxyhydroxide, and reduction of this compound poses a challenge, as cell envelopes are impermeable to the poorly soluble electron acceptor. Much of the work on dissimilatory Fe(III) reduction that has been done to date has focused on species from two mesophilic genera, Geobacter and Shewanella (9, 10). While these studies have found that both species utilize c-type cytochromes for Fe(III) respiration, they seem to have evolved different mechanisms for contact with the Fe(III) oxide particle; while Geobacter species primarily rely on direct electron transfer from cells to Fe(III) oxides, Shewanella species have the ability to produce soluble electron shuttles (i.e., flavin) that can account for 75% of the insoluble Fe(III) reduction (11, 12).
Similar to their mesophilic bacterial counterparts, it also appears that hyperthermophilic archaea have evolved different mechanisms for Fe(III) reduction. For example, Pyrobaculum aerophilum seems to release an electron-shuttling compound(s) that transfers electrons from the cell surface to the surface of Fe(III) oxides not in direct contact with the cells (13), while Geoglobus ahangari and Geoglobus acetivorans require direct contact with Fe(III) hydroxides in the absence of chelated iron or reduced electron shuttles (14, 15). The involvement of c-type cytochrome proteins in Fe(III) reduction also seems to vary among hyperthermophilic archaea. While c-type cytochrome proteins appear to be involved in electron transfer to Fe(III) by G. ahangari (14), they are not required for Fe(III) respiration by Pyrobaculum species (13, 16).
In this study, Fe(III) respiration was examined in Ferroglobus placidus, a hyperthermophilic archaeon that was isolated from hydrocarbon- and iron-rich sediments associated with a hydrothermal system in Vulcano Island (17, 18). F. placidus is a member of the family Archaeoglobaceae, which is composed of three genera: Ferroglobus, Archaeoglobus, and Geoglobus. Although some Archaeoglobus species are capable of chemolithotrophic iron reduction, Ferroglobus and Geoglobus species are the only members of the family known to couple the complete oxidation of organic compounds with Fe(III) reduction (19–21). F. placidus also is unique among the Archaeoglobaceae in that it can transfer electrons from the oxidation of aromatic compounds to Fe(III) (22–24). Although genes involved in aromatics degradation were identified in the G. acetivorans genome, attempts to grow G. acetivorans on such compounds have been unsuccessful (15). Therefore, any information regarding Fe(III) reduction by an aromatics-degrading hyperthermophile such as F. placidus is significant. In this study, genomic, transcriptomic, and proteomic approaches were used to show that F. placidus has more multiheme c-type cytochromes than any other hyperthermophilic archaeon, and that many of its electron transfer proteins have homologues in mesophilic Fe(III)-reducing bacteria. We also were able to identify some of the proteins likely to be involved in electron transfer to Fe(III) and show that the availability of soluble Fe(III) affects expression of these proteins.
MATERIALS AND METHODS
Microbial strains and culturing conditions.
Ferroglobus placidus strain AEDII12DO (DSM 10642) was obtained from the type culture collection of the Deutsche Sammlung von Mikroorganismen and Zellkulturen (DSMZ), Braunschweig, Germany. Strict anaerobic culturing and sampling techniques were used throughout (25, 26). Ferroglobus placidus cells were grown with acetate (10 mM) as the electron donor and Fe(III) citrate (56 mM) or amorphous Fe(III) oxyhydroxide (100 mM) as the electron acceptor.
Ferroglobus placidus medium was prepared as previously described (27). After autoclaving, FeCl2 (1.3 mM), Na2SeO4 (30 μg/liter), Na2WO4 (40 μg/liter), APM salts (1 g/liter MgCl2, 0.23 g/liter CaCl2) (28), DL vitamins (29), and all electron donors were added to the sterilized medium from anaerobic stock solutions. Cultures were incubated under N2 and CO2 (80:20) at 85°C in the dark.
Amorphous Fe(III) oxyhydroxide was prepared by dissolving Fe(III) chloride hexahydrate (Sigma-Aldrich, St. Louis, MO, USA) in water, slowly adding sodium hydroxide (Thermo Scientific, Rockford, IL, USA) to a stable pH of 7, centrifuging the mixture in a Sorvall SLA-3000 rotor at 4,225 × g for 20 min at 4°C, and repeatedly resuspending the pellet in water and centrifuging it until the supernatant was brownish red. The Fe(III) oxide gel was resuspended in water to a concentration of approximately 1 M total iron and stored at 4°C. Total iron concentration was determined by ferrozine assay: 0.1 ml Fe(III) gel suspension was reacted overnight with 4.7 ml 0.5 M hydrochloric acid and 0.2 ml 6.25 M hydroxylamine and then diluted 50-fold in 0.5 M hydrochloric acid prior to assay (30).
Alginate bead assays.
Amorphous Fe(III) oxyhydroxide, prepared as described above, was incorporated into microporous alginate beads (diameter, 5 mm) with a nominal molecular mass cutoff of 12 kDa as previously described (11). Beads were added to F. placidus medium to provide Fe(III) at 50 mM. The production of Fe(II) was determined with the ferrozine assay (30) after the beads were incubated in 0.5 N HCl at room temperature for 12 h.
Protein extraction and quantification.
Proteins were extracted from 200 ml of cells grown until stationary phase [when 60 mM Fe(III) was reduced] with acetate as the electron donor and either Fe(III) citrate or Fe(III) oxide as the electron acceptor. Cells first were centrifuged at 17,700 × g for 20 min and washed with 10 ml TPE buffer (100 mM Tris HCl, 10 mM EDTA, 300 mM KH2PO4, pH 7.0) and 4 ml oxalate solution (197 mM ammonium oxalate and 119 mM oxalic acid). The pellet then was resuspended in 5 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 1% SDS) with protease inhibitor (Complete Mini EDTA-free protease inhibitor cocktail tablet; Roche Diagnostics, Indianapolis, IN, USA) and added to 5 different screw-cap tubes containing lysing matrix B (MP Biomedicals, Santa Ana, CA, USA). Cells were lysed in the FastPrep-24 tissue and cell homogenizer (MP Biomedicals, Santa Ana, CA, USA) for 45 s at 6 m/s. Cellular debris then was removed by centrifugation at 16,100 × g for 30 min at 4°C, and the supernatant from all 5 lysis tubes was combined and transferred to an Amicon Ultra centrifugal filter unit with a molecular mass cutoff of 3,000 g/mol (Millipore, Billerica, MA, USA). Proteins were concentrated at room temperature by centrifugation at 2,000 × g for 1 h.
Proteins were quantified with the BCA (bicinchoninic acid) protein assay kit (Thermo Scientific, Rockford, IL, USA) according to the manufacturer's instructions. Equal amounts of protein (20 μg) from Fe(III) citrate-grown and Fe(III) oxide-grown cells were boiled for 5 min in 4× sample loading buffer (Amresco, Solon, OH, USA) and separated by electrophoresis in glycine-buffered 12.5% polyacrylamide gels. Total proteins were stained with Coomassie blue solution (0.2% Coomassie blue, 7.5% acetic acid, 50% ethanol) and destained in a solution consisting of 30% ethanol and 10% acetic acid. Heme groups were identified by peroxidase activity detected with H2O2 and 3,3′,5,5′-tetramethylbenzidine (TMBZ) as previously described (31, 32).
RNA extraction.
RNA was extracted from six batch cultures (100 ml), three grown with acetate as the electron donor and Fe(III) oxide as the electron acceptor and three grown with acetate as the electron donor and Fe(III) citrate as the electron acceptor, during the exponential phase [Fe(II) concentrations were 30 mM]. Cells were split into 50-ml conical tubes (BD Biosciences, San Jose, CA, USA), mixed with RNA Protect (Qiagen, Valencia, CA, USA) in a 1:1 ratio, and pelleted by centrifugation at 3,000 × g for 15 min at 4°C. Pellets then were immediately frozen in liquid nitrogen and stored at −80°C.
