Background: ER stress induces cell surface translocation of GRP78/BiP.
Results: GRP78 translocation to the cell surface requires its substrate binding domain and exists majorly as a peripheral protein.
Conclusion: GRP78 anchors on the cell surface via interaction with other proteins, and the translocation mechanism is cell context-dependent.
Significance: Learning how GRP78 exists on the cell surface is crucial for understanding its signaling regulatory functions.
Keywords: Cell Surface Protein, Chaperone, Endoplasmic Reticulum Stress (ER Stress), Imaging, Translocation, GPI-anchored Proteins, GRP78/BiP, Cancer Cell Lines
Abstract
Glucose-regulated protein (GRP78)/BiP, a major chaperone in the endoplasmic reticulum, is recently discovered to be preferably expressed on the surface of stressed cancer cells, where it regulates critical oncogenic signaling pathways and is emerging as a target for anti-cancer therapy while sparing normal organs. However, because GRP78 does not contain classical transmembrane domains, its mechanism of transport and its anchoring at the cell surface are poorly understood. Using a combination of biochemical, mutational, FACS, and single molecule super-resolution imaging approaches, we discovered that GRP78 majorly exists as a peripheral protein on plasma membrane via interaction with other cell surface proteins including glycosylphosphatidylinositol-anchored proteins. Moreover, cell surface GRP78 expression requires its substrate binding activity but is independent of ATP binding or a membrane insertion motif conserved with HSP70. Unexpectedly, different cancer cell lines rely on different mechanisms for GRP78 cell surface translocation, implying that the process is cell context-dependent.
Introduction
The glucose-regulated protein, GRP783 (also referred to as BiP or HSPA5), is a member of the heat shock protein 70 (HSP70) superfamily and is evolutionarily conserved from yeast to human (1, 2). GRP78 contains a signal peptide that targets it to the endoplasmic reticulum (ER) and a carboxyl KDEL motif for retrieval from the Golgi apparatus leading to ER retention (3). The ER is an essential organelle for the synthesis and processing of plasma membrane and secretory proteins. As a major ER chaperone protein with ATPase activity, GRP78 complexes with nascent polypeptides and is critical for their folding and maturation in the ER compartment. Under ER stress conditions, when malfolded proteins accumulate in the ER, GRP78 is up-regulated and prevents protein aggregation as well as facilitates degradation of misfolded proteins (1, 4). GRP78 is a key regulator of the unfolded protein response (UPR) such that it binds and maintains the transmembrane ER stress sensors (PERK, IRE1, and ATF6) in their inactive forms, and upon ER stress, GRP78 is released resulting in the activation of these signaling pathways, impacting both cell survival and apoptosis (4, 5). Analogously, in non-stressed cells, GRP78 forms a complex with ER-associated pro-apoptotic signaling machineries and blocks their activation (2).
Although traditionally GRP78 has been regarded as an ER lumenal protein, evidence is emerging that GRP78 can also be detected in other cellular locations including the cell surface, cytosol, mitochondria, and the nucleus and assume novel functions that control signaling, proliferation, invasion, apoptosis, inflammation, and immunity (2, 4, 6). Of particular importance is that a subfraction of GRP78 can relocalize to the surface of specific cell types, such as cancer cells, and this process is actively enhanced by ER stress (7–9). At the cell surface, in complexes with specific cell surface proteins, GRP78 exerts functions beyond the ER (6). For example, GRP78 serves as co-receptor for the proteinase inhibitor α2-macroglobulin-induced signal transduction for cancer survival and metastasis (10). Cell surface GRP78 is also an obligatory binding partner for Cripto, a GPI-anchored protein on the cell surface for its activity in regulating stem cell regeneration and tumorigenesis (11, 12). Cell surface GRP78 can also mediate endothelial cell survival, TRAIL (TNF-related apoptosis-inducing ligand)-induced apoptosis as well as viral entry into host cells. The recent discovery that GRP78 is preferably expressed on the surface of tumor cells but not normal organs in vivo opens a unique opportunity for specific tumor targeting with minimal harmful effects on normal cells. As cell surface GRP78 is further detected in some tumor-initiating cells and increased in metastatic and cancer cells that have developed therapy resistance as well as in hypoxic endothelial cells that support tumor cells, cytotoxic agents including peptide-drug conjugates and monoclonal antibodies targeting against cell surface GRP78 has shown great promise in cancer therapy in multiple settings and are currently under development (2, 7, 8, 13–18).
Considering the significance of cell surface GRP78 from both the basic cell biology and therapeutic targeting perspective, it is important to understand how GRP78 exists stably on the cell surface and how it reaches the cell surface. This is particularly intriguing because the primary amino acid sequence of the mature GRP78 contains only a few weak hydrophobic domains, and GRP78 containing the intact KDEL ER retrieval motif is capable of localizing on the cell surface (9, 15). Global profiling of cell surface proteome of tumor cells clearly revealed relative abundance of cytosolic heat shock and ER lumen chaperones, including GRP78 (19), suggesting relocating these stress-inducible chaperones to the cell surface could represent a common adaptive mechanism for cells to respond to stress-perturbing protein homeostasis. In this study, using a combination of biochemical, mutational, FACS, and super-resolution microscopy approaches, we address these issues in a panel of cancer cells. Our studies reveal previously unidentified physical and biochemical properties of cell surface GRP78, which have important implications for its function as a novel regulator of cell signaling outside the ER and its therapeutic targeting.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cervical cancer cell line HeLa and breast cancer cell line MCF-7 were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) (Life Technologies) and 1% penicillin/streptomycin. Human colon cancer cell line HCT-116 was cultured in McCoy's 5A medium containing 10% FBS and 1% penicillin/streptomycin. Human prostate cancer cell line C4-2B was cultured in RPMI 1640 medium containing 10% FBS and 1% penicillin/streptomycin. Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. For stress treatment, the cells were treated with thapsigargin (Tg) at 300 nm, tunicamycin (Tu) at 1.5 μg/ml for 16 h, or 2-deoxy-d-glucose (2-DG) at 10 mm for 24 h. For brefeldin A (BFA) treatment, the cells were incubated with 0.2–5 μg/ml BFA for 16 h before harvest. For cyclohexamide treatment, the cells were incubated with 0.2 or 2 μg/ml cyclohexamide for 16 h. For MG-115 treatment, the cells were incubated with 20 μm for 16 h before harvest. All the agents mentioned above were purchased from Sigma.
Expression Vector Construction
The construction of expression plasmid for FLAG-GRP78 (WT) has been described previously (9). The mutants of GRP78 were generated using FLAG-GRP78 as template and following the protocol of QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). The construction of expression plasmid for GRP78 substrate binding domain (SBD) with KDEL motif at the C terminus was generated by PCR amplification from FLAG-GRP78 (WT) expression plasmid using TaqDNA polymerase (M0273S, New England Biolabs, Ipswich, MA) and primers 5′-TATTATCCCGGGGTCCAGGCTGGTGTGCTCTCTG-3′ and 5′-TTATATGTCGACCTATTACAACTCATCTTTGGTGACTTCAATCTGTGGGAC-3′. The PCR product was inserted in-frame into pDisplay expression vector (Life Technologies) between XmaI and SalI sites. The construction of bacterial expression plasmid for GST-HA fusion protein was generated by insertion of annealed oligonucleotides 5′-GATCCCCGAAGCTTTACCCATACGATGTTCCAGATTACGCTTAGC-3′ and 5′-TCGAGCTAAGCGTAATCTGGAACATCGTATGGGTAAAGCTTCGGG-3′ into the BamHI and XhoI sites of pGEX 4T1 plasmid (GE Healthcare). The caveolin-1-SNAP expression plasmid was produced by PCR amplification of the SNAP coding sequence from a pSNAP-tag(m) plasmid (New England Biolabs) using forward primer 5′-AACAAACCGCGGATGGACAAA-3′ and reverse primer 5′-AACAAATCTAGATCAGG TACC-3′. The SNAP coding sequence was then inserted in-frame with caveolin-1 in a pCav1-GFP 1–10(h)-N1 mammalian expression vector (20) after SacII and XbaI extraction of the GFP 1–10(h) coding sequence. The construct was verified by sequencing.
Transfection and Collection of Secreted Proteins
Transfection was performed as described using BioT (Bioland Scientific, Paramount, CA) following the manufacturer's instructions (9). The secreted proteins were collected as described (9).
Cell Surface Protein Biotinylation and Avidin Pulldown
After treatment, the cells were washed with cold PBS three times. EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific, Waltham, MA) in PBS at 0.5 mg/ml was added, and the cells were gently shaken at 4 °C for 30 min. To stop the biotinylation reaction, the biotin solution was removed, and the cells were rinsed 3 times with the quenching buffer containing Tris-Cl, pH 7.4, in cold PBS. Then cells were rinsed with cold PBS and subjected to either sodium carbonate extraction (see below) or radioimmune precipitation (RIPA) lysis. The RIPA lysis buffer was supplemented with protease and phosphatase inhibitor mixture (Thermo Scientific). Protein concentrations were determined by the Bradford assay (Bio-Rad). Part of the lysate was saved as whole cell lysate for Western blots to measure the total level of the target proteins. To purify the surface proteins, the remaining lysates were mixed with High Capacity NeutrAvidin Agarose Resin (Thermo Scientific) at room temperature for 1 h, and the resin was washed by RIPA buffer and centrifuged at 3000 × g for 1 min 10 times. The cell surface proteins were released by the addition of 50 μl of 2× SDS-PAGE sample buffer followed by heating at 95 °C for 5 min and centrifuged at 6000 × g for 5 min to collect the supernatant.
Sodium Carbonate Extraction
The cells were lysed in 100 mm sodium carbonate by three freeze-thaw cycles and ultracentrifuged for 1 h at 195,000 × g at 4 °C as described (21). The supernatant was concentrated with Vivaspin 6 concentrator (10-kDa cut-off, GE Healthcare) and buffer-exchanged to RIPA buffer to obtain soluble protein fraction. The pellet was resuspended and washed in cold 100 mm sodium carbonate 5 times and solubilized in RIPA buffer to obtain membrane protein fraction. The volume of membrane protein fraction corresponded to the final volume of soluble protein fraction.
