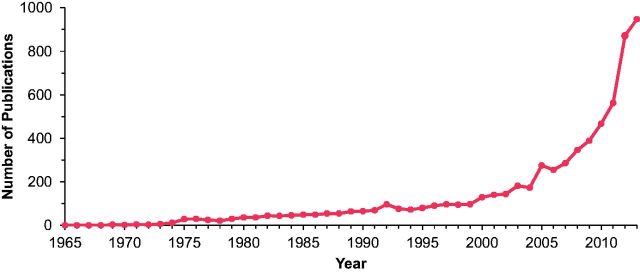
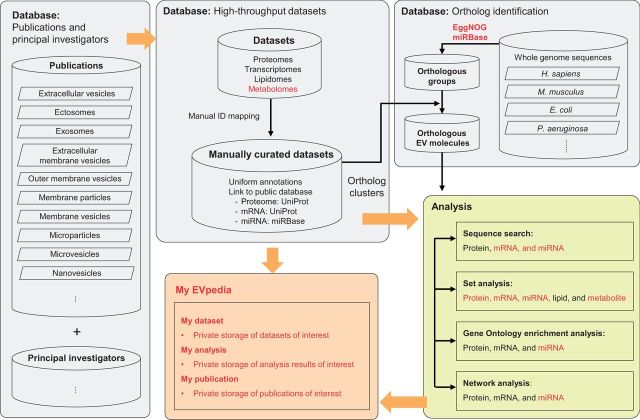
Dae-Kyum Kim
Dae-Kyum Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,†,‡,
Jaewook Lee
Jaewook Lee
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,†,‡,
Sae Rom Kim
Sae Rom Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Dong-Sic Choi
Dong-Sic Choi
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Yae Jin Yoon
Yae Jin Yoon
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
2,‡,
Ji Hyun Kim
Ji Hyun Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Gyeongyun Go
Gyeongyun Go
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Dinh Nhung
Dinh Nhung
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Kahye Hong
Kahye Hong
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Su Chul Jang
Su Chul Jang
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Si-Hyun Kim
Si-Hyun Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Kyong-Su Park
Kyong-Su Park
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Oh Youn Kim
Oh Youn Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Hyun Taek Park
Hyun Taek Park
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Ji Hye Seo
Ji Hye Seo
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,
Elena Aikawa
Elena Aikawa
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
3,
Monika Baj-Krzyworzeka
Monika Baj-Krzyworzeka
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
4,
Bas W M van Balkom
Bas W M van Balkom
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
5,
Mattias Belting
Mattias Belting
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
6,
Lionel Blanc
Lionel Blanc
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
7,
Vincent Bond
Vincent Bond
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
8,
Antonella Bongiovanni
Antonella Bongiovanni
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
9,
Francesc E Borràs
Francesc E Borràs
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
10,
Luc Buée
Luc Buée
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
11,
Edit I Buzás
Edit I Buzás
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
12,
Lesley Cheng
Lesley Cheng
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
13,
Aled Clayton
Aled Clayton
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
14,
Emanuele Cocucci
Emanuele Cocucci
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
15,
Charles S Dela Cruz
Charles S Dela Cruz
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
16,
Dominic M Desiderio
Dominic M Desiderio
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
17,
Dolores Di Vizio
Dolores Di Vizio
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
18,
Karin Ekström
Karin Ekström
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
19,20,
Juan M Falcon-Perez
Juan M Falcon-Perez
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
21,
Chris Gardiner
Chris Gardiner
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
22,
Bernd Giebel
Bernd Giebel
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
23,
David W Greening
David W Greening
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
24,
Julia Christina Gross
Julia Christina Gross
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
25,
Dwijendra Gupta
Dwijendra Gupta
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
26,
An Hendrix
An Hendrix
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
27,
Andrew F Hill
Andrew F Hill
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
13,
Michelle M Hill
Michelle M Hill
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
28,
Esther Nolte-'t Hoen
Esther Nolte-'t Hoen
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
29,
Do Won Hwang
Do Won Hwang
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
30,
Jameel Inal
Jameel Inal
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
31,
Medicharla V Jagannadham
Medicharla V Jagannadham
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
32,
Muthuvel Jayachandran
Muthuvel Jayachandran
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
33,
Young-Koo Jee
Young-Koo Jee
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
34,
Malene Jørgensen
Malene Jørgensen
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
35,
Kwang Pyo Kim
Kwang Pyo Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
36,
Yoon-Keun Kim
Yoon-Keun Kim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
37,
Thomas Kislinger
Thomas Kislinger
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
38,
Cecilia Lässer
Cecilia Lässer
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
39,
Dong Soo Lee
Dong Soo Lee
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
30,
Hakmo Lee
Hakmo Lee
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
40,
Johannes van Leeuwen
Johannes van Leeuwen
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
41,
Thomas Lener
Thomas Lener
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
42,43,
Ming-Lin Liu
Ming-Lin Liu
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
44,45,
Jan Lötvall
Jan Lötvall
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
39,
Antonio Marcilla
Antonio Marcilla
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
46,
