Abstract
Loop-mediated isothermal amplification (LAMP) is a method for enzymatically replicating DNA that has great utility for clinical diagnosis at the point of care (POC), given its high sensitivity, specificity, speed, and technical requirements (isothermal conditions). Here, we adapted LAMP for measuring protein analytes by creating a protein-DNA fusion (referred to here as a “LAMPole”) that attaches oligonucleotides (LAMP templates) to IgG antibodies. This fusion consists of a DNA element covalently bonded to an IgG-binding polypeptide (protein L/G domain). In our platform, LAMP is expected to provide the most suitable means for amplifying LAMPoles for clinical diagnosis at the POC, while quantitative PCR is more suitable for laboratory-based quantification of antigen-specific IgG abundance. As proof of concept, we measured serological responses to a protozoan parasite by quantifying changes in solution turbidity in real time. We observed a >6-log fold difference in signal between sera from vaccinated versus control mice and in a clinical patient sample versus a control. We assert that LAMPoles will be useful for increasing the sensitivity of measuring proteins, whether it be in a clinical laboratory or in a field setting, thereby improving acute diagnosis of a variety of infections.
INTRODUCTION
Numerous applications require sensitive measurement of biological analytes to provide actionable information in real time. In considering biomarker measurement, no other molecule is as easily or sensitively quantified as nucleic acid because of facile means to enzymatically replicate sequence-specific templates in vitro. The detection limit of specific DNA templates has been reported to be at or below 10 molecules when measured by real-time PCR (quantitative PCR [qPCR]) (1, 2). This level of sensitivity is useful in the context of a laboratory, but there is need for diagnostics that can be used at the point of care (POC) without thermocycling equipment. An alternative method for amplifying DNA is loop-mediated isothermal amplification (LAMP), which requires four different oligonucleotides to promote Bst polymerase-catalyzed DNA synthesis at a constant 65°C. The end products of LAMP (pyrophosphate and high-molecular weight DNA) can be monitored by measuring changes in turbidity or color with the addition of Mg2+ (3) or dyes (4), respectively.
Highly sensitive measurement of non-DNA analytes (e.g., proteins, toxins, lipids, and carbohydrates) at the POC can be achieved by combining the molecular recognition of immunoassays with the signal amplification of LAMP. We previously developed means to label protein and small-molecule ligands with unique oligonucleotides measurable by PCR (5). We labeled molecular targets using a protein-DNA fusion known as a “Tadpole,” which binds ligand with high specificity so as to attach an identifying oligonucleotide. When we labeled antibodies or protein antigens with oligonucleotides, we created toxin and biomarker assays that were hundreds of times more sensitive than matched enzyme-linked immunosorbent assay (ELISA) (5–7). Compared to PCR, however, LAMP is more amenable to POC use because of reaction simplicity and product detection by visual readouts (8–11). In this regard, we previously advocated (12) that LAMP and Tadpoles be combined such that the DNA element of a Tadpole is amplified by LAMP instead of PCR. To investigate this concept, we created a protein-DNA fusion consisting of a polypeptide that binds mammalian IgG and a DNA element encompassing 1 PCR and 2 LAMP amplicons. During the construction of this fusion, we explored the activities of 10 different primer pair combinations for conducting LAMP and several of chemistries for conjugating protein and DNA. We further investigated the specificity of the LAMPole fusion for recognizing human IgG and, as a benchmark of this approach, applied it to measuring antiparasite antibodies in humans and mice. Our assay was inspired by an ELISA for detecting host antibodies to Trypanosoma brucei gambiense variable surface glycoproteins (VSGs), which detected titers of 1:30,000 and 1:40 from serum and saliva, respectively (13). This work indicated that detection of relatively low titers of antibodies in noninvasive specimen types is possible provided the method is sufficiently sensitive. Here, we present our findings. Rapid, accurate diagnosis of other proteins, such as antigen and IgM antibody, may enable sensitive and specific diagnosis of the etiology of acute illness at the POC.