The pellets were resuspended in 10 ml of cold (4°C) TPE buffer (100 mM Tris HCl, 10 mM EDTA, 100 mM KH2PO4; pH 7.6), and 1-ml aliquots were dispensed into 2-ml screw-cap tubes. RNA was extracted from these screw-cap tubes with a hot acidic phenol extraction protocol as previously described (22). Nucleic acids were precipitated at −30°C for 1 h and pelleted by centrifugation at 16,100 × g for 30 min. The pellet was cleaned with cold (−20°C) 70% ethanol, dried, and resuspended in sterile diethylpyrocarbonate (DEPC)-treated water (Ambion, Austin, TX, USA). The resuspended pellets were combined and cleaned with the RNeasy RNA cleanup kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. The RNA cleanup product was treated with DNA-free DNase (Ambion, Austin, TX, USA) according to the manufacturer's instructions.
High-quality RNA was extracted from these culture samples. All samples had A260/A280 ratios of 1.8 to 2.0, indicating that they were of high purity (33). In order to ensure that RNA samples were not contaminated with DNA, PCR amplification with primers targeting the 16S rRNA gene was conducted on RNA samples that had not undergone reverse transcription (RT).
Microarray analysis.
Whole-genome microarray hybridizations were carried out by Roche NimbleGen, Inc. (Madison, WI, USA). The TransPlex whole-transcriptome amplification kit (Sigma-Aldrich, St. Louis, MO, USA) was used to amplify RNA and generate cDNA prior to microarray analyses. Three biological and technical replicates were conducted for microarray analyses; three cDNA samples were generated from each Fe(III) citrate and Fe(III) oxide RNA template, and all 18 samples were labeled with Cy3 and hybridized by NimbleGen. The oligonucleotide microarrays used in this study were designed based on preliminary genome sequence data of F. placidus (accession number NC_013849), which was obtained from the DOE Joint Genome Institute (JGI) website (www.jgi.doe.gov).
Results from microarray hybridizations were analyzed with the software Array 4 Star (DNASTAR, Madison, WI, USA). P values were determined with Student's t test analysis. Multiple oligonucleotide probes (3, 4) were analyzed for each gene. For each of the six technical replicates in each experiment, a probe was considered valid if its signal intensity was above the average signal from three probes for the rgy gene (Ferp_0787, reverse gyrase) for either the control or the experimental condition. A probe was considered valid for a biological replicate if it was valid for one or both technical replicates thereof. A gene was considered expressed only if at least two probes were valid for at least two biological replicates each. A gene was considered differentially expressed only if at least two probes had P values less than or equal to 0.01.
qRT-PCR.
Genome sequence data obtained from the DOE JGI website www.jgi.doe.gov was used to design quantitative RT-PCR (qRT-PCR) primers. All qRT-PCR primers were designed according to the manufacturer's specifications (amplicon size, 100 to 200 bp), and representative products from each of these primer sets were verified by sequencing clone libraries constructed with a TOPO TA cloning kit, version M (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. One hundred plasmid inserts from each clone library were sequenced with the M13F primer at the University of Massachusetts Sequencing Facility. All primer pairs used in this study are provided in Table S1 in the supplemental material.
The housekeeping gene rgy (Ferp_0787), which encodes the DNA topoisomerase reverse gyrase, was used as an internal control. This gene was selected because it appears to be constitutively expressed by F. placidus under a variety of growth conditions and was not differentially expressed in any of the microarray experiments (22, 23).
First-strand cDNA for qPCR was generated with the enhanced avian reverse transcriptase kit (Sigma-Aldrich, St. Louis, MO, USA) using 3′ antisense-specific primers (see Table S1 in the supplemental material). The reaction mixture consisted of 0.25 μg total RNA, 1 μl deoxynucleotide mix (10 mM mixed stock), 1 μl 3′ antisense-specific primer (20 μM stock), 2 μl 10× buffer for avian myeloblastosis virus (AMV)-RT, 1 μl RNase inhibitor (1 U/μl stock), enhanced avian RT (1 U/μl stock), and enough nuclease-free water to bring the final volume up to 20 μl.
Once the appropriate cDNA fragments were generated by reverse transcription (3 cDNAs from each of the 6 conditions), quantitative PCR amplification and detection were performed with the 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). Each reaction mixture consisted of a total volume of 25 μl and contained 1.5 μl of the appropriate primers (stock concentrations, 1.5 μM), 5 ng of cDNA, and 12.5 μl of Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA). Standard curves covering 8 orders of magnitude were constructed with serial dilutions of known amounts of purified cDNA quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at an absorbance of 260 nm. Transcript abundances and qPCR efficiencies (90% to 99%) were calculated from appropriate standard curves, and all qPCR experiments followed MIQE guidelines (34).
Optimal thermal cycling parameters consisted of an activation step at 50°C for 2 min and an initial 10-min denaturation step at 95°C, followed by 50 cycles of 95°C for 15 s and 58 to 60°C for 1 min. After 50 cycles of PCR amplification, dissociation curves were made for all qPCR products by increasing the temperature from 58°C to 95°C at a ramp rate of 2%. The curves all yielded a single predominant peak, further supporting the specificity of the PCR primer pairs.
TEM.
For transmission electron microscopy (TEM), F. placidus cells were grown with acetate (10 mM) as the electron donor and either Fe(III) citrate (50 mM) or Fe(III) oxide (100 mM) as the electron acceptor until stationary phase [Fe(II) concentrations were 60 mM]. Cells (4 ml) were fixed for 1 h with 500 μl of 16% paraformaldehyde and 250 μl of 8% gluteraldehyde. After fixation, 4 ml of oxalate solution (197 mM ammonium oxalate and 119 mM oxalic acid) was added to the solution to dissolve all Fe(III) particles. Fixed cells then were pelleted by centrifugation at 3,000 × g for 20 min, washed twice in 1 ml of 50 mM PIPES [piperazine-N,N′-bis-(2-ethanesulfonic acid)] buffer by centrifugation at 2,300 × g for 5 min, and resuspended in 20 μl of water.
After resuspension in water, cells were placed on 400-mesh (400 holes per inch) carbon-coated copper grids, incubated for 5 min, and then stained with 2% uranyl acetate. Cell appendages were observed using a Tecnai 12 transmission electron microscope at an accelerating voltage of 100 kV. Images were taken digitally with the Teitz TCL camera system.
Phylogenetic analysis.
Putative c-type cytochrome genes were identified in the F. placidus genome by the presence of the Cys-Xaa-Xaa-Cys-His peptide motif and examined for the presence of signal peptides with the SignalP 3.0 server (35). The subcellular location of each c-type cytochrome gene then was predicted with PSORT-B (36), PRED-SIGNAL (37), and Tmpred (38).
Homologues of putative c-type cytochrome proteins were identified by comparing F. placidus sequences to the GenBank protein database with the BLASTp algorithm (39, 40). Protein alignments were made in ClustalX (40) and corrected with ProSeq v2.9 (41) before phylogenetic trees were constructed with Mega v6 (42) and FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). The maximum likelihood algorithm (43) was used to construct all phylogenetic trees. All evolutionary distances were computed with the Jones-Taylor-Thornton model (44) with 100 bootstrap replicates.
Nucleotide sequence accession number.
A complete record of all oligonucleotide sequences used and raw and statistically treated data files is available in the NCBI Gene Expression Omnibus database (GEO data series number GSE59467).
