Abstract
Tracking fast-swimming bacteria in three dimensions can be extremely challenging with current optical techniques and a microscopic approach that can rapidly acquire volumetric information is required. Here, we introduce phase-contrast holographic video microscopy as a solution for the simultaneous tracking of multiple fast moving cells in three dimensions. This technique uses interference patterns formed between the scattered and the incident field to infer the three-dimensional (3D) position and size of bacteria. Using this optical approach, motility dynamics of multiple bacteria in three dimensions, such as speed and turn angles, can be obtained within minutes. We demonstrated the feasibility of this method by effectively tracking multiple bacteria species, including Escherichia coli, Agrobacterium tumefaciens, and Pseudomonas aeruginosa. In addition, we combined our fast 3D imaging technique with a microfluidic device to present an example of a drug/chemical assay to study effects on bacterial motility.
Introduction
In nature, many prokaryotic cells exhibit self-propelling motions. The cost of motility is significant for these microorganisms. Precious resources are spent in metabolizing and assembling motility apparatus and energies are consumed in maintaining and fueling the respective molecular motors. The cost is balanced by the many benefits of motility including translocation to preferred host or colonization sites, increased nutrient acquisition, and avoidance of potential hazardous or toxic substances (1). Therefore, motility behaviors have been widely used to study biological responses to external stimuli, such as chemotaxis (2) and phototaxis (3). Optical microscopy is an ideal noninvasive method for studying motility of individual microorganisms.
Due to the small physical size of bacteria, imaging them requires high magnification and large numerical aperture objectives. The limited working distance and shallow depth of field associated with such lenses make tracking microscopic particles in three dimensions very challenging (4). Currently, most quantitative studies on bacterial motility are based on either two-dimensional (2D) analysis of bacterial motion using an optical microscope (5,6) or from swarming assays of their collective motion in agar dishes (7) or capillary tubes (2). Although 2D studies can provide information on bacterial locomotion, such motility is strongly influenced by interactions between cells (7,8) and the cells with the surface (9). In their natural habitat, bacteria are not confined to 2D motility. Many species of bacteria explore the three-dimensional (3D) space of their habitat by rotating their flagella located on their cell surfaces. Therefore, for a complete study on bacterial motility, a high-magnification microscopy technique that can rapidly capture large depths of field for 3D dynamics measurements of individual motile bacteria is essential. In this work, we will introduce phase-contrast holographic video microscopy for capturing the 3D dynamics of bacterial motility.
For tracking 3D dynamics of microscopic particles, two noteworthy approaches are mechanical scanning and the use of optical techniques. For mechanical scanning methods, there is confocal microscopy (10), scanning stage microscopy (11–13) and scanning objectives microscopy (14). In these methods, the microscope stage or objective lens is moved to follow a single particle’s motion or to scan the volume of interest. These methods are limited by their scanning speed in providing detailed spatial and temporal information simultaneously. The movement of the stage or objective lenses also makes some of these methods difficult to integrate with other assays, such as those employing microfluidic devices.
For 3D optical tracking, there is intensity correlation microscopy (15), defocused microscopy (16), parallax or stereoscopic microscopy (17), and holographic microscopy (18–22). Most of these methods need special optical alignments or complex calibrations. Among these 3D imaging methods, the holographic imaging method has been proven to be easy to implement (18,19), has a large depth of field (21), and is efficient in providing high-throughput data (23–25).
Earlier approaches for inline holographic microscopy use a laser point source for sample illumination on a lensless camera (18,24). Although point illumination overcomes the issue of limited spatial resolution for digital cameras, the imaging depth of field is still very limited (19). Our approach uses a coherent plane wave laser as an illumination source and a microscope objective to expand the resultant inline holograms (19,20). Our previous attempts in holographic video microscopy successfully yielded quantitative measurements of 3D positions and the physical quantities of individual scatterers such as microspheres (19,21,26), emulsion droplets (26), and nanorods (27). Successful applications of this imaging approach include holographic microrheology (28), particle-image velocimetry (25,29), particle-resolved porosimetry (30), microrefractometry (31), and label-free molecular binding assays (25).
With our approach, we are able to improve the contrast in the volumetric reconstructions by an order of magnitude compared to previous attempts. With this improvement in technique, tracking bacterial locomotion in three dimensions becomes easier. This improvement also reduces computational resources and time in analyzing holographic video. In addition, we combined our fast 3D imaging technique with a microfluidic device to present an example of a drug/chemical assay to study effects on bacterial motility. This has potential applications not only in studying fast-moving microorganisms such as bacteria and sperm cells, but also in detecting pathogens in water and in quantifying the efficacy of chemical and drugs developed to target such pathogens.
Materials and Methods
Single colonies of Escherichia coli K12 (ATCC MG1655; American Type Culture Collection, Manassas, VA), Agrobacterium tumefaciens (Luria Broth (LB) 4404), and Pseudomonas aeruginosa (PAO1) cells were separately cultured overnight in 10 mL of LB or M9 minimum media at 37 and 30°C, respectively. The E. coli, A. tumefaciens, and P. aeruginosa cultures were then diluted with fresh medium to OD600 = 0.1 and grown to late-exponential growth phase. The prepared cell cultures were then diluted one part per thousand (∼1 × 105 cells/mL) before dispersing into a glass chamber consisting of a microscope slide and No. 1.5 cover slide for imaging. The prepared sample was allowed to sit on the microscope stage for 10 min to reach thermal equilibrium before imaging.