Immunoblot Analysis
Protein samples were subjected to 10% or 15% SDS-PAGE and Western blot analysis as previously described (9). Primary antibodies used are as follows: anti-ANXA2 mouse antibody (610068, BD Biosciences), 1:2500; anti-β-actin mouse antibody (A5316, Sigma), 1:5000; anti-calreticulin rabbit antibody (catalogue no. 2891, Cell Signaling, Danvers, MA), 1:5000; anti-calnexin rabbit antibody (ADI-SPA-860, Enzo Life Sciences), 1:1000; anti-EphB4 mouse antibody (mAb131) (22), 1:1000; anti-FLAG M2 mouse antibody (F1804, Sigma), 1:1000; anti-GAPDH mouse antibody (sc-32233, Santa Cruz Biotechnology, Inc., Dallas, TX), 1:1000; anti-GRP78 mouse antibody (mAb159) (16), 1:1000; anti-GRP78 rat antibody (sc-13539, Santa Cruz Biotechnology, Inc.), 1:1000; anti-GRP94 rat antibody (SPA-851, Enzo Life Sciences), 1:1000; anti-HA rabbit antibody (sc-805, Santa Cruz Biotechnology, Inc.), 1:1000; anti-HSP70 mouse antibody (sc-66048, Santa Cruz Biotechnology, Inc.) 1:500; anti-HTJ1 rabbit antibody (GTX103858, GeneTex, Inc., Irvine, CA) 1:500; anti-integrin β1 rabbit antibody (EP1041Y, Millipore, Billerica, MA), 1:500; anti-PDI rabbit antibody (ADI-SPA-890, Enzo Life Sciences), 1:1000; anti-PDI rabbit antibody (sc-20132, Santa Cruz Biotechnology, Inc.), 1:500; anti-uPAR rabbit antibody (GTX100467, GeneTex, Inc.), 1:500. The secondary antibodies used in this study are as follows: horseradish peroxidase conjugate goat anti-mouse, anti-rabbit, and anti-rat antibodies (sc-2005, sc-2004, sc-2006, Santa Cruz Biotechnology, Inc.) and goat anti-mouse IRDye® 800CW, 1:7500 and goat anti-rabbit IRDye® 680RD secondary antibodies (LI-COR Biosciences, Lincoln, NE), 1:7500. The experiments were repeated 2–4 times. Protein levels were visualized and quantitated by ChemiDoc™ XRS+ Imager (Bio-Rad) or LI-COR Odyssey (LI-COR Biosciences).
Recombinant Protein Preparation
The GST-GRP78 expression plasmid was generated as described previously (23). Recombinant GST-GRP78 and GST-HA were purified using glutathione-Sepharose 4B (GE Healthcare) following the manufacturer's instructions.
Cell Permeabilization Assay
The assay was performed as previously described (24) with the exception that the cells were permeabilized by 0.005% digitonin for 5 min on ice.
Phosphatidylinositol-specific Phospholipase C (PI-PLC) Treatment
The cells were treated with 0.5 units/ml PI-PLC (Life Technologies) in PBS for 1 h at 37 °C as described (25).
Surface Protein Cross-linking
Cells were treated with membrane-impermeable cross-linker DTSSP (Thermo Scientific) at 1 mm in cold PBS following manufacturer's instruction.
FACS Analysis
Cell surface proteins were analyzed using flow cytometry as described previously (15). Surface GRP78 was detected by the mouse anti-GRP78 (mAb159) (16) and ephrin type-B receptor 4 (EphB4) by the anti-EphB4 (mAb131) antibody (22).
Cell Culture and Staining for Super-resolution Imaging
HCT-116 cells were seeded at 5 × 104 cells per well in 6-well plates containing Marienfeld-Superior precision cover glasses (thickness no. 1.5H) coated with fibronectin (50 μg/ml, Sigma), grown for 24 h, then transfected with the caveolin-1-SNAP expression plasmid (2 μg/well) using BioT transfection reagent (Bioland Scientific). Six hours after transfection, media were changed to the normal culture media. To induce ER stress, Tg (300 nm, Sigma) was added into the culture media 30 h after transfection. Sixteen hours after thapsigargin treatment, cells were washed with warm PBS and then fixed with 4% paraformaldehyde in PBS for 15 min at RT. Cell immunofluorescence staining was performed as previously described with some modifications (15). In brief, cells were blocked with 4% BSA in PBS for 1 h at RT and then incubated overnight at 4 °C in a humidified chamber with Alexa488 dye conjugated mouse anti-GRP78 monoclonal antibody (0.4 μg, mAb159) diluted in blocking solution. Alexa488 conjugation to anti-GRP78 antibody was performed using an APEX™ Alexa Fluor® 488 antibody labeling kit (Life Technologies) and resulted in a degree of labeling of 1.23. Cells were then washed with PBS and permeabilized by 0.5% saponin in PBS for 15 min at RT followed by blocking with 4% BSA in PBS/saponin (0.01% saponin in PBS) for 1 h at RT. The cells were incubated with SNAP-Surface® Alexa Fluor® 647 (New England Biolabs) at a concentration of 1 μm in PBS/saponin for 30 min at RT followed by 4 washes with 0.5% BSA in PBS/saponin for 15 min each at RT and then a final wash with PBS before imaging.
Single Molecule Super-resolution Microscopy
Dual color three-dimensional super-resolution imaging was performed by direct stochastic optical reconstruction microscopy (STORM) (26, 27) of fixed HCT-116 cells on an inverted Nikon Eclipse Ti-E microscope equipped with TIR optics, an axial stabilizing system (Perfect Focus System, Nikon, Melville, NY), 405-, 488-, and 647-nm fiber-coupled excitation lasers, a ×100 1.49 NA objective, a dual camera light path splitter (Andor Technology, South Windsor, CT), and two iXon EMCCD cameras (Andor Technology). A quad-band pass ZET405/488/561/647× excitation filter (Chroma Technology, Bellows Falls, VT), a quad-band ZT 405/488/561/647 dichroic mirror (Chroma Technology), an emission FF640-FDi01 dichroic mirror (Semrock, Rochester, NY), and two emission filters at 525/50 (Semrock) and 700/75 (Chroma Technology) were used for the detection of photoswitchable Alexa488 and Alexa647 fluorophores, respectively. Three-dimensional imaging was achieved with an astigmatic lens inserted before the dual camera light path splitter. Channel alignment and generation of astigmatic calibration curves for both detection channels were achieved by imaging 40-nm diameter TransFluoSphere beads (488/685 nm, Life Technologies) by total internal reflection fluorescence (TIRF) on a z-piezo stage (Mad City Labs, Madison, WI) using 10-nm incremental steps across a maximum point spread function distortion range of +1 μm and −1 μm above and below the focal plan. Single molecule switching and imaging of fluorophores on labeled cells were performed in oxygen scavenging PBS buffer composed of 10% (w/v) glucose, 20 mm cysteamine (Sigma), 0.5 mg/ml glucose oxidase (Sigma), and 40 μg/ml catalase (Sigma) (28) and using 405-, 488-, and 647-nm laser lines in TIRF illumination. Both channels were acquired sequentially.
Single molecule localizations and image reconstructions were performed using the open source program rapidSTORM (Version 3.3.1) (29). The z position of each detected single molecule was determined using z-calibration curves corresponding to each respective detection channels. The localization precisions for GRP78 in x, y, and z were 9, 12, and 21 nm, respectively. The localization precisions for caveolin-1 (cav-1) in x, y, and z was 8-, 10-, and 16 nm, respectively. Super-resolved z-stacks from rapidSTORM analyses were saved as TIFF stacks in ImageJ, imported into UCSF Chimera three-dimensional rendering software (30), and displayed using surface modeling and color-coded z-depth to visualize structural information. z-Sections are displayed in 60-nm intervals, with z = 0 nm set at the estimated position of the cell plasma membrane and with positive and negative z values corresponding to localization above and below the plasma membrane, respectively.
Statistical Analysis
A two-tailed Student's t test was applied for all pairwise comparisons. Data are expressed as the mean ± S.D.