MATERIALS AND METHODS
To construct the LAMPole fusion, we needed to design (i) the sequence of the DNA element and primer sets that specifically recognize this template, even in the presence of contaminating human or pathogen DNA, and (ii) a conjugation chemistry that bonds the DNA element to the polypeptide without inhibiting IgG-binding activity or interfering with the ability to promote LAMP. We designed 10 LAMP primer sets (4 different primers in each set) predicted to hybridize with elements in the Arabidopsis thaliana chalcone synthase (CHS)-coding sequence (GenBank accession number M20308.1) using the software PrimerExplorer V4 (Fujitsu Limited). We identified primers according to the nomenclature originally described by Notomi et al. (14) as F3, B3, FIP, and BIP. The predicted melting temperature for the F3 and B3 primers was fixed close to 60°C in each set to promote LAMP at the optimal temperature for Bst activity. The LAMP reactions for selecting our primer sets (Fig. 1) and for subsequently measuring IgG serology were performed as previously described (15–17). Briefly, the reaction mixture contained 12.5 μl of 2× LAMP buffer [40 mM Tris-HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M betaine, 2.8 mM each deoxyribonucleoside triphosphate], 1.0 μl CHS LAMP primer mix (5 pmol each of F3 and B3 and 40 pmol each of FIP and BIP), 8 units Bst DNA polymerase (New England BioLabs, Tokyo, Japan), and template DNA. Final volumes were adjusted to 25 μl with distilled water. All reactions were conducted in duplicate and were monitored in real time in a Loopamp LA320C real-time turbidimeter (Teramecs, Tokyo, Japan). For assaying production of high-molecular-weight DNA ladders, we fractionated 10 μl of reaction products by 1% agarose gel electrophoresis in 1× Tris-borate-EDTA (TBE) and visualized with ethidium bromide (EtBr).
FIG 1.
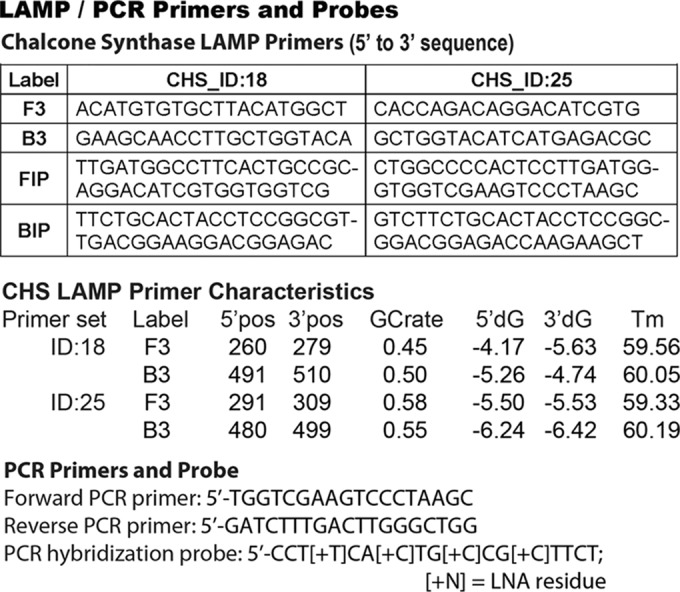
Primers sets developed for sequence-specific LAMP and real-time PCR. The LAMP primer set ID:18 and ID:25 sequences are shown going from 5′ to 3′ with the F3, B3, FIP, and BIP nomenclature derived from Notomi et al. (14). The annealing positions corresponding to the A. thaliana chalcone synthase (CHS) complete coding sequence (GenBank accession number M20308.1), GC content (GCrate), 5′ and 3′ gene positions, free energy (5′dG/3′dG), and melting temperatures (Tm) are indicated. Also shown are the forward and reverse primer sequences (5′ to 3′), along with the fluorescent hybridization probe sequence, used for real-time PCR.
To create the recombinant 253-bp double-stranded DNA (dsDNA) oligonucleotide (LAMP template) (Fig. 2), we digested pTRX-CHS plasmid DNA (18) with PciI and NaeI to liberate an element with a single 3′ recessed terminus (PciI) and a single blunt end terminus (NaeI). We incorporated an amino-allyl dUTP nucleotide at the 3′ recessed terminus using the 3′-to-5′ exo-Klenow fragment (New England BioLabs) for 30 min according to the manufacturer's instructions. We converted this amino residue to an azide using 10:1 excess N-hydroxysuccinimide-azide (Pierce) in phosphate-buffered saline (PBS) for 1 h at 25°C and purified this element using a NAP-5 column (Life Technologies). To create our protein L/G chimera, we directed expression of the L/G mammalian antibody-binding polypeptide (19) fused to polyhistidine and the Methanobacterium thermoautotrophicum RIR1 intein (20) Rosetta-gami-B (DE3) pLysS (Novagen) as previously described (21). Briefly, we cultured selected transformants in Luria-Bertani broth to an optical density (OD) of 0.6 at 600 nm, induced L/G expression with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h, and collected cells at 3,000 × g for 10 min at 4°C. We purified full-length L/G-intein fusion protein by immobilized metal affinity chromatography from cell-free lysates and cleaved the intein domain from L/G using 1 mM 2-mercaptoethanesulfonic acid in vitro (21). We labeled the purified L/G protein with phosphine-N-hydroxysuccinimide (NHS) at a 1:5 protein/phosphine stoichiometry in 0.1 M HEPES (pH 8.5) for 2 h at 25°C and buffer exchanged the phosphine-labeled L/G protein into phosphate-buffered saline. We conjugated azide-modified CHS dsDNA to phosphine-modified L/G protein for 6 h at 25°C to yield an L/G protein-DNA fusion (LAMPole) and purified fusions by cation exchange (Q-Sepharose) and hydrophobic chromatography (phenyl-Sepharose) as described previously (5, 21).