For measuring the reactive oxygen species (ROS) production in bacteria, E. coli MG1655 wild-type was labeled with 25 μM of dihydrorhodamine-123 (D633; Life Technologies, Carlsbad, CA) (32). The labeled cells were confined onto a cover-glass slide by a thin film of agar gel. After irradiation with a 405-nm laser, the change in fluorescence signal for each individual cell was measured with excitation at 500 nm and emission at 530 nm.
The microvalve-controlled microfluidic device (Fig. S1 in the Supporting Material) was fabricated from PDMS (poly(dimethysiloxane)) using multilayer soft lithography (33). The device contains two microchambers connected by a microchannel where the connection between the chambers can be controlled using a microvalve (34). One chamber contains bacteria while the other chamber contains gentamicin (15 mg/mL) or bleach solution (10%). The microfluidic devices were precoated with 1% (V/V) BSA (bovine serum albumin) to prevent bacterial adhesion to the internal surfaces. Laser powers were measured with a hand-held laser power meter (Edmund Optics, Barrington, NJ).
Phase-contrast holographic video microscopy
Most bacteria can swim 10–100 times their length per second. For reliable tracking of these fast-moving cells, we applied inline holographic video microscopy for tracking their 3D motions. Digital inline holographic video microscopy records 3D spatial information of matter that scatters coherent light at video frame rates (22). As schematically described in Fig. 1 a, holographic video microscopy uses a coherent light source to illuminate the sample. The interference between scattered light and the unscattered incident beam (reference light) forms an inline hologram on the focal plane. An objective lens then magnifies and projects the hologram onto an imaging device. The dynamics of the scatterers is captured in holographic video. Each 2D holographic video encodes the full 3D spatial and temporal information of individual microscopic scatterers.
Figure 1.
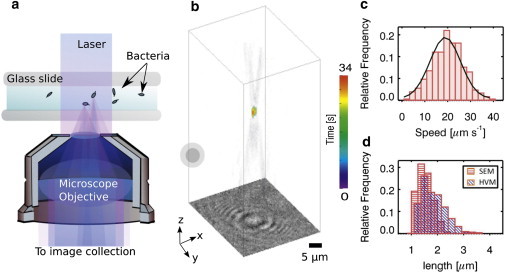
(a) Schematic of our phase-contrast holographic video microscope. A laser replaces conventional incandescence illumination in a conventional inverted microscope. Inline holograms are generated from the interference between the incident light and the scattered light at a focal plane below the scatterers. (b) Volumetric reconstruction of E. coli from its inline hologram. The 3D trajectory represents an example of single-cell swimming dynamics in M9 minimum media. (Concentrated circles) Start of the trajectory. (c) Distribution of the instantaneous speed for this bacterial swimming in three dimensions. The bacterium traveled with a mean speed of 18.9 ± 6.7 μm/s. (d) Length distribution of the bacteria measured by phase-contrast HVM (blue slanted lines) and by scanning-electron microscopy (SEM; red horizontal lines). To see this figure in color, go online.
Our inline holographic video microscopy, as illustrated in Fig. 1 a, consists of an inverted microscope (Eclipse TE2000; Nikon Instruments, Melville, NY) outfitted with high numerical aperture (NA) objectives (CFI Apo TIRF 60× H, NA = 1.49; Plan Apochromat VC 100× H, NA = 1.4; CFI Plan Apochromat DM 40× C, NA = 0.95; Nikon Instruments). The collimated illumination is from a diode laser (CVI; Melles Griot, Albuquerque, NM) operating at a vacuum wavelength of λ = 405 nm and a maximum power measured at 43 mW. A rotatable neutral density filter wheel (NDM4; Thorlabs, Newton, NJ) was used to adjust the intensity of the laser. The holographic videos were captured with a low-noise grayscale CMOS camera (DC 1645; Thorlabs) that can record uncompressed digital video stream at tens to hundreds of frames per second. The camera’s millisecond exposure time was also fast enough to avoid motion-blurring effects.
Each holographic video capture was then analyzed frame-by-frame to extract the size of the bacteria and its center of mass. A sample inline hologram of a single E. coli K12 bacterium cell is illustrated in Fig. 1 b. The position of the center of mass, xi, was extracted from the volumetric reconstruction of this hologram by identifying the highest intensity points. Our reconstruction and tracking algorithms were coded in the software language IDL 8.1 (Exelis Visual Information Solutions, Boulder, CO).
With little prior knowledge of the sample, image analysis of the captured holograms was performed with numerical reconstruction of the light field. Such volumetric reconstruction (22,27,35,36) of biological samples is done at high speed, is label-free and shape-independent, and can be used for measuring multiple bacteria positions in three dimensions.