RESULTS
Cellular Distribution of Endogenous GRP78 after ER Stress Induction
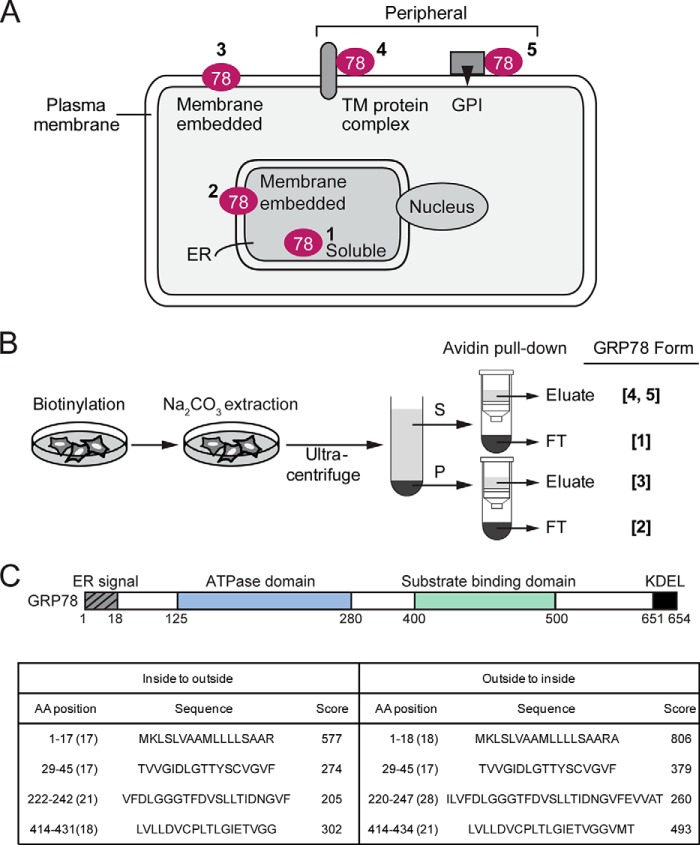
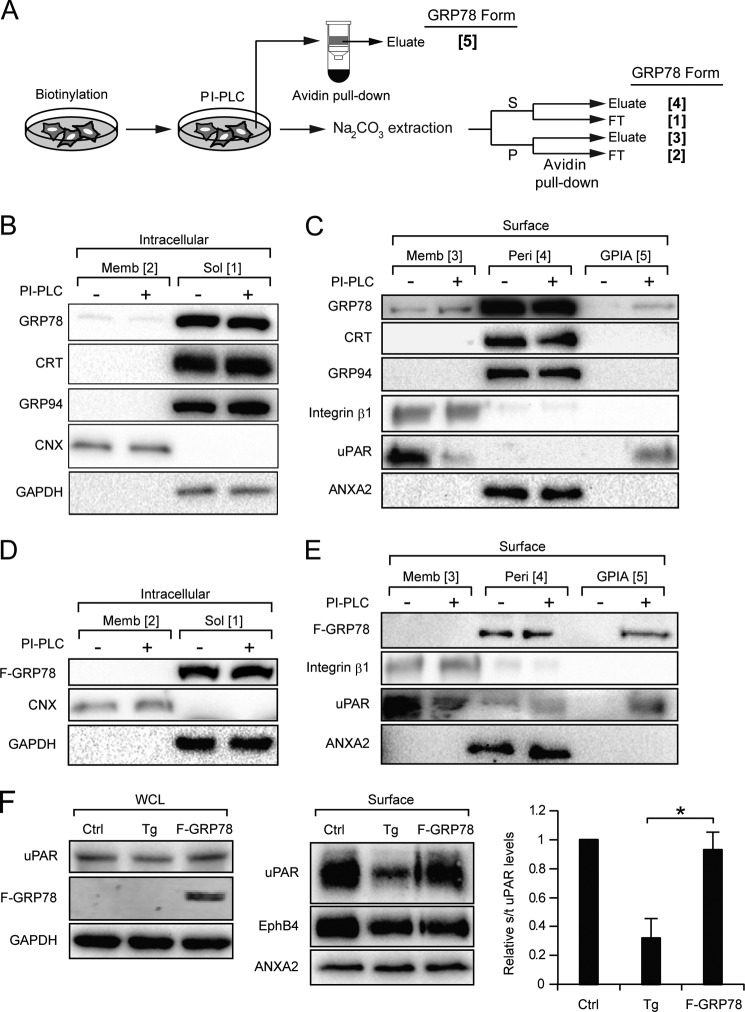
Previous studies showed that although GRP78 is primarily an ER lumenal protein, a subpopulation of GRP78 from isolated microsomes was resistant to sodium carbonate extraction and existed as a partially protease-resistant protein (31, 32), suggesting that some form of GRP78 is membrane-embedded. Thus, as depicted in Fig. 1A, in principle, GRP78 can be a soluble ER lumenal protein (Form 1) or an ER membrane-embedded protein (Form 2). Likewise, as a cell surface protein, GRP78 can be a membrane-embedded protein (Form 3), a peripheral protein associated with transmembrane protein (Form 4) or GPI-anchored protein (Form 5). To perform a comprehensive analysis of the soluble and membrane-embedded form of intracellular and cell surface GRP78, we devised a biochemical fractionation scheme to fractionate GRP78 into the five forms described above (Fig. 1B). Cell membrane-impermeable biotinylation reagent was used to specifically label the cell surface form of GRP78, which can then be purified from intracellular GRP78 by avidin pulldown. Sodium carbonate extraction was used to precipitate membrane-embedded form of GRP78 in the pellet, whereas the soluble ER lumenal GRP78 as well as peripheral form of cell surface GRP78 remain in the supernatant.
FIGURE 1.
Hypothetical cellular subpopulations of GRP78 and their fractionation scheme. A, schematic drawing of GRP78 localization in the ER lumen (Form 1), the ER membrane (Form 2), or the plasma membrane as an embedded protein (Form 3) associated with a transmembrane protein complex (Form 4), or a GPI-anchored protein (Form 5). B, fractionation scheme for the various GRP78 subpopulations depicted in A. S denotes supernatant, P denotes the pellet, and FT denotes flow-through fractions. Na2CO3 denotes sodium carbonate. C, schematic drawing of the functional domains of human GRP78 and their amino acid spans. Transmembrane helix predictions of GRP78 by TMPred are shown below. Scores >500 are considered significant.
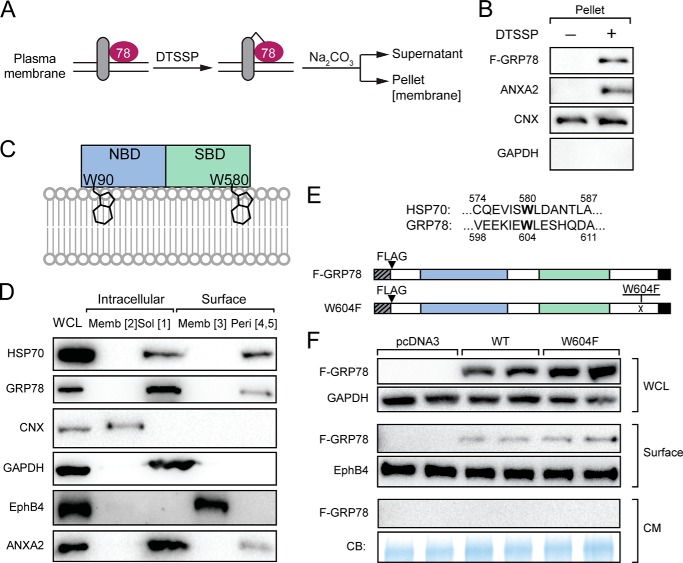
GRP78 contains several well established functional motifs/domains that include the ER signal peptide at the N terminus, the ATPase domain, the substrate binding domain, and the ER retention signal KDEL at the C terminus (2). Subjection of the primary GRP78 amino acid sequence to the TMpred program for possible transmembrane (TM) helices yielded one sequence (amino acid 1–18) containing the ER signal peptide with a significant score (>500); however, within the mature GRP78 protein, the scores for the three predicted hydrophobic regions were below the threshold for significance (Fig. 1C). Thus, the primary amino acid sequence of GRP78 does not predict a traditional TM configuration, suggesting the membrane association of GRP78 may majorly involve association with other transmembrane proteins, and this could be influenced by cellular changes mediated by ER stress.
To examine this, we utilized several mammalian cancer cell lines. HeLa cells were chosen for adherent properties suited for the fractionation protocol and high transfection efficiency, and they were used in previous proof-of-principle studies for cell surface GRP78 (9, 15). The other cell lines include MCF-7 breast adenocarcinoma cells, which expressed elevated cell surface GRP78 upon therapeutic resistance (15), and HCT-116, a colon adenocarcinoma cell line that is highly adherent. All these cell lines express suitable markers for cell surface peripheral and transmembrane proteins. Additionally, the androgen-insensitive, metastatic LNCaP-derivative C4-2B prostate adenocarcinoma cell line was also examined based on its elevated expression of cell surface GRP78 compared with the LNCaP cells (15). The cells were either cultured in normal conditions to obtain the basal expression levels of the five forms of GRP78 in non-stressed cells or subjected to three classic stimuli of ER stress including Tg, which inactivates the ER calcium ATPase thereby resulting in depletion of ER calcium, Tu, which blocks N-linked glycosylation, and 2-DG, which mimics glucose starvation; both Tu and 2-DG treatment lead to accumulation of underglycosylated proteins in the ER. After treatment, the cells were subjected to the scheme described in Fig. 1B, and the levels of GRP78 in the various fractions were determined by Western blot.
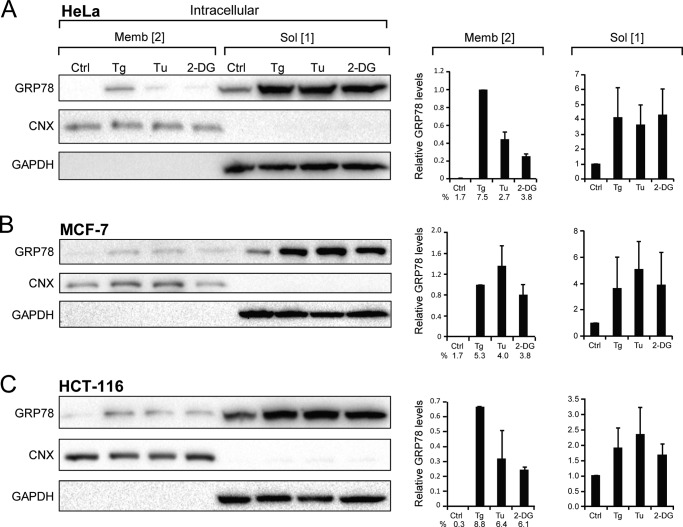
First, we examined the intracellular forms of GRP78, which were not labeled by biotinylation and collected as flow-through in the avidin pulldown assays (Fig. 1, A and B). Calnexin (CNX), a well established TM ER protein (1, 33) was the control for the purity of the membrane fraction, and GAPDH, a soluble protein, was the control for the purity of the soluble fraction containing non-membrane-embedded protein (Fig. 2, A–C). The relative GRP78 levels as the soluble and membrane-embedded forms were quantitated, normalized to the respective controls, and graphed alongside the Western blots. It is well established that the intracellular forms of GRP78 reside majorly in the ER. As expected, all three cell lines (HeLa, MCF-7, and HCT-116 cells) showed basal expression of the soluble (Form 1) of GRP78 in non-stressed cells, and upon ER stress there was an ∼4–5-fold increase for HeLa and MCF-7 cells and ∼2-fold increase for HCT-116 cells, which expressed higher basal level of GRP78. In contrast, the basal level of membrane-embedded (Form 2) intracellular GRP78 was below the detection limit in HeLa cells and very low in MCF-7 and HCT-116 cells. Strikingly, upon ER stress, membrane-embedded GRP78 (Form 2) markedly increased, in particular after Tg treatment, in all three cancer cell lines (Fig. 2, A–C).
