Background: miR-3189-3p is a putative mirtron of growth differentiation factor 15 (GDF15).
Results: MiR-3189-3p is down-regulated in glial tumors.
Conclusion: MiR-3189-3p has tumor suppressor activities in glioblastoma cells.
Significance: MiR-3189-3p has potential therapeutic properties.
Keywords: Cell Migration, Cell Proliferation, Glioblastoma, MicroRNA (miRNA), Tumor Suppressor Gene
Abstract
Glioblastoma is one of the most aggressive brain tumors. We have previously found up-regulation of growth differentiation factor 15 (GDF15) in glioblastoma cells treated with the anticancer agent fenofibrate. Sequence analysis of GDF15 revealed the presence of a microRNA, miR-3189, in the single intron. We then asked whether miR-3189 was expressed in clinical samples and whether it was functional in glioblastoma cells. We found that expression of miR-3189-3p was down-regulated in astrocytoma and glioblastoma clinical samples compared with control brain tissue. In vitro, the functionality of miR-3189-3p was tested by RNA-binding protein immunoprecipitation, and miR-3189-3p coimmunoprecipitated with Argonaute 2 together with two of its major predicted gene targets, the SF3B2 splicing factor and the guanine nucleotide exchange factor p63RhoGEF. Overexpression of miR-3189-3p resulted in a significant inhibition of cell proliferation and migration through direct targeting of SF3B2 and p63RhoGEF, respectively. Interestingly, miR-3189-3p levels were increased by treatment of glioblastoma cells with fenofibrate, a lipid-lowering drug with multiple anticancer activities. The attenuated expression of miR-3189-3p in clinical samples paralleled the elevated expression of SF3B2, which could contribute to the activation of SF3B2 growth-promoting pathways in these tumors. Finally, miR-3189-3p-mediated inhibition of tumor growth in vivo further supported the function of this microRNA as a tumor suppressor.
Introduction
Glioblastomas are deadly cancers characterized by rapid cell proliferation, high invasiveness, and resistance to radio- and chemotherapy (1). Patients with this aggressive tumor, which accounts for nearly 50% of all adult brain tumors, have a median survival of ∼15 months (2). The standard treatment for glioblastoma involves invasive surgery and radiotherapy, which is often followed by chemotherapy with temolozomide (3). Because novel therapeutic treatments for glioblastoma are needed desperately, it is essential to understand the molecular mechanisms supporting growth and survival of this highly malignant and practically incurable brain tumor.
MicroRNAs are short single-stranded non-coding RNAs that regulate gene expression by incomplete base pairing with mRNAs. Alterations in microRNA expression have been associated with a variety of pathologies, including cancer. In addition to their effect on carcinogenesis, microRNAs have been investigated for their potential values in diagnosis, prognosis, and cancer therapy. In this study, we found miR-3189-3p down-regulated in astrocytoma and glioblastoma clinical samples in comparison with unaffected brain tissue. Genomic sequence analysis revealed that the miR-3189 stem loop is located in the intron of the growth differentiation factor 15 (GDF15) gene. Interestingly, we have previously reported increased expression of GDF15 in the LN-229 glioblastoma cell line following the treatment with fenofibrate (4), an agonist of peroxisome proliferator-activated receptor α (PPARα),4 commonly used as a lipid-lowering drug, which has strong antiglioblastoma properties (5, 6). GDF15, also known as nonsteroidal anti-inflammatory drug-activated gene 1 (NAG-1) or macrophage inhibitory cytokine 1 (MIC-1), is a member of the TGF-β superfamily (7, 8). The GDF15 gene is encoded by two exons that produce a precursor protein cleaved further to generate a 112-amino acid mature GDF15 peptide that is secreted into the extracellular matrix as a biologically active dimer (9). GDF15 can be induced by anti-inflammatory drugs, cytotoxic agents, PPAR agonists, and anticancer drugs (4, 10). Increased GDF15 mRNA expression has been reported in patients during malignant progression to glioblastomas (11), and others have reported that expression levels of GDF15 are up-regulated in glioblastoma cells in response to cytotoxic stimuli during chemotherapy treatment (4, 12, 13). The precursor sequence encoded by miR-3189 contains two mature microRNA sequences within the stem loop: miR-3189-3p and miR-3189-5p, 21 and 25 nucleotides in length, respectively. The biological function of these microRNAs has never been described. Here we show that ectopic expression of miR-3189-3p inhibited glioblastoma cell growth and migration through down-regulation of SF3B2 and p63RhoGEF, respectively. In comparison with normal brain tissue, we also found that astrocytoma and glioblastoma clinical samples have increased levels of GDF15 and decreased levels of miR-3189-3p and that these changes correlated with increased expression of SF3B2 and p63RhoGEF. Finally, the subcutaneous and intracranial growth of glioblastoma cells expressing miR-3189-3p was reduced significantly when compared with the parental control cells, further supporting the role of this microRNA as a tumor suppressor.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Reagents
LN-229 and U87MG cells were obtained from the ATCC and cultured under standard growth conditions. Fenofibrate was from Sigma and was used in all experiments at a concentration of 50 μm. The miR-3189-3p mirVana miRNA mimic and miR-3189-3p mirVana miRNA inhibitor (anti-miR-3189-3p) were purchased from Life Technologies. For transfection experiments, cells were seeded at a density of 4 × 105 cells/60-mm dish or 2.5 × 104 cells/well in 12-well plates and transfected using Lipofectamine 2000 (Life Technologies) according to the instructions of the manufacturer. SF3B2 and p63RhoGEF siRNAs and controls were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at a final concentration of 10 nm. Human PPARα siRNA was the ON-TARGET Plus SMARTpool from Thermo Scientific Dharmacon (Lafayette, CO), and it was used at a final concentration of 100 nm. The PPARα inhibitor GW 9662 was from Cayman Chemical (St. Louis, MO). For the transient transfection experiments, cells were seeded at a density of 4 × 105 cells/60-mm dish or 2.5 × 104 cells/well in 12-well plates and transfected using Lipofectamine 2000 reagent (Life Technologies) according to the instructions of the manufacturer.