FIG 2.

The 5′-to-3′ sequence of the LAMP template. The annealing sites for the ID:18 F3 and B3 primers are underlined, while the annealing sites for the ID:25 F3 and B3 primers are shaded. The annealing site for the locked nucleic acid (LNA) hybridization probe for use in quantitative PCR (qPCR) is indicated in italics and gray. The adenosine residue complementary to amino-allyl dUTP nucleotide (Roche) incorporation (PciI site) is indicated in bold and underlined. PciI targets and cuts 5′...ACATGT...3′ sequences to 5′-CATGT...3′ products. As a result, the recombinant fragment digested is 1 bp shorter at the 5′ end than the CHS plasmid sequence used to design the LAMP primers. LAMP recognizes and amplifies the recombinant construct and parent CHS plasmid equally well.
Human African trypanosomiasis (HAT) is caused by Trypanosoma brucei gambiense (22). Our prototype LAMPole assay is based on a published ELISA that detects host antibodies against T. b. gambiense LiTat 1.3, 1.5, and 1.6 variable surface glycoproteins (VSGs) (13). To create the solid-phase substrate for use in our assays, we conjugated 2.8-μm tosyl-activated beads (Invitrogen) with either purified LiTat 1.3 VSG (23) or purified mouse total IgG (Jackson Immunologicals) in 0.1 M borate (pH 9.5)–1 M (NH4)2SO4–0.1 M NaCl for 16 h at 37°C. We capped unreacted tosyl residues with 0.1 M ethanolamine for 2 h at 37°C and blocked unoccupied surface area with PBS containing 0.1% dephosphorylated casein and 0.05% Tween 20 (blocking buffer). The mouse sera used to optimize assay conditions were prepared from animals vaccinated with 20 μg purified VSG once a week for 3 weeks. No adjuvant was used. Sham (naive) controls received vehicle alone (PBS). For proof of concept, we also used archived serum obtained from a patient with stage 2 T. b. gambiense HAT confirmed by a positive card agglutination test for trypanosomiasis (CATT) and with trypanosomes identified on blood and cerebrospinal fluid (CSF) smears.
For our serological assays, we serially diluted sera from test samples and negative controls. We incubated 106 VSG-coated beads with 50-μl serial dilutions of blocking buffer for 1 h at 25°C, washed the beads 3 times in 1 ml of casein-Tween-PBS by vortexing for 10 s, and then resuspended the beads with 50 μl of protein L/G LAMPole. After incubating for 1 h, we washed the beads 3 times with vortexing (as described above), resuspended the beads in 10 μl 10 mM Tris-HCl (pH 8.0)–0.05% Tween 20, and used 1 μl as the template for amplifying CHS templates by LAMP (25-μl total volume) at 63°C. Here, we monitored the production of insoluble Mg2+-pyrophosphate in real time using an LA-320C turbidimeter according to the manufacturer's instructions. For instances in which we used real-time PCR to quantify the amount of LAMPole bound to beads (affinity measurements), we used a fluorescent hydrolysis probe containing locked nucleic acid (LNA) nucleotides (5′-CCT[+T]CA[+C]TG[+C]CG[+C]TTCT, where [+N] is a LNA residue). For these measurements, we thermocycled bead-bound templates for 15 s at 95°C, 20 s at 60°C, and 60 s at 72°C. The quantities of LAMPole present in these samples were calibrated by measurement of known numbers of CHS template with an r2 value of 0.9988 over a 6-log range in concentration. We used the four-parameter logistic equation in the software package Prism v4.0 (GraphPad Software, Inc.) to fit a curve describing LAMPole activity to calculate the binding affinity constant according to previously published methods (5, 21).
RESULTS
We designed primer sets that specifically annealed and amplified a 253-nucleotide (nt) element of the A. thaliana chalcone synthase (CHS) gene (i) even in the presence of contaminating human or pathogen DNA and (ii) conjugated to the protein L/G polypeptide without inhibiting IgG-binding activity or interfering with the ability to promote LAMP. We identified 2 primer sets out of 10 that promoted LAMP of the A. thaliana CHS template at 63°C by assaying production of high-molecular-weight DNA ladders via agarose gel electrophoresis (data not shown). The LAMP primer set ID:18 and ID:25 sequences in Fig. 1 are shown going from 5′ to 3′, with F3, B3, FIP, A. thaliana CHS complete coding sequence (GenBank accession number M20308.1), GC contents, 5′ and 3′ gene positions, free energy, and melting temperatures (Tm) indicated. Also shown are the forward and reverse primer sequences (5′ to 3′) that allow for more accurate DNA quantitation by real-time PCR, along with the fluorescent hybridization probe sequence used to determine antibody-binding affinities as well as for DNA quantitation.