Suresh Mathivanan
Suresh Mathivanan
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
24,
Andreas Möller
Andreas Möller
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
47,
Jess Morhayim
Jess Morhayim
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
41,
François Mullier
François Mullier
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
48,49,
Irina Nazarenko
Irina Nazarenko
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
50,
Rienk Nieuwland
Rienk Nieuwland
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
51,
Diana N Nunes
Diana N Nunes
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
52,
Ken Pang
Ken Pang
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
53,54,
Jaesung Park
Jaesung Park
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
55,
Tushar Patel
Tushar Patel
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
56,
Gabriella Pocsfalvi
Gabriella Pocsfalvi
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
57,
Hernando del Portillo
Hernando del Portillo
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
58,
Ulrich Putz
Ulrich Putz
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
59,
Marcel I Ramirez
Marcel I Ramirez
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
60,
Marcio L Rodrigues
Marcio L Rodrigues
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
61,62,
Tae-Young Roh
Tae-Young Roh
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,2,
Felix Royo
Felix Royo
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
21,
Susmita Sahoo
Susmita Sahoo
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
63,
Raymond Schiffelers
Raymond Schiffelers
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
64,
Shivani Sharma
Shivani Sharma
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
65,
Pia Siljander
Pia Siljander
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
66,
Richard J Simpson
Richard J Simpson
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
24,
Carolina Soekmadji
Carolina Soekmadji
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
67,
Philip Stahl
Philip Stahl
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
68,
Allan Stensballe
Allan Stensballe
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
69,
Ewa Stępień
Ewa Stępień
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
70,
Hidetoshi Tahara
Hidetoshi Tahara
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
71,
Arne Trummer
Arne Trummer
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
72,
Hadi Valadi
Hadi Valadi
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
73,
Laura J Vella
Laura J Vella
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
74,
Sun Nyunt Wai
Sun Nyunt Wai
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
75,
Kenneth Witwer
Kenneth Witwer
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
76,
María Yáñez-Mó
María Yáñez-Mó
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
77,
Hyewon Youn
Hyewon Youn
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
30,
Reinhard Zeidler
Reinhard Zeidler
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
78,
Yong Song Gho
Yong Song Gho
1Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea, 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang, Republic of Korea, 3Cardiovascular Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 4Department of Clinical Immunology, Polish-American Institute of Paediatrics, Jagiellonian University Medical College, Cracow, Poland, 5Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands, 6Section of Oncology, Department of Clinical Sciences, Lund University, Lund, Sweden, 7The Feinstein Institute for Medical Research, Manhasset, NY, USA, 8Department of Microbiology, Biochemistry, and Immunology, Morehouse School of Medicine, Atlanta, GA, USA, 9Institute of Biomedicine and Molecular Immunology (IBIM), National Research Council (CNR), Palermo, Italy, 10Innovation in Vesicles and Cells for Application in Therapy, Germans Trias i Pujol Research Institute, Germans Trias i Pujol University Hospital, Badalona, Spain, 11INSERM, UMR837 JEAN-PIERRE Aubert Research Centre, Lille, France, 12Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary, 13Department of Biochemistry and Molecular Biology, BIO21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC, Australia, 14Institute of Cancer & Genetics, School of Medicine, Velindre Cancer Centre, Cardiff University, Cardiff, UK, 15Program in Cellular and Molecular Medicine at Boston Children’s Hospital and Department of Cell Biology, Harvard Medical School, Boston, MA, USA, 16Section of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, USA, 17Department of Neurology, College of Medicine, University of Tennessee Health Science Center, Memphis, TN, USA, 18Cancer Biology Program, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA, 19Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden, 20BIOMATCELL VINN Excellence Center of Biomaterials and Cell Therapy, Gothenburg, Sweden, 21Metabolomics Unit, CIC bioGUNE, CIBERehd, IKERBASQUE Research Foundation, Technology Park of Bizkaia, Derio, Bizkaia, Spain, 22Nuffield Department of Obstetrics and Gynaecology, John Radcliffe Hospital, University of