Abstract
A member of the TTP family of proteins, TIS11d binds RNA with high specificity using a pair of CCCH-type tandem zinc fingers separated by a 18 residue long linker. Our previous work showed that the formation of hydrogen bonds between the C-terminal residue E220 and the residues of the linker region stabilized a compact structure of TIS11d in the absence of RNA. To investigate the role of the C-terminal residues in the structure of unbound TIS11d, the E220A mutant and the truncation mutant lacking the last two residues (D219/E220) were studied using molecular dynamics, NMR spectroscopy, and biochemical methods. This study confirmed the importance of the charged residues D219 and E220 in maintaining structural stability in unbound TIS11d and elucidated the underlying physical mechanisms. We observed a greater structural heterogeneity for the residues of the linker in the molecular dynamics trajectories of both mutant proteins relative to the wild-type. This heterogeneity was more pronounced in the D219/E220 deletion mutant than in the E220A mutant, indicating that a greater reduction of the charge of the C-terminus results in greater flexibility. In agreement with the increased flexibility and the reduced number of negatively charged residues of the D219/E220 deletion mutant, we measured more unfavorable entropic and a more favorable enthalpic contribution to the free energy of RNA binding in the mutant than in the wild-type protein. The relative orientation of the zinc fingers was stabilized by the electrostatic interaction between E220 and positively charged residues of the linker in TIS11d. In the E220A mutant, the relative orientation of the zinc fingers was less constrained, whereas in the D219/E220 deletion mutant, little orientational preference was observed. We posit that favorable electrostatic interactions provide a mechanism to promote preferential orientation of separate domains without imposing structural rigidity.
Introduction
The crucial functions of gene replication, transcription, and translation can be controlled at transcriptional and posttranscriptional levels through interactions between proteins and nucleic acids. The stability and/or translational efficiency of mRNA must to be regulated to control gene expression at the posttranscriptional stage. Tristetraprolin (TTP) regulates mRNA stability by promoting deadenylation and degradation of RNA transcripts in a highly specific manner (1–5). TTP binds AU-rich elements (ARE) in the 3′-untranslated region of mRNAs that encode for tumor necrosis factor-α and other proinflammatory cytokines (1,6,7). The TTP family of CCCH-type tandem zinc finger (TZF) proteins contains two additional human proteins: TIS11b and TIS11d. Although the biological function of TIS11b/d is not yet fully understood, their function has been linked to thymocyte development and their dysregulation to cancer (8–10). In vitro, these proteins promote degradation of the same RNAs recognized by TTP (11), but TIS11b/d have also been shown to regulate other oncogenic transcription factors such as Notch-1 (8). The RNA-binding domains (RBD) of these proteins have high sequence identity: TIS11d and TIS11b are 91% identical to each other and 71% identical to TTP (Fig. 1). The CCCH zinc finger motifs in the TTP family are characterized by a strict spacing between the zinc binding residues of CX8CX5CX3H (Fig. 1) and are separated by a 18 residue long linker. Furthermore, mutation of any of the zinc binding residues abrogates RNA binding (12).
Figure 1.
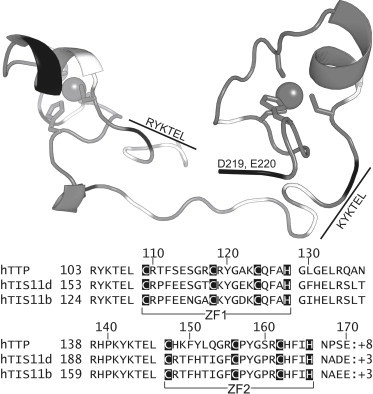
Structure and sequence alignment of the RBD of TIS11d. Top: representation of TIS11d (PDB access code 1RGO, RNA not shown) colored by charge (black: negative residues, white: positive residues). Gray shows backbone (tube), Zn2+ ions (spheres). Bottom: sequence alignment of the RBD of TIS11 family members with total charge.
The structure of the TZF domain of TIS11d (residues 151–220) was determined by NMR spectroscopy while bound to the RNA sequence (5′-UUAUUUAUU-3′) (13). Cys-159, Cys-168, Cys-174, and His-178 coordinate the zinc ion in zinc finger 1 (ZF1) and Cys-197, Cys-206, Cys-212, and His-216 coordinate the zinc ion in zinc finger 2 (ZF2) (Fig. 1). TIS11d is characterized by relatively few secondary structural elements. An α-helix starts immediately after the first zinc-coordinating cysteine, spanning residues 160–165 (198–203) in ZF1 (ZF2). A single turn of a 310-helix is present between the second and third zinc-coordinating cysteine residues, spanning residues 171–173 (209–211) in ZF1 (ZF2). The only secondary structural element in the linking residues is a single turn of a 310-helix consisting of three residues of the linker immediately following ZF1. RNA binding is achieved through hydrogen bonding and the base intercalation of aromatic residues: Tyr-170 and Phe-176 (Tyr-208 and Phe-214) in ZF1 (ZF2). Hydrogen bonds form between the RNA bases and the backbone of the zinc finger domains and highly conserved (R/K)YKTEL motifs preceding each zinc finger (see Fig. 1), with the exception of the side chains of Glu-157 and Cys-174 (Cys-212) in ZF1 (ZF2) (13).
Although the three-dimensional structure of TIS11d was determined in the presence of RNA, the 2D NMR spectrum of apo-TIS11d shows that the protein is structured in solution (13). Previous work explored the changes in the structure of TIS11d after the removal of RNA by molecular dynamics (MD) simulations (14). In agreement with the NMR spectroscopic study of apo-TIS11d, the unbound form of TIS11d maintained the structure of the two zinc finger domains. However, a conformational change was seen in the linking residues, enabling ZF2 to rotate to form hydrogen bonds between the C-terminal residue E220 and residues of the linker and a hydrogen bond between ZF1 and ZF2. The rotation of ZF2 relative to ZF1 and formation of these hydrogen bonds resulted in a more compact structure compared to the RNA-bound form of TIS11d (14).
The terminal glutamic acid, indicated to be important for stabilizing a more compact structure in unbound TIS11d (14), is conserved in TTP CCCH domain homologs for both human and nonhuman proteins (15). To further explore the importance of the charged C-terminus in the stabilization of the structure of unbound TIS11d, MD simulations, NMR spectroscopy, and biochemical studies were performed for wild-type (WT) TIS11d, the mutation of E220A (TIS11d-E220A), and a truncation of the last two residues (TIS11d-Δ D219/E220). In this work, we used longer trajectories (120 ns) than previous works on TZF domains (14,16,17) to more fully explore the flexibility and dynamics of unbound TIS11d and the two C-terminal mutant proteins. We found that the WT protein is very flexible in the residues of the linker and samples many conformations over longer time periods. The two mutations in the C-terminus affected the structural stability of the mutant proteins, resulting in dramatic structural extensions in the residues of the linker while preserving most of the TZF structure. We explored the sources of this increased flexibility and concluded that the negatively charged residues of the C-terminus play a critical role in stabilizing the more compact structure of TIS11d through attractive electrostatic interactions.