Quantitative RT-PCR
Total RNA was isolated using the miRVana miRNA extraction kit (Ambion, Austin, TX). RNA was reverse-transcribed using the high-capacity cDNA reverse transcription kit for mRNAs or TaqMan assays for microRNAs (Applied Biosystems, Carlsbad, CA). Quantitative real-time PCR was performed in duplicate using a Roche LightCycler 480 real-time PCR system (Indianapolis, IN). Each sample was normalized using GAPDH or RNU6B control (ΔCt), and the relative quantification of gene expression was calculated using the comparative Ct (2−ΔΔCt) method as described in our previous publications (14–16). For the clinical samples, relative quantification was represented as 1/ΔCt to maintain real differences in Ct values between samples.
Western Blot Analysis
Cells were collected by gently scraping in the presence of PBS (Gibco, Life Technologies) followed by centrifugation and disruption of the cell pellet in lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA (pH 8.4), 10% glycerol, 1% Triton X-100, 1 mm PMSF, 1 mm sodium orthovanadate (NA3VO4), and phosphatase and protease inhibitor mixtures (Sigma). Whole-cell lysates (30–70 μg) were separated on a 4–15% SDS-PAGE gel (Bio-Rad). GDF15 and E2F-1 antibodies were purchased from Cell Signaling Technology (Beverly, MA). The antibody to detect p63RhoGEF was obtained from GeneTex (Irvine, CA). The GDF15 antibody was purchased from R&D Systems (Minneapolis, MN), and anti-GRB2 was from BD Transduction Laboratories (San Jose, CA). SF3B2 and 14-3-3ζ antibodies were obtained from Santa Cruz Biotechnology.
ELISA Assay
Mature GDF15 was detected in the cell culture medium using the GDF15 Quantikine ELISA kit from R&D Systems according to the instructions of the manufacturer. Sample absorbance was measured at 450 nm using a Bio-Rad Benchmark Plus microplate reader.
RNA-binding Protein Immunoprecipitation (RIP)
RIP assay was performed using Argonaute 2 (Ago2) antibody from Millipore (RIPAb+Ago2 RIP). This kit includes a negative control mouse IgG antibody and control primers specific for human FOS, which were utilized for the optimization of Ago2-IP in our cellular model. The RIP Ago-IP was performed essentially as described previously (17). In brief, 10 × 106 LN-229 cells were used for Ago2-IP and for the IgG isotype control. 2 × 106 cells were used for the extraction of total RNA and for total protein lysates. The cell monolayer was washed twice with cold PBS and lysed in 500 μl of lysis buffer (150 mm KCl, 25 mm Tris-HCl (pH 7.4), 5 mm EDTA, 0.5% Nonidet P-40, and 5 mm DTT) supplemented with 10 mm protease inhibitor mixture, 10 mm PMSF, 10 mm phosphatase inhibitor, 10 mm Na3VO4, and 100 units/ml RNase inhibitor (Applied Biosystems). After 30 min of incubation on ice, lysates were centrifuged for 30 min at 14,000 rpm at 4 °C in a microcentrifuge. 30 μl of protein A/G magnetic beads (Pierce, Thermo Scientific) were washed three times with blocking solution (0.5% BSA dissolved in Dulbecco's PBS+/+) and incubated for 1 h at 4 °C with 5 μg of anti-Ago2 mouse monoclonal IgG1κ or with isotype IgG1κ control antibodies (Millipore). The immunocomplexes were washed three times with blocking solution. Immunoprecipitations with specific lysates were carried out overnight at 4 °C. Next day, the immunocomplexes were washed three times with lysis buffer supplemented with the inhibitors. Leftover IP samples before the first wash were collected to determine the efficiency of Ago2 depletion from the cellular lysate. After the last wash, immunocomplexes were resuspended in 40 μl of lysis buffer, of which 20 μl was used for the RNA extraction and 20 μl for Western blot analysis. RNA extraction from the beads and from the leftover samples was followed by reverse transcription and quantitative real-time PCR, which were performed as described above.
Cloning of the p63RhoGEF and SF3B2 Open Reading Frames
Total RNA was isolated from LN-229 cells and reverse-transcribed to cDNA using a high-capacity cDNA reverse transcription kit containing random hexamers. The cDNA sequence corresponding to the ORF of p63RhoGEF was PCR-amplified. The primers used were as follows: forward, 5′-GGTGGAATTCTGCAGATATGCGGGGGGGGCACAAA; reverse, 5′-CCACTGTGCTGGATTTACAGCTCATCTTCATCCAGCTTGG. Sequences compatible with pcDNA3.1(+) are underlined. Next, the pcDNA3.1(+) vector was digested with EcoRV. This vector and the PCR product (above) were digested with the Klenow fragment of DNA polymerase I to generate single-stranded 3′ overhangs compatible between the two DNA molecules. These products were annealed by incubation at incremental reducing temperatures from 95 °C to 45 °C using a PCR cycler (Bio-Rad). The ORF sequence corresponding to the SF3B2 gene was also cloned into the pcDNA3.1(+) vector using the approach described above. The primers used were as follows: forward, 5′-GGTGGAATTCTGCAGATATGGCGACGGAGCATCCC; reverse, 5′-CCACTGTGCTGGATCTAAAACTTGAACTCCTTATATTTCTTGCTGCC. Sequences compatible with pcDNA3.1(+) are underlined.