FIGURE 2.
Distribution of intracellular forms of GRP78 under basal and ER stress conditions. HeLa (A), MCF-7 (B), and HCT-116 (C) cells were either untreated (Ctrl) or subjected to Tg, Tu, or 2-DG treatment followed by fractionation to obtain the intracellular soluble (Sol, Form 1) or membrane-embedded (Memb, Form 2). The levels of GRP78 were analyzed by Western blots, with CNX and GAPDH serving as controls for the purity of the membrane and soluble fractions, respectively. The band intensities were quantitated and normalized against the respective loading controls. In the graph denoted on the right, the level of GRP78 in the untreated cells in the membrane and soluble fractions was set as 1. For the membrane-embedded (Form 2), the percentage of GRP78 under each condition is also indicated. These were derived from dividing the intensity of (Form 2) by sum of intensities of (Form 1 and 2).
Cell Surface GRP78 Exists Majorly as a Peripheral Protein
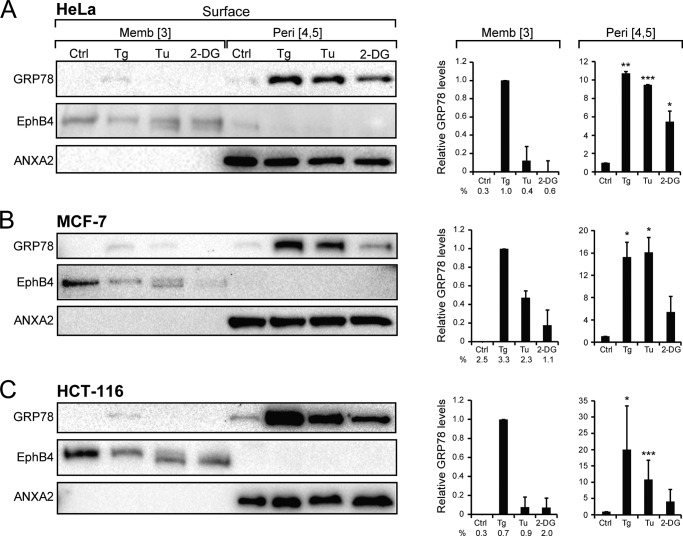
Next we examined the cell surface forms of GRP78, which were labeled by biotinylation and purified through avidin pulldown followed by elution (Fig. 1, A and B). The GRP78 level was monitored by Western blot, with EphB4, a cell surface TM receptor (34), and annexin A2 (ANXA2), a cell surface peripheral protein (35), serving as controls for the successful fractionation of the target proteins (Fig. 3, A–C). The relative GRP78 levels in the membrane and peripheral forms from multiple experiments were quantitated and graphed.
FIGURE 3.
Distribution of cell surface forms of GRP78 and other ER chaperones under basal and ER stress conditions. HeLa (A), MCF-7 (B), HCT-116 (C) cells were either untreated (Ctrl) or subjected to Tg, Tu, or 2-DG treatment followed by fractionation to obtain the plasma membrane-embedded (Form 3) or the peripheral GRP78 (Forms 4 and 5). The levels of GRP78 were analyzed by Western blots, with EphB4 and ANXA2 serving as controls for the respective fractions. The band intensities were quantitated from multiple experiments and graphed, with the level of GRP78 in the untreated cells in the membrane (Memb) and peripheral (Peri) fractions set as 1. *, p < 0.05; **, p < 0.01; ***, p < 0.005. For the membrane-embedded (Form 3), the percentage of GRP78 under each condition is also indicated. These were derived from dividing the intensity of (Form 3) by sum of intensities of (Forms 3, 4, and 5).
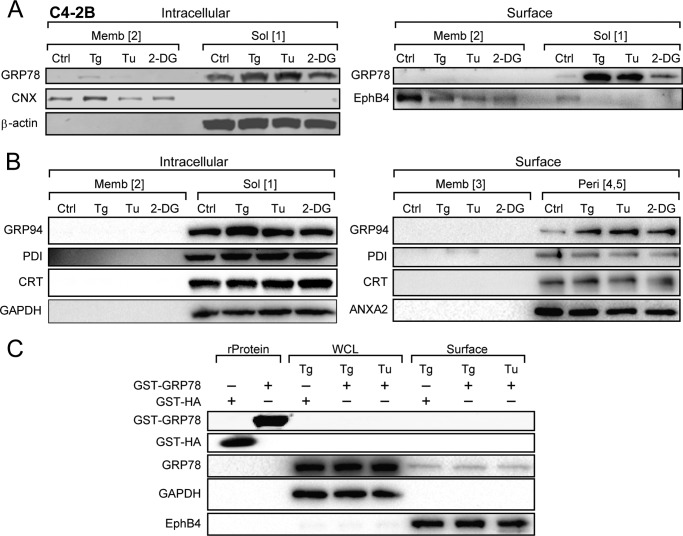
In all three cell lines, ER stress inducer Tg and Tu dramatically increased cell surface expression of GRP78 by ∼10–15-fold, with the effect of 2-DG being more variable (Fig. 3, A–C). Importantly, almost all biotinylated cell surface GRP78 was sensitive to sodium carbonate extraction, suggesting that they are peripherally associated with plasma membrane (Form 4 or 5). Similar to intracellular membrane-embedded GRP78 (Form 2), there was hardly any plasma membrane-embedded GRP78 (Form 4) in non-stressed cells; however, upon ER stress, in particular Tg treatment, membrane-embedded GRP78 was detectable in all three cell lines, albeit still at a very low level (Fig. 3, A–C). In the case of C4-2B cells, a large increase in cell surface GRP78 was detected when the cells were treated with Tg or Tu, and a moderate increase was detected with 2-DG (Fig. 4A). Interestingly, another ER lumenal chaperone, GRP94, which was ER stress-inducible, also showed substantially increased expression at the cell surface upon Tg, Tu, or 2-DG treatment. On the other hand, cell surface expression of protein disulfide isomerase (PDI) and calreticulin (CRT) was readily detected in the non-stressed cells and maintained after ER stress (Fig. 4B). GRP94, PDI, and CRT all exist as peripheral protein on the cell surface, and no membrane-embedded form was detected even after ER stress. Previously we reported that exogenously added recombinant GRP78 did not bind to the surface of non-stressed cells (9). Here, we further determined that recombinant GST-GRP78 or GST-HA were also unable to bind to the surface of ER stressed cells (Fig. 4C).
FIGURE 4.
Distribution of intracellular and cell surface forms of GRP78 in C4-2B cells and other chaperones under basal and ER stress conditions in HeLa cells. A, C4-2B cells were either untreated (Ctrl) or treated with Tg, Tu, or 2-DG treatment and subjected to biotinylation and fractionation to isolate the various forms of GRP78. The levels of GRP78 were analyzed by Western blots, with β-actin, CNX, and EphB4 serving as controls for the respective fractions. B, Western blot analysis of GRP94, CRT, and PDI in each fraction under the conditions indicated. C, HeLa cells were treated with Tg or Tu, and 10 μg of GST-GRP78 or GST-HA was added to the cell culture at the same time. After 16 h, whole cell lysate (WCL) and cell surface proteins were isolated, and endogenous GRP78, recombinant GST-GRP78, and GST-HA were analyzed by Western blots along with GAPDH and EphB4 serving as controls for the respective fractions. Purified recombinant protein (rProtein) GST-HA and GST-GRP78 (1 μg) were loaded in the first two lanes as controls.
To determine whether GRP78 binds to the cell surface via association with GPI-anchored protein (Form 5), we utilized PI-PLC to cleave off the GPI anchor, thereby releasing the GPI-anchored protein and its associating peripheral proteins from the cell surface. The scheme for the separation of the GPI-anchored protein-associated (Form 5) GRP78 from the other forms (Forms 1–4) was summarized in Fig. 5A. In this series of experiments, the HeLa cells were treated with Tg to enhance GRP78 cell surface expression, and urokinase plasminogen activator receptor (uPAR), a known GPI-anchored protein, was used as the control for its release after PI-PLC treatment. For comparison, GRP94 and CRT were also monitored in parallel. First, we established that treatment of cells with PI-PLC in PBS for 1 h had no effect on the intracellular levels of these ER proteins (Fig. 5B). Upon PI-PLC treatment, uPAR was detectable in the released fraction as expected, and GRP78 was also detectable, albeit at a low level (∼4.5% of total peripheral GRP78), whereas GRP94 and CRT were either not released or were below our detection limit. Integrin β1 and ANXA2, not known to associate with GPI-anchored proteins, were not released (Fig. 5C).
FIGURE 5.
A fraction of cell surface GRP78 complexes with GPI-anchored proteins. A, fractionation scheme for isolating the GPI-anchored protein-associated cell surface GRP78 (Form 5) from the other forms of GRP78 (Forms 1–4). PI-PLC was used to cleave the GPI anchor, thereby releasing the GPI-anchored proteins and their associating proteins (GPIA) into the PBS buffer. S denotes the supernatant, P denotes the pellet, and FT denotes the flow-through. B and C, HeLa cells treated with Tg to promote cell surface GRP78 localization were subjected to biotinylation, PI-PLC treatment, and fractionation as shown in A. The levels of GRP78, CRT, GRP94, and CNX in each fraction under the conditions indicated were determined by Western blots. B, for the intracellular forms, CNX and GAPDH served as controls for the purity of the membrane (Memb) and soluble (Sol) fractions, respectively. C, for the cell surface forms, integrin β1 served as marker for the membrane-embedded fraction, ANXA2 for the peripheral (Peri) fraction, and uPAR, a GPI-anchored protein, served as positive control for PI-PLC cleavage and release, and GAPDH served as negative control for release of cytosolic protein into the buffer. D and E, same as B and C except the HeLa cells were transfected with the F-GRP78 expression vector, and the fraction of ectopic F-GRP78 as GPIA was assayed as above. F, HeLa cells were either untreated (Ctrl), treated with Tg or transfected with FLAG-GRP78-expressing vector (F-GRP78). Whole cell lysate (WCL) and surface proteins were isolated, and uPAR levels were analyzed by Western blot. Quantitation of cell surface uPAR level to total uPAR level is shown in the right panel. *, p < 0.05.