RESULTS AND DISCUSSION
Comparison of c-type cytochromes found in F. placidus to other Fe(III)-reducing archaea and bacteria.
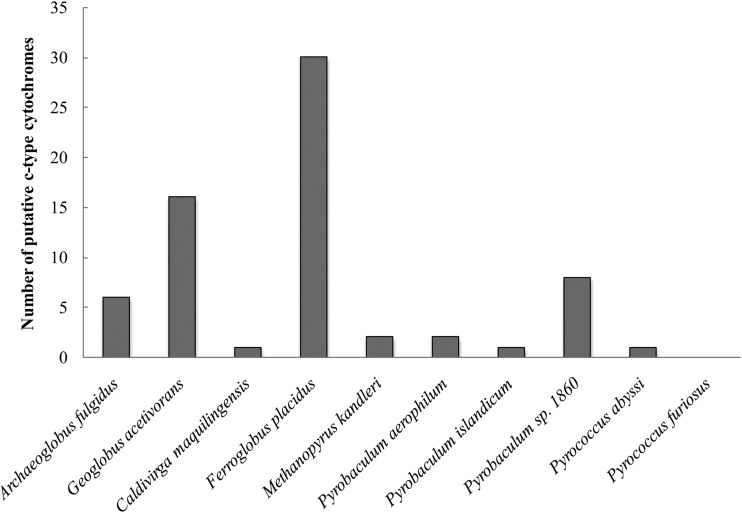
When the genomes of Fe(III)-respiring hyperthermophilic archaea were scanned, it was apparent that F. placidus had significantly more genes coding for putative c-type cytochrome proteins (Fig. 1). While the majority of hyperthermophilic archaea contained a few putative monoheme c-type cytochromes (see Table S2 in the supplemental material), only 3 of the genomes (F. placidus, G. acetivorans, and Pyrobaculum sp. strain 1860) contained putative c-type cytochrome proteins with more than 5 heme groups, and F. placidus was the only organism that contained genes coding for c-type cytochromes with more than 20 heme groups (Table 1; also see Table S2).
FIG 1.
Number of genes coding for putative c-type cytochrome proteins found in genomes from hyperthermophilic archaea capable of Fe(III) respiration.
TABLE 1.
Putative c-type cytochrome genes found in the F. placidus genome and comparison to c-type cytochromes from other archaeal and bacterial species
| Locus IDa | No. of heme groups | Top orthologous cytochrome protein in archaeal or bacterial isolate (organism and gene locus ID) | % identity | % similarity |
|---|---|---|---|---|
| Ferp_0264 | 8 | Ignavibacterium album (IALB_2703) | 40 | 56 |
| Ferp_0310 | 2 | Geobacter metallireducens (Gmet_0328) | 40 | 60 |
| Ferp_0648 | 4 | Geoglobus acetivorans (Gace_0102) | 35 | 51 |
| Ferp_0649 | 8 | Geoglobus acetivorans (Gace_0430) | 47 | 61 |
| Ferp_0660 | 6 | Geoglobus acetivorans (Gace_1853) | 53 | 71 |
| Ferp_0668 | 4 | Geoglobus acetivorans (Gacet_1846) | 51 | 63 |
| Ferp_0669 | 5 | Geoglobus acetivorans (Gacet_1846) | 50 | 63 |
| Ferp_0670 | 35 | Geoglobus acetivorans (Gace_1847) | 43 | 56 |
| Ferp_0672 | 31 | Geoglobus acetivorans (Gace_1847) | 41 | 56 |
| Ferp_0673 | 4 | Archaeoglobus veneficus (Arcve_0353) | 42 | 59 |
| Ferp_0674 | 4 | Archaeoglobus veneficus (Arcve_0353) | 37 | 57 |
| Ferp_0675 | 7 | Geoglobus acetivorans (Gacet_1843) | 44 | 61 |
| Ferp_0676 | 24 | Geoglobus acetivorans (Gacet_1847) | 33 | 48 |
| Ferp_0711 | 8 | Geoglobus acetivorans (Gacet_1826) | 85 | 91 |
| Ferp_0712 | 6 | Geobacter sulfurreducens (GSU2737) | 34 | 46 |
| Ferp_0802 | 1 | Archaeoglobus veneficus (Arcve_2078) | 63 | 75 |
| Ferp_0937 | 1 | Geobacter sp. strain M18 (GM18_0298) | 35 | 54 |
| Ferp_1198 | 8 | Pyrolobus fumarii (Pyrfu_1339) | 58 | 74 |
| Ferp_1255 | 2 | No homologue | ||
| Ferp_1267 | 4 | Geoglobus acetivorans (Gace_1344) | 41 | 61 |
| Ferp_1270 | 10 | Desulfocapsa sulfexigens (UWK_01207) | 28 | 40 |
| Ferp_1336 | 2 | Geoglobus acetivorans | 69 | 78 |
| Ferp_1338 | 1 | Anaerolinea thermophila (ANT_19420) | 33 | 55 |
| Ferp_1341 | 1 | Beggiatoa sp. strain PS (BGP_5602) | 38 | 56 |
| Ferp_1361 | 1 | Geoglobus acetivorans (Gace_2071) | 60 | 72 |
| Ferp_1439 | 4 | Geoglobus acetivorans (Gace_0102) | 74 | 84 |
| Ferp_1813 | 5 | Geoglobus acetivorans (Gacet_1361) | 64 | 73 |
| Ferp_1814 | 5 | Geoglobus acetivorans (Gacet_1360) | 42 | 55 |
| Ferp_2064 | 5 | Anaeromyxobacter dehalogenans (Adeh_1422) | 41 | 56 |
| Ferp_2107 | 12 | Geoglobus acetivorans (Gace_0099) | 66 | 81 |
ID, identifier.
More than half (57%) of the F. placidus c-type cytochrome sequences were most similar to G. acetivorans (Table 1; also see Fig. S1 in the supplemental material), another hyperthermophilic archaeon that is able to couple the oxidation of organic compounds with Fe(III) respiration. A large proportion (27%) of the c-type cytochrome proteins had best BLAST hits to cytochromes of bacteria (Table 1), and almost all of the c-type cytochromes had homologues in known Fe(III)-reducing bacteria from the genera Geobacter, Anaeromyxobacter, Thermincola, and Rhodoferax (see Fig. S1).
Three of the c-type cytochrome genes (Ferp_0648, Ferp_1267, and Ferp_1439) are in arrangements commonly found in Fe(III)-reducing Deltaproteobacteria (see Fig. S2 in the supplemental material). This operon consists of genes coding for a tetraheme c-type cytochrome, an iron-sulfur cluster-binding protein (Ferp_0647, Ferp_1268, and Ferp_1438), and a putative cytoplasmic membrane-associated b-type cytochrome (Ferp_0646, Ferp_1269, and Ferp_1437) with quinone-binding site motifs (45). It is remarkable that F. placidus possesses these three putative menaquinol:ferricytochrome c oxidoreductases of the type known as Cbc4 in Geobacter species (46). These Cbc4 complexes may be the primary interfaces for electrons from the oxidation of organic compounds in the cytoplasm, carried by menaquinone, to reach periplasmic c-type cytochromes in both Bacteria and Archaea en route to extracellular electron acceptors such as Fe(III).
Another cytochrome found solely in both F. placidus (cbcZ; Ferp_0937) and Geobacter species is an integral membrane protein that appears to be a fusion of c-type and b5-type cytochromes (Table 2). This membrane-associated c-type cytochrome also is likely to be involved in electron transfer across the cytoplasmic membrane. However, further investigation is required.
TABLE 2.