Cloning for MicroRNA Functional Analysis
The genomic sequence corresponding to the 3′UTR of p63RhoGEF was PCR-amplified from LN-229 cells. This PCR product was ligated into multiple cloning sites downstream of the Renilla luciferase reporter gene in the psiCHECK-2 vector (Promega, Madison, WI). This vector also contains a firefly luciferase reporter sequence that allows for normalization of transfection efficiency. The primers used were as follows: forward, 5′-CCGCTCGAGCTGGTGAAAACCATGGGGGTG containing the XhoI restriction site; reverse, 5′-ATAAGAATGCGGCCGCGCAGCCTCGGTGATATAACAAAACC containing the NotI restriction site. The genomic sequence corresponding to the 3′UTR of SF3B2 was also cloned into the psiCHECK-2 vector. The primers used were as follows: forward, 5′-CCGCTCGAGTTCAA GTTTTAGGTCCCCTCAC containing the XhoI restriction site; reverse, 5′-ATAAGAATGCGGCCGCGGAGGCTCAGGAGTGTTAAATATTCATCTC containing the NotI restriction site. The XhoI and NotI restriction sites are underlined.
Mutations of the miR-3189-3p putative binding sites in the p63RhoGEF and SF3B2 3′UTR sequences were generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) using the respective psiCHECK2/3′UTR plasmids as a template. The oligonucleotides for the mutagenesis of p63RhoGEF sites were as follows: site 1 forward, 5′-TCAGCCGCCTATTCCCCTTCCAG CTTCAGGGCAGTCCT; site 2 forward, 5′-TGGAGGAGAACACCTAGACCCTTCCACTTTTTTCTGCCCAAGGAAC; and site 3 forward, 5′-CCCAAGGACTTTTTTCTGCCCTTCCAACACAGTTTCCTTCAGCTCC. The oligonucleotides for the mutagenesis of SF3B2 sites were as follows: site 1 forward, 5′-GAACCACCTCTCCCGCAGTTCC CTTCCACTTGTCATTTCATGTTCTTATT; and site 2 forward, 5′-GACCTGTTTTGTAAATAAA GCTGTTTCCCTTCCAAAGAGATGAATATTTAACACT CCTGAGC. Mutated bases in the miR-3189-3p binding sites are underlined. The reverse oligonucleotide primers were complementary to the forward primers.
Dual Luciferase Assay
LN-229 cells were plated at a density of 8 × 104 cells/well in a 12-well plate and transfected with psiCHECK-2 vector expressing the target 3′UTR (160 ng/well) alone, the target 3′UTR with the miR-3189-3p mimic (30 nm), or the target 3′UTR with the miRNA mimic and anti-miR-3189-3p using Lipofectamine 2000. After 24 h, cells were harvested, and lysates were assayed for luciferase activity with the Dual-Luciferase reporter assay system (Promega) using a Synergy 2 microplate reader (BioTek Instruments, Inc., Winooski, VT). Relative units of Renilla luciferase activity were normalized to the firefly luciferase internal control in each sample. Experiments were performed in duplicate.
Cell Proliferation Assay
LN-229 or U87MG cells were plated at a density of 2.5 × 104 cells/well in a 12-well plate and transfected with mock or miR-3189-3p mimic ± anti-miR-3189-3p. 72 h after transfection, cells were incubated with medium containing 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent (Promega) diluted according to the instructions of the manufacturer. Cells were then incubated at 37 °C for 30–60 min, and absorbance was measured at a wavelength of 490 nm using a Bio-Rad Benchmark Plus microplate reader.
Cell Cycle Analysis
Cells were collected 48 h after transfection and fixed in 70% ethanol overnight at −20 °C. Cells were then centrifuged at 300 × g, resuspended in 150 μl of Guava cell cycle reagent (Guava Technologies, Hayward, CA), and stained for 45 min at 25 °C while protected from light. Cells were counted by flow cytometry using a FACSAria (BD Biosciences) in the Comprehensive Alcohol Research Center Analytical Core Laboratory at the Louisiana State University Health Sciences Center. Cell cycle distribution was evaluated using the ModFit LT program (Verity Software House, Topsham, ME).
Scratch Assay
LN-229 or U87MG cells were transfected with miR-3189-3p and plated in a 35 mm glass bottom dish (MatTek Corporation, Ashland, MA) at a density of 1.8 × 105 cells/dish. The scratch assay was performed by moving a pipette tip across the cell monolayer. Migration into the cell-free area was monitored for up to 24 h using live cell time-lapse imaging (VivaView FL incubator fluorescent microscope, Olympus, Center Valley, PA).
Subcutaneous Injection of Mice with LN-229/mCherry Glioblastoma Cells
Female Fox1nu athymic nude mice at 6–8 weeks of age (Harlan Laboratories, Inc., Indianapolis, IN) were injected subcutaneously with 2 × 106 LN-229 glioblastoma cells stably expressing the mCherry fluorescent protein and mock-transfected, or transfected with miR-3189-3p mimic. All experiments were performed in accordance with institutional ethical guidelines.
Intracranial Injection of Mice with U87MG/luciferase Glioblastoma Cells
Intracranial implantation of mice was performed as described previously (6, 18). Briefly, female nude-Foxn1nu athymic mice 6–8 weeks of age (Harlan Laboratories) were anesthetized with 4% isoflurane and secured in a stereotaxic head frame (Harvard Apparatus, Holliston, MA). The tumor cells (3 × 104 in 2 μl of artificial cerebrospinal fluid) were injected into the left striatum through a burr hole in the skull using a 10-μl Hamilton syringe.