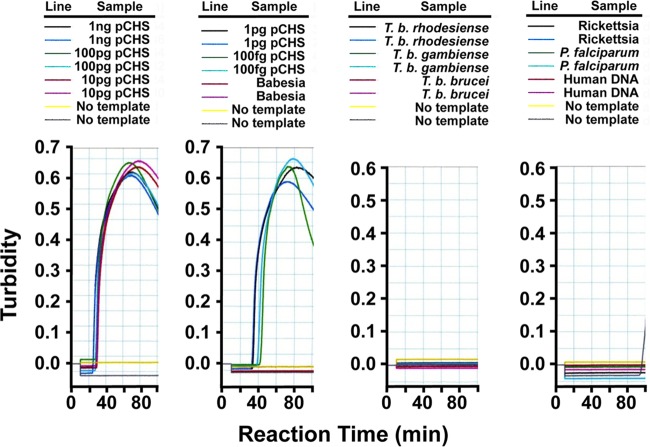
While both ID:18 and ID:25 enabled detection of at least 100 fg pET32(a)CHS as positive control (roughly 12,000 template molecules), ID:25 generated a more rapid LAMP reaction. We confirmed the primers to be 100% specific for A. thaliana CHS when tested against A. thaliana chalcone isomerase (CHI) plasmid and genomic DNAs from several protozoan parasites as negative controls, including Babesia microti, Plasmodium falciparum, African trypanosomes (T. b. brucei, T. b. rhodesiense, and T. b. gambiense) (Fig. 3), and Toxoplasma gondii (not shown). DNA from Rickettsia prowazekii, normal human blood (Fig. 3), and Escherichia coli and blood from bacteremic patients (not shown) and also gave negative results. We considered precipitate occurring after 60 min to be an artifact. Importantly, the amount of time needed to develop a visual turbidity of ≥0.1 OD unit in these experiments was inversely proportional to the amount of pET32(a)CHS template, demonstrating that the signal is quantitatively proportional to the abundance of template (Fig. 4). Fortuitously, the ID:18 and ID:25 amplicons overlap in sequence, which allowed us to finalize our LAMPole DNA design to include both overlapping amplicons and an internal PCR amplicon containing a hybridization site for a locked nucleic acid (LNA) (24) hydrolysis probe (see Materials and Methods).
FIG 3.
Sensitivity and specificity of LAMP primers for pET32(a)CHS template. We serially diluted plasmid encoding A. thaliana chalcone synthase (18) [pET32(a)CHS] or genomic DNA from Trypanosoma brucei subspecies, Babesia microti, Plasmodium falciparum, Toxoplasma gondii, E. coli, Rickettsia parkeri, bacteremic human patients, and normal human blood and assessed LAMP by monitoring turbidity in real-time. We used primer set ID:18 to direct LAMP in these reactions.
FIG 4.
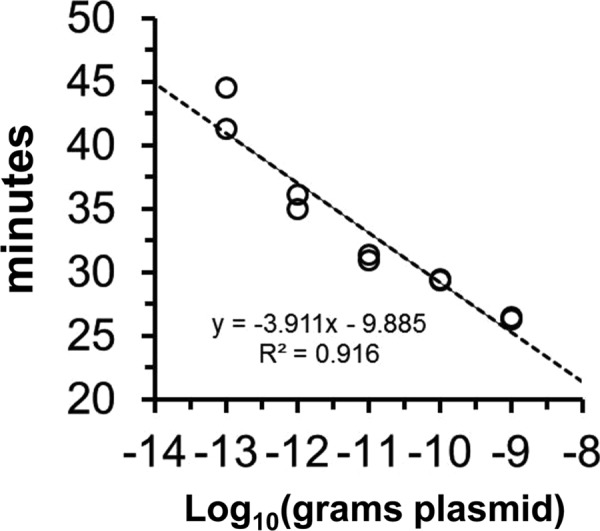
Quantitative LAMP of CHS templates. Based on data in Fig. 1, the amount of time necessary to accumulate turbidity above the threshold of detection (0.1 OD unit) is inversely proportional to the amount (100 fg to 1 ng) of starting A. thaliana CHS plasmid DNA template. The individual data points (n = 2) are shown.