In our holographic video microscopy, the incident illumination can be described by a plane wave, as , which propagates along the optical axis, z, with a real-valued amplitude Uo(r) that depends on position r = (x, y) in the transverse plane, and a uniform polarization . After scattering by the object, the scattered wave, , propagates in three dimensions with complex amplitude Es(r,z) and spatially varying polarization . Their superposition in the focal plane yields the holographic pattern captured by camera, as
| (1) |
Defocusing the focal plane below the scatterer ensures both that the polarization change is minimum, and that the scattered wave is substantially less intense than the illumination, . Expanding Eq. 1, the following relation is obtained:
Normalizing the measured hologram in each frame by the incident illumination’s intensity distribution (where the sample is not in the field of view) then yields
| (2) |
where is the reduced scattered field.
The complex scattered field above the focal plane can be fully reconstructed at height z as the convolution of the scattered amplitude in the focal plane with the Rayleigh-Sommerfeld propagator, , where , and yields
| (3) |
Equation 3 may be rewritten with the Fourier convolution theorem as , where the Fourier transform of and h(r, −z) is given by and , respectively.
To relate the above formalism with our normalized holograms, we note that the Fourier transform of b(r) is .
Therefore, may be recognized as the superposition of the scattered field at height z above the focal plane and a spurious field due to the object’s twin image in the focal plane. Fig. 2 a shows the twin image’s influence on the reconstructed field.
Figure 2.
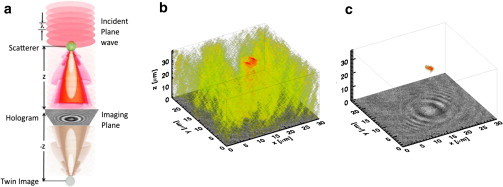
(a) Schematic of the light fields (and virtual image) contributions in the formation of an inline hologram with our experimental setup. Volumetric reconstructions of (b) amplitude-only scattered intensity and (c) phase-contrast intensity reconstruction, from a single E. coli MG1655 bacterium, colored by intensity with the same threshold cutoff. (Colored regions) Iso-surface of the brightest 70% of the reconstructed voxels. To see this figure in color, go online.
Using inverse Fourier transformation, will yield
| (4) |
At height z above the focal plane, the reduced scattered light’s intensity and the associated phase are then estimated, respectively, by
and
Using this relation, we can numerically reconstruct the volumetric intensity distribution of the scattered light propagating back through the bacterium. In this work, we associated the voxel with intensity above a threshold (70% of the peak intensity) as an estimate for the scatterer’s position at r and , . Ideally, the whole bacterium can be identified with the associate volume that matches the selected threshold value. However, most of the signals related to the bacterium are often masked by upstream artifacts and the spurious field of the virtual twin image as shown in Fig. 2 b.
To reduce these distractions, we can either use more stringent intensity threshold (22) or deconvolute the reconstructed volume with a point-spread function (35). For most biological samples, which have refractive indexes very close to their embedding medium, intensity signal alone offers limited contrast. Therefore, phase-contrast microscopy, rather than conventional bright-field microscopy, is used in visualizing biological specimens. Phase-contrast microscopy converts phase shift into intensity signal for visualizing unstained biological samples. It increases the imaging contrast for identifying the specimen from the background. A similar concept can be implemented into digital inline holographic video microscopy (HVM) to track bacteria.
For reconstructing a phase-contrast intensity profile in our HVM, we revisit the interference relationship between the scattered light intensity and the unscattered light intensity (37). Total intensity of light, at the point of scattering, can be written as
where ∅ is the phase-shifted term between the incident light intensity, and the scattered light intensity, Is(r,z). The value ξ(r,z) describes the phase shift due to the scatterer.
At the biological samples position (r,zo), there is no emission or absorption of light by the sample. By conservation of energy, all illuminating light at (r,zo) must therefore be scattered. Hence, I0(r,zo) ≅ Is(r,zo). Thus, the intensity signal of the feature, Ifeature, and background signal, Ibackground, can be written as
| (5) |
| (6) |
By digitally assigning ∅ = π, the background signals are . Also, the signal is related to the specimen as ). By choosing ∅ = π, the deviation between Ifeature and Ibackground will be the largest.
For small phase shift, ξ(r,zo), by the bacterium, small-angle approximation can be used to further simplify Eq. 5 to obtain the following:
| (7) |
In our case, when a bacterium is illuminated by the laser, both intensity and phase of the incident light shift by a small amount. Therefore, it is difficult to identify the bacterium’s 3D position using reconstruction with intensity contribution alone, as shown in Fig. 2 b. By including the corresponding phase contribution, ξR(r,z), for the intensity in the volumetric reconstruction, the bacterium’s 3D position and orientation can easily be quantified from the background intensity, as shown in Fig. 2 c and in the Movies S1, S2, S3, and S4 in the Supporting Material.
This approach is computationally more economical than using deconvolution approach (35). Using the point-spread function of the particular objects in an effort to optimize detection resolution requires additional computational steps and resources in deconvoluting the reconstructed data. In addition, calibration measurement for the point-spread function is also eliminated in our proposed approach. The effort to identify and track particles with HVM is thereby significantly simplified with our proposed approach.