In Vivo Imaging of Tumor Xenografts
In vivo growth of LN-229/mCherry and U87MG/luciferase tumors was monitored by biophotonic imaging using a Xenogen IVIS 200 system (Xenogen, Palo Alto, CA). Prior to imaging, mice bearing U87MG/luciferase tumors received an intraperitoneal injection of 100 μl of d-luciferin (30 mg/ml solution) and were anesthetized by 3% isoflurane inhalation. Anesthesia was sustained at 1.5% isoflurane inside the imaging chamber using nose cones. Images were captured and quantified with Xenogen Living Image 4.1 software on the basis of equivalent regions of interest over the lower dorsal flank or cranium of the mouse. Image intensities were expressed as photon flux per second per square centimeter and steradian.
Statistical Analysis
Data are presented as mean ± S.D. Comparison between two experimental groups was performed using Student's t test. One way ANOVA was used to compare three or more groups. p ≤ 0.05 were considered statistically significant.
RESULTS
MiR-3189-3p Is Down-regulated in Glial Tumors and Regulates Growth and Migration of Glioblastoma Cells
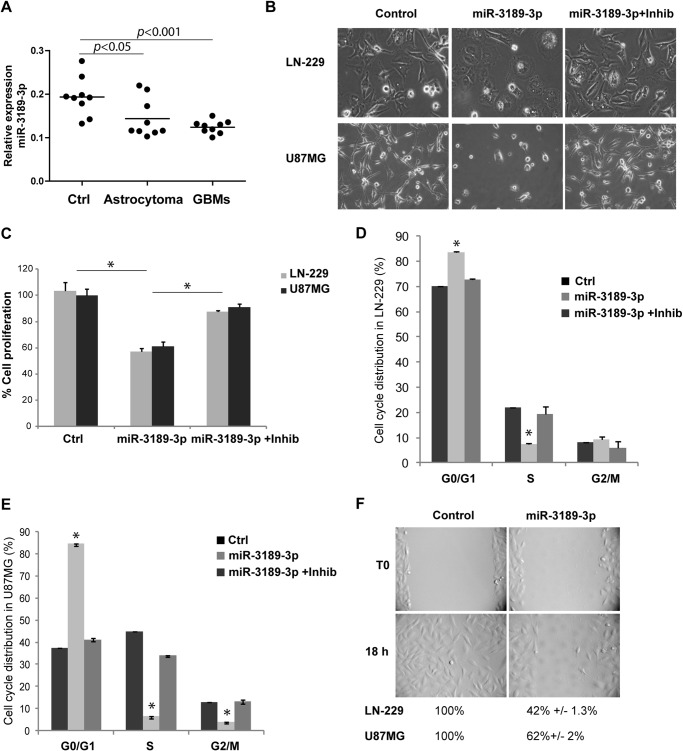
Formalin-fixed paraffin-embedded tissue samples from astrocytomas, glioblastomas, and normal brains were utilized for total RNA isolation and were subjected to quantitative real-time PCR. Fig. 1A shows significantly less miR-3189-3p levels in astrocytoma and glioblastoma tissue samples compared with controls (p < 0.05 and p < 0.001, respectively), with a trend of down-regulation that correlated with the tumor progression. We next evaluated the function of miR-3189-3p in cell growth and migration of the human glioblastoma cell lines LN-229 and U87MG. 48 h post-transfection morphology of miR-3189-3p-expressing cells was visibly altered, presenting a more polygonal shape compared with the typical spindle-shaped cells under normal growth conditions or when both miR-3189-3p and anti-miR-3189-3p were coexpressed (Fig. 1B). In addition, the expression of miR-3189-3p resulted in a 50% reduction in cell proliferation (percent decrease in cell number over control, Fig. 1C) accompanied by a significant, 40% reduction in S phase, as determined by cell cycle distribution analysis (Fig. 1, D and E). No significant changes were observed when miR-3189-3p was cotransfected with anti-miR-3189-3p, and the cells expressing this inhibitor behaved essentially as the control, mock-transfected cells. Similarly, transfection with miR-3189–5p did not elicit changes in cell morphology, nor did it inhibit cell proliferation and migration (data not shown). To minimize possible off-target effects, we repeated this set of experiments using another commercially available miR-3189-3p mimic (mission miRNA mimic, Sigma) and obtained the same results (data not shown).
FIGURE 1.
MiR-3189-3p is down-regulated in glial tumors and affects growth and migration of glioblastoma cells in culture. A, relative expression of miR-3189-3p in controls (Ctrl), astrocytomas, and glioblastomas (GBM) (n = 9/group). B, phase-contrast images showing the morphology of LN-229 and U87MG cells following transfection with miR-3189-3p or miR-3189-3p + anti-miR-3189-3p (original magnification, ×10). Images were acquired 48 h post-transfection. Inhib, inhibitor. C, cell proliferation assay performed 72 h post-transfection of the indicated cell lines with mock, miR-3189-3p, or miR-3189-3p + inhibitor and quantified using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium reagent. Results are expressed as percent of growth/mock-treated control. D and E, cell cycle analysis of LN-229 (D) and U87MG (E) cells transfected with mock (ctrl), miR-3189-3p, and anti-miR-3189-3p (Inhib). Cells were stained with Guava cell cycle reagent and cell cycle distribution (%) was quantified by flow cytometry using a FACSAria. F, representative images of a scratch assay to monitor migration of controls (mock-transfected) and miR-3189-3p-transfected LN-229 cells (original magnification, ×10). Migration into the cell-free area was monitored by time-lapse imaging in a VivaView fluorescent microscope. The same experiment was performed in U87MG cells, and data are shown below the images in F. *, p < 0.05.