To construct the antibody-binding protein domain, we used a recombinant protein originally created from the immunoglobulin-binding domains of Streptococcus sp. protein G (which binds mammalian immunoglobulin G [IgG] Fc chains) and Peptostreptococcus sp. protein L (which binds mammalian immunoglobulin κ and λ light chains) (19). We previously used this “L/G” domain to construct a probe with PCR readout and quantified as few as 21,559 and 1,016 molecules of an E. coli transcriptional initiation factor 1 fusion protein (INFA-TAP) (25) and interleukin-6 (26) in 50 μl, equivalent to detection limits of 716 and 34 aM, respectively (21). Here, we directly adapted this IgG-binding domain to synthesizing LAMPoles. We constructed our original Tadpoles exclusively from recombinant protein and synthetic oligonucleotides (60 to 85 bp) using “intein-mediated ligation” (27) to covalently bond a single DNA element per protein domain. However, the minimum size of templates that are amplifiable by LAMP averages two hundred nucleotides (3). We ultimately want large quantities of LAMPoles to be synthesized inexpensively so that they can be used as field diagnostics. The cost per yield for using synthetic DNA to manufacture LAMPoles would be prohibitive. This conclusion led us to develop an alternate synthetic route utilizing the “Staudinger ligation” (28) to covalently bond recombinant DNA to the L/G domain in limiting stoichiometry. Here, we conjugated azide-modified DNA to phosphine-modified L/G protein to create the IgG-binding LAMPole fusion. We verified that the L/G domain remained active after conjugation to DNA by using a hybridization LNA probe in qPCR to measure the Kd (dissociation constant) of the LAMPole for binding purified mouse polyclonal IgG as previously described (21). We measured a Kd of 250 nM for mouse polyclonal IgG (Jackson Immunologicals) covalently linked to M280 magnetic beads (Invitrogen). This affinity is less than the Kd of 3.4 nM measured for this protein linked to an 82-nt DNA element (21).
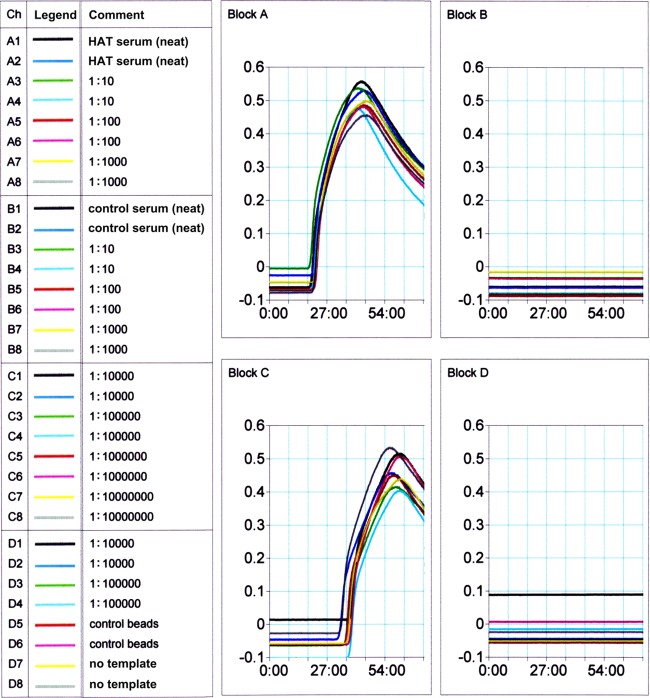
As proof of concept that our LAMPole could be used to assay antigen-specific antibodies, we measured levels of IgG that recognize a specific T. b. gambiense antigen, variable surface glycoprotein (VSG), in the sera of vaccinated mice and a stage 2 HAT patient. Various VSGs have traditionally been selected as antigenic targets in serological assays for HAT (13, 22). For all these assays, we scavenged antiparasite IgG from sera using T. b. gambiense LiTat 1.3 VSG-coated magnetic beads and detected bound IgG by forming a sandwich with our anti-IgG LAMPole (Fig. 5). Using spike and recovery assays, we previously confirmed that no substances in human or mouse serum interfered with the use of L/G-DNA fusions to measure IgG (21), and we found the same to be true in these experiments. We accounted for cross-reactive binding of IgG to the beads and nonspecific binding of the LAMPole by subtracting the baseline signal obtained in replicates with neither IgG nor antigen, and we confirmed equivalently low levels of nonspecific signal, even in negative-control serum. We formulated assays with 0.576 mg/ml LAMPole (10-fold Kd) to ensure that at least 90% of available epitopes are occupied during measurement. We initially formulated assay conditions using sera from naive and VSG-vaccinated mice. We incubated 106 VSG-coated magnetic beads with mouse sera and measured the amount of bound antibody using the ID:25 primers (Fig. 1) to amplify the LAMPole DNA element. When tested neat, only sera from VSG-vaccinated mice (n = 4) yielded LAMP-dependent turbidity, in contrast to naive controls (n = 2). We also serially diluted sera from one immune mouse and one nonimmune mouse (in 1:10 steps) to 1:10−5 and found no loss of anti-VSG antibody signal at maximal dilution (Table 1). However, after further dilution, anti-VSG antibodies were detected in sera diluted to at least 1:10−6. Based on these findings, we incubated VSG-coated beads with the sera of a patient with HAT and a healthy control (Fig. 6). We detected anti-VSG antibodies in maximally diluted (1:10−7) serum from a patient with stage 2 HAT but not in serum from the healthy control. Thus, we observed a >6- to 7-log fold difference in signal between sera from vaccinated versus control mice and from a stage 2 HAT patient versus a control.