To quantify the physical length, center of mass, and orientation of bacteria in every frame of the holographic video, we make use of the single-value decomposition procedure on the threshold selected points in the reconstructed volume (27). Intensity-weight skeletonization of Is(r,zo) using the single-value decomposition procedure is an efficient computational approach to estimate the bacterium axis. Points lying on the axis of interest are then fitted by linear regression to a line segment that can provide a good estimate of the bacterium’s orientation. The center of mass and the length of the bacterium are then retrieved from the median position and the maximum distance among the points along the identified axis, respectively.
Results and Discussion
Three-dimensional trajectories of the bacterium were obtained by linking its center-of-mass position dynamics over every single frame in Movie S1. Fig. 1 b also shows a 34-s three-dimensional trajectory plot of a single E. coli bacterium swimming in M9 minimal media. The 3D instantaneous speed was calculated using vi = dxi/dt, where dxi is the change in the center of mass between frames and dt is the time between each frame. The mean speed of this cell was obtained from the probability distribution of the speeds, as illustrated in Fig. 1 c. In this case, this E. coli cell was swimming at an average speed of 19 ± 7 μms−1. This result corresponds to the reported swim speed of E. coli measured using a microscope that can track in three dimensions (12).
The mean-length distribution measured with HVM for this batch of bacteria was ∼1.5 μm. The cell-length distribution of the same batch of cultured E. coli measured with a scanning-electron microscope (SEM; model No. JSM-6010LV; JEOL, Peabody, MA) was 1.5 ± 0.3 μm. This result corroborated with our holographic results as shown in Fig. 1 d. Such in situ holographic characterization of bacterial physical size is useful in differentiating actual bacteria from unidentified micron-size debris or colloidal particles.
With a few minutes of holographic video capture, we can sample hundreds of bacteria swimming continually into the field of view. As illustrated in Fig. 2 a and Movies S1, S2, S3, and S4, a phase-contrast holographic video microscope can track multiple bacteria simultaneously at the designated video frame rates. Using a 40× objective lens, this current setup can view a volume of ∼200 × 200 × 100 μm3. From just 5 min of data acquisition, ∼300 ± 100 independent E. coli cells swimming freely within the volume can be tracked.
Most bacteria swim using their flagella. In the flagellar system, a molecular motor at the basal body of the flagellum structure drives the semirigid filament structure (38). Different bacteria arrange their flagella distribution differently. E. coli is peritrichously flagellated (39), distributing their flagella over the entire cell body. In contrast, A. tumefaciens flagella are distributed at the poles (40,41) and monotrichous bacterium, P. aeruginosa has only one flagellum located at the pole. Differences in the flagellar position and numbers are reflected in the 3D motility of individual species.
The ease in obtaining multiple trajectories of bacteria in a large field of view makes this approach suitable to obtain statistics of the locomotion of various species far from the influence of the wall. An individual bacterium swimming close to the glass wall swims slower and makes circular trajectories due mainly to hydrodynamics coupling between the cell and solid surfaces (9,13). To minimize such effects, only tracks that were located >10 μm from the planar glass wall were chosen.
Following the description used in Berg and Brown (12), swim speed is defined as the mean swimming speed of a bacterium during runs. The turn angle is obtained from the angle supported by the trajectories between smooth runs. For E. coli, their population-average mean swim speed was measured to be 17 ± 5 μm/s (Fig. 3 a), with a most probable turn angle to be at 71° ± 34° (Fig. 3 b and Movie S1) (12). Notably, this speed and turn angle are consistent with previous reports of E. coli’s run-and-tumble mechanism (12). During smooth runs, the flagella rotate counterclockwise and bundle together to propel the bacterium forward. When the motors reverse, the flagellar bundle dissociates, causing tumbling and random reorientation to a new direction before the flagella bundle again for the next run. Without any favorable conditions, E. coli switch between run-and-tumble actions almost every second. Peritrichous bacteria employ such a run-and-tumble strategy efficiently for sensing its surroundings (12).
Figure 3.
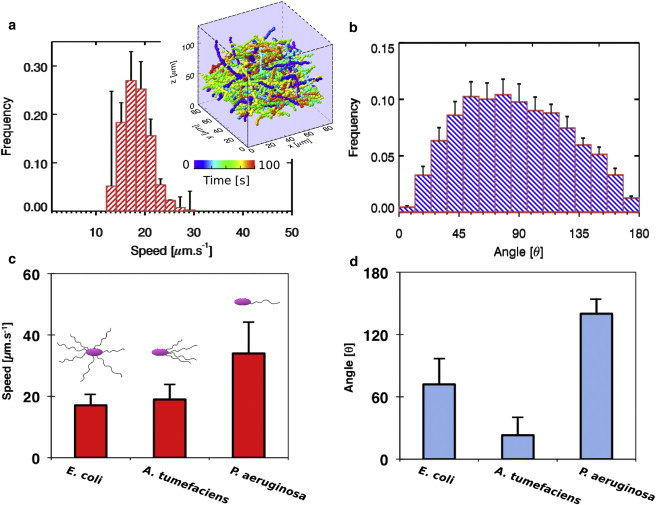
(a) Trajectory-averaged, 3D swimming-speed distribution plot for E. coli MG1655. (Inset) Trajectory plots of free-swimming E. coli MG1655 (n = 611 cells). The plot is color-coded according to the time in the holographic video micrographs. (b) Turn-angle distributions plot for E. coli MG1655. (c) Plot of most probable 3D swimming speed of three different species of bacteria. (d) Plot of most probable turn angles of three different species of bacteria. All experiments were conducted in LB media. Experiments for each bacterial species were repeated at least five times with samples obtained from at least two different colonies. To see this figure in color, go online.