Genes in F. placidus and Geobacter genomes that code for a protein (cbcZ) consisting of a c-type cytochrome fused to a cytochrome b5-type heme/steroid binding domain

Differential expression of c-type cytochromes during growth with soluble or insoluble Fe(III) as electron acceptor.
Previous studies with Fe(III)-reducing bacteria, including Geobacter and Shewanella species, have shown that these organisms express different c-type cytochrome and electron transport proteins when grown on soluble and insoluble electron acceptors (9, 46–48). Proteomic and transcriptomic studies were done to see if F. placidus also differentially expresses c-type cytochromes and other electron transport proteins when electron acceptors are varied.
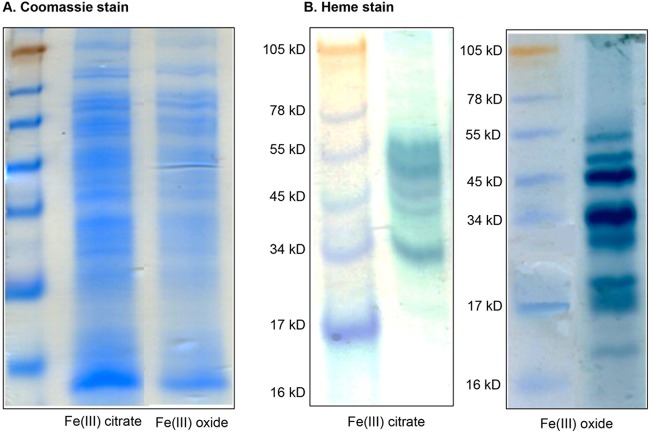
SDS-PAGE experiments revealed that 5 different c-type cytochromes were faintly expressed during growth with soluble Fe(III) citrate (55 kDa, 50 kDa, 45 kDa, 43 kDa, and 34 kDa), one of which (43 kDa) was unique to Fe(III)-citrate-grown cells (Fig. 2). On the other hand, eight different c-type cytochrome proteins were highly expressed in Fe(III)-oxide grown cells, four of which were unique to this condition (28 kDa, 20 kDa, 17 kDa, and 16.5 kDa).
FIG 2.
SDS-PAGE of proteins extracted from F. placidus cells grown with acetate as an electron donor and either insoluble Fe(III) oxide or soluble Fe(III) citrate as the electron acceptor. Total proteins were stained with Coomassie blue (A) or heme stain (B).
The largest c-type cytochrome protein that was expressed by F. placidus under both conditions had an apparent molecular mass of 55 kDa. There are two c-type cytochrome genes with predicted protein products of this size: Ferp_0649 and Ferp_1198. The next band corresponds to a molecular mass of approximately 50 kDa, and there are two genes coding for c-type cytochrome proteins of this size: Ferp_0711 and Ferp_0712. There also are two c-type cytochrome genes that would encode a protein around 45 kDa: Ferp_0264 and Ferp_0675. The proteins encoded by Ferp_0937 and Ferp_2107 are ∼34 kDa, and the proteins encoded by Ferp_0310, Ferp_1336, and Ferp_2064 are ∼28 kDa. The protein band that migrated with the 20-kDa marker likely is the protein product of Ferp_1814, the 17-kDa band is likely to be Ferp_1267, and the 16.5-kDa band most likely is Ferp_1255.
In order to further analyze the putative c-type cytochromes that were differentially expressed during growth on either soluble or insoluble Fe(III), a whole-genome DNA microarray was done comparing F. placidus cells grown with acetate as the electron donor and either Fe(III) oxide or Fe(III) citrate as the electron acceptor. A total of 405 genes were differentially expressed by F. placidus during growth on Fe(III) oxide compared to growth on Fe(III) citrate (218 upregulated and 187 downregulated). Genes encoding proteins involved in energy production and conversion, amino acid metabolism and transport, and translation had increased transcription in cells grown with insoluble Fe(III) oxide, while the majority of genes that were downregulated in Fe(III) oxide-grown cells coded for hypothetical proteins.
Among the putative c-type cytochromes, 11 genes were differentially expressed (8 upregulated, 3 downregulated) in Fe(III) oxide-grown cells (Table 3). All of the genes encoding c-type cytochrome proteins that had higher transcript abundance in Fe(III) oxide-grown cells had multiple heme-binding motifs, and two had more than 30 heme-binding motifs (Ferp_0672 and Ferp_0670). Ferp_0670 has homologues in G. sulfurreducens (GSU2898 and GSU2884) and G. metallireducens (Gmet_0571, Gmet_0580, and Gmet_0581) that transcriptomic studies have shown are involved in electron transfer to insoluble Fe(III) oxides (46, 47).
TABLE 3.
Fold difference in mRNA transcripts for c-type cytochrome proteins
| Locus ID | No. of heme-binding motifs | Predicted size of protein (kDa) | Resulta by: |
|
|---|---|---|---|---|
| Microarray | qRT-PCR | |||
| Upregulated genes | ||||
| Ferp_0670 | 35 | 189.28 | 3.67 | 13.24 ± 3.31 |
| Ferp_0668 | 4 | 25.59 | 2.69 | 7.88 ± 2.55 |
| Ferp_1336 | 2 | 31.91 | 2.55 | 5.26 ± 1.38 |
| Ferp_1267 | 4 | 17.73 | 2.41 | 17.80 ± 5.36 |
| Ferp_0672 | 31 | 180.93 | 2.28 | 6.94 ± 2.42 |
| Ferp_1814 | 5 | 20.80 | 2.13 | 3.56 ± 1.22 |
| Ferp_1255 | 2 | 15.67 | 2.08 | 8.65 ± 2.34 |
| Ferp_1813 | 5 | 21.20 | 2.08 | 4.56 ± 2.10 |
| Downregulated genes | ||||
| Ferp_0660 | 6 | 22.89 | −2.20 | −4.21 ± 1.98 |
| Ferp_2064 | 5 | 31.01 | −2.34 | −5.94 ± 2.67 |
| Ferp_1341 | 1 | 25.21 | −2.77 | −6.33 ± 2.35 |
Shown are fold differences in mRNA transcripts for c-type cytochrome proteins that were significantly upregulated or downregulated in F. placidus cells grown with acetate as an electron donor and Fe(III) oxide compared to Fe(III) citrate as an electron acceptor.
The protein products of several of the genes that had higher transcript levels during growth on Fe(III) oxide are predicted to be similar in size to some of the c-type cytochrome proteins that were observed by SDS-PAGE analysis: Ferp_1814 corresponds to the 20-kDa band, Ferp_1336 would migrate with the 28-kDa band, and the products of Ferp_1267 and Ferp_1255 could be the 17-kDa and 16.5-kDa c-type cytochromes, respectively.
The membrane-associated polyheme c-type cytochrome protein encoded by Ferp_1267 has a homologue in G. sulfurreducens (GSU0068) that genetic studies have shown to be important for insoluble Fe(III) reduction (46). This protein could be similarly important for Fe(III) respiration in F. placidus. However, until a genetic system is developed for F. placidus, the significance of any of these proteins cannot be determined.
Other genes involved in electron transport that were differentially expressed during growth on Fe(III) oxide.
In addition to c-type cytochrome proteins, 40 other genes coding for proteins involved in energy production and conversion were significantly upregulated in Fe(III) oxide-grown cells, 25 of which encoded ferredoxins or subunits of oxidoreductases or flavoproteins (Table 4). A cupredoxin superfamily protein gene (Ferp_0125), predicted to belong to an operon divergently transcribed from the operon for nitrous oxide reductase, which also contains a cupredoxin domain (49), was upregulated in F. placidus cells during growth with Fe(III) oxide. However, the two genes predicted to be cotranscribed with Ferp_0125, one of which encodes another protein of the cupredoxin superfamily (Ferp_0127), were not differentially regulated, and neither was nitrous oxide reductase (nosZ; Ferp_0128).