Oxford, Oxford, UK, 23Institute for Transfusion Medicine, University Hospital Essen, University of Duisburg-Essen, Essen, Germany, 24La Trobe Institute for Molecular Science (LIMS), La Trobe University, Melbourne, VIC, Australia, 25Division of Signaling and Functional Genomics, German Cancer Research Center, Heidelberg, Germany, 26Jai Prakash University, Chapra, Bihar, India, 27Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Ghent, Belgium, 28The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, QLD, Australia, 29Department of Biochemistry & Cell Biology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, 30Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea, 31Cellular and Molecular Immunology Research Centre, Faculty of Life Sciences and Computing, London Metropolitan University, London, UK, 32CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Andhra Pradesh, India, 33Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, MN, USA, 34Department of Internal Medicine, Dankook University College of Medicine, Cheonan, Republic of Korea, 35Department of Clinical Immunology, Aalborg University Hospital, Aalborg, Denmark, 36Department of Applied Chemistry, Kyung Hee University, Yongin, Republic of Korea, 37Ewha Womans University Medical Center, Seoul, Republic of Korea, 38Ontario Cancer Institute, Toronto, ON, Canada, 39Krefting Research Centre, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 40Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea, 41Department of Internal Medicine, Erasmus University Rotterdam Medical Centre, Rotterdam, The Netherlands, 42Department of Blood Group Serology and Transfusion Medicine, University Hospital of Salzburg, Paracelsus Medical University, Salzburg, Austria, 43Spinal Cord Injury & Tissue Regeneration Center Salzburg (SCI-TReCS), Paracelsus Medical University, Salzburg, Austria, 44Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA, 45Department of Medicine, Temple University School of Medicine, Philadelphia, PA, USA, 46Área de Parasitología, Departamento de Biología Celular y Parasitología, Universitat de València, Burjassot, Valencia, Spain, 47Tumour Microenvironment Laboratory, QIMR Berghofer Medical Research Institute, Herston, QLD, Australia, 48Hematology Laboratory, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), CHU Dinant Godinne UCL Namur, Université Catholique de Louvain, Belgium, 49Department of Pharmacy, NARILIS, Namur Thrombosis and Hemostasis Center (NTHC), University of Namur, Naumr, Belgium, 50Institute of Environmental Health Sciences and Hospital Infection Control, Medical Center-University of Freiburg, Freiburg, Germany, 51Department of Clinical Chemistry, Academic Medical Center, Amsterdam, The Netherlands, 52Laboratory of Medical Genomics, A.C. Camargo Cancer Center, São Paulo, Brazil, 53Inflammation Division, The Walter and Eliza Hall Institute for Medical Research, Parkville, VIC, Australia, 54Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia, 55Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Republic of Korea, 56Departments of Transplantation and Cancer Biology, Mayo Clinic, Jacksonville, FL, USA, 57Mass Spectrometry and Proteomics, Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy, 58ICREA, Barcelona Centre for International Health Research, Barcelona, Spain, 59The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Melbourne, VIC, Australia, 60Laboratory of Molecular Biology of Parasites and Vectors, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 61Paulo de Góes Microbiology Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 62Centre for Technological Development in Health (CDTS), Oswaldo Cruz Foundation, Rio de Janeiro, Brazil, 63Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA, 64Department of Clinical Chemistry and Hematology, University Medical Center Utrecht, Utrecht, The Netherlands, 65California Nanosystems Institute, University of California Los Angeles, Los Angeles, CA, USA, 66Division of Biochemistry and Biotechnology, Department of Biosciences, University of Helsinki, Helsinki, Finland, 67Australian Prostate Cancer Research Centre-Queensland, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, 68Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, USA, 69Department of Health Science and Technology, Aalborg University, Aalborg, Denmark, 70Department of Medical Physics, Jagiellonian University, Cracow, Poland, 71Department of Cellular and Molecular Biology, Graduate School of Biomedical Science, Hiroshima University, Hiroshima, Japan, 72Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany, 73Department of Rheumatology and Inflammation Research, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 74Ludwig Institute for Cancer Research Melbourne, Austin Hospital, Heidelberg, VIC, Australia, 75Department of Molecular Biology, Umeå University, Umeå, Sweden, 76Department of Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA, 77Unidad de Investigación, Hospital Santa Cristina, Instituto de Investigación Sanitaria Princesa, Madrid, Spain, 78Department of Otorhinolaryngology and Research Group Gene Vectors, University of Muenchen, Helmholtz-Zentrum München, Munich, Germany
1,‡,*