FIG 5.
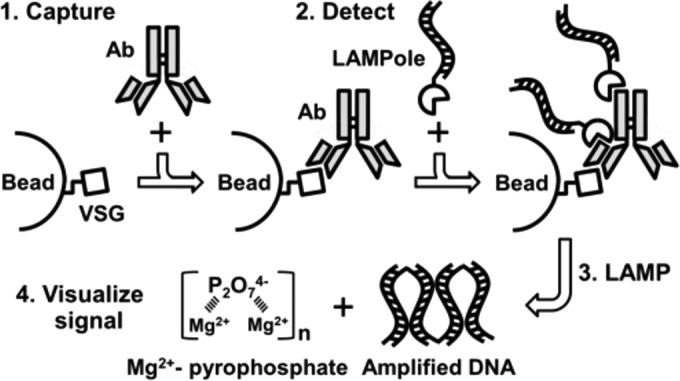
LAMPole-based measurement of antigen-specific IgG. Measuring antigen-specific serology is a four-step process. (1) Capture. Antigen-coated beads (LiTat 1.3 VSG) are incubated with serum samples containing anti-VSG IgG (Ab). (2) Detect. After nonspecific IgG is washed away, bound IgG is detected by adding LAMPoles to create a molecular sandwich. (3) LAMP. The LAMPole sandwich is subjected to Bst polymerase-catalyzed LAMP to yield amplified high-molecular-weight DNA and insoluble pyrophosphate in complex with Mg2+. Here, the abundance of LAMPole retained on the bead surface is proportional to the amount of specific anti-VSG IgG. (4) Visualize signal. Insoluble precipitate may be monitored in real time, or DNA may be directly visualized using intercalating dyes or gel electrophoresis.
TABLE 1.
LAMPole detection of VSG antibodies in immune mouse serum
| Expta | Serum source | LAMP primer set | Resultb with serum dilution: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Neat | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | 10−7 | |||
| A | Vaccinated mouse | CHS_ID:25 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ND | |
| Control mouse | −/− | −/− | −/− | −/− | −/− | −/− | ND | |||
| B | Vaccinated mouse | CHS_ID:25 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | −/− |
| Control mouse | −/− | −/− | −/− | −/− | −/− | ND | ND | ND | ||
The same serum samples were used in experiments A and B.
Duplicate test samples were used. −/−, negative result; +/+, positive result; ND, not done.
FIG 6.
Measurement of anti-VSG in a patient with HAT. Normal and HAT patient sera were incubated in duplicate with VSG-coated beads, followed by exposure to L/G LAMPole. We assayed the production of turbidity in real time. Samples incubated with HAT-positive serum were specifically amplified by CHS LAMP (blocks A and C), while normal serum and no-serum and no-template controls produced no signal (blocks B and D). We used primer set ID:25 to direct LAMP in these reactions.
DISCUSSION
Our strategy to measure protein analytes (e.g., pathogen-specific host antibodies) by LAMP is to use an adaptor probe that attaches DNA oligonucleotides to the protein analyte to enable sensitive and specific molecular detection of the protein-DNA LAMPole. We found that we could measure protein analytes using LAMP as a signal amplifier. Importantly, our goal was not to formulate a better test for HAT, per se, but to demonstrate the LAMPole concept as a technological advance applicable to improving the measurement of a wide variety of protein analytes or pathogens with high sensitivity and low cost. This is the first instance that we are aware of in which a protein-DNA fusion has been used to combine LAMP with immunoassays to measure protein analytes.
In its fully developed form, we expect that this technology may be used in field situations without electricity, but for the sake of this benchmark, we chose to detect turbidity using a turbidimeter for increased precision. We envisioned the “field-deployed” assay to be a two-step process in which a relevant biomarker of interest (e.g., antigen) present in a patient's sample (e.g., blood, serum, saliva, urine, etc.) is captured onto magnetic beads coated with a specific capture agent (e.g., capture antibody). The bound biomarker is then detected by addition of LAMPole to the beads, and a visual signal is developed by supplementation with primers and Bst polymerase. The inclusion of overlapping amplicons in the DNA element of the LAMPole enables the use of two separate LAMP primer sets to generate signal output and may be confirmed using an orthogonal amplification processes such as qPCR for more precise quantification when needed. However, our LAMPole fusion may also be embedded into existing and future microfluidic platforms to provide signal amplification in situ. While a colorimetric or turbidity output visible to the naked eye could be used instead of microtiter plate readers in resource-poor settings, this visual readout could also be useful in developed countries for POC diagnosis. When quantitative analysis is important, real-time LAMP is also available.