To confirm these results with epitope-tagged GRP78, we transfected HeLa cells with the expression vector for FLAG-GRP78 (F-GRP78) and performed the same fractionation. As shown in Fig. 5, D and E, F-GRP78, which exists majorly as peripheral cell surface protein as endogenous GRP78, was clearly detectable in the released fraction after PI-PLC treatment, constituting ∼30% of total peripheral F-GRP78. Although collectively these results establish that a subpopulation of peripheral GRP78 is bound on the cell surface by association with GPI-anchored proteins, the fraction of peripheral GRP78 binding to GPI-anchored proteins in cells subjected to Tg treatment is substantially less. In view of the recent report that ER stress induced acute clearance of mutant, misfolded GPI-anchored proteins via the secretory pathway (36), we investigated whether wild-type GPI-anchored proteins such as uPAR was also affected. Our results showed that upon Tg treatment, the amount of uPAR at the cell surface was also decreased (Fig. 5F), Thus, the reduction of GPI-anchored proteins on the cell surface after ER stress could contribute to their reduced interaction with peripheral GRP78.
Differential Mechanism for GRP78 Cell Surface Translocation in Different Cancer Cell Lines
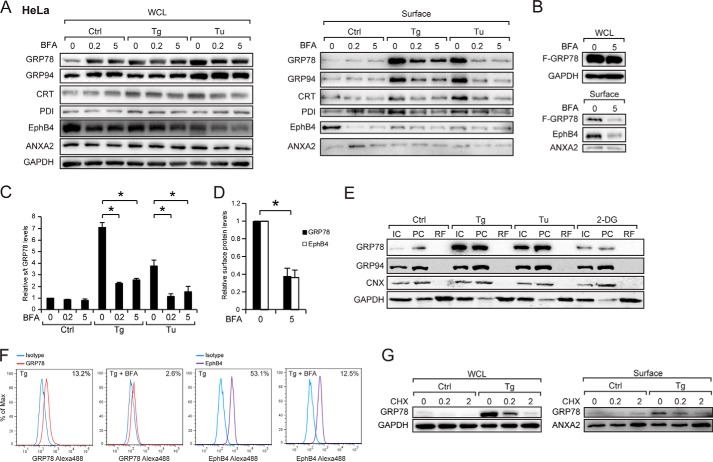
To determine whether the translocation of GRP78 to cell surface utilizes the traditional secretory ER to Golgi traffic, we treated the cells with BFA, which specifically blocks protein transport from the ER to the Golgi apparatus and causes resorption of Golgi membrane (37). Interestingly, BFA, which causes accumulation of proteins in the ER, is itself an ER stress inducer and has been reported to induce GRP78 expression (38). In the following series of experiments, we compared the whole cell and cell surface expression levels of GRP78 and other ER proteins in HeLa cells treated with increasing doses of BFA in the absence or presence of ER stress stimuli (Tg and Tu). Our results showed treatment of cells alone with BFA induced intracellular GRP78 and GRP94 but not CRT or PDI (Fig. 6A). BFA suppressed cell surface expression of GRP78, GRP94, CRT, and PDI, most notably after Tg and Tu treatment (Fig. 6A), as well as ectopically expressed F-GRP78 at the cell surface (Fig. 6B). As controls, BFA suppressed the cell surface expression of EphB4 known to traffic through the ER-Golgi apparatus but has no effect on cell surface expression of ANXA2 known to traffic to the cell surface via a mechanism independent of the classical ER-Golgi pathway (39). Quantitation of the Western blot results showed that BFA reduced Tg- and Tu-induced surface GRP78 by ∼60–70% (Fig. 6C) and EphB4 by similar amounts (Fig. 6D).
FIGURE 6.
Stress-induced cell surface GRP78 expression in HeLa cell requires Golgi integrity and new protein synthesis. A, HeLa cells were either untreated (Ctrl) and treated with Tg or Tu as well as increasing amounts of BFA, either alone or in combination as indicated. Whole cell lysate (WCL) and cell surface proteins were isolated, and the indicated ER chaperone levels were analyzed by Western blot along with EphB4 and ANXA2 as BFA-sensitive and -insensitive controls respectively. B, same as A except the HeLa cells were transfected with the FLAG-GRP78 expression vector and treated with the indicated amount of BFA. C, the experiments in A were repeated three times, and the levels of cell surface GRP78 relative to total GRP78 levels under each condition were graphed with the level of the untreated cells set as 1. *, p < 0.05. D, the experiments in B were repeated two times, the relative levels of cell surface F-GRP78 and EphB4 were normalized against ANXA2 were graphed, with the levels of the untreated cells were set as 1. *, p < 0.05. E, HeLa cells treated as indicated were subjected to cell permeabilization assay. The levels of GRP78, GRP94, CNX, and GAPDH were determined by Western blot. IC denotes intact cell, PC denotes permeabilized cells, and RF released fraction. F, FACS analysis of cell surface GRP78 and EphB4 in HeLa cells with or without BFA treatment. Blue line, IgG1 isotype control; red line, anti-GRP78 antibody; purple line, anti-EphB4 antibody. The percentage of cells expressing cell surface GRP78 or EphB4 is indicated at the right upper corner of each FACS profile. G, HeLa cells were either untreated (Ctrl) or treated with Tg as well as increasing amounts of cyclohexamide, either alone or in combination as indicated. Whole cell lysate and cell surface proteins were isolated. The levels of GRP78, ANXA2, and GAPDH were analyzed by Western blot.
Another potential mechanism for ER proteins to transit to the cell surface is through translocation into the cytosol followed by exit to the cell membrane. To test whether ER stress induces GRP78 release into the cytosol, we performed a cell permeabilization assay where upon treatment of intact cells with 0.005% digitonin, cytosolic proteins were recovered in the “released fraction” and non-cytosolic proteins in the “permeabilized cell” fraction (24, 40). GAPDH served as a cytosolic protein control, and the ER transmembrane protein CNX served as the non-cytosolic control. Our results showed that GRP78, as well as GRP94, were not detected in the released fraction either before or after Tg, Tu, or 2-DG treatment (Fig. 6E). FACS assays were performed to independently confirm that in HeLa cells surface GRP78 and EphB4 expression were suppressed by BFA after Tg treatment (Fig. 6F). Furthermore, Tg-induced translocation of GRP78 to the cell surface was suppressed by treatment of cells with cycloheximide (Fig. 6G). In contrast, cell surface ANXA2 was up-regulated by cyclohexamide treatment as previously reported (39). Collectively, these results demonstrated that in HeLa cells GRP78 trafficking to the cell surface is at least in part dependent on the integrity of the ER and Golgi apparatus and new protein synthesis.
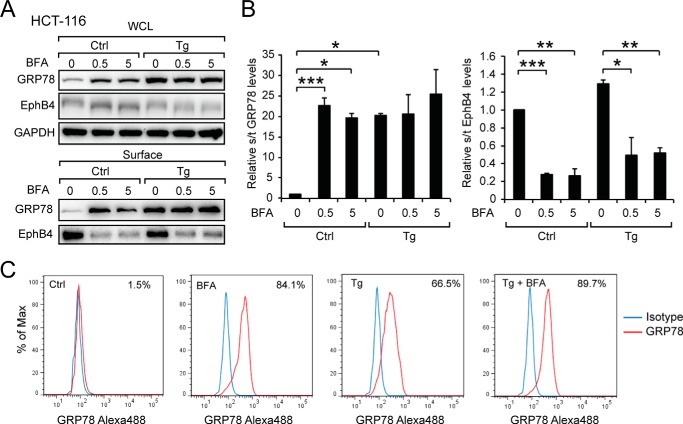
To test whether this finding also applies to other cancer cell lines, we tested the effect of BFA in the HCT-116 colon cancer cell line. At the intracellular level, BFA induced GRP78 expression as was observed for HeLa cells (Fig. 6, A and B). Cell surface GRP78 expression in HCT-116 cells cultured in the presence of BFA was readily detectable biochemically and by FACS (Fig. 7, A–C). In contrast to HeLa cells, Tg-induced cell surface GRP78 expression was not affected by BFA even at high dose, whereas BFA effectively blocked EphB4 surface translocation (Fig. 7, A and B). Thus, HCT-116 appears to be able to utilize a BFA-independent mechanism to transport GRP78 to the cell surface before and after ER stress.
FIGURE 7.
Stress-induced cell surface GRP78 expression in HCT-116 cell is insensitive to brefeldin A. A, HCT-116 cells were either untreated (Ctrl) or treated with Tg as well as increasing amounts of BFA, either alone or in combination as indicated. Whole cell lysate (WCL) and cell surface proteins were isolated, and the levels of GRP78, EphB4, and GAPDH were analyzed by Western blot. B, quantitation of cell surface GRP78 level to total GRP78 level and surface EphB4 level to total EphB4 level. The experiments were repeated four times. *, p < 0.05; **, p < 0.01; ***, p < 0.005. C, cell surface GRP78 was detected by FACS. The percentage of cells expressing cell surface GRP78 under each condition is indicated at right upper corner of each FACS profile. The blue line denotes IgG1 isotype control, and the red line denotes anti-GRP78 antibody.