Materials and Methods
Simulation protocol
We modeled the unknown structure of ligand-free TIS11d, starting from the lowest energy model of the NMR solution structure of TIS11d bound to ARE (UUAUUUAUU) (13), by removal of the RNA followed by energy minimization and equilibration. The structure was solvated and neutralized in a TIP3P water box of 9261 water molecules using VMD (18). Structural optimization and MD simulations were subsequently carried out using the NAMD2.9 molecular modeling package (19) and the CHARMM27 force field (20). A nonbonded model that included charge transfer and local polarization effects (21) was used to model the zinc ions, with van der Waals parameters taken from set A (Table 2) of Sakharov and Lim (21).
After energy minimization, particle velocities were randomly assigned from the Maxwell distribution and the system was simulated in the isothermal-isobaric ensemble. A constant temperature of 298 K and pressure of 1 atm were maintained using Langevin dynamics and the Nosé-Hoover Langevin piston method. The SHAKE constraint algorithm (22) was used, thereby allowing the equations of motion to be integrated using a 2 fs time step. Nonbonded interactions were calculated every time step with a cut-off distance of 12 Å and a switching distance of 10 Å. The particle mesh Ewald method was used to treat electrostatic interactions using periodic boundary conditions. After equilibration in the isothermal-isobaric ensemble, an additional stage of equilibration was performed in the microcanonical ensemble for three independent trajectories, with velocity reassignment to ensure that the temperature stabilized to the desired value. Although the thermodynamic values are equilibrated at this point, the simulations were run in the microcanonical ensemble for an additional 20 ns for structural equilibration. After this point, we collected 100 ns for each of the three independent trajectories that were used for data collection and analysis.
The mutation of residue E220 to alanine was performed using the Mutator plugin of VMD (18), starting from the lowest energy model of the NMR solution structure of TIS11d bound to ARE after removal of the RNA sequence. The D219/E220 deletion mutant was modeled directly using VMD. Both structures were also solvated and neutralized in a TIP3P water box of 9261 water molecules using VMD. Structural optimization, minimization, and equilibration were all carried out in the same manner as for WT TIS11d, with the exception of an additional 20 ns that was allowed for structural equilibration, resulting in 80 ns of three independent trajectories for both TIS11d-E220A and TIS11d-Δ D219/E220 that were used for data collection and analysis.
Two independent trajectories were collected of WT TIS11d bound to ARE (UUAUUUAUU) (13) using the minimization and thermodynamic equilibration procedures described previously (14). Structural equilibration was reached at 6 ns; an additional 8 ns were used for data analysis.
Structural characterization of MD trajectories
The quantities used to characterize the structural changes and dynamics were calculated using VMD (18) to process the trajectories and generate the values used to calculate the reported statistics. Visualization of structures was also done in VMD, using the STRIDE method (23). The data generated was processed and plotted using Tableau Software v8.0 (Seattle, WA).
ZF1 was defined as residues 159–178 and ZF2 was defined as residues 197–216. The distance between the Cα of residues 179 and 196 defines the linker distance. Radius of gyration, as implemented in VMD, follows the standard form (25). The Zn-Zn distance is defined as the distance between the zinc ions of ZF1 and ZF2. Root mean-square displacement (RMSD) was calculated after applying a r.m.s. fit to minimize the contribution from global rotations by using the average structure as the point of reference.
Definition of hydrogen bonding
The distance between the donor and the acceptor is <4.0 Å and the angle between the donor and acceptor groups must be within 113–180° (26). This was the most stringent cut-off possible such that the mean distance between the hydrogen and the acceptor of the bonds reported by Wright et al. (13) were within the cut-off for the twenty original NMR structures, excluding hydrogen bonds involving sulfur.
Relative orientation of the tandem zinc fingers
Zn1 is the vector defined by the zinc atom of the first finger and the coordinating nitrogen atom of His-178. Zn2 is the vector defined by the zinc atom of the second finger and the coordinating nitrogen atom of His-216. The normalized dot product of these two vectors characterizes the relative orientation of the two zinc fingers.
Dihedral angles
The dihedral angles were calculated by the implementation in VMD (18). Because dihedral angles are defined in the range of (–180°, 180°), corrections were applied to minimize artificial transitions at the boundaries, which affect both the mean and the SD.
Distance maps
The backbone distance maps show the average distance between the Cα atoms of each residue. The color scale is truncated at 10 Å to highlight close contacts, similar to the technique used in contact maps.
Cross correlation
The correlation between residues was calculated using the dot product of the change in the position vectors
| (1) |
normalized by the mean square fluctuations of Ri and Rj, such that the cross correlation is (27):
| (2) |
The position vectors ΔRi and ΔRj are defined as the change in position of the Cα atoms of residues i and j with respect to the average position. To remove the overall molecular rotation from the internal motions of the protein, every coordinate frame was translated and rotated until the r.m.s. displacement of residues 179–190 relative to a reference configuration (the average structure) was minimized. The linker residues were chosen for the alignment to explore correlations in the motion of the two zinc finger domains.
Interaction energy
The interaction energy (electrostatic + van der Waals) was calculated for each residue pair using the NamdEnergy plugin of VMD (18). The data were plotted using Tableau Software v8.0 where the size of the circles is determined by the square of the interaction energy to highlight strong interactions. In addition, (N, N+1) interactions were excluded from the plot for clarity.
Empirical chemical shifts
The empirical chemical shifts were predicted using SPARTA+ (28) using the combined conformations from all three trajectories of WT TIS11d. The errors for the calculated shifts are the estimated prediction errors given by SPARTA+. An additional unknown error is present because existing chemical shift prediction programs do not include the effects of the zinc ions. Linear regression was performed on the experimental versus calculated chemical shifts. The correlation coefficients were 0.98 for Cα, 0.99 for Cβ, 0.56 for H, and 0.92 for N.
Cloning, expression, and purification of the TZF of TIS11d and mutants
The RBD of human TIS11d (residues 152–220) was synthesized by Genescript and cloned into the pHMTC vector between BamHI and HindIII restriction sites. TIS11d-E220A and TIS11d-Δ D219/E220 (152–218) constructs were generated using the QuikChange Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). TIS11d and mutated constructs were expressed in BL21(DE3) Escherichia coli competent cells. Isotopic labeling with 15N was performed by growing the cells in M9 enriched with 1 g of 15NH4Cl per liter. Carbon labeling was performed by growing cells in M9 containing 13C-glucose. The cells were grown at 37°C to an OD600 of 0.8 and then induced for 4 h with 1 mM Isopropyl β-D-1 thiogalactopyranoside (IPTG) and 0.1 mM ZnSO4 at the same temperature. Harvested cells were then passed through a cell disrupter in 50 ml lysis buffer containing 50 mM Tris HCl, pH 8.0, 100 mM NaCl, 100 μM ZnOAc, 2 mM dithiothreitol (DTT), and 1 EDTA free Complete protease inhibitor tablet (Roche, Indianapolis, IN). Lysate was centrifuged at 19,500 RPM for 1 h at 4°C and passed through a 20 ml Amylose column preequilibrated with wash buffer (50 mM Tris HCl, pH 8.0, 100 mM NaCl, 100 μM ZnOAc, and 2 mM DTT). The column was washed with 10 column volumes of wash buffer. The protein was eluted with 10 mM maltose in wash buffer. The eluent was then placed over a 5 ml Hi TRAP Q column (GE Healthcare Life Science, Piscataway, NJ) preequilibrated with the same wash buffer used previously. The column was washed with 3 column volumes of wash buffer. The protein was eluted with a gradient of NaCl from 0 to 1 M; the peptides typically eluted at 0.3–0.5 M NaCl. Fractions containing our protein were combined, concentrated and dialyzed in 50 mM Tris HCl, pH 8.0, 300 mM NaCl, 100 μM ZnOAc, and 2 mM DTT in the presence of TEV at 1:10 ratio and left to undergo proteolysis for 24 h. The protein was further purified using a 1.6 X 60-cm Superdex-75 size exclusion column (Amersham Bio/Science, Piscataway, NJ) equilibrated with 50 mM Tris HCl pH 7, 300 mM NaCl, 2 mM BME to separate the 8.5 kDa fragments. For NMR analysis, fractions containing TIS11d or mutant forms of TIS11d were buffer exchanged into 10 mM Tris pH 6.2, 20 mM KCl, 2 mM DTT, 0.1 mM ZnSO4. Protein concentrations were determined via Ninhydrin assays.