TABLE 4.
Genes encoding electron transport proteins that are not c-type cytochromes and that were differentially expressed in F. placidus cellsa
| Locus ID | Gene product | Fold upregulated |
|---|---|---|
| Ferp_0125 | Cupredoxin superfamily protein | 2.49 |
| Ferp_0140 | Heterodisulfide oxidoreductase, iron-sulfur cluster-binding subunit B | 2.10 |
| Ferp_0349 | Ferredoxin | 2.16 |
| Ferp_0602 | Formylmethanofuran dehydrogenase, bis-(molybdopterin guanine dinucleotide)-oxotungsten-binding subunit B | 2.11 |
| Ferp_0603 | Formylmethanofuran dehydrogenase, subunit A | 3.54 |
| Ferp_0604 | Formylmethanofuran dehydrogenase, subunit C | 2.32 |
| Ferp_0728 | Formylmethanofuran dehydrogenase, iron-sulfur cluster-binding subunit G | 2.19 |
| Ferp_1088 | Oxidoreductase, membrane protein subunit | 2.30 |
| Ferp_1256 | Phenylacetyl-coenzyme A dehydrogenase, iron-sulfur cluster-binding subunit | 2.02 |
| Ferp_1268 | Menaquinol oxidoreductase complex Cbc4, iron-sulfur cluster-binding subunit, putative | 2.43 |
| Ferp_1269 | Menaquinol oxidoreductase complex Cbc4, cytochrome b subunit, putative | 2.72 |
| Ferp_1300 | 2-Oxoacid:ferredoxin oxidoreductase, small subunit | 2.14 |
| Ferp_1437 | Menaquinol oxidoreductase complex Cbc4, cytochrome b subunit, putative | 2.17 |
| Ferp_1687 | Electron transfer flavoprotein, beta subunit | 2.27 |
| Ferp_1688 | Electron transfer flavoprotein, alpha subunit | 2.63 |
| Ferp_1705 | Coenzyme F420:quinone oxidoreductase, membrane protein subunit M, putative | 2.09 |
| Ferp_1707 | Coenzyme F420:quinone oxidoreductase, membrane protein subunit N, putative | 2.23 |
| Ferp_1709 | Coenzyme F420:quinone oxidoreductase, subunit BC, putative | 3.58 |
| Ferp_1710 | Coenzyme F420:quinone oxidoreductase, subunit D, putative | 2.38 |
| Ferp_1711 | Coenzyme F420:quinone oxidoreductase, membrane protein subunit H, putative | 2.49 |
| Ferp_1712 | Coenzyme F420:quinone oxidoreductase, iron-sulfur cluster-binding subunit I, putative | 2.44 |
| Ferp_1940 | Electron transfer flavoprotein, alpha subunit | 2.13 |
| Ferp_2216 | Sulfate adenylyltransferase | 2.32 |
| Ferp_2218 | Adenosine-5′-phosphosulfate reductase, FAD-binding catalytic alpha subunit | 3.01 |
| Ferp_2220 | Oxidoreductase, iron-sulfur cluster-binding subunit | 2.83 |
| Ferp_2377 | Oxidoreductase domain protein | 2.32 |
Shown are genes encoding electron transport proteins that are not c-type cytochromes and that were differentially expressed in F. placidus cells grown with acetate as the electron donor and either insoluble Fe(III) oxide or soluble Fe(III) citrate as the electron acceptor.
As seen in Geobacter sulfurreducens (46), significantly more transcripts from genes coding for iron sulfur cluster-binding proteins were made by Fe(III) oxide-grown cells (Table 4). These proteins may participate in electron transport to Fe(III) oxides. One of the upregulated genes coding for an iron-sulfur cluster-binding domain protein in F. placidus, Ferp_1268, is homologous to a subunit of the Cbc4 complex from G. sulfurreducens (GSU0069; 42% identical) that was upregulated in G. sulfurreducens during growth on Fe(III) oxide (46). Both the c-type cytochrome (Ferp_1267) and the b-type cytochrome (Ferp_1269) in this cluster also were upregulated during growth on Fe(III) oxide, which suggests that these three proteins form a complex that are involved in extracellular electron transfer to Fe(III).
Genes coding for proteins of the coenzyme F420:quinone oxidoreductase complex (Ferp_1705, Ferp_1707, Ferp_1709, Ferp_1710, Ferp_1711, and Ferp_1712) and the formylmethanofuran dehydrogenase complex (Ferp_0602, Ferp_0603, Ferp_0604, and Ferp_0728) also were significantly upregulated in Fe(III) oxide-grown cells (Table 4). Although both of these protein complexes are involved in methane production by methanogenic archaea, genomes of species from the family Archaeoglobaceae are missing the genes for coenzyme M biosynthesis and coenzyme F430 biosynthesis, as well as methyl-coenzyme M reductase genes from the pathway, and they are not capable of methanogenesis (50). Studies of these proteins in A. fulgidus show that they are part of the energy-conserving electron transport chain that connects the oxidation of lactate with the reduction of sulfate, sulfite, and thiosulfate (51, 52). The coenzyme F420:quinone oxidoreductase complex in A. fulgidus plays a role similar to that of NADH:quinone oxidoreductase, except that coenzyme F420H2 rather than NADH acts as an electron carrier for the enzyme (51). NADH dehydrogenase homologues also were significantly upregulated in Geobacter cells grown with insoluble Fe(III) oxide or with current-harvesting electrodes compared to soluble Fe(III) citrate (46, 47, 53), and membrane-bound NADH dehydrogenase proteins have been shown to transfer electrons to c-type cytochromes during Fe(III) reduction (54).
Genes coding for adenylsulfate reductase (Ferp_2218) and sulfate adenylyltransferase (Ferp_2216), proteins involved in dissimilatory sulfate reduction, also had more mRNA transcripts in Fe(III) oxide-grown cells. Two genes coding for alpha subunits of electron transfer flavoproteins (Ferp_1688 and Ferp_1940), for which a homologue was upregulated in G. sulfurreducens (GSU2796) (46), also had higher transcript abundance in Fe(III) oxide-grown cells of F. placidus.
Expression of archaella and other extracellular appendages during growth on Fe(III).
Previous analysis of the F. placidus genome with the software program FlaFind (55) identified 31 different genes that could code for archaellin/pilin preproteins with type IV-like signal peptides. In this study, genes encoding putative archaellum/pilus-associated proteins were further analyzed (Table 5). Three of the type IV-like signal peptide-bearing preproteins (Ferp_0458, Ferp_1118, and Ferp_1554) contained a DUF1628 domain that is associated with previously described archaeal pilin proteins (56), one (Ferp_0926) contained a DUF361 domain that is characteristic of a class III signal peptide processed by a unique archaeal prepilin peptidase, EppA (55), and two were homologous to bacterial type IV pilin proteins (Ferp_1456 and Ferp_0066). Five of the predicted archaellum/pilus-associated genes had more transcripts in Fe(III) oxide-grown cells, including two type IV pilin-like proteins (Ferp_1456 and Ferp_1554).
TABLE 5.