Because glioblastoma cells are characterized by high migratory potentials, we evaluated the possible effects of this microRNA on cell migration using a scratch assay and by monitoring both cell locomotion and cell division using time-lapse imaging. Fig. 1F shows representative images for LN-229 cells. Although control cells populated the entire scratched area in 18 h, LN-229 cells transfected with miR-3189-3p covered only 42% of the scratched surface (Fig. 1F) in the same time. Although with decreased efficiency compared with LN-229 cells, miR-3189-3p expression also inhibited U87MG cell migration, and, after 18 h of transfection, cells covered only 62% of the available surface. Note that both decreased cell motility and decreased cell proliferation contributed to delayed invasion of the cell-free space.
SF3B2 and p63RhoGEF Are Targets of miR-3189-3p
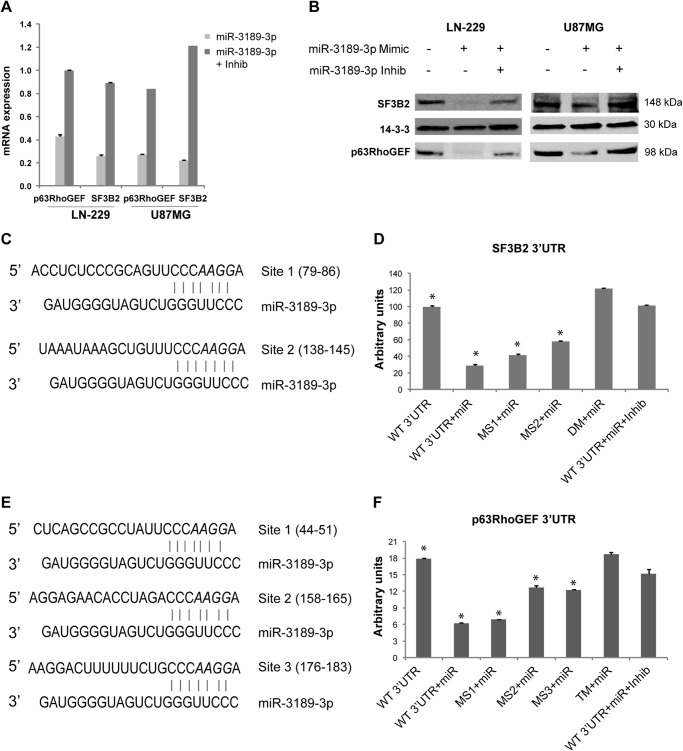
According to microRNA gene target predictions, the splicing factor SF3B2 and the Rho guanine nucleotide exchange factor 25, ARHGEF25 (also known as p63RhoGEF) are the top targets of miR-3189-3p and were chosen for validation studies. The direct contribution of miR-3189-3p to SF3B2 and p63RhoGEF mRNA and protein levels was evaluated in miR-3189-3p-transfected LN-229 and U87MG cells by quantitative real-time PCR and Western blot analyses. In comparison with controls, nearly 5-fold and 2.5-fold lower levels of SF3B2 mRNA and p63RhoGEF mRNA were detected in miR-3189-3p-expressing cells, respectively (Fig. 2A). Importantly, we were able to counteract down-regulation of these two transcripts by overexpressing the anti-miR-3189-3p (miR-3189-3p inhibitor), further supporting the presence of miR-3189-3p-specific regulation. A remarkable down-regulation of SF3B2 and p63RhoGEF at the translational level was confirmed by Western blots in cells transfected with miR-3189-3p compared with control or cells cotransfected with the anti-miR-3189-3p (Fig. 2B). Notably, the down-regulation of SF3B2 and p63RhoGEF was more pronounced in LN-229 cells, and this cell line was selected for the next experiments.
FIGURE 2.
MiR-3189-3p directly targets the 3′UTR sequences of SF3B2 and p63RhoGEF. A, quantitative real-time PCR showing expression of p63RhoGEF and SF3B2 mRNAs in mock-transfected cells transfected with miR-3189-3p and cells transfected with miR-3189-3p + anti-miR-3189-3p. Results are expressed as -fold change compared with mock-treated cells. Inhib, inhibitor. B, Western blots for SF3B2 and p63RhoGEF proteins performed on lysates from LN-229 and U87MG cells mock-transfected or transfected with miR-3189-3p or miR-3189-3p + anti-miR-3189-3p (Inhib) for 48 h. C–E, predicted binding sites for miR-3189-3p in the SF3B2 (C) and p63RhoGEF (E) 3′UTR sequences. The bases mutated in the microRNA binding site are shown in italics. D, luciferase assays of LN-229 cells cotransfected with psiCHECK2/SF3B2 3′UTR and the mutants in the miR-3189-3p putative binding sites (MS1 and MS2) and miR-3189-3p ± anti-miR-3189-3p (Inhib). F, luciferase assays of LN-229 cells cotransfected with psiCHECK2/p63RhoGEF 3′UTR and mutants (MS1, MS2, and MS3) and miR-3189-3p with or without anti-miR-3189-3p (Inhib). MS1–3, specific microRNA binding site mutants; DM, double-binding site mutants; TM, triple-binding site mutants. *, p < 0.05.