The cost of LAMPole synthesis makes it possible to create a wide array of probes for detecting all antigenic varieties expressed by infectious agents that undergo antigenic variation, such as the trypanosome parasites that cause HAT. Using our originally published method for making Tadpoles that employed chemically synthesized DNA elements (5), we estimate a reagent cost of about $1.00 per assay. However, using the Staudinger ligation as outlined here, we estimate a reagent cost of about $0.0075 per assay. Although this cost consideration is less important in developed countries that can afford higher-priced methods, this issue is significant in resource-limited countries where cost alone may prevent test availability. Moreover, the modular nature of LAMPole synthesis allows new probes to be easily reconfigured, which may accommodate a multitude of antigenic variants corresponding to geographic location.
Several multiplex LAMP methods are available (29, 30), which may be applied to amplifying mixtures of our LAMPole probes and to simultaneously measure different target biomarkers in a clinical sample. Using technologies that we describe here, the creation of antibody LAMPoles for antigen detection should now also be possible. Together with intelligent LAMPole tail and LAMP primer design, the creation of a single diagnostic LAMP-based test for detection of host antipathogen antibodies (i.e., protein L/G LAMPoles), pathogen antigens (antibody LAMPoles), and pathogen DNA or RNA (LAMP or RT-LAMP) in a single assay format should now be possible to increase the likelihood of identifying the cause of the acute infection (increase sensitivity). For example, an ideal LAMPole diagnostic test for acute dengue might detect NS1 antigen, viral RNA, and IgM antibody; if it also detected IgG, it could rapidly distinguish secondary from primary dengue, which could have prognostic implications (higher risk of severe dengue).
By merging LAMP and Tadpoles, we created a method that may improve pathogen detection at the POC (7, 21). We previously showed that the usefulness of traditional LAMP assays that recognize multicopy gene targets for pathogen DNA detection can be dramatically enhanced by sample pretreatment to lyse or solubilize the pathogen(s) and facilitate release of its DNA prior to assay, especially when the sample size is limited (31). Similar modifications could also be applied to the LAMPole-based assays described here. Furthermore, we predict that LAMPoles will be a suitable platform for the development of noninvasive saliva- and potentially lachrymal (tear) fluid-based assays (32, 33), with sensitivities equivalent to those found with blood or CSF. Therefore, the use of LAMPoles could provide cost-effective, ultrasensitive antigen/antibody diagnostic assays with low technical requirements, which could improve acute diagnosis of neglected tropical and other infections that plague the developing world and thereby related clinical outcomes.
ACKNOWLEDGMENTS
We are grateful to J. Stephen Dumler, from Johns Hopkins University, for reading and editing the manuscript.
This research was supported in part by grants from the National Institutes of Health (5R01AI082695 and 1R21AI079282) to D.J.G. and support from the French Ministry of Foreign Affairs in Gabon, Angola, and Central African Republic awarded to S.B.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
D.J.G. conceived of the project. I.E.B., S.T.P., S.M., and D.J.G. discussed and designed the experiments. D.J.G., O.V.N., K.Y., S.T.P., S.M., and I.E.B. performed the experiments. I.E.B., K.Y., O.V.N., S.T.P., S.M., S.B., M.E.R., and D.J.G. analyzed the data and made comments on the manuscript. I.E.B. and D.J.G. wrote the manuscript.
We declare no competing financial interests.