RNA-binding activity measured by fluorescence polarization
All RNA binding assays were performed with uncleaved maltose binding protein (MBP) tagged protein for accurate determination of protein concentration via absorbance at 280 nm. Optimal binding of the TZF domains to RNA was measured in the presence of 10 mM Tris HCl, pH 8.0, 100 mM NaCl, 0.1 mM Zn(OAc)2, 0.01% IGEPAL CA630, and 2 mM DTT. The affinities of TIS11d, TIS11D-E220A, and TIS11d-Δ D219/E220 (152–218) for the sequence: 5′−UUUUAUUUAUUUU−3′ (ARE13) were measured by direct titration of 3 nM labeled RNA with increasing concentrations of protein using fluorescence polarization (29). The Kd,app were determined by fitting to the quadratic binding isotherm (29).
Thermodynamics of protein/RNA binding
The previously mentioned fluorescence anisotropy-based assay was then employed to generate binding curves for the ARE13 RNA at various temperatures using a VICTOR X Light Luminescence Plate Reader (PerkinElmer, Waltham, MA). The Kd,app measured at 26, 29, 32, 35, and 38°C were determined by fitting to the quadratic equation. The Kd,app obtained from fitting the data at each temperature are reported in Table S1 in the Supporting Material. Enthalpic (ΔH) and entropic (TΔS) contributions to the free energy of TIS11d and TIS11d-Δ D219/E220 binding to RNA substrates were estimated by a global fit of all the binding data measured at the five different temperatures using the quadratic binding isotherm expressed as a function of ΔH and ΔS:
| (3) |
where θ is the fraction of RNA bound, cRNA is the labeled RNA concentration, cp is the protein concentration, m and b are the maximum and base signals, respectively.
NMR spectroscopy
Folding of TIS11d, TIS11D-E220A, and TIS11d-Δ D219/E220 (152–218) was monitored using NMR spectroscopy. To determine whether amino acid changes within the C-terminus have an effect on the structure of the TZF domain of TIS11d, 15N-1H HSQC spectra were recorded at 25°C on a Varian Inova 600MHz NMR spectrometer (Palo Alto, CA) for each mutant protein. The concentration of TIS11d was 0.1 mM. Due to sample precipitation, the spectra for TIS11d-E220A and TIS11d-Δ D219/E220 were collected at a protein concentration of 50 μM. Experiments were processed with NMRpipe (30) and the spectra were compared to the WT TIS11d spectrum using Sparky (Goddard, T. D. and D. G. Kneller, SPARKY 3. UCSF, San Francisco, CA).
Results
NMR spectroscopic and biochemical studies of TIS11d and C-terminus mutants
TIS11d and the two mutants were studied using 15N–1H heteronuclear single-quantum coherence (HSQC) experiments. Absent from the 15N–1H HSQC spectrum of TIS11d are proline residues (161, 190, 207, and 210) and the crosspeaks of residues 181 and 188, which are severely overlapped and cannot be resolved. The HSQC spectrum of TIS11d-E220A (Fig. 2 A) shows loss of signal for residues 154, 169, 219, and 220 relative to the spectrum of WT TIS11d. The 15N–1H HSQC spectrum of TIS11d-Δ D219/E220 (Fig. 2 B) shows a loss of signal for significantly more residues: 153, 154, 169, 172, 185–194, 198, 199, 206, 213, and 216–218. Arg-153 and Tyr-154 are located on the RYKTEL RNA-binding motif preceding ZF1. Lys-169 is before the 310-helix turn on ZF1 (residues 171–173) and Glu-172 is in the middle of the 310-helix turn. Residues 185–194 are located in the unstructured linker region. Arg-198 and Thr-199 are located at the N-terminus of the α-helix in ZF2. Cys-206 is the second zinc-coordinating cysteine on ZF2. His-213 is adjacent to the third zinc-coordinating cysteine. His-216 coordinates Zn2+ on ZF2, and Asn-217 and Ala-218 are the two following residues. In addition, the crosspeaks of residues 180, 208, and 211 have reduced intensity than in the WT protein, they are 5.5%, 44%, and 13.9% of the WT signal, respectively. The loss of signal for residues in the linker in both mutants and several residues of ZF2 in the spectrum of TIS11d-Δ D219/E220 suggests that the mutant proteins have different flexibilities from the WT. Residues that exhibit a loss of signal in the two mutants relative to WT TIS11d are mapped onto the structures in Fig. 3.
Figure 2.
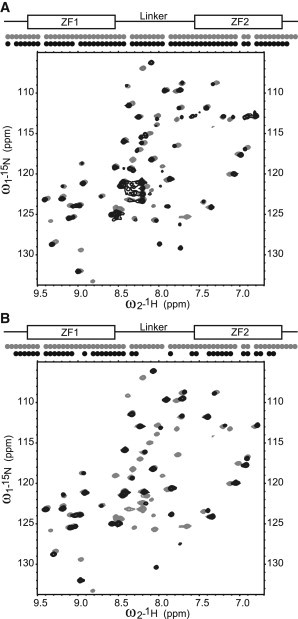
Comparison of 15N-1H HSQC spectra. Dots on top of the spectrum indicate whether a crosspeak was observed for each residue. (A) TIS11d (gray) and TIS11d-E220A (black), (B) TIS11d (gray) and TIS11d-Δ D219/E220 (black).
Figure 3.
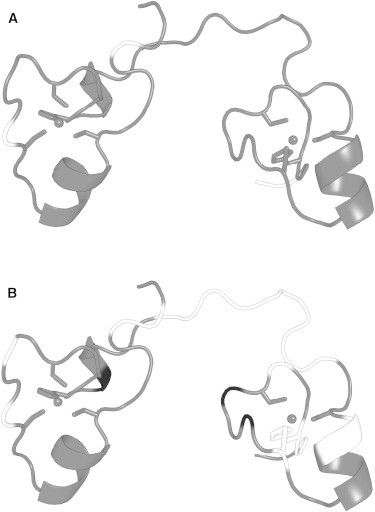
Crosspeaks with lost or reduced signal in the 15N-1H HSQC spectra of TIS11-E220A and TIS11d-Δ D219/E220 relative to WT TIS11d mapped on the initial NMR structure. Gray: residues with no significant change, black: residues with reduced signal, white: residues with no signal above the noise. (A) TIS11d-E220A and (B) TIS11d-Δ D219/E220.