On the other hand, lophotrichous flagellated bacteria such as A. tumefaciens have their flagella localized asymmetrically at one pole of the cell (41,42). With most of the torque generated from a pole, A. tumefaciens makes small turn angles. From the swim speed distribution, the most probable swim speed of A. tumefaciens (LB 4404) was at 19 ± 5 μm/s (Fig. 3 c) with the most probable turn angle at ∼23° ± 16° (Fig. 3 d and Movie S2). In contrast to E. coli, A. tumefaciens flagella rotate in a clockwise direction to propel the cell forward. Their flagella do not rotate counterclockwise. Instead, asynchronous flagellar rotation occurs to create the tumbling effect (40).
P. aeruginosa possess only single flagellum and do not apply the run-and-tumble mechanism for motility. Monotrichous bacteria use a run-and-reverse mechanism where the single flagellum either pushes or pulls the cell by rotating clockwise or counterclockwise (38). P. aeruginosa can swim at an average speed of 34 ± 10 μm/s (Fig. 3 c) with the most probable turn angle at ∼140° × 14° (Fig. 3 d and Movie S3). From this result, we can estimate that the P. aeruginosa molecular motor provides ∼0.3 pN force to overcome viscous drag and propel the cell forward or backward.
A phase-contrast holographic video microscope can work with any wavelength of light. A shorter wavelength laser (λ = 405 nm) gives a better signal because it scatters much more strongly than other visible wavelengths. Because our approach is based on scattering theory, our spatial resolution is still limited to approximately one-half the wavelength of incident light. In our case, we can only differentiate objects larger than 200 nm. And hence, we are not able to see individual flagellum of the bacteria in our studies.
Using 405-nm lasers can have some disadvantages. As shown in Fig. 4 a, A. tumefaciens 3D motility was affected at a 405-nm laser intensity >100 mW/cm2. When we illuminated A. tumefaciens with lasers of different wavelengths (λ = 532 and 660 nm), we did not see any correlation between laser intensity and bacterial 3D motility. These results suggest that photoinduced effects on cells were dependent on the applied laser wavelengths and intensity.
Figure 4.
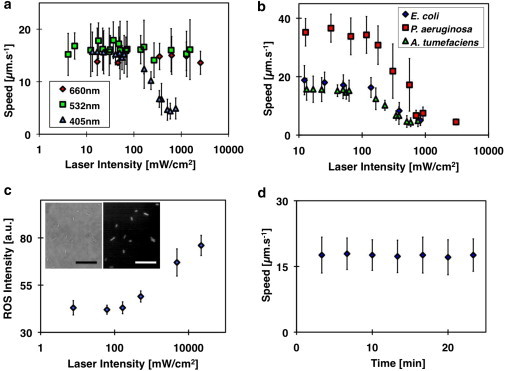
(a) Plot of average speeds of A. tumefaciens at different applied intensities of 405-nm, 532-nm, and 660-nm lasers in LB. There was a significant decrease in average speed for bacteria swimming at 405-nm laser intensity, with incident intensity larger than 100 mW/cm2. There was no obvious change in the mean 3D dynamics of the bacteria when they were illuminated with a low-intensity laser. (b) Plot of swim speed of three different species of bacteria at different applied intensities of the 405-nm laser. (c) Plot of ROS level induced in E. coli at different applied laser (λ = 405 nm) intensities. (Inset) Bright-field image of E. coli and the corresponding ROS-induced florescence image (scale bar =10 μm). (d) Plot of swim speed of E. coli against time. To see this figure in color, go online.
We further investigated this photoinduced effect on the other bacteria species. Similar responses to a 405-nm laser were observed in E. coli MG1655 and P. aeruginosa PAO1, as shown in Fig. 4 b. Their motility decreased when the laser intensity was higher than 100 mW/cm2. At <5 s exposure, we did observe that some bacteria recovered their locomotion. When bacteria were exposed to sufficiently high intensity light (I > 1 W/cm2), the paralyzed bacteria were unable to recover their motility even at an exposure of a few seconds.