Cell Surface GRP78 Expression Requires Its Substrate Binding Property
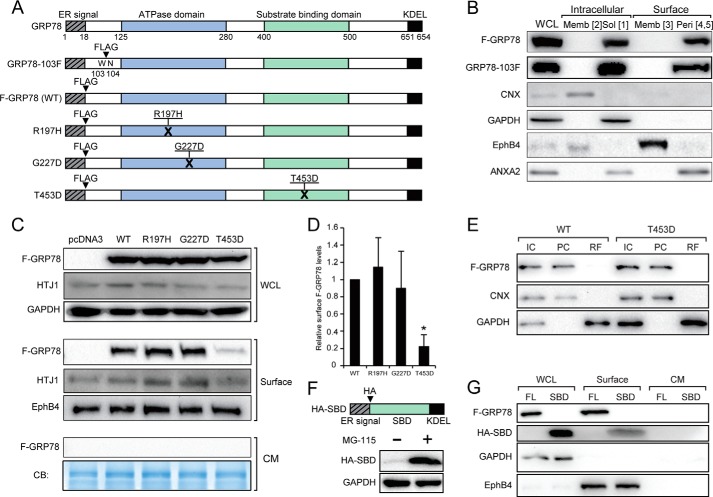
GRP78 contains an ATP binding domain required for its ATPase catalytic activity in protein folding and a substrate binding domain required for interaction with its client proteins (Fig. 8A). To probe whether either one or both of these domains are required for cell surface localization, we utilized the F-GRP78 as the parental construct for subsequent mutant creations. First, we showed that the N-terminal location of the FLAG tag in F-GRP78 did not affect its property as a cell surface peripheral protein, as both F-GRP78 and GRP78–103F, where the FLAG tag was internally inserted, showed similar expression and localization profiles for intracellular soluble (Form 1) and peripheral cell surface protein (Forms 4 and 5) (Fig. 8B). We noted that in contrast to endogenous GRP78, ectopically expressed F-GRP78 and GRP78–103F were not detected in the membrane-embedded form either intracellularly (Form 2) or at the cell surface (Form 3).
FIGURE 8.
Mutational analysis of GRP78 for cell surface translocation. A, the locations of the ER signal peptide, ATPase domain, substrate binding domain, and KDEL motifs of human GRP78 are indicated. FLAG (F) epitope was either tagged at the N terminus or inserted between Trp-103 and Asn-104. For the endoplasmic reticulum DnaJ homologue interaction-defective mutant (R197H), Arg-197 was replaced by His. For ATP binding-defective mutant (G227D), Gly-227 was replaced by Asp. For substrate binding-defective mutant (T4530), Thr-453 was replaced by Asp. B, HeLa cells were transfected with FLAG-GRP78 or GRP78-F103 vector and subjected to biotinylation and fractionation to isolate the various forms of GRP78. The levels of indicated proteins in various fractions were determined by Western blot. WCL, whole cell lysate; Memb, membrane; Sol, soluble; Peri, peripheral. C, HeLa cells were transfected with the indicated vectors. After 24 h the cells were switched to serum-free medium and cultured for another 24 h. The whole cell lysate, the cell surface proteins, and the conditioned medium (CM) were subjected to Western blot analysis for the indicated proteins. The conditioned medium was further subjected to Coomassie Blue (CB) staining. D, quantification of relative cell surface expression of wild-type F-GRP78 and the mutants normalized against EphB4. *, p < 0.05. E, HeLa cells transfected with indicated vectors were subjected to cell permeabilization assay. The indicated proteins were analyzed by Western blot. IC denotes intact cell, PC denotes permeabilized cell, and RF denotes released fraction. F, schematic drawing of HA-SBD with the locations of the ER signal peptide, HA tag, SBD, and KDEL motif indicated. HA-SBD could be expressed in the presence of proteasome inhibitor MG-115. G, HeLa cells were transfected with FLAG-GRP78 (FL) or HA-SBD. Whole cell lysates, cell surface proteins, and the conditioned medium were subjected to Western blot analysis for the indicated proteins.
We then created the R197H mutant, which renders GRP78 unable to associate with co-chaperone DnaJ proteins (41), the G227D mutant unable to bind ATP (42), and the T453D mutant unable to bind protein substrates (43) (Fig. 8A). In all these mutants, the KDEL retention motif is retained. Upon transfection into HeLa cells, these constructs expressed the GRP78 protein mutants, as shown by equivalent expression levels as the wild-type protein in the whole cell lysates, and none of the proteins were secreted into the culture medium (Fig. 8C). Strikingly, although the F-GRP78(WT), R197H, and G227D were able to localize to the cell surface, T453D uniquely exhibited an 80% reduction in surface expression (Fig. 8D). Thus, the substrate binding domain of GRP78 is paramount in its cell surface expression. We further determined that as in the case for F-GRP78(WT), T453D was not released in the cytosol (Fig. 8E) and that HTJ-1, the human homologue of MTJ-1, which is one of the substrate proteins required for GRP78 surface expression in macrophages (44), was expressed in HeLa cells and detectable on the cell surface (Fig. 8C).
We next tested whether the SBD domain alone is able to translocate from the ER to the cell surface. To achieve this, we constructed an expression vector for the GRP78 SBD, which contains an ER signal peptide followed by an HA epitope at its N terminus and a KDEL motif at its C terminus (Fig. 8F). After transfection, HA-SBD expression could be stabilized by treatment of cells with the proteasome inhibitor MG-115 and was detectable at the cell surface at ∼30% that of the level of full-length F-GRP78 (Fig. 8, F and G).
GRP78 Cell Surface Localization Is Independent of the “Membrane Insertion” Motif Conserved with HSP70
Potential mechanisms for peripheral GRP78 to retain on the cell membrane may rely on complex formation with TM proteins or direct binding to the lipid bilayer of the plasma membrane. To test these, HeLa cells were transfected with F-GRP78 expression vector and treated with membrane-impermeable DTSSP, which cross-links proteins by reacting with the primary amines of lysine or the N terminus of proteins in proximity followed by sodium carbonate extraction (Fig. 9A). Upon cross-linking of peripheral GRP78 with its partner TM proteins, it would become sodium carbonate-resistant and retained in the membrane. As shown in Fig. 9B, we observed that F-GRP78 was recovered in the membrane (pellet) fraction after DTSSP treatment, which was also observed for ANXA2 known to bind to TM proteins (45). For CNX, which is a TM protein, it was present in the pellet fraction before and after DTSSP cross-link as expected. GAPDH, an intracellular soluble protein, was not detected at all in the pellet fraction. These results provide additional evidence that GRP78 formed a complex with TM proteins.
FIGURE 9.
GRP78 association with plasma membrane was enhanced by protein cross-linking but unaffected by W604F substitution mutation. A, schematic drawing of protein cross-linking by DTSSP and fractionation. Peripheral proteins cross-linked with transmembrane protein complexes would become sodium carbonate (Na2CO3)-resistant during extraction and recovered in the pellet. B, HeLa cells transfected with F-GRP78 vector were subjected to DTSSP treatment as described in A. The levels of F-GRP78 and the indicated marker proteins were determined by Western blot analysis. C, schematic drawing of HSP70 association with membrane via tryptophan insertions (Trp-90 and Trp-580) into the lipid bilayer. NBD and SBD denote nucleotide and substrate binding domains, respectively. D, HeLa cells were subjected to biotinylation and Na2CO3 extraction, and the various fractions of intracellular and cell surface proteins as indicated were isolated. The levels of the indicated proteins were analyzed by Western blot. Memb, membrane; Sol, soluble; Peri, peripheral. E, Trp-580 of HSP70 is conserved and corresponds to Trp-604 of GRP78. To mutate this amino acid, Trp-604 of F-GRP78 was replaced by Phe, creating the W604F mutant. F, HeLa cells were transfected with the indicated vectors. After 24 h, the cells were switched to serum-free medium and cultured for another 24 h. The whole cell lysate (WCL), the cell surface proteins, and the conditioned medium (CM) were subjected to Western blot analysis for the indicated proteins. The conditioned medium was further subjected to Coomassie Blue (CB) staining.
GRP78 belongs to the HSP70 superfamily, and the two proteins contain both a nucleotide binding domain and a substrate binding domain. Recently, it has been proposed based on in vitro assays that HSP70 can peripherally associate with the membrane via direct insertion into the lipid bilayer mediated by tryptophan residue Trp-90 in the nucleotide binding domain and Trp-580 in the substrate binding domain (46) (Fig. 9C). In HeLa cells, we confirmed that as observed for GRP78, HSP70 exists as a peripheral protein on the cell surface (Fig. 9D). Protein sequence alignment showed that Trp-580, but not Trp-90, of HSP70 is conserved and corresponds to Trp-604 of GRP78 (Fig. 9E). Therefore, to test whether GRP78 and HSP70 share this phospholipid insertion mechanism for membrane association, Trp-604 in GRP78 was replaced by phenylalanine to create the GRP78 W604F mutant, which corresponds to the HSP70 W580F mutant that abolishes its lipid insertion property (46) (Fig. 9E). HeLa cells were transfected with wild-type F-GRP78 or W604F, and surface protein fractions were pulled down, and conditioned medium were collected. Our results showed that as compared with the F-GRP78, the amount of GRP78 associating with the cell membrane remained unchanged in W604F, and no secretion of W604F was detected (Fig. 9F). This suggests that Trp-604 with potential lipid insertion property is dispensable for GRP78 cell surface expression.
Single Molecule Super-resolution Imaging of Cell Surface GRP78
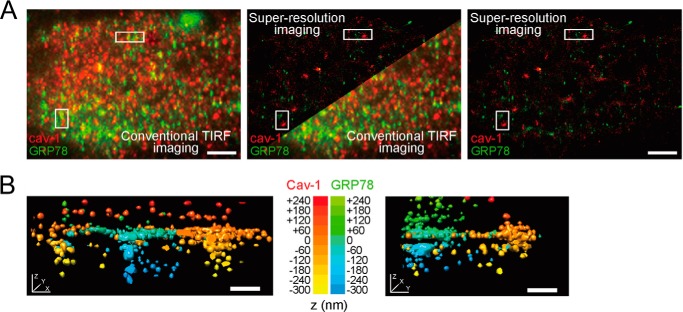
To detect the presence of GRP78 at the plasma membrane of intact cells with extremely high sensitivity, we imaged Tg-stressed HCT-116 cells by single molecule super-resolution TIRF microscopy. We performed three-dimensional and dual-color STORM imaging (26, 27) of endogenous GRP78 and compared its cell surface distribution with that of cav-1. cav-1 is a well characterized integral membrane protein that can form ∼150-nm-deep caveolae invaginations at the plasma membrane (47) and is overexpressed in HCT-116 cells (48). Consistent with our biochemical data, GRP78 could be readily detected at the plasma membrane of stressed HCT-116 cells by both conventional TIRF microscopy and single molecule super-resolution imaging (Fig. 10A). In super-resolved three-dimensional renderings, cav-1 was associated with membrane-lined caveolar invaginations a few hundreds of nanometers below the cell surface and with flatter plasma membrane areas (Fig. 10B and supplemental Movies 1 and 2). Although GRP78 very rarely co-localized with cav-1 (Fig. 10A), it was positioned in the same z-plane as flat cav-1-rich plasma membrane areas as well as within intracellular structures adjacent to the plasma membrane (Fig. 10B and supplemental Movies 1 and 2). Although the exact nature of GRP78-rich subplasma membrane structures remains to be determined, these results provide direct evidence that GRP78 localizes to the plasma membrane of HCT-116 cells after ER stress.