Genes encoding archaeallar biogenesis proteins that were upregulated in F. placidus cells growing with acetate as electron donor and Fe(III) oxidea
| Locus ID | Gene product | Gene | Fold upregulated |
|---|---|---|---|
| Ferp_0061 | Type IV prepilin-like proteins leader peptide processing enzyme | flaK | NDb |
| Ferp_0252 | Type II secretion system ATPase PulE | pulE-1 | ND |
| Ferp_0253 | Type II secretion system inner membrane protein PulF | pulF | ND |
| Ferp_0283 | Twitching motility pilus retraction ATPase | pilT | ND |
| Ferp_0458 | Type IV pilin, putative (DUF1628-containing) | ND | |
| Ferp_0578 | Type II secretion system ATPase domain protein | ND | |
| Ferp_0783 | Type II secretion system ATPase domain protein | ND | |
| Ferp_0926 | DUF361 domain like pattern EppA-pilin protein | ND | |
| Ferp_1118 | Type IV pilin, DUF1628-containing | ND | |
| Ferp_1431 | Type II secretion system ATPase GspE | gspE-1 | ND |
| Ferp_1456 | Archaellin | flaB | 3.54 |
| Ferp_1457 | Archaellar accessory protein FlaC | flaC | 2.11 |
| Ferp_1458 | Archaellar accessory protein FlaD/FlaE | flaDE | 2.51 |
| Ferp_1459 | Archaellar accessory protein FlaF | flaF | 2.02 |
| Ferp_1460 | Archaellar accessory protein FlaG | flaG | ND |
| Ferp_1461 | Archaellar accessory ATPase FlaH | flaH | ND |
| Ferp_1462 | Archaellar accessory ATPase FlaI | flaI | ND |
| Ferp_1463 | Archaellar membrane protein FlaJ | flaJ | ND |
| Ferp_1533 | VirB11-like ATPase | flaI-2 | ND |
| Ferp_1554 | Type IV pilin, DUF1628-containing | 2.46 |
Shown are genes encoding archaeallar biogenesis proteins that were upregulated in F. placidus cells growing with acetate as the electron donor and Fe(III) oxide compared to Fe(III) citrate as the electron acceptor according to microarray analysis.
ND, no difference.
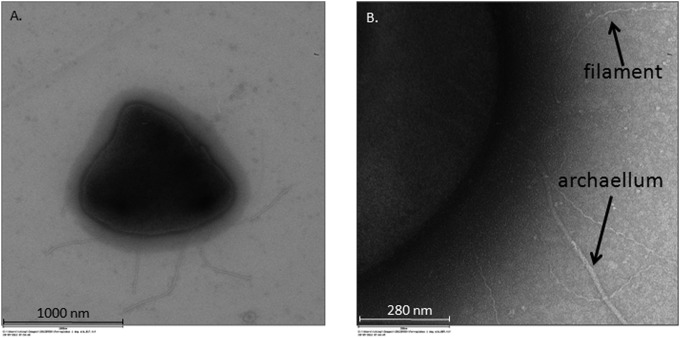
TEM microscopy was done to further examine the archaellum and/or pilus appendages during growth on Fe(III). Multiple archaellar extensions were apparent on the F. placidus cell surface (Fig. 3A). This result differs significantly from the original isolation study in which a single archaellum was detected during growth with nitrate as the electron acceptor (18). In addition to archaella, smaller filament-like structures (putative pili) also were observed in these images (Fig. 3B).
FIG 3.
Transmission electron microscopy of F. placidus cells grown with insoluble Fe(III) oxide as the electron acceptor and acetate as the electron donor. (A) Cell with multiple archaella; (B) cell with archaellum and significantly smaller filaments.
The archaellum (archaeal flagellum) is a surface structure that shares many similarities with the bacterial type IV pilus system (57, 58). Archaellar proteins are made as preproteins with N-terminal signal peptides that are cleaved by a prepilin peptidase (FlaK) and assembled by the addition of archaellin subunits to the base of the archaellum. Type IV pili associated with dissimilatory Fe(III)-reducing bacteria are known to be involved in biofilm formation and electron transfer to extracellular Fe(III) compounds and current-harvesting electrodes (59, 60). Although electron transfer by archaella has not been observed, studies have shown that archaella help with attachment to surfaces and biofilm formation (58). It is possible that the archaellin proteins in F. placidus perform a role similar to that of type IV pilins in Fe(III)-reducing bacteria. However, further investigation into this possibility is required.
Evidence of direct electron transfer to insoluble Fe(III) oxides.
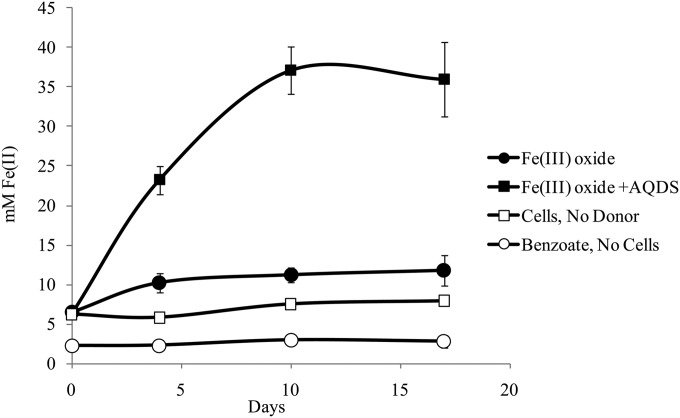
F. placidus cells were grown in medium with insoluble Fe(III) oxides entrapped in microporous alginate beads to determine whether this organism utilizes an electron shuttle to reduce insoluble Fe(III) indirectly. Pores in the beads were too small for cells to make direct contact with the Fe(III) oxides. Therefore, only Fe(III) oxide exposed on the bead surface (ca. 3.5 mM) was directly accessible to the cells. F. placidus was unable to reduce Fe(III) oxides in the beads unless the electron shuttle anthraquinone 2,6,-disulfonate (AQDS) was provided (Fig. 4). These results suggest that F. placidus does not produce electron mediators that are small enough to fit through the alginate bead pores.
FIG 4.
F. placidus grown in medium with benzoate (0.5 mM) provided as an electron donor and insoluble Fe(III) oxides entrapped in alginate beads provided as the electron acceptor. As a control, 50 μM anthraquinone 2,6,-disulfonate (AQDS) was added to the medium and served as a shuttle for Fe(III) reduction. ■, Fe(III) oxide; ●, Fe(III) oxide plus AQDS; ☐, cells, no donor; ○, donor, no cells.
Conclusions.
There are a variety of mechanisms used for Fe(III) respiration by prokaryotes, and these do not appear to be domain specific. Direct contact with Fe(III) oxides and reliance on electron-shuttling or chelating compounds for reduction of Fe(III) are traits seen in both bacterial and archaeal species (1, 11–16, 61). In fact, the majority of electron transport proteins that transcriptomic and proteomic studies showed to be important for reduction of insoluble Fe(III) oxide by F. placidus have homologues in mesophilic Fe(III)-reducing bacteria.
For example, three multiheme c-type cytochrome proteins (Ferp_0670, Ferp_0672, and Ferp_1267) that appear to be important for insoluble Fe(III) respiration by F. placidus have homologues in Geobacter that are known to be involved in electron transfer to Fe(III) (46, 47). In addition to similarities in expression patterns of c-type cytochromes, other genes encoding important electron transport proteins found in Geobacter also had higher transcript abundance in Fe(III) oxide-grown F. placidus cells (46). A menaquinol oxidoreductase Cbc4 complex (Ferp_1267-1269) and a unique periplasmic c-type cytochrome fused to a cytochrome b5-type heme/steroid binding domain (CbcZ; Ferp_0937) seem to be involved in electron transfer to insoluble Fe(III) oxide in both F. placidus and Geobacter species. It is also significant that, similar to Geobacter species, F. placidus seemed to express more pilin-like structures during growth on insoluble Fe(III) oxide.