There are two putative binding sites for miR-3189-3p in the 3′UTR sequence of SF3B2 mRNA, one conserved (MS2) and one non-conserved (MS1) (Fig. 2C), and their expression was tested by a luciferase-based reporter assay (Fig. 2D). A reduction of ∼75% of luminescence was observed in cells expressing miR-3189-3p, and, again, this inhibition was almost completely alleviated in the presence of anti-miR-3189-3p. In addition, site-directed mutagenesis of the microRNA binding sequences in the 3′UTR showed a slightly different, although cumulative, inhibitory activity. The conserved microRNA binding site (MS2) appeared to be slightly more effective in microRNA-induced transcript degradation because mutation of this site significantly reversed a decrease in luciferase signal (compare MS1 and MS2). As expected, mutation of both microRNA binding sequences (double mutation) abrogated inhibition by miR-3189-3p. The p63RhoGEF 3′UTR contains three putative binding sites for miR-3189-3p, one conserved and two non-conserved (Fig. 2E). Similarly to SF3B2 3′UTR, we tested the p63RhoGEF 3′UTR (Fig. 2F). Here the inhibition was also reversed efficiently, either in the presence of anti-miR-3189-3p or by mutating the three binding sequences (triple mutation) of the p63RhoGEF 3′UTR. Quantitatively, one of the non-conserved binding sites did not appear to be required for miR-3189-3p-induced gene regulation because mutation of this site (MS1) failed to revert the expected inhibition. Conversely, mutation of the other two microRNA binding sites (MS2 and MS3), one conserved and one non-conserved, showed a stronger and cumulative reversal of inhibition by miR-3189-3p (Fig. 2F).
Role of SF3B2 and p63RhoGEF in the Inhibition of Cellular Proliferation and Migration Induced by miR-3189-3p
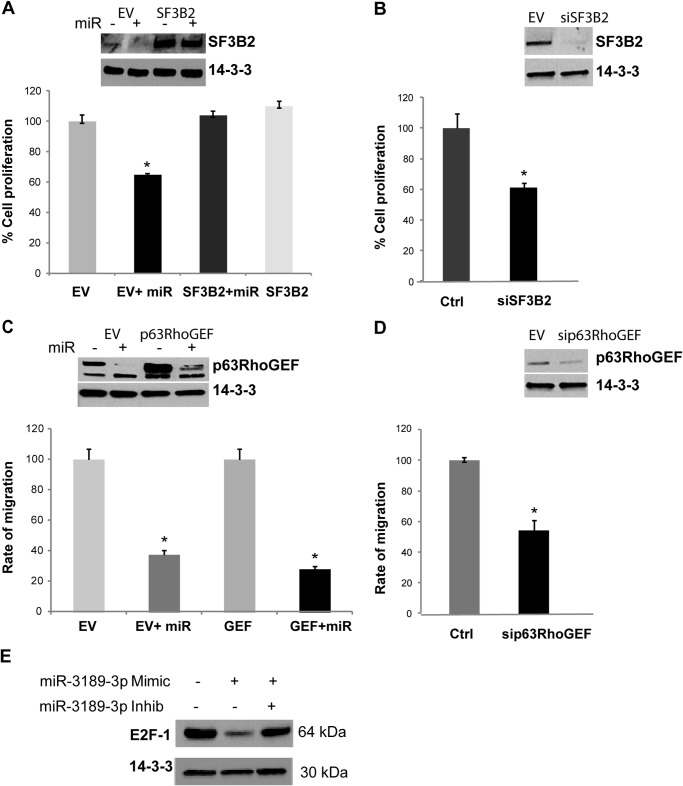
Because SF3B2 expression is strongly down-regulated by miR-3189-3p, we hypothesized that forced expression of SF3B2 might reverse the microRNA-mediated effects on cell proliferation. The results from the growth rate analysis in Fig. 3A confirm this assumption. Moreover, we measured that constitutive expression of SF3B2 was enough to rescue cell growth to steady-state levels despite the addition of miR-3189-3p. Conversely, down-regulation of SF3B2 by siRNA mimics the biological effects of miR-3189-3p expression by inducing a 55% reduction in cell growth (Fig. 3B). When tested on a migration assay, cells in which SF3B2 was silenced behaved essentially as the control cells, indicating that SF3B2 is involved in the proliferation but not in the migration of these cells (data not shown). The impaired growth by SF3B2 is likely mediated by E2F-1, a known molecule downstream of SF3B2 that is important for cell cycle progression (19). The Western blot in Fig. 3E shows a marked down-regulation of E2F-1 protein levels in LN-229 cells transfected with miR-3189-3p, and this effect was reversed in the presence of the anti-miR-3189-3p. Therefore, these data suggest that E2F-1 is a potential downstream target of miR-3189-3p/SF3B2 in coordinating delayed cell growth in our model.
FIGURE 3.
MiR-3189-3p regulates LN-229 cell growth and migration through the down-regulation of SF3B2 and p63RhoGEF. A, cell growth assay performed 72 h after transient transfection of LN-229/pcDNA3.1 (EV) or LN-229/SF3B2 with miR-3189-3p. The inset shows levels of expression of SF3B2 under the indicated experimental conditions. Results are expressed as percent growth/mock-treated control. B, cell growth assay performed with mock-treated control (Ctrl) or siSF3B2-treated LN-229 cells 72 h post-transfection. Results are expressed as percent growth/mock-treated control. C, results of scratch assays to monitor the migration of cells expressing the p63RhoGEF gene (GEF). The inset shows the levels of expression of p63RhoGEF protein under the indicated conditions. D, plot of a scratch assay to monitor the migration of mock-treated control or sip63RhoGEF-treated LN-229 cells. The inset shows levels of expression of p63RhoGEF protein under the indicated conditions. E, Western blots for E2F-1 protein expression performed on lysates from mock-transfected cells or cells transfected with miR-3189-3p or miR-3189-3p + anti-miR-3189-3p (Inhib). 14-3-3ζ antibody was used to show equal loading of cellular lysates. *, p < 0.05.