The dependence of motility on laser wavelength suggests the possible involvement of intrinsic photoreceptors. Most bacteria have blue light receptors that regulate bacterial lifestyle decisions (43), including the transition from a free-swimming planktonic state to a multicellular state. Light alters protein-protein interactions, altering signal transduction (44), and bacterial motility. Another possible explanation is that endogenous porphyrin absorbs the photon energy to generate the ROS that results in photokinesis (32,45). In Fig. 4 c, the amount of ROS generated within E. coli increased exponentially when exposed to a 405-nm laser intensity larger than 100 mW/cm2. This intensity coincided with the laser intensity, where there was a decrease in bacterial motility in Fig. 4 b. This result suggested that ROS play a critical role in influencing bacterial motility. At low light dosage, the cell can mediate the amount of ROS generated with antioxidants expressed in each bacterial strain. However, as ROS production increases with laser intensity, a threshold is reached and ROS toxicity may result in cell death and immobility. Taken together, our observation of reduced motility at high 405-nm laser intensity can be attributed to either lifestyle transitions or cell toxicity.
Laser intensities <100 mW/cm2 do not cause long-term effects on the bacteria strains examined. We monitored the time-dependence on motility by continually exposing E. coli to 405-nm laser intensity at ∼20 mW/cm2. No change in the 3D motility of E. coli was observed after >20-min (Fig. 4 d) exposure to a 405-nm laser. Therefore, for the rest of this work, we maintained the laser intensity at ∼20 m W/cm2, which was well below the measured laser toxicity threshold for this species.
As with many inline holographic microscopy methods, the density of objects to be resolved must be limited to ensure accurate reconstruction. In our image analysis, we have not accounted for the effects of secondary scattering by the bacteria. Therefore, the errors from overlapping signals are significant when the concentration of cells is too high. We have tested various cell densities experimentally, and a concentration of <1 × 105 cell/mL is recommended.
Application
One application of this phase-contrast HVM is to observe the effects of antibiotics on bacterial motility. Microfluidic platforms can be incorporated with this imaging method for systematic studies of bacterial 3D locomotion under the influence of chemicals or drugs. Microvalve-controlled microfluidic platforms provide precise control of the fluid in microliter volumes for biological studies (33,34). The advantage of not moving the stage or the optics makes phase-contrast HVM an ideal 3D imaging method for use with a microfluidic platform. As a proof of principle, we demonstrated the effectiveness of such a platform on determining the effect of the amino-glycoside antibiotic gentamicin on motile E. coli, as illustrated in Fig. 5 a.
Figure 5.
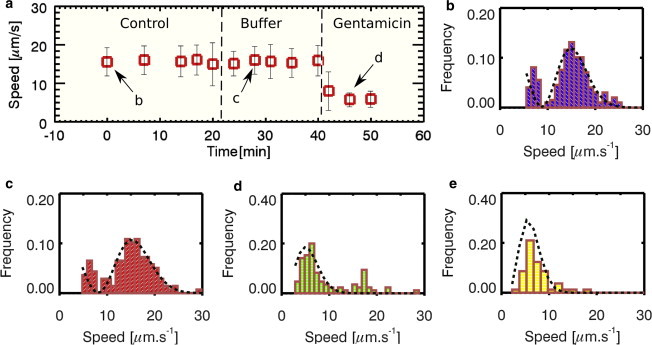
(a) Plot of average speeds of E. coli at different times. (b–d) Speed distribution plots of E. coli swimming speed at different time points, respectively. (Dotted line plot) Least-square fits of the 3D speed distribution to a Maxwell-Boltzmann distribution. (b) Distribution plot of the bacterial speed before the introduction of the amino-glycoside antibiotic gentamicin (number of cells, n = 191 cells). The average swim speed of bacteria was 16 μm/s (Movie S4). The secondary peaks at a lower velocity could be from bacteria swimming near the top of the microfluidic chamber. (c) The 3D speed distribution after addition of buffer (n = 177). (d) The 3D speed distribution after addition of gentamicin. Most of the bacteria populations were swimming with speeds <6 μm/s (n = 57 cells). (e) The 3D speed distribution after addition of 10% bleach solution (n = 80 cells). To see this figure in color, go online.
In this experiment, the effects of gentamicin on the motility of E. coli were studied with a microvalve-controlled microfluidic device (33). Before introduction of gentamicin, the population of bacteria was swimming at 15.8 ± 4 μm/s, as shown in Fig. 5 b. As control, 10 μL of LB (Fig. 5 c) was added. As expected, there was no observable change in the mean swim speed of the bacteria. After 20 min, 10 μL of gentamicin (150 ng/mL) was added to the bacteria and within minutes, most of the bacteria were moving at <6 μm/s (Fig. 5 d). This result was similar to a separate experiment where we added 10 μL of a 10% bleach solution (Clorox Liquid Bleach, The Clorox Company, Oakland, CA) (Fig. 5 e). Because bleach solution can effectively kill the E. coli cells (46), speed of <6 μm/s is indicative of nonflagellated motion (such as Brownian motion or residue sample drift).
Conclusions
Phase-contrast HVM can be widely applicable for observing the kinetics of fast moving microorganisms in three dimensions. HVM records videos of inline holograms of scatterers above the focal plane. We incorporate phase information in our intensity calculation to increase the contrast of the reconstructed images for 3D tracking. We have applied this imaging method successfully in tracking E. coli (Movie S1), A. tumefaciens (Movie S2), and P. aeruginosa (Movie S3). Because this method is based solely on scattering and interference of coherent light, it should work with any laser wavelength on most bacterial species.