It is also interesting that F. placidus has more c-type cytochromes than any other hyperthermophilic Fe(III)-reducing archaeon, and 83.3% of its cytochromes have more than one heme group. The genomes of most other hyperthermophilic archaea that are capable of Fe(III) reduction have few to no c-type cytochrome genes and can only reduce Fe(III) chemolithotrophically (16, 62–65). On the other hand, Ferroglobus and Geoglobus, both hyperthermophilic archaea that are known to effectively couple the complete oxidation of organic compounds with Fe(III) reduction (20, 21), contain numerous multiheme c-type cytochromes. More than half of these multiheme c-type cytochromes have homologues in Fe(III)-reducing bacteria from the genera Geobacter, Shewanella (48), Rhodoferax (66), and Thermincola (67), organisms that also are efficient at electron transfer from the oxidation of organic compounds to Fe(III). Many of the multiheme cytochromes found in Ferroglobus, Geoglobus, and Fe(III)-reducing bacteria are class III, which means that they have heme groups with reduction potentials that can vary significantly (68). For example, a 50-kDa cytochrome purified from Desulfuromonas acetoxidans, a relative of the Geobacteraceae, contains 6 heme groups that have reduction potentials ranging from +100 mV to −375 mV (69). This range of reduction potentials may help facilitate the flow of electrons released from the oxidation of organic compounds through the periplasm and out to extracellular Fe(III). However, further investigation into this possibility is required.
This study clearly shows that there is a considerable amount of overlap in the mechanisms for Fe(III) reduction employed by archaea and bacteria, and to fully understand Fe(III) respiration, one needs to examine organisms from both domains of life.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the Office of Science (BER), U.S. Department of Energy, cooperative agreement no. DE-FC02-02ER63446.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04038-14.
REFERENCES
- 1.Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol 49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 2.Thamdrup B. 2000. Bacterial manganese and iron reduction in aquatic sediments. Adv Microb Ecol 16:41–84. doi: 10.1007/978-1-4615-4187-5_2. [DOI] [Google Scholar]
- 3.Roling WF, Head IM, Larter SR. 2003. The microbiology of hydrocarbon degradation in subsurface petroleum reservoirs: perspectives and prospects. Res Microbiol 154:321–328. doi: 10.1016/S0923-2508(03)00086-X. [DOI] [PubMed] [Google Scholar]
- 4.Slobodkin AI. 2005. Thermophilic microbial metal reduction. Mikrobiologiia 74:581–595. doi: 10.1007/s11021-005-0096-6. [DOI] [PubMed] [Google Scholar]
- 5.Kashefi K, Holmes DE, Lovley DR, Tor JM. 2004. Potential importance of dissimilatory Fe(III)-reducing microorganisms in hot sedimentary environments. Geophys Monogr Ser 144:199–211. doi: 10.1029/144GM13. [DOI] [Google Scholar]
- 6.Holm NG, Cairns-Smith AG, Daniel RM, Ferris JP, Hennet RJ, Shock EL, Simoneit BR, Yanagawa H. 1992. Marine hydrothermal systems and the origin of life: future research. Life Evol Biosph 22:181–242. [DOI] [PubMed] [Google Scholar]
- 7.Kim JD, Yee N, Nanda V, Falkowski PG. 2013. Anoxic photochemical oxidation of siderite generates molecular hydrogen and iron oxides. Proc Natl Acad Sci U S A 110:10073–10077. doi: 10.1073/pnas.1308958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yucel M, Gartman A, Chan CS, Luther GW. 2011. Hydrothermal vents as a kinetically stable source of iron-sulphide-bearing nanoparticles to the ocean. Nat Geosci 4:367–371. doi: 10.1038/ngeo1148. [DOI] [Google Scholar]
- 9.Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP. 2011. Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100. doi: 10.1016/B978-0-12-387661-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 10.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 11.Nevin KP, Lovley DR. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl Environ Microbiol 66:2248–2251. doi: 10.1128/AEM.66.5.2248-2251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotloski NJ, Gralnick JA. 2013. Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4:e00553–12. doi: 10.1128/mBio.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg LF, Holden JF. 2006. Characterization of dissimilatory Fe(III) versus NO3− reduction in the hyperthermophilic archaeon Pyrobaculum aerophilum. J Bacteriol 188:525–531. doi: 10.1128/JB.188.2.525-531.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzella MP, Reguera G, Kashefi K. 2013. Extracellular electron transfer to Fe(III) oxides by the hyperthermophilic archaeon Geoglobus ahangari via a direct contact mechanism. Appl Environ Microbiol 79:4694–4700. doi: 10.1128/AEM.01566-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardanov AV, Slododkina GB, Slobodkin AI, Beletsky AV, Gavrilov SN, Kublanov IV, Bonch-Osmolovskaya EA, Skryabin KG, Ravin NV. 2014. The genome of Geoglobus acetivorans: Fe(III) reduction, acetate utilization, autotrophic growth and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl Environ Microbiol 81:1003–1012. doi: 10.1128/AEM.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childers SE, Lovley DR. 2001. Differences in Fe(III) reduction in the hyperthermophilic archaeon, Pyrobaculum islandicum, versus mesophilic Fe(III)-reducing bacteria. FEMS Microbiol Lett 195:253–258. doi: 10.1111/j.1574-6968.2001.tb10529.x. [DOI] [PubMed] [Google Scholar]
- 17.Rogers KL, Amend JP, Gurrieri S. 2007. Temporal changes in fluid chemistry and energy profiles in the Vulcano island hydrothermal system. Astrobiology 7:905–932. doi: 10.1089/ast.2007.0128. [DOI] [PubMed] [Google Scholar]
- 18.Hafenbradl D, Keller M, Dirmeier R, Rachel R, Rossnagel P, Burggraf S, Huber H, Stetter KO. 1996. Ferroglobus placidus gen nov, sp nov, a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch Microbiol 166:308–314. doi: 10.1007/s002030050388. [DOI] [PubMed] [Google Scholar]
- 19.Kashefi K, Tor JM, Holmes DE, Gaw Van Praagh CV, Reysenbach AL, Lovley DR. 2002. Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int J Syst Evol Microbiol 52:719–728. doi: 10.1099/ijs.0.01953-0. [DOI] [PubMed] [Google Scholar]
- 20.Slobodkina GB, Kolganova TV, Querellou J, Bonch-Osmolovskaya EA, Slobodkin AI. 2009. Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 59:2880–2883. doi: 10.1099/ijs.0.011080-0. [DOI] [PubMed] [Google Scholar]
- 21.Tor JM, Kashefi K, Lovley DR. 2001. Acetate oxidation coupled to Fe(III) reduction in hyperthermophilic microorganisms. Appl Environ Microbiol 67:1363–1365. doi: 10.1128/AEM.67.3.1363-1365.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes DE, Risso C, Smith JA, Lovley DR. 2012. Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J 6:146–157. doi: 10.1038/ismej.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes DE, Risso C, Smith JA, Lovley DR. 2011. Anaerobic oxidation of benzene by the hyperthermophilic archaeon Ferroglobus placidus. Appl Environ Microbiol 77:5926–5933. doi: 10.1128/AEM.05452-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aklujkar M, Risso C, Smith JA, Beaulieu D, Dubay R, Giloteaux L, DiBurro K, Holmes D. 2014. Anaerobic degradation of aromatic amino acids by the hyperthermophilic archaeon, Ferroglobus placidus. Microbiology 160:2694–2709. doi: 10.1099/mic.0.083261-0. [DOI] [PubMed] [Google Scholar]
- 25.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller TL, Wolin MJ. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol 27:985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tor JM, Lovley DR. 2001. Anaerobic degradation of aromatic compounds coupled to Fe(III) reduction by Ferroglobus placidus. Environ Microbiol 3:281–287. doi: 10.1046/j.1462-2920.2001.00192.x. [DOI] [PubMed] [Google Scholar]