Next, we evaluated the contribution of p63RhoGEF in reversing the inhibitory effects of miR-3189-3p expression on cell migration. Silencing p63RhoGEF in LN-229 resulted in a 54% (±7%) inhibition of migration (Fig. 3D), confirming data reported previously (20). To test the contribution of p63RhoGEF on the inhibition of migration because of miR-3189-3p, we overexpressed p63RhoGEF in LN-229 cells. When tested using the scratch assay, p63RhoGEF-expressing cells behaved essentially similar as the control cells (transfected with pcDNA3.1 empty vector). However, transient transfection with miR-3189-3p was still capable of reducing cell migration by 60% (Fig. 3C). Knocking down p63RhoGEF did not alter the proliferation of LN-229 cells (data not shown). Together, these results indicate that down-regulation of SF3B2 by miR-3189-3p is necessary and sufficient for the miR-mediated impairment of cell growth, whereas down-regulation of p63RhoGEF might be required, but is not sufficient, for the inhibition of migration by miR-3189-3p.
MiR-3189-3p Has Tumor Suppressor Activity in Mice
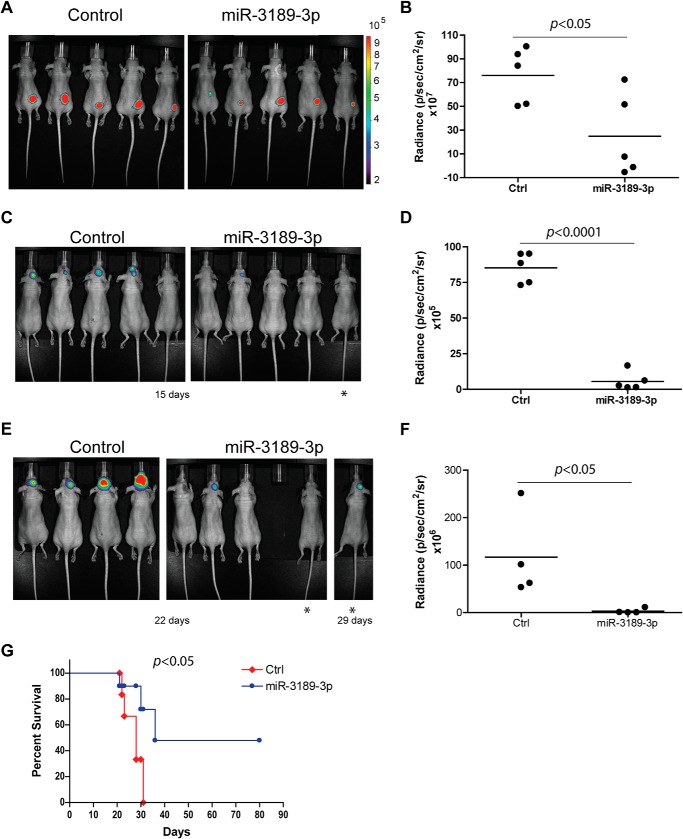
Next, we evaluated the ability of miR-3189-3p to inhibit tumor growth in vivo. LN-229 cells, bearing the pmCherry plasmid to facilitate detection of the tumor by fluorescence, were mock-transfected or transfected with a miR-3189-3p mimic. The next day, 2 × 106 cells of either transfection were injected subcutaneously in the flank of nude mice (n = 5/group). Beginning at 1 week post-injection, mice were visualized via in vivo biophotonic epifluorescence, and the mean fluorescence radiance for each tumor was collected. We found that mice bearing LN-229/miR-3189-3p cells had a nearly 75% smaller tumors compared with LN-229/Mock (Fig. 4, A and B, p < 0.05) or with cells expressing both miR-3189-3p and its anti-miR (data not shown). The anti-tumor effect of miR-3189-3p was also confirmed in nude mice bearing U87MG glioblastoma cells (data not shown). In another experimental setting, U87MG cells bearing the luciferase gene were transfected with miR-3189-3p prior to their intracranial injection in nude mice. Tumor growth was monitored using biophotonic epifluorescence as described above. The results in Fig. 4, C–F, show a strong inhibition of tumor growth in animals bearing the cells transfected with the microRNA at both 15 and 22 days post-injection (Fig. 4, C–F, respectively). Indeed, more than 40 days after the control mice had died because of tumor growth, two of the animals injected with cells transfected with miR-3189-3p never developed the tumor and were still alive. Fig. 4G shows the survival data for the two groups of mice. All mice in the control group died because of the intracranial tumor growth. One mouse in the miR-3189-3p group died after 21 days without apparent signs of tumor formation. Two mice developed intracranial tumors and were sacrificed at days 31 and 36. Of note, the mouse that died after 36 days had a delay in developing the tumor, which was visible at day 29 but not at days 22 or 15 (Fig. 4E, mouse labeled with an asterisk). The delay in tumor formation may be attributable to the loss of cells bearing the microRNA over several days combined with the presence of some untransfected cells that later took over and formed the tumor. In fact, the levels of miR-3189-3p in this tumor were comparable with the controls, indicating loss of miR-3189-3p (data not shown).
FIGURE 4.