To investigate the effect of the mutations on the activity of the proteins, we measured the RNA binding activity of all the proteins and determined the thermodynamics of binding for TIS11d and TIS11d-Δ D219/E220 (Fig. 4, A–C). In vitro binding data collected at 22°C shows nanomolar RNA binding affinity for TIS11d, TIS11d-E220A, and TIS11d-Δ D219/E220: (7.0 ± 0.9)×10−9 M, (2.9 ± 0.6)×10−9 M, and (3.2 ± 0.7)×10−9 M, respectively (Fig. 4, A–C). Despite the different flexibility and the different net charge of the mutant proteins relative to the WT, all three proteins bind RNA with similar affinity. These results are not surprising, because experimental evidence demonstrates that TTP is less structured than TIS11d in the absence of RNA (32), yet TTP and TIS11d bind RNA with similar affinity (33). To understand why these two mutations do not affect the RNA-binding affinity, we determined the thermodynamics of binding for TIS11d and TIS11d-Δ D219/E220 by measuring the binding constants at different temperatures (see Table S1 for Kd,app by temperature). The range of temperatures that were sampled was limited by the instrument and the limited thermal stability of these proteins. The enthalpic (ΔH0) and entropic (TΔS0) contributions to the free energy of binding for TIS11d and TIS11d-Δ D219/E220 were estimated from a global fit, where we simultaneously fit all of the binding data collected at different temperatures using Eq. 3. The results of the fitting are shown in Table 1 and Fig. S1, and indicate that for both proteins, RNA-binding is enthalpically driven. The mutant protein has a more favorable enthalpy of binding and a less favorable entropy of binding than the WT protein. Given the small difference between the enthalpy and entropy of binding between the WT and mutant proteins relative to the errors, we performed a t-test to determine if the observed differences in entropy and enthalpy of binding are statistically significant (34). In addition, we also used the F-test to directly compare the binding data of the WT and mutant proteins, and determine if the mutation affects the thermodynamics of binding RNA (34). From the t-test, p-values of 2 × 10−9 and 5 × 10−13 were determined for ΔH0 and ΔS0, respectively. Thus, the difference between the enthalpy and entropy of binding between the WT and mutant proteins is statistically significant. The same conclusion is reached from the F-test: the difference between the binding curves obtained for the WT and D219/E220 truncation mutant protein is statistically significant (p-value of 3 × 10−13), which shows that the mutation affects the thermodynamics of RNA-binding.
Figure 4.
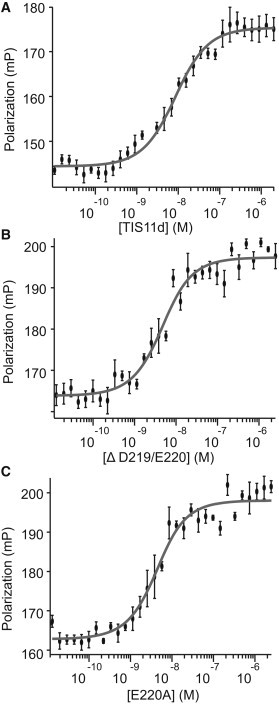
Thermodynamics of protein/RNA binding. Fluorescence polarization data used to calculate Kd,app of RNA-binding for (A) TIS11d, (B) TIS11d-Δ D219/E220, (C) TIS11d-E220A.
Table 1.
Thermodynamics of protein/RNA-binding for TIS11d and TIS11d-Δ D219/E220
| ΔH0 (kcal/mol) | TΔS0 (kcal/mol), 298 K | |
|---|---|---|
| TIS11d | −29.7 ± 1.7 | −18.5 ± 1.7 |
| TIS11d-Δ D219/E220 | −32.3 ± 0.6 | −21.6 ± 0.6 |
Structural dynamics of TIS11d and C-terminus mutants
To investigate how the charged C-terminus affected the structure and dynamics of unbound TIS11d with atomistic details, MD simulations were performed of TIS11d and the C-terminus mutants. Validation of MD trajectories is achieved by comparing chemical shifts calculated from MD trajectories with experimental data. The good agreement between the experimental chemical shifts and the empirical chemical shifts predicted by SPARTA+ (28) supports the validity of our MD simulation model for WT TIS11d (Fig. S2).
Structural parameters (Zn-Zn distance, linker end-to-end distance, radius of gyration, and RMSD from the average structure) were used to quantify the differences between TIS11d and the two mutants (Fig. 5). These quantities are plotted as a function of time for each trajectory in Fig. S3. Although the data are not normally distributed, mean and SD can be used to quantify differences when the histograms are used to interpret the aggregate statistics. The modes (most frequently populated values) are used instead of the mean in the case of bimodal distributions.
Figure 5.
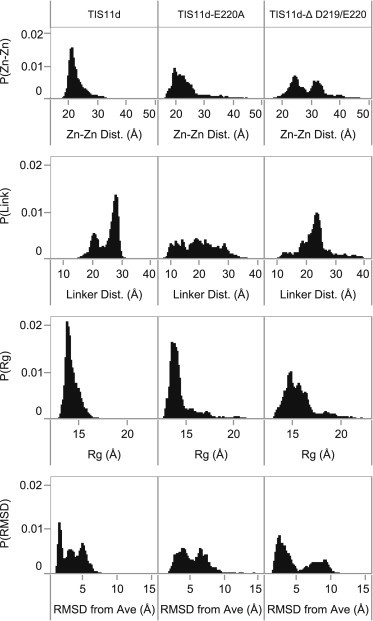
Normalized histograms of combined trajectories of structural metrics for each system (as labeled). From top to bottom: distance between zinc ions, linker end-to-end distance, radius of gyration, and RMSD from average structure.
The Zn-Zn distance distribution for WT TIS11d has a single maximum and is skewed with a long tail toward larger distances; the mean and SD of the Zn-Zn distance are 22.7 ± 2.6 Å. The distribution of the linker end-to-end distances is bimodal, suggesting that the WT protein fluctuates between two states characterized by linker end-to-end distances of 20 Å (less populated mode) and 28 Å (more populated mode), with a SD of 3.4 Å. Plotted as a function of time (Fig. S3), the linker end-to-end distance and radius of gyration show a dynamic picture of unbound TIS11d with occasional transitions between two states, one less compact than the other.
In contrast, the histograms of the structural parameters of TIS11d-E220A show that this mutant protein samples a significantly increased range of conformations (Fig. 5, middle). The distribution of the Zn-Zn distances has a much longer tail toward larger distances than in TIS11d. The mean Zn-Zn distance of 23.5 Å in this mutant is slightly increased over the WT protein, whereas the SD is increased to 4.9 Å, reflecting that this mutant protein samples a broader range of conformations than TIS11d. The increased flexibility of this protein relative to the WT is also highlighted by the distribution of the linker end-to-end distance that presents many small maxima between a wider range of values than in TIS11d. The mean linker end-to-end distance is TIS11d-E220A to 19.9 Å, similar to the less populated mode in TIS11d, however the SD is increased to 6.2 Å. Trajectories of these quantities (Fig. S3) show that these differences in the histograms are associated with transitions between conformational states not accessible to TIS11d, and that are characterized by greater dynamic changes in the calculated structural parameters.
For TIS11d-Δ D219/E220, the histograms of the structural parameters show that this protein samples an even more enhanced range of conformations (Fig. 5, right). The Zn-Zn distance is bimodal with maxima centered ∼24 and 32 Å. Both modes in this mutant are larger than the mean Zn-Zn distance in TIS11d and the first mode is similar to the mean Zn-Zn distance of TIS11d-E220A. The SD of the Zn-Zn distance is 5.3 Å, larger than in the two other proteins. The distribution of the linker end-to-end distances in TIS11d-Δ D219/E220 exhibits a significant maximum near the mean value of 23.0 Å, midway between the two linker end-to-end distances most frequently populated by TIS11d, and the SD of 4.8 Å is increased relative to the WT protein. Examining these quantities as a function of time shows that in one of the trajectories, the protein undergoes a larger number of transitions (Fig. S3, blue). This trajectory also loses the coordination of Cys-212 with the zinc ion of ZF2 at 58 ns and this coordination is not regained. The remaining two trajectories show transitions between many different conformations that are more compact than those sampled by the trajectory where Cys-212 becomes uncoordinated with the Zn2+ ion, thus are characterized by smaller radii of gyration.
The extent of these large-scale structural transitions observed in TIS11d-E220A and TIS11d-Δ D219/E220 is shown in Fig. 6. Representative structures are shown for before (Fig. 6 A) and during (Fig. 6 B) the linker extension seen in the third trajectory of TIS11d-E220A (Fig. S3, green). An example of the wide array of backbone conformations sampled by the most dynamic trajectory of TIS11d-Δ D219/E220 is shown in Fig. 6, C–F, including structures before and after the loss of coordination of Cys-212 with Zn2+. Fig. 6 F shows a unique conformation where the linker becomes twisted, not seen in any other trajectory of TIS11d-Δ D219/E220, indicating that the loss of Zn2+ coordination by one of the residues in ZF2 allows for greater flexibility in not just the structure of ZF2, but of the linker region as well.
Figure 6.
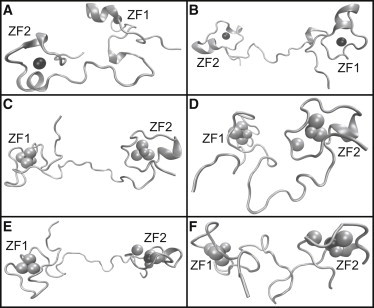
Example structures of TIS11d-E220A (A and B) and TIS11d-Δ D219/E220 (C–F). Black spheres: zinc ions, gray spheres: sulfur atoms of coordinating cysteine residues (large spheres) and nitrogen residues (small spheres), gray ribbon: backbone. TIS11d-E220A: (A) before extension, (B) during extension. TIS11d-Δ D219/E220: (C) extension with zinc ion of ZF2 still coordinated to Cys-212, (D) after extension with loss of Cys-212 coordination, (E) extended structure with loss of Cys-212 coordination, (F) more compact structure with crossed linker structure.
Flexibility is localized to the residues of the linker
Although the MD simulations of TIS11d showed that it is a dynamic protein, those of TIS11d-E220A and TIS11d-Δ D219/E220 showed markedly increased fluctuations (Fig. S3). To determine the source of the increased flexibility, we calculated the mean backbone distance between residues and the backbone dihedral angles.
The backbone distance maps shown in Fig. 7 (above the diagonal) quantify contacts between different residues of the protein and allow us to visualize structural features from the examination of the off-diagonal contacts and the identification of long-range interactions. The long-range contacts in TIS11d show a characteristic fingerprint of the tandem zinc finger domains, with an additional area of long-range contacts between ZF1 and the linking residues. The backbone contacts are preserved in both mutants with the exception of a slight decrease in contacts in ZF2 (Fig. 7, B and C, above the diagonal). This indicates that the structural transitions that occur, despite the size of the fluctuations observed in the radius of gyration or the Zn-Zn distance, do not greatly disrupt the fundamental structure of the zinc finger domains. However, backbone distance maps do not allow us to characterize differences in regions of a protein that are flexible and extended, and thus form few and transient contacts. For this reason, we calculated the backbone dihedral angles and observed that there is little difference of the mean values in the zinc finger regions between the three studied systems, but differences are seen in the linker region indicating that the linker is sampling different conformations in the three proteins (Fig. S4).
Figure 7.
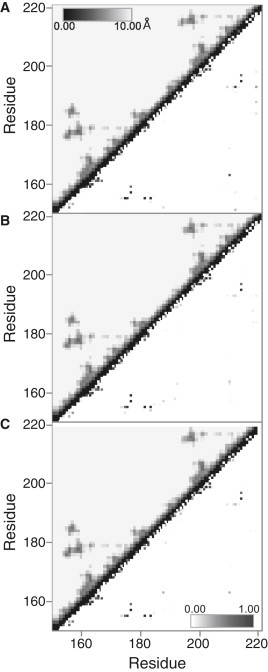
Above the diagonal: backbone distance maps by system averaged over all trajectories. The mean distance between Cα atoms of each residue pair is shown from dark (0 Å) to light (10 Å cutoff). Below the diagonal: hydrogen bond probability maps by system averaged over all trajectories. (A) TIS11d, (B) TIS11d-E220A, (C) TIS11d-Δ D219/E220.
When we analyzed the SD of the dihedral angles (Fig. 8), we observed that TIS11d is flexible at residues 178, 179, and 184–188 (Fig. 8, light gray). The ψ dihedral angle of His-178 and the φ dihedral angle of Gly-179 have much larger amplitudes in their oscillations than any other residue, resulting in the larger SD. This indicates that an inherent flexibility is present in the structure of TIS11d at the juncture between ZF1 and the linker due to the glycine residue at position 179. The increased SD of residues 184–188 is the result of infrequent dihedral transitions.
Figure 8.
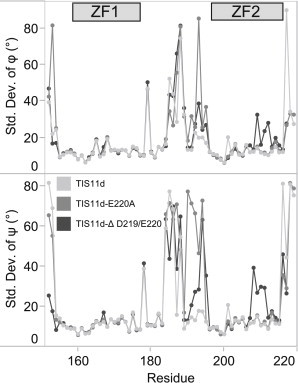
SD of backbone dihedral angles over all trajectories of TIS11d (light gray), TIS11d-E220A (medium gray), and TIS11d-Δ D219/E220 (dark gray).
Both mutants show the same increased SD of the linker dihedral angles as TIS11d, but with additional areas of increased flexibility (Fig. 8). The increased SD in residues 191–194 is also the result of infrequent transitions in the dihedral angles, as with residues 184–188. The increased flexibility seen throughout the linker region in the mutants is absent at Pro-190, which maintains its trans conformation. TIS11d-E220A (Fig. 8, medium gray) shows greater variability in the linker residues than TIS11d-Δ D219/E220 (Fig. 8, dark gray). However, TIS11d-Δ D219/E220 shows a region of increased variability in ZF2 of residues 207–212, which spans the region after the second coordinating cysteine (Cys-206) to the third coordinating cysteine (Cys-212), showing the effect of the loss of coordination of Cys-212.
Long-range hydrogen bonds are heterogeneous
The hydrogen bond probabilities are plotted for each of the three systems in Fig. 7 (lower diagonal). In all three systems, four long-range (between residues that are far apart in primary sequence) hydrogen bonds are characteristic of ZF1: Lys-155-Gln-175 (backbone), Lys-155-Phe-176 (side chain/backbone), Glu-157-Ala-177 (backbone), and Cys-159-Ala-177 (backbone). A similar pattern is seen in ZF2 for TIS11d: Lys-193-His-213 (backbone), Lys-193-Phe-214 (side chain/backbone), Glu-195-Ile-215 (backbone), and Cys-197-Ile-215 (backbone), although the Lys-193-Phe-214 hydrogen bond is very weak, unlike its equivalent in ZF1. Two additional long-range hydrogen bonds are seen between ZF1 and the linker, Lys-155-Glu-182 (side chain) and Lys-155-Arg-184 (backbone), which correspond to the smaller contact area adjacent to ZF1 in the pairwise backbone distance maps (Fig. 7 A, upper diagonal).
In TIS11d, long-range hydrogen bonds are involved in stabilizing the relative positions of ZF1 and ZF2 through direct hydrogen bonds between the two zinc fingers or interactions between the C-terminus and the linker (Fig. 7 A, below the diagonal). These bonds are ionic (35) in nature, but are distinct from salt bridges in that the hydrogen bond geometry is also satisfied. The stabilizing ionic hydrogen bonds vary by trajectory, but their general effect on the structure is similar (for hydrogen bond probabilities by trajectory, see Fig. S5). Between ZF1 and ZF2, the ionic hydrogen bonds can form between the side chain of Glu-163 or Glu-172 and the side chain of Arg-211. The C-terminus and linker interact through ionic hydrogen bonds between the side chains of Arg-184, Arg-188, and Lys-193 with the side chain of Glu-220.
In both TIS11d-E220A and TIS11d-Δ D219/E220, the hydrogen bonds of ZF1 are maintained but in ZF2 the Lys-193-His-213 backbone hydrogen bond is greatly weakened (Fig. 7, B and C, below the diagonal). The weakening of this ZF2 hydrogen bond in both mutants explains the increase in the mean distance between Lys-193 and Phe-214 and neighboring residues seen in the backbone distance maps (Fig. 7, B and C, above the diagonal). Similarly, the RMSD of ZF2 increases for the two mutants (Table S2). As expected, the side-chain hydrogen bonds of residue 220 with the linker are greatly weakened by the E220A mutation. Interestingly, the ionic hydrogen bonds between ZF1 and ZF2 are also weakened as a result of the E220A mutation. These observations suggest that E220 can promote the formation of additional stabilizing hydrogen bonds between ZF1 and ZF2 by bringing the two zinc fingers into closer proximity through a strong electrostatic interaction. With the deletion of residues D219/E220, no C-terminal hydrogen bonds can form. As a result, similar to TIS11d-E220A, the ZF1/ZF2 ionic hydrogen bonds are reduced (Fig. 7 C, below the diagonal). In only one trajectory was a moderate interaction formed between the side chains of Glu-163 and Arg-211 (Fig. S5, column 3, row 2).
Although the majority of the hydrogen bonds are preserved across trajectories, subtle variations are observed among the trajectories for each system (Fig. S5). This heterogeneity is the result of the flexibility of the protein (Fig. S3), particularly of the linker region, as shown by the large fluctuations and the different mean values of the backbone dihedral angles of the linker region (Figs. 8 and S4).
Lack of favorable electrostatic interaction causes increased flexibility
To identify the origin of the increased flexibility of the mutant proteins relative to the WT, we calculated the interaction energy between each residue pair for each of the three proteins (Fig. 9 A). Several strong interactions are seen in all three systems. The two zinc fingers are characterized by unfavorable interactions between the three deprotonated cysteine residues of ∼60–64 kcal/mol each (Fig. 9 A, dark blue circles). In the absence of zinc, the strong repulsion of the charged cysteine residues likely would initiate the unraveling of the zinc finger structure, as seen by the decrease of two of these unfavorable interaction energies in one trajectory of TIS11d-Δ D219/E220 where Cys-212 becomes uncoordinated (Fig. S6, column 3, row 1). ZF1 has a favorable electrostatic interaction between one of its preceding RYKTEL residues (Lys-155) and one of the residues of the linker (Glu-182) of ∼–90 kcal/mol. The analogous interaction is lacking for ZF2, suggesting that ZF1 may have greater structural stability.
Figure 9.
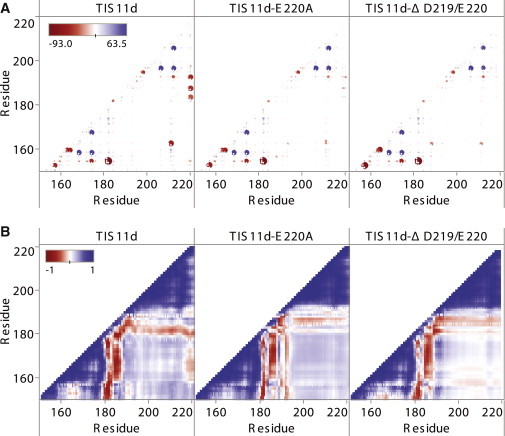
(A) Interaction energy between residue pairs by system (as labeled) averaged over all trajectories. Red: negative (favorable) interaction energies, blue: positive (unfavorable) interaction energies. Size of circles is a function of the interaction strength to highlight strong interactions, the larger the circle the stronger the interaction. (B) Cross correlation between residues when aligned on residues 179–190 by system (as labeled) averaged over all trajectories. Blue is positively correlated, red is anticorrelated.
A prominent feature among the interaction energies in TIS11d is the favorable interactions between the C-terminus (D219 and E220) and the linker residues (Arg-188, Lys-191, and Lys-193) and between ZF1 and ZF2 (Fig. 9 A, left). The interaction energies range between –85 and –115 kcal/mol and are associated with the ionic hydrogen bonds seen in TIS11d, which promote stabilization of the compact structure. These favorable interactions are greatly reduced or absent in TIS11d-E220A (Fig. 9 A, middle) and only a weak ZF1/ZF2 favorable interaction is present in TIS11d-Δ D219/E220 (Fig. 9 A, right). Although averaging over the trajectories allows us to see clearly the differences between the three systems, it obscures interesting details of the interactions. The strong electrostatic interactions vary between trajectories in TIS11d (Fig. S6) according to the hydrogen bonds that are present (Fig. S5). TIS11d forms very strong interactions either between the C-terminus and the linker residues (Fig. S6, column 1, row 2) or between ZF1 and ZF2 (Fig. S6, column 1, rows 1 and 3). The weak interaction between ZF1 and ZF2 in TIS11d-Δ D219/E220, seen in Fig. 9 A (right), is observed in only one trajectory (Fig. S6, column 3, row 2). Interestingly, this is the same trajectory where the protein samples conformations that are more similar to the WT protein, as shown by the fact that the Zn-Zn and linker distances are most like those of TIS11d (Fig. S3, orange). This result highlights the importance of the interactions between ZF1 and ZF2 in stabilizing the structure of TIS11d in the absence of RNA.
In TIS11d, the attraction between the negatively charged C-terminus and the positively charged linker residues brings the C-terminus closer to the linker and rotates the orientation of ZF2, allowing the formation of stabilizing ionic hydrogen bonds between the C-terminus and the linker as well as between ZF1 and ZF2. Reducing the negative charge in the C-terminus by the mutation E220A weakens this attraction and these stabilizing ionic hydrogen bonds are diminished. The truncation of the last two residues (D219 and E220) removes this attractive potential completely. As a result, both mutants sample more extended structures and a greater range of conformational states than WT TIS11d.
Global effects of electrostatic interactions
The long-range interactions present in TIS11d have effects not only in stabilizing a more compact structure but also in the dynamics of the protein. In the cross correlation matrix of the Cartesian coordinate fluctuations (Fig. 9 B), only TIS11d has a distinct band of anticorrelation between the motion of the C-terminal residues and ZF1. Thus, throughout the trajectories, the C-terminal residues and ZF1 are either moving toward each other, or away. Anticorrelation is consistent with an attractive potential limited by steric restrictions. This correlation is lacking in the mutants because the negative charge of the C-terminus is reduced or lost. In addition, the localized band of anticorrelation of the motions seen in TIS11d is broadened in the trajectories of the mutants between ZF1 and the linking residues, and weakened between ZF2 and the linking residues. The D219/E220 truncation has the further effect that the positive correlation between the motion of ZF1 and ZF2 is decreased and become in some regions slightly anticorrelated (Fig. 9 B, right). These additional losses of correlation in the mutants indicate that the electrostatic interactions promoted by the C-terminus enable a stronger communication between the two zinc finger domains in TIS11d.
The most striking and possibly the most functionally relevant effect of the reduction or loss of these long-range interactions is seen in the relative orientation of the zinc fingers. To estimate the relative orientation of the zinc fingers, we calculated an orientation vector for each zinc finger defined by the Zn2+ ion and its coordinating nitrogen of His-178 or His-216, respectively. We took the normalized dot product of the two vectors (when |Zn1·Zn2| = 1 the two zinc finger have the same orientation and when |Zn1·Zn2| = −1 the two zinc fingers have opposite orientation) and calculated the probability, P(>0), that the dot product was greater than zero (the zinc finger domains are aligned with an angle between them of <90°, hence they face the same side of the molecule). Because the zinc finger domains bind RNA with the zinc coordinating residues oriented toward the RNA, the probability P(>0) is the fraction of the trajectories where the two zinc fingers are in a conformation compatible with RNA-binding.
Snapshots from the trajectories illustrating the change in the relative orientation of the zinc finger domains and the normalized histograms of the relative orientation of the zinc fingers are shown in Fig. 10. Additionally, Fig. S7 shows the relative orientation of the zinc fingers as a function of time for each of the three systems by trajectory and the probability of each trajectory being in an RNA-binding compatible conformation. Fig. 10 (top) shows that the relative orientation of the two zinc fingers is predominantly in an RNA-binding compatible conformation in TIS11d (P(>0) = 0.76 over the three trajectories). The charged residues of the C-terminus in TIS11d help maintain much of the same relative orientation of the two zinc fingers, however one trajectory does change its relative orientation to be antiparallel (Fig. S7, top). Although the WT is able to sample RNA-binding incompatible conformations, the two mutants do so with much higher frequency (Fig. 10). Reducing the strength of the C-terminus interactions in TIS11d-E220A results in a higher population of orientations incompatible with RNA-binding, although the two zinc finger domains primarily face the same side of the molecule, with P(>0) = 0.66 over the three trajectories (Fig. 10, middle). Deleting the last two residues in TIS11d-Δ D219/E220 removes the preferred orientation of the two zinc fingers, shown by the more uniform distribution of |Zn1·Zn2| in Fig. 10 (bottom) and P(>0) = 0.47 over the three trajectories. The increased population of intermediate orientations (–0.5 < |Zn1·Zn2| < 0.5) compared to the WT is the result of frequent changes in the relative orientation of the zinc fingers observed in both mutants compared to the single transition observed in the WT (Fig. S7).
Figure 10.
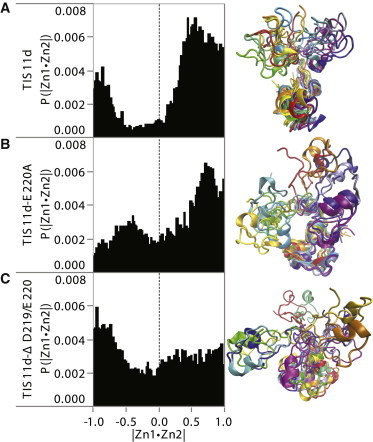
Characterization of zinc finger relative orientation for each system (as labeled). Left: normalized histograms of zinc finger domain relative orientation. Right: representative structures colored by time from trajectory 1 for each system aligned on residues 179–190. Red-yellow structures are early time points in the trajectories, orange-green structures are mid trajectory, and blue-purple structures are toward the end of the trajectories.
Discussion
Previous work has indicated the importance of E220 in maintaining a compact TIS11d structure in solution (14). To test this finding, we studied the following C-terminal mutants: TIS11d-E220A, where the last charged residue is mutated to a small nonpolar amino acid and TIS11d-Δ D219/E220 where the last two charged residues were deleted. MD simulations of TIS11d in the unbound state show that this protein is very flexible in solution, however both TIS11d-E220A and TIS11d-Δ D219/E220 sample more conformations and can adopt a fully extended linker conformation, indicating that the charged residues of the C-terminus are important to the overall stability of the structure of TIS11d. The deletion of D219 and E220 also was shown to permit the loss of zinc coordination with Cys-212 in ZF2 in one trajectory. This trajectory showed the most heightened flexibility and variation in structural conformations. The MD simulations were validated using NMR spectroscopy, by comparing the chemical shifts calculated from MD trajectories with experimental data. In addition, the line broadening and loss of signal observed in the 2D 15N-1H HSQC spectra of the mutant proteins indicates that both mutant proteins have different flexibilities from the WT, in agreement with the simulation data.
Given the intrinsically flexible nature of the TIS11d structure, the long-range attractive Coulomb interactions between the positively charged linker residues and the negatively charged C-terminus serve an important function in stabilizing the structure of TIS11d in a more compact conformation even when the ionic hydrogen bonds fluctuate. This arrangement allows for conformational flexibility and structural heterogeneity while still maintaining a compact structure and avoiding unfolding.