FIGURE 10.
Dual color super-resolution TIRF imaging of GRP78 and caveolin-1 at the plasma membrane of an ER-stressed HCT-116 cell. A, conventional TIRF (left) and super-resolved (right) dual color images of GRP78 (green) and caveolin-1 (cav-1, red) displayed as overlaid two-dimensional projections (x and y planes) of all z-planes. Scale bars: 2 μm. B, three-dimensional renderings of cav-1-rich (red-to-yellow z-gradient) and GRP78-rich (green-to-blue z-gradient) membrane structures within the super-resolved image (left, upper white rectangle in A; right, lower white rectangle in A). Scale bars: 200 nm.
DISCUSSION
The discovery that traditional ER chaperone lumenal proteins are capable of expressing on the cell surface and assuming critical functions in health and disease represents a paradigm shift on our view on this class of proteins (4). Although the regulation of their expression at the transcriptional and translational level is important, their relocalization to different cellular compartments under physiological and pathological conditions could also dictate how they control vital biological pathways and how these can be exploited for therapeutic intervention. For example, upon presentation on the surface of apoptotic cells, CRT serves as the “eat me” signal, triggering specific T cell-mediated immune response against these apoptotic cells leading to phagocytosis (49).
Here in this study we focus on cell surface GRP78 where a large volume of recent studies provide compelling evidence that it acts as a crucial regulator of multiple cellular signaling pathways implicated in survival and apoptosis as well as a vital co-receptor for viral entry into host cells (1, 2, 4, 6, 8, 18). Cell surface GRP78, through its interaction with α-synuclein, is also linked to regulating the dynamics of actin cytoskeleton integrity in neurons and onset of neurodegeneration (50). A role of cell surface GRP78 in tissue factor-mediated initiation of coagulation cascade has also been reported in endothelial and cancer cells (51, 52). Therefore, understanding how GRP78 is presented on the cell surface can lead to novel approaches for therapeutic interventions for cancer and other human diseases. Here, in this report, we uncovered salient features of GRP78 relocalization that are unconventional, and our findings provide new insights into the cell response to ER stress targeting protein homeostasis.
A major discovery is that cell surface GRP78 as well as other chaperones exist primarily as a peripheral protein and a small subpopulation of cell surface GRP78 is tethered by GPI-anchored proteins. This is consistent with the TMpred analysis that GRP78 is unlikely to possess TM configuration under normal physiologic conditions and explains the accessibility of both the N and C termini as well as previously predicted internal cytosolic domains to antibodies, peptides, and toxins applied externally to the cells targeting GRP78 (2, 8, 9, 13, 16, 18, 53). The observation that a fraction of GRP78 can be released from the cell surface after PI-PLC treatment is in agreement with the association of GRP78 with GPI-anchored proteins such as Cripto, and as such, regulates stem cell viability and oncogenesis (12, 54). Nonetheless, our study showed that only a very small fraction of endogenous GRP78 is bound to the cell surface through association with GPI-anchored proteins upon ER stress, and this could be in part due to the reduction of some GPI-anchored proteins from the plasma membrane in stressed cells as was observed with uPAR in this study.
This raises the interesting question of how the majority of GRP78s stay bound to the cell surface, as we were unable to detect any amount of GRP78 spontaneously secreted into the medium either before and after ER stress in the cell lines being examined. Through mutational analysis, we discovered that the GRP78 substrate binding activity is required for cell surface translocation of GRP78, whereas the ATP binding did not appear to be obligatory. This implies that GRP78 traffics to the cell surface via association with its client proteins. Previous studies have indicated that specific cell types may utilize different proteins for transporting GRP78 to the cell surface. For example, the ER TM protein, MTJ-1, has been reported as a GRP78 carrier in macrophages (44) and the tumor suppressor Par-4 in PC-3 cells (55). Thus, different substrate proteins are likely to partner with GRP78 in a cell context-dependent manner for its translocation to the cell surface and maintain it on the cell surface. Recently, it has been proposed based on in vitro assays that HSP70, a major cytosolic chaperone that shares 60% sequence homology with GRP78, could be anchored on the cell surface as peripheral protein by inserting the tryptophan residue contained within its substrate binding domain into the lipid bilayer (46). Here we confirmed that HSP70 exists as a peripheral protein on the surface of HeLa cells, and this amino acid was conserved in GRP78. Nonetheless, mutation of this conserved tryptophan has no impact on GRP78 cell localization, suggesting that this residue is not required for GRP78 cell surface expression. In contrast, cross-linking with membrane-impermeable agent resulted in substantial increase in GRP78 cell surface expression, providing additional evidence that GRP78 anchoring on the cell surface is protein-dependent.
In determining the route(s) whereby GRP78 translocates to the cell surface, we observed that ER stress-induced increase in cell surface GRP78 was substantially reduced when the ER to Golgi transport was blocked by BFA in HeLa but not in HCT-116 cells. This suggests that there are alternative routing mechanisms for the translocation of GRP78 to the cell surface independent of ER to Golgi transport, and the transport routes could be cell type- and context-dependent.
Although present at a very low level, membrane-embedded GRP78, which is sodium carbonate-resistant, was detected both intracellularly (Form 2) and at the cell surface (Form 3), and these forms were actively promoted by subjecting the cells to various ER stress-inducing agents, in particular, by Tg, which caused ER calcium efflux, in all three cell lines that we tested. The trapping of GRP78 in the ER membrane could be a consequence of translational pausing (56) or that under ER stress conditions, GRP78 is misfolded and exists as an atypical form trapped in the membrane, the composition or structure of which may also be altered under ER stress. Lastly, through the use of ultrasensitive super-resolution microscopy, we provided nanometer precision and three-dimensional mapping of distinct GRP78 patterns on the cell surface with respect to the well characterized plasma membrane protein cav-1. Compared with conventional TIRF imaging, the increased optical resolution afforded by super-resolution microscopy allowed us to confirm that ER-stressed HCT-116 cells showed robust localization of GRP78 at the plasma membrane. Because the ER membrane serves as a source for the plasma membrane and ER may fuse with the plasma membrane, it is tempting to speculate that the cell surface GRP78 membrane-embedded form (Form 3) originates from its intracellular counterpart (Form 2), and this awaits future validation. The presence of some GRP78 just below the plasma membrane is consistent with active trafficking of intracellular GRP78 to the cell surface as well as the ability of Tg to induce ER-plasma membrane contact sites (57). In summary, our studies provide the first evidence that cell surface GRP78 exists primarily as a peripheral protein and establishes the critical parameters for its cell surface localization. As cell surface GRP78 is increasingly recognized as a target for anti-cancer therapy while sparing normal organs, its mode of cell surface translocation has great implications for therapeutic interventions.
Supplementary Material
Acknowledgments
We thank Drs. Parkash Gill and Ren Liu for the gift of antibodies and reagents and James Ou, Tobias Ulmer, Erik Snapp, and Linda Hendershot for helpful discussions. Flow cytometry was performed in the USC Flow Cytometry Core Facility supported in part by the National Cancer Institute Cancer Center Shared Grant P30 CA014089 and the USC Provost Office, Dean's Development Funds, Keck School of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA027607, R21 CA179273, and P01 AG034906 (to A. S. L.).

This article contains supplemental Movies 1 and 2.
- GRP78
- 78-kDa glucose-regulated protein
- F-GRP78
- FLAG-GRP78
- HSP70
- heat shock protein 70
- ER
- endoplasmic reticulum
- GPI
- glycosylphosphatidylinositol
- Tg
- thapsigargin
- Tu
- tunicamycin
- 2-DG
- 2-deoxy-d-glucose
- SBD
- substrate binding domain
- RIPA
- radioimmune precipitation assay buffer
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- RT
- room temperature
- STORM
- stochastic optical reconstruction microscopy
- TIRF
- total internal reflection fluorescence
- CNX
- calnexin
- EphB4
- ephrin type-B receptor 4
- ANXA2
- annexin A2
- PDI
- protein disulfide isomerase
- CRT
- calreticulin
- uPAR
- urokinase plasminogen activator receptor
- cav-1
- caveolin-1
- BFA
- brefeldin A
- TM
- transmembrane
- DTSSP
- 3,3′-dithiobis[sulfosuccinimidylpropionate].