Biophotonic measurements of orthotopic glioblastoma xenografts in vivo show significant tumor suppressor activity of miR-3189-3p. A, fluorescent images of LN-229/mCherry cells mock-transfected (Control) or transfected with miR-3189-3p implanted subcutaneously into nude mice (n = 5/group, p < 0.05). B, plot of tumor burden 3 weeks post-injection with control (Ctrl) or miR-3189-3p-transfected LN-229/mCherry cells. Relative fluorescence values are represented as photon flux per second per square centimeter and steradian. C–F, U87MG/luciferase cells mock-transfected (control) or transfected with miR-3189-3p were injected intracranially into nude mice (n = 5/group). Images were taken after 15 (C) and 22 days (E). The asterisks in E indicate the same mouse at day 29. The mouse missing in the miR-3189-3p group at day 21 died of tumor-unrelated reasons (see details under “Results”). Relative plots of radiance are shown in D and F. G, Kaplan-Meier curves comparing mice bearing mock-transfected (ctrl) or miR-3189-3p-transfected U87MG/luciferase cells. Statistical analysis revealed a significantly longer survival (p < 0.05) of mice injected with miR-3189-3p-transfected cells.
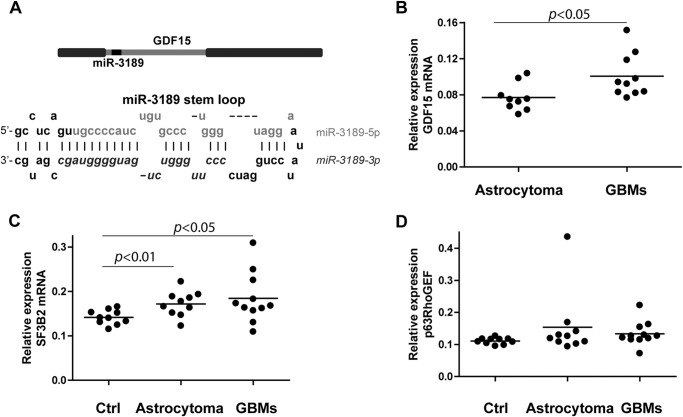
Because GDF15 and miR-3189 originate from the same transcript (Fig. 5A) and because miR-3189-3p is down-regulated in clinical samples, we asked whether GDF15 expression would also be down-regulated in human brain tumor extracts compared with controls. The results shown in Fig. 5, B–D represent the relative expression (1/ΔCt) of the indicated RNA species normalized using GAPDH as reference gene. Interestingly, although GDF15 was not detected by real-time PCR in normal brain tissues, its expression yielded a trend specific to tumor type with higher up-regulation in glioblastomas than in astrocytomas (Fig. 5B, p < 0.05). These data may indicate that GDF15 and miR-3189 are differentially regulated and that conditions or treatments that affect the expression of GDF15 may not alter the expression of the microRNA. Of the two major targets of miR-3189-3p, SF3B2 and p63RhoGEF, only SF3B2 showed a statistically significant increase in expression in both astrocytomas and glioblastomas compared with the control group (p < 0.01 and p < 0.05, respectively) (Fig. 5C), whereas p63RhoGEF mRNA expression analysis did not result in significant changes between the three groups (Fig. 5D).
FIGURE 5.
Evaluation of GDF15, SF3B2, and p63RhoGEF mRNA in clinical samples. A, schematic of the GDF15 gene encoding miR-3189 within the exon and of the stem loop of miR-3189 containing -3p and -5p mature microRNAs. Note that this location arrangement is typical of mirtrons. B–D, relative expression of GDF15, SF3B2, and p63RhoGEF mRNAs in human astrocytoma and glioblastoma (GBM) clinical samples, calculated as 1/ΔCt. Note that GDF15 mRNA was undetectable in control brain samples. Student's t test results (p values) are shown in the graphs.
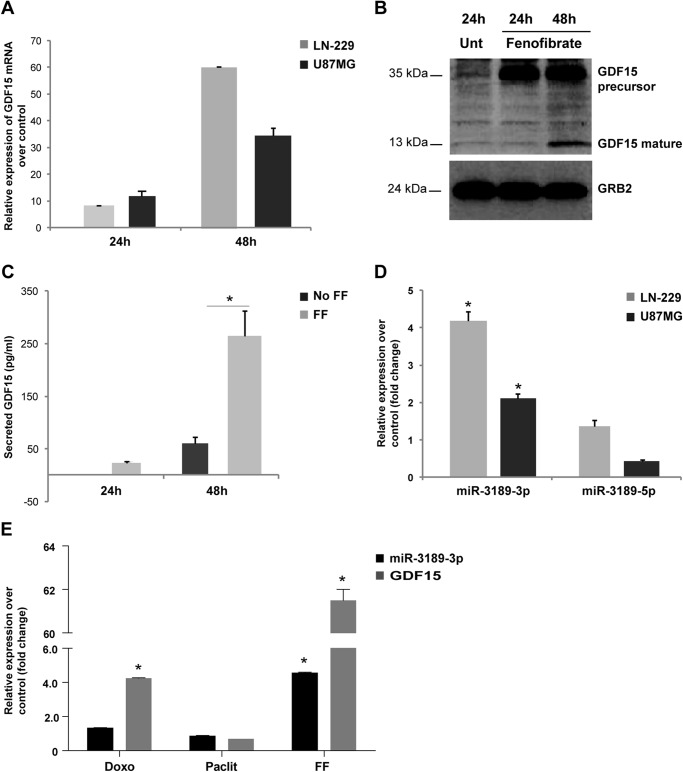
Expression of GDF15 and miR-3189-3p Is Increased after Fenofibrate Treatment
Previous reports have demonstrated that GDF15 expression is induced following treatment by a variety of chemotherapeutic