REFERENCES
- 1.Higuchi R, Dollinger G, Walsh PS, Griffith R. 1992. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY) 10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi R, Fockler C, Dollinger G, Watson R. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 3.Mori Y, Nagamine K, Tomita N, Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun 289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 4.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. 2009. Colormetric detection of loop-mediated isothermal amplification reaction by using htdroxy naphthol blue. Biotechniques 46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 5.Burbulis I, Yamaguchi K, Gordon A, Carlson R, Brent R. 2005. Using protein-DNA chimeras to detect and count small numbers of molecules. Nat Methods 2:31–37. doi: 10.1038/nmeth729. [DOI] [PubMed] [Google Scholar]
- 6.Evans J. 2005. Tadpoles stick to protein analysis. Chem World 18. [Google Scholar]
- 7.Nolan GP. 2005. Tadpoles by the tail. Nat Methods 2:11–12. doi: 10.1038/nmeth0105-11. [DOI] [PubMed] [Google Scholar]
- 8.Matovu E, Kazibwe AJ, Mugasa CM, Ndungu JM, Njiru ZK. 2012. Towards point-of-care diagnostic and staging tools for human African trypanosomiaisis. J Trop Med 2012:340538. doi: 10.1155/2012/340538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bearinger JP, Dugan LC, Baker BR, Hall SB, Ebert K, Mioulet V, Madi M, King DP. 2011. Development and initial results of a low cost, disposable, point-of-care testing device for pathogen detection. IEEE Trans Biomed Eng 58:805–808. doi: 10.1109/TBME.2010.2089054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori Y, Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother 15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wastling SL, Picozzi K, Kakembo AS, Welburn SC. 2010. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl Trop Dis 4:e865. doi: 10.1371/journal.pntd.0000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grab DJ, Lonsdale-Eccles J, Inoue N. 2005. Lamp for tadpoles. Nat Methods 2:635 (Author reply, 2: 635–636.) doi: 10.1038/nmeth0905-635a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lejon V, Jamonneau V, Solano P, Atchade P, Mumba D, Nkoy N, Bebronne N, Kibonja T, Balharbi F, Wierckx A, Boelaert M, Buscher P. 2006. Detection of trypanosome-specific antibodies in saliva, towards non-invasive serological diagnosis of sleeping sickness. Trop Med Int Health 11:620–627. doi: 10.1111/j.1365-3156.2006.01620.x. [DOI] [PubMed] [Google Scholar]
- 14.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thekisoe OM, Kuboki N, Nambota A, Fujisaki K, Sugimoto C, Igarashi I, Yasuda J, Inoue N. 2007. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop 102:182–189. doi: 10.1016/j.actatropica.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Thekisoe OM, Inoue N, Kuboki N, Tuntasuvan D, Bunnoy W, Borisutsuwan S, Igarashi I, Sugimoto C. 2005. Evaluation of loop-mediated isothermal amplification (LAMP), PCR and parasitological tests for detection of Trypanosoma evansi in experimentally infected pigs. Vet Parasitol 130:327–330. doi: 10.1016/j.vetpar.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol 41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelletier MK, Burbulis IE, Winkel-Shirley B. 1999. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol Biol 40:45–54. doi: 10.1023/A:1026414301100. [DOI] [PubMed] [Google Scholar]
- 19.Kihlberg BM, Sjobring U, Kastern W, Bjorck L. 1992. Protein LG: a hybrid molecule with unique immunoglobulin binding properties. J Biol Chem 267:25583–25588. [PubMed] [Google Scholar]
- 20.Evans TC Jr, Benner J, Xu MQ. 1999. The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J Biol Chem 274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 21.Burbulis I, Yamaguchi K, Yu R, Resnekov O, Brent R. 2007. Quantifying small numbers of antibodies with a ‘near-universal’ protein-DNA chimera. Nat Methods 4:1011–1013. doi: 10.1038/nmeth1127. [DOI] [PubMed] [Google Scholar]
- 22.Chappuis F, Loutan L, Simarro P, Lejon V, Büscher P. 2005. Options for field diagnosis of human african trypanosomiasis. Clin Microbiol Rev 18:133–146. doi: 10.1128/CMR.18.1.133-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, Senter P, Revets H, De Baetselier P, Muyldermans S, Magez S. 2004. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem 279:1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 24.Braasch DA, Liu Y, Corey DR. 2002. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res 30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 26.Hirano T, Yasukawa K, Harada H, Taga T, Watanabe Y, Matsuda T, Kashiwamura S, Nakajima K, et al. 1986. Complementary DNA for a novel interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature 324:73–76. doi: 10.1038/324073a0. [DOI] [PubMed] [Google Scholar]
- 27.Muir T, Sondhi D, Cole P. 1998. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A 95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staudinger H, Meyer J. 1919. Über neue organische Phosphorverbindungen. III. Phosphinmethylenderivate und Phosphinimine. Helv Chim Acta 2:635. doi: 10.1002/hlca.19190020164. [DOI] [Google Scholar]
- 29.Crum NF, Aronson NE, Lederman ER, Rusnak JM, Cross JH. 2005. History of U.S. military contributions to the study of parasitic diseases. Mil Med 170:17–29. [DOI] [PubMed] [Google Scholar]
- 30.Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue N, Nambota A, Yasuda J, Igarashi I. 2007. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods 71:281–287. doi: 10.1016/j.mimet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Grab DJ, Nikolskaia OV, Inoue N, Thekisoe MM, Morrison L, Gibson W, Dumler JS. 2011. Using detergent to enhance detection sensitivity of African trypanosomes in human CSF and blood by Loop-mediated isothermal amplification (LAMP). PLoS Negl Trop Dis 5:e1249. doi: 10.1371/journal.pntd.0001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lolli F, Franciotta D. 2010. Oligoclonal bands in tears. Mult Scler 16:760. doi: 10.1177/1352458510367663. [DOI] [PubMed] [Google Scholar]
- 33.Wartofsky L, Handelsman DJ. 2010. Standardization of hormonal assays for the 21st century. J Clin Endocrinol Metab 95:5141–5143. doi: 10.1210/jc.2010-2369. [DOI] [PubMed] [Google Scholar]