Using phase-contrast HVM, we have shown that the free-swimming behavior of bacteria depends on the wavelength and intensity of the light. Although bacteria scattered shorter wavelengths of light more than longer wavelengths, they are affected by short-wavelength light. Hence, when using a 405-nm laser, we recommend the intensity should be kept to less than the threshold intensity of 100 mW/cm2. Alternatively, when illuminating bacteria at intensity higher than 100 mW/cm2, antioxidants, such as peroxidase, can be added to the sample to control the level of ROS produced (47). The best solution is still to choose a wavelength of light that has minimum interaction with the species under investigation. The flexibility to select any wavelength is one important advantage of this optical technique.
Coupling this technique with microfluidic devices, we can achieve high throughput 3D quantitative microscopic measurements for studying the 3D behavior of fast moving cells. In this work, we have demonstrated the usefulness of coupling this imaging technique with a microfluidic device to analyze the effect of antibiotics on bacterial motility. The phase-contrast HVM method will complement contemporary 3D tracking techniques as an important imaging method for tracking fast moving cells for biological and physical studies.
Acknowledgments
We thank Professor Chong Haur Sow for the loan of his lasers.
This work was supported by the Mechanobiology Institute, Singapore (a Research Centre of Excellence under the Ministry of Education), under grant No. VA-10BX000372.
Editor: Jeffrey Fredberg.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supporting Material
References
- 1.Berg H.C. Bacterial behavior. Nature. 1975;254:389–392. doi: 10.1038/254389a0. [DOI] [PubMed] [Google Scholar]
- 2.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 3.Armitage J.P. Chapter 5. Three hundred years of bacterial motility. In: Ord M.G., Stocken L.A., editors. Further Milestones in Biochemistry, Vol. 3. Foundations of Modern Biochemistry. Elsevier; New York: 1997. pp. 107–171. [Google Scholar]
- 4.Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 5.Larsen S.H., Reader R.W., Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 6.Vaituzis Z., Doetsch R.N. Motility tracks: technique for quantitative study of bacterial movement. Appl. Microbiol. 1969;17:584–588. doi: 10.1128/am.17.4.584-588.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg H.C. Symmetries in bacterial motility. Proc. Natl. Acad. Sci. USA. 1996;93:14225–14228. doi: 10.1073/pnas.93.25.14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S., Wolanin P.M., Austin R.H. Motion to form a quorum. Science. 2003;301:188. doi: 10.1126/science.1079805. [DOI] [PubMed] [Google Scholar]
- 9.DiLuzio W.R., Turner L., Whitesides G.M. Escherichia coli swim on the right-hand side. Nature. 2005;435:1271–1274. doi: 10.1038/nature03660. [DOI] [PubMed] [Google Scholar]
- 10.Prasad V., Semwogerere D., Weeks E.R. Confocal microscopy of colloids. J. Phys. Condens. Matter. 2007;19:113102. [Google Scholar]
- 11.Berg H.C. How to track bacteria. Rev. Sci. Instrum. 1971;42:868–871. doi: 10.1063/1.1685246. [DOI] [PubMed] [Google Scholar]
- 12.Berg H.C., Brown D.A. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 13.Frymier P.D., Ford R.M., Cummings P.T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA. 1995;92:6195–6199. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corkidi G., Taboada B., Darszon A. Tracking sperm in three-dimensions. Biochem. Biophys. Res. Commun. 2008;373:125–129. doi: 10.1016/j.bbrc.2008.05.189. [DOI] [PubMed] [Google Scholar]
- 15.Soni G.V., Ali B.M., Shivashankar G.V. Single particle tracking of correlated bacterial dynamics. Biophys. J. 2003;84:2634–2637. doi: 10.1016/s0006-3495(03)75068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M., Roberts J.W., DeLisa M.P. Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique. Appl. Environ. Microbiol. 2006;72:4987–4994. doi: 10.1128/AEM.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman R., Gibson G., Padgett M. Particle tracking stereomicroscopy in optical tweezers: control of trap shape. Opt. Express. 2010;18:11785–11790. doi: 10.1364/OE.18.011785. [DOI] [PubMed] [Google Scholar]
- 18.Xu W., Jericho M.H., Kreuzer H.J. Digital in-line holography for biological applications. Proc. Natl. Acad. Sci. USA. 2001;98:11301–11305. doi: 10.1073/pnas.191361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng J., Malkiel E., Katz J. Digital holographic microscope for measuring three-dimensional particle distributions and motions. Appl. Opt. 2006;45:3893–3901. doi: 10.1364/ao.45.003893. [DOI] [PubMed] [Google Scholar]
- 20.Lee S.H., Grier D.G. Holographic microscopy of holographically trapped three-dimensional structures. Opt. Express. 2007;15:1505–1512. doi: 10.1364/oe.15.001505. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.H., Roichman Y., Grier D.G. Characterizing and tracking single colloidal particles with video holographic microscopy. Opt. Express. 2007;15:18275–18282. doi: 10.1364/oe.15.018275. [DOI] [PubMed] [Google Scholar]