REFERENCES
- 1. Ni M., Lee A. S. (2007) ER chaperones in mammalian development and human diseases. FEBS Lett. 581, 3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee A. S. (2014) Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat. Rev. Cancer 14, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Munro S., Pelham H. R. (1986) An Hsp70-like protein in the ER: identity with the 78-kDa glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 4. Luo B., Lee A. S. (2013) The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 32, 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332 [DOI] [PubMed] [Google Scholar]
- 6. Ni M., Zhang Y., Lee A. S. (2011) Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J. 434, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arap M. A., Lahdenranta J., Mintz P. J., Hajitou A., Sarkis A. S., Arap W., Pasqualini R. (2004) Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell 6, 275–284 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalez-Gronow M., Selim M. A., Papalas J., Pizzo S. V. (2009) GRP78: a multifunctional receptor on the cell surface. Antioxid. Redox Signal. 11, 2299–2306 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Liu R., Ni M., Gill P., Lee A. S. (2010) Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 285, 15065–15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misra U. K., Gonzalez-Gronow M., Gawdi G., Hart J. P., Johnson C. E., Pizzo S. V. (2002) The role of Grp 78 in α2-macroglobulin-induced signal transduction. Evidence from RNA interference that the low density lipoprotein receptor-related protein is associated with, but not necessary for, GRP 78-mediated signal transduction. J. Biol. Chem. 277, 42082–42087 [DOI] [PubMed] [Google Scholar]
- 11. Shani G., Fischer W. H., Justice N. J., Kelber J. A., Vale W., Gray P. C. (2008) GRP78 and Cripto form a complex at the cell surface and collaborate to inhibit transforming growth factor β signaling and enhance cell growth. Mol. Cell. Biol. 28, 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spike B. T., Kelber J. A., Booker E., Kalathur M., Rodewald R., Lipianskaya J., La J., He M., Wright T., Klemke R., Wahl G. M., Gray P. C. (2014) CRIPTO/GRP78 signaling maintains fetal and adult mammary stem cells ex vivo. Stem Cell Reports 2, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miao Y. R., Eckhardt B. L., Cao Y., Pasqualini R., Argani P., Arap W., Ramsay R. G., Anderson R. L. (2013) Inhibition of established micrometastases by targeted drug delivery via cell surface-associated GRP78. Clin. Cancer Res. 19, 2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasche L., Duell J., Morgner C., Chatterjee M., Hensel F., Rosenwald A., Einsele H., Topp M. S., Brändlein S. (2013) The natural human IgM antibody PAT-SM6 induces apoptosis in primary human multiple myeloma cells by targeting heat shock protein GRP78. PLoS ONE 8, e63414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Tseng C. C., Tsai Y. L., Fu X., Schiff R., Lee A. (2013) Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PLoS ONE 8, e80071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu R., Li X., Gao W., Zhou Y., Wey S., Mitra S. K., Krasnoperov V., Dong D., Liu S., Li D., Zhu G., Louie S., Conti P. S., Li Z., Lee A. S., Gill P. S. (2013) Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin. Cancer Res. 19, 6802–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Ridder G. G., Ray R., Pizzo S. V. (2012) A murine monoclonal antibody directed against the carboxyl-terminal domain of GRP78 suppresses melanoma growth in mice. Melanoma Res. 22, 225–235 [DOI] [PubMed] [Google Scholar]
- 18. Sato M., Yao V. J., Arap W., Pasqualini R. (2010) GRP78 signaling hub a receptor for targeted tumor therapy. Adv. Genet. 69, 97–114 [DOI] [PubMed] [Google Scholar]
- 19. Shin B. K., Wang H., Yim A. M., Le Naour F., Brichory F., Jang J. H., Zhao R., Puravs E., Tra J., Michael C. W., Misek D. E., Hanash S. M. (2003) Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 278, 7607–7616 [DOI] [PubMed] [Google Scholar]
- 20. Pinaud F., Dahan M. (2011) Targeting and imaging single biomolecules in living cells by complementation-activated light microscopy with split-fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E201–E210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okamoto T., Schwab R. B., Scherer P. E., Lisanti M. P. (2001) Analysis of the association of proteins with membranes. Curr. Protoc. Cell Biol. 10.1002/0471143030.cb0101s05 [DOI] [PubMed] [Google Scholar]
- 22. Liu R., Ferguson B. D., Zhou Y., Naga K., Salgia R., Gill P. S., Krasnoperov V. (2013) EphB4 as a therapeutic target in mesothelioma. BMC Cancer 13, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu Y., Li J., Lee A. S. (2007) GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen-starvation induced apoptosis. Cancer Res. 67, 3734–3740 [DOI] [PubMed] [Google Scholar]
- 24. Ni M., Zhou H., Wey S., Baumeister P., Lee A. S. (2009) Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS ONE 4, e6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tamilselvam B., Daefler S. (2008) Francisella targets cholesterol-rich host cell membrane domains for entry into macrophages. J. Immunol. 180, 8262–8271 [DOI] [PubMed] [Google Scholar]
- 26. Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M. (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 [DOI] [PubMed] [Google Scholar]
- 27. Huang B., Wang W., Bates M., Zhuang X. (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dempsey G. T., Vaughan J. C., Chen K. H., Bates M., Zhuang X. (2011) Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolter S., Löschberger A., Holm T., Aufmkolk S., Dabauvalle M. C., van de Linde S., Sauer M. (2012) rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 [DOI] [PubMed] [Google Scholar]
- 30. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 31. Reddy R. K., Mao C., Baumeister P., Austin R. C., Kaufman R. J., Lee A. S. (2003) Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 278, 20915–20924 [DOI] [PubMed] [Google Scholar]
- 32. Rao R. V., Peel A., Logvinova A., del Rio G., Hermel E., Yokota T., Goldsmith P. C., Ellerby L. M., Ellerby H. M., Bredesen D. E. (2002) Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 514, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ellgaard L., Helenius A. (2001) ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13, 431–437 [DOI] [PubMed] [Google Scholar]
- 34. Flanagan J. G., Vanderhaeghen P. (1998) The ephrins and Eph receptors in neural development. Annu. Rev. Neurosci. 21, 309–345 [DOI] [PubMed] [Google Scholar]
- 35. Gerke V., Creutz C. E., Moss S. E. (2005) Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6, 449–461 [DOI] [PubMed] [Google Scholar]
- 36. Satpute-Krishnan P., Ajinkya M., Bhat S., Itakura E., Hegde R. S., Lippincott-Schwartz J. (2014) ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell 158, 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu E. S., Ou J. H., Lee A. S. (1992) Brefeldin A as a regulator of grp78 gene expression in mammalian cells. J. Biol. Chem. 267, 7128–7133 [PubMed] [Google Scholar]
- 39. Deora A. B., Kreitzer G., Jacovina A. T., Hajjar K. A. (2004) An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J. Biol. Chem. 279, 43411–43418 [DOI] [PubMed] [Google Scholar]
- 40. Afshar N., Black B. E., Paschal B. M. (2005) Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell. Biol. 25, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Awad W., Estrada I., Shen Y., Hendershot L. M. (2008) BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc. Natl. Acad. Sci. U.S.A. 105, 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei J., Gaut J. R., Hendershot L. M. (1995) In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270, 26677–26682 [DOI] [PubMed] [Google Scholar]
- 43. Guo F., Snapp E. L. (2013) ERdj3 regulates BiP occupancy in living cells. J. Cell Sci. 126, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Misra U. K., Gonzalez-Gronow M., Gawdi G., Pizzo S. V. (2005) The role of MTJ-1 in cell surface translocation of GRP78, a receptor for α2-macroglobulin-dependent signaling. J. Immunol. 174, 2092–2097 [DOI] [PubMed] [Google Scholar]
- 45. Nakayama H., Fukuda S., Inoue H., Nishida-Fukuda H., Shirakata Y., Hashimoto K., Higashiyama S. (2012) Cell surface annexins regulate ADAM-mediated ectodomain shedding of proamphiregulin. Mol. Biol. Cell 23, 1964–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahalka A. K., Kirkegaard T., Jukola L. T., Jäättelä M., Kinnunen P. K. (2014) Human heat shock protein 70 (Hsp70) as a peripheral membrane protein. Biochim. Biophys. Acta 1838, 1344–1361 [DOI] [PubMed] [Google Scholar]
- 47. Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G. (1992) Caveolin, a protein component of caveolae membrane coats. Cell 68, 673–682 [DOI] [PubMed] [Google Scholar]
- 48. Patlolla J. M., Swamy M. V., Raju J., Rao C. V. (2004) Overexpression of caveolin-1 in experimental colon adenocarcinomas and human colon cancer cell lines. Oncol. Rep. 11, 957–963 [PubMed] [Google Scholar]
- 49. Obeid M., Panaretakis T., Tesniere A., Joza N., Tufi R., Apetoh L., Ghiringhelli F., Zitvogel L., Kroemer G. (2007) Leveraging the immune system during chemotherapy: moving calreticulin to the cell surface converts apoptotic death from “silent” to immunogenic. Cancer Res. 67, 7941–7944 [DOI] [PubMed] [Google Scholar]
- 50. Bellani S., Mescola A., Ronzitti G., Tsushima H., Tilve S., Canale C., Valtorta F., Chieregatti E. (2014) GRP78 clustering at the cell surface of neurons transduces the action of exogenous α-synuclein. Cell Death Differ. 21, 1971–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bhattacharjee G., Ahamed J., Pedersen B., El-Sheikh A., Mackman N., Ruf W., Liu C., Edgington T. S. (2005) Regulation of tissue factor-mediated initiation of the coagulation cascade by cell surface grp78. Arterioscler. Thromb. Vasc. Biol. 25, 1737–1743 [DOI] [PubMed] [Google Scholar]
- 52. Al-Hashimi A. A., Caldwell J., Gonzalez-Gronow M., Pizzo S. V., Aboumrad D., Pozza L., Al-Bayati H., Weitz J. I., Stafford A., Chan H., Kapoor A., Jacobsen D. W., Dickhout J. G., Austin R. C. (2010) Binding of anti-GRP78 autoantibodies to cell surface GRP78 increases tissue factor procoagulant activity via the release of calcium from endoplasmic reticulum stores. J. Biol. Chem. 285, 28912–28923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ray R., de Ridder G. G., Eu J. P., Paton A. W., Paton J. C., Pizzo S. V. (2012) The Escherichia coli subtilase cytotoxin A subunit specifically cleaves cell-surface GRP78 protein and abolishes COOH-terminal-dependent signaling. J. Biol. Chem. 287, 32755–32769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gray P. C., Vale W. (2012) Cripto/GRP78 modulation of the TGF-β pathway in development and oncogenesis. FEBS Lett. 586, 1836–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burikhanov R., Zhao Y., Goswami A., Qiu S., Schwarze S. R., Rangnekar V. M. (2009) The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell 138, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Képès F. (1996) The “+70 pause”: hypothesis of a translational control of membrane protein assembly. J. Mol. Biol. 262, 77–86 [DOI] [PubMed] [Google Scholar]
- 57. Wu M. M., Buchanan J., Luik R. M., Lewis R. S. (2006) Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.