- 28.Coates JD, Lonergan DJ, Philips EJ, Jenter H, Lovley DR. 1995. Desulfuromonas palmitatis sp. nov., a marine dissimilatory Fe(III) reducer that can oxidize long-chain fatty acids. Arch Microbiol 164:406–413. doi: 10.1007/BF02529738. [DOI] [PubMed] [Google Scholar]
- 29.Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley DR, Phillips EJ. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl Environ Microbiol 53:1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francis RT, Becker RR. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal Biochem 136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 32.Thomas PE, Ryan D, Levin W. 1976. Improved staining procedure for detection of peroxidase-activity of cytochrome-P-450 on sodium dodecyl-sulfate polyacrylamide gels. Anal Biochem 75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 2001. Current protocols in molecular biology, vol 3 John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 34.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 35.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 36.Gardy JL, Spencer C, Wang K, Ester M, Tusnady GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FSL. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res 31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. 2009. Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35. doi: 10.1093/protein/gzn064. [DOI] [PubMed] [Google Scholar]
- 38.Hofmann K, Stoffel W. 1993. TMbase–a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166. [Google Scholar]
- 39.Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filatov DA. 2002. PROSEQ: a software for preparation and evolutionary analysis of DNA sequence data sets. Mol Ecol Notes 2:621–624. doi: 10.1046/j.1471-8286.2002.00313.x. [DOI] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aldrich J. 1997. R.A. Fisher and the making of maximum likelihood 1912-1922. Statist Sci 12:162–176. [Google Scholar]
- 44.Jones D, Taylor W, Thornton J. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 45.Fisher N, Rich PR. 2000. A motif for quinone binding sites in respiratory and photosynthetic systems. J Mol Biol 296:1153–1162. doi: 10.1006/jmbi.2000.3509. [DOI] [PubMed] [Google Scholar]
- 46.Aklujkar M, Coppi MV, Leang C, Kim BC, Chavan MA, Perpetua LA, Giloteaux L, Liu A, Holmes DE. 2013. Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159:515–535. doi: 10.1099/mic.0.064089-0. [DOI] [PubMed] [Google Scholar]
- 47.Smith JA, Lovley DR, Tremblay PL. 2013. Outer cell surface components essential for Fe(III) oxide reduction by Geobacter metallireducens. Appl Environ Microbiol 79:901–907. doi: 10.1128/AEM.02954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, Squier TC, Zachara JM, Fredrickson JK. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol Microbiol 65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Gao C, Zhang A, Jin P, Wang L, Feng L. 2008. The nos gene cluster from Gram-positive bacterium Geobacillus thermodenitrificans NG80-2 and functional characterization of the recombinant NosZ. FEMS Microbiol Lett 289:46–52. doi: 10.1111/j.1574-6968.2008.01362.x. [DOI] [PubMed] [Google Scholar]
- 50.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, Richardson DL, Kerlavage AR, Graham DE, Kyrpides NC, Fleischmann RD, Quackenbush J, Lee NH, Sutton GG, Gill S, Kirkness EF, Dougherty BA, McKenney K, Adams MD, Loftus B, Peterson S, Reich CI, McNeil LK, Badger JH, Glodek A, Zhou LX, Overbeek R, Gocayne JD, Weidman JF, McDonald L, Utterback T, Cotton MD, Spriggs T, Artiach P, Kaine BP, Sykes SM, Sadow PW, D'Andrea KP, Bowman C, Fujii C, Garland SA, Mason TM, Olsen GJ, Fraser CM, Smith HO, Woese CR, Venter JC. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 51.Bruggemann H, Falinski F, Deppenmeier U. 2000. Structure of the F420H2:quinone oxidoreductase of Archaeoglobus fulgidus identification and overproduction of the F420H2-oxidizing subunit. Eur J Biochem 267:5810–5814. doi: 10.1046/j.1432-1327.2000.01657.x. [DOI] [PubMed] [Google Scholar]
- 52.Schmitz RA, Linder D, Stetter KO, Thauer RK. 1991. N5,N10-methylenetetrahydromethanopterin reductase (coenzyme-F420-dependent) and formylmethanofuran dehydrogenase from the hyperthermophile Archaeoglobus fulgidus. Arch Microbiol 156:427–434. doi: 10.1007/BF00248722. [DOI] [Google Scholar]
- 53.Holmes DE, Chaudhuri SK, Nevin KP, Mehta T, Methe BA, Liu A, Ward JE, Woodard TL, Webster J, Lovley DR. 2006. Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ Microbiol 8:1805–1815. doi: 10.1111/j.1462-2920.2006.01065.x. [DOI] [PubMed] [Google Scholar]
- 54.Magnuson TS, Hodges-Myerson AL, Lovley DR. 2000. Characterization of a membrane-bound NADH-dependent Fe(3+) reductase from the dissimilatory Fe(3+)-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol Lett 185:205–211. doi: 10.1111/j.1574-6968.2000.tb09063.x. [DOI] [PubMed] [Google Scholar]
- 55.Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJM, Pohlschroder M. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 189:772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esquivel RN, Xu R, Pohlschroder M. 2013. Novel archaeal adhesion pilins with a conserved N terminus. J Bacteriol 195:3808–3818. doi: 10.1128/JB.00572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pohlschroder M, Ghosh A, Tripepi M, Albers SV. 2011. Archaeal type IV pilus-like structures-evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol 14:357–363. doi: 10.1016/j.mib.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 58.Ng SYM, Zolghadr B, Driessen AJM, Albers SV, Jarrell KF. 2008. Cell surface structures of archaea. J Bacteriol 190:6039–6047. doi: 10.1128/JB.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 60.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nevin KP, Lovley DR. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl Environ Microbiol 68:2294–2299. doi: 10.1128/AEM.68.5.2294-2299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vargas M, Kashefi K, Blunt-Harris EL, Lovley DR. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65–67. doi: 10.1038/25720. [DOI] [PubMed] [Google Scholar]
- 63.Itoh T, Suzuki K, Sanchez PC, Nakase T. 1999. Caldivirga maquilingensis gen. nov., sp nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Bacteriol 49:1157–1163. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 64.Kashefi K, Lovley DR. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100 degrees C by Pyrobaculum islandicum. Appl Environ Microbiol 66:1050–1056. doi: 10.1128/AEM.66.3.1050-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kletzin A. 2007. General characteristics and important model organisms, p 14–92. In Cavicchioli R. (ed), Archaea: molecular and cellular biology. ASM Press, Washington, DC. [Google Scholar]
- 66.Risso C, Sun J, Zhuang K, Mahadevan R, Deboy R, Ismail W, Shrivastava S, Huot H, Kothari S, Daugherty S, Bui O, Schilling CH, Lovley DR, Methe BA. 2009. Genome-scale comparison and constraint-based metabolic reconstruction of the facultative anaerobic Fe(III)-reducer Rhodoferax ferrireducens. BMC Genomics 10:447. doi: 10.1186/1471-2164-10-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlson HK, Iavarone AT, Gorur A, Yeo BS, Tran R, Melnyk RA, Mathies RA, Auer M, Coates JD. 2012. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc Natl Acad Sci U S A 109:1702–1707. doi: 10.1073/pnas.1112905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mowat CG, Chapman SK. 2005. Multi-heme cytochromes–new structures, new chemistry. Dalton Trans 21:3381–3389. doi: 10.1039/b505184c. [DOI] [PubMed] [Google Scholar]
- 69.Pereira IAC, Pacheco I, Liu MY, Legall J, Xavier AV, Teixeira M. 1997. Multiheme cytochromes from the sulfur-reducing bacterium Desulfuromonas acetoxidans. Eur J Biochem 248:323–328. doi: 10.1111/j.1432-1033.1997.00323.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.