- 22.Cheong F.C., Krishnatreya B.J., Grier D.G. Strategies for three-dimensional particle tracking with holographic video microscopy. Opt. Express. 2010;18:13563–13573. doi: 10.1364/OE.18.013563. [DOI] [PubMed] [Google Scholar]
- 23.Su T.W., Erlinger A., Ozcan A. Compact and light-weight automated semen analysis platform using lensfree on-chip microscopy. Anal. Chem. 2010;82:8307–8312. doi: 10.1021/ac101845q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su T.W., Xue L., Ozcan A. High-throughput lensfree 3D tracking of human sperms reveals rare statistics of helical trajectories. Proc. Natl. Acad. Sci. USA. 2012;109:16018–16022. doi: 10.1073/pnas.1212506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheong F.C., Sun B., Grier D.G. Flow visualization and flow cytometry with holographic video microscopy. Opt. Express. 2009;17:13071–13079. doi: 10.1364/oe.17.013071. [DOI] [PubMed] [Google Scholar]
- 26.Cheong F.C., Xiao K., Grier D.G. Technical note: characterizing individual milk fat globules with holographic video microscopy. J. Dairy Sci. 2009;92:95–99. doi: 10.3168/jds.2008-1361. [DOI] [PubMed] [Google Scholar]
- 27.Cheong F.C., Grier D.G. Rotational and translational diffusion of copper oxide nanorods measured with holographic video microscopy. Opt. Express. 2010;18:6555–6562. doi: 10.1364/OE.18.006555. [DOI] [PubMed] [Google Scholar]
- 28.Cheong F.C., Duarte S., Grier D.G. Holographic microrheology of polysaccharides from Streptococcus mutans biofilms. Rheol. Acta. 2008;48:109–115. [Google Scholar]
- 29.Dixon L., Cheong F.C., Grier D.G. Holographic particle-streak velocimetry. Opt. Express. 2011;19:4393–4398. doi: 10.1364/OE.19.004393. [DOI] [PubMed] [Google Scholar]
- 30.Cheong F.C., Xiao K., Grier D.G. Holographic characterization of individual colloidal spheres’ porosities. Soft Matter. 2011;7:6816–6819. [Google Scholar]
- 31.Shpaisman H., Krishnatreya B.J., Grier D.G. Holographic microrefractometer. Appl. Phys. Lett. 2012;101:091102. [Google Scholar]
- 32.Krisko A., Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA. 2010;107:14373–14377. doi: 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam S.W., Van Noort D., Park S. A biological sensor platform using a pneumatic-valve controlled microfluidic device containing Tetrahymena pyriformis. Lab Chip. 2007;7:638–640. doi: 10.1039/b617357h. [DOI] [PubMed] [Google Scholar]
- 34.Unger M.A., Chou H.P., Quake S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 35.Dixon L., Cheong F.C., Grier D.G. Holographic deconvolution microscopy for high-resolution particle tracking. Opt. Express. 2011;19:16410–16417. doi: 10.1364/OE.19.016410. [DOI] [PubMed] [Google Scholar]
- 36.Kapfenberger D., Sonn-Segev A., Roichman Y. Accurate holographic imaging of colloidal particle pairs by Rayleigh-Sommerfeld reconstruction. Opt. Express. 2013;21:12228–12237. doi: 10.1364/OE.21.012228. [DOI] [PubMed] [Google Scholar]
- 37.Born M., Wolf E. 4th Ed. Pergamon; Oxford, UK: 1970. Principles of Optics. [Google Scholar]
- 38.Berry R.M., Armitage J.P. The bacterial flagella motor. Adv. Microb. Physiol. 1999;41:291–337. doi: 10.1016/s0065-2911(08)60169-1. [DOI] [PubMed] [Google Scholar]
- 39.Berg H.C. Motile behavior of bacteria. Phys. Today. 2000;53:24–29. [Google Scholar]
- 40.Chesnokova O., Coutinho J.B., Kado C.I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol. Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 41.Shaw C.H. Swimming against the tide: chemotaxis in Agrobacterium. BioEssays. 1991;13:25–29. doi: 10.1002/bies.950130105. [DOI] [PubMed] [Google Scholar]
- 42.Merritt P.M., Danhorn T., Fuqua C. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 2007;189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomelsky M., Hoff W.D. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011;19:441–448. doi: 10.1016/j.tim.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Zoltowski B.D., Gardner K.H. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipovsky A., Nitzan Y., Lubart R. Visible light-induced killing of bacteria as a function of wavelength: implication for wound healing. Lasers Surg. Med. 2010;42:467–472. doi: 10.1002/lsm.20948. [DOI] [PubMed] [Google Scholar]
- 46.Merritt K., Hitchins V.M., Brown S.A. Safety and cleaning of medical materials and devices. J. Biomed. Mater. Res. 2000;53:131–136. doi: 10.1002/(sici)1097-4636(2000)53:2<131::aid-jbm1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 47.Cui Y., Oh Y.J., Park S. AFM study of the differential inhibitory effects of the green tea polyphenol (−)-epigallocatechin-3-gallate (EGCG) against Gram-positive and Gram-negative bacteria. Food Microbiol. 2012;29:80–87. doi: 10.1016/j.fm.2011.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