Furthermore, general electrostatic interactions arising from the charged C-terminus residues help maintain the relative orientation of the two zinc fingers in a conformation that is suitable for RNA-binding (P(>0) = 0.76) while allowing for flexibility. Reducing the strength of the interaction by mutation of one charged residue results in greater flexibility and variation in the orientation of the zinc fingers (P(>0) = 0.66). Removing the two charged resides results in significantly higher flexibility than the single mutation and the relative orientation of the zinc fingers approaches random (P(>0) = 0.47).
In agreement with the increased flexibility that was observed in the MD simulations of TIS11d-Δ D219/E220 relative to the WT protein, we found that the thermodynamics of RNA-binding of these two proteins differ from one another (Table 1, Fig. S1). Although these proteins bind RNA with similar affinity, their entropic and enthalpic contributions to the free energy of binding are different. MD simulations of TIS11d bound to ARE (14) show that electrostatic interactions between D219/E220 and RNA can have an unfavorable contribution to the interaction energy (Fig. S8). The interaction energy between Glu-220 and RNA has a bimodal distribution due to opposing electrostatic forces. An attractive potential exists between Lys-193 and Glu-220, however, Lys-193 is part of the KYKTEL RNA-binding motif that forms backbone hydrogen bonds with RNA. Thus, when Glu-220 interacts with the side chain of Lys-193, it is brought into closer proximity to the negatively charged RNA backbone, which results in a repulsive potential (and unfavorable interaction energies) between RNA and Glu-220. When Glu-220 is not interacting with Lys-193, the C-terminus extends away from the RNA backbone. Deleting the last two negatively charged residues in TIS11d-Δ D219/E220 changes the overall charge of the protein from +3 to +5 and removes the unfavorable interactions of the RNA with the C-terminus of the protein, thus increasing the affinity for the negatively charged RNA. The thermodynamics of binding for TIS11d-Δ D219/E220 compared to TIS11d (Table 1) show that a more favorable enthalpy of binding (likely due to the change in the overall charge of the protein from +3 to +5) compensates for a less favorable entropy change (likely due to the increased flexibility of the mutant protein relative to the WT), and explains the similar binding free energy observed for the WT and mutant proteins.
These observations suggest an intriguing explanation for the difference in charge in the C-terminal residues of the TTP family of proteins. In TIS11b/d, where both zinc fingers are structured in the unbound form, the last two residues of the C-terminus are both negatively charged (Fig. 1). This may enable TIS11b/d to maintain a preferential relative orientation of the two zinc fingers that is RNA-binding compatible (a similar orientation as the RNA-bound state), thereby minimizing the unfavorable entropy change associated with binding. In contrast, TTP, where the C-terminal zinc finger is unstructured or partially unstructured in solution (32), has only a single negative residue in the C-terminus, perhaps because there is no second structured finger and thus no relative orientation to maintain in the absence of RNA. The total charge of TTP is more positive than that of TIS11d/b (+8 vs. +3), hence has increased affinity for the negatively charged RNA. Thus, the more favorable enthalpic term might compensate for the less favorable entropic term, due to the folding of the second zinc finger, in the free energy of binding. The thermodynamics of binding show that a similar entropic/enthalpic compensation occurs in TIS11d-Δ D219/E220 relative to TIS11d.
Conclusions
We have shown that reduction or loss of these charged residues in the C-terminus increases the flexibility of the TIS11d mutants. Our results suggest an important mechanism that relies on the long-range nature of electrostatic interactions to maintain two domains, connected by a flexible linker, in a conformation that is compatible with ligand binding while allowing them to remain highly flexible in solution. Furthermore, the electrostatic nature of the interactions between the negatively charged C-terminus of TIS11d with the linker guarantees that the transition from the more compact free state to the bound state can be easily initiated upon interaction with the highly negatively charged RNA.
Author Contributions
B.R.M., L.M.D., and F. M. designed the research; B.R.M. performed the molecular dynamics simulations and computational analysis; L.M.D. prepared the samples, performed the NMR experiments, backbone resonance assignment, and binding experiments; B.R.M. and F.M wrote the paper.
Acknowledgments
The authors thank Troy W. Whitfield for helpful discussion.
This work was supported by the National Institutes of Health Grant GM098763.
Supporting Material
References
- 1.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 2.Chen C.Y., Gherzi R., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 3.Lai W.S., Carballo E., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackshear P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 5.Brewer B.Y., Malicka J., Wilson G.M. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 6.Ogilvie R.L., Abelson M., Bohjanen P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 7.Carrick D.M., Lai W.S., Blackshear P.J. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodson D.J., Janas M.L., Turner M. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stumpo D.J., Broxmeyer H.E., Blackshear P.J. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpo D.J., Byrd N.A., Blackshear P.J. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell. Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai W.S., Carballo E., Blackshear P.J. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 12.Lai W.S., Kennington E.A., Blackshear P.J. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 13.Hudson B.P., Martinez-Yamout M.A., Wright P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 14.Morgan B.R., Massi F. A computational study of RNA binding and specificity in the tandem zinc finger domain of TIS11d. Protein Sci. 2010;19:1222–1234. doi: 10.1002/pro.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worthington M.T., Amann B.T., Berg J.M. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc. Natl. Acad. Sci. USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin F., Chen Y., Chen H.F. Induced fit for mRNA/TIS11d complex. J. Chem. Phys. 2009;131:115103. doi: 10.1063/1.3224126. [DOI] [PubMed] [Google Scholar]
- 17.Lai W.S., Perera L., Blackshear P.J. Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin. J. Biol. Chem. 2014;289:565–580. doi: 10.1074/jbc.M113.466326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 19.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 21.Sakharov D.V., Lim C. Zn protein simulations including charge transfer and local polarization effects. J. Am. Chem. Soc. 2005;127:4921–4929. doi: 10.1021/ja0429115. [DOI] [PubMed] [Google Scholar]
- 22.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1997;23:327–341. [Google Scholar]
- 23.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 24.Reference deleted in proof.
- 25.Berne B.J., Pecora R. Wiley-Interscience; New York, NY: 1976. Dynamic Light Scattering. [Google Scholar]
- 26.Simmerling C., Elber R., Zhang J. Kluwer; Netherlands: 1995. A Program for Visualization of Structure and Dynamics of Biomolecules and STO-a Program for Computing Stochastic Paths. [Google Scholar]
- 27.Haliloglu T., Bahar I. Coarse-grained simulations of conformational dynamics of proteins: application to apomyoglobin. Proteins. 1998;31:271–281. [PubMed] [Google Scholar]
- 28.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagano J.M., Clingman C.C., Ryder S.P. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted in proof.
- 32.Brewer B.Y., Ballin J.D., Wilson G.M. Substrate dependence of conformational changes in the RNA-binding domain of tristetraprolin assessed by fluorescence spectroscopy of tryptophan mutants. Biochemistry. 2006;45:13807–13817. doi: 10.1021/bi061320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amann B.T., Worthington M.T., Berg J.M. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 34.Motulsky H., Christopoulos A. Oxford University Press; Oxford: 2004. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. [Google Scholar]
- 35.Meot-Ner M. The ionic hydrogen bond. Chem. Rev. 2005;105:213–284. doi: 10.1021/cr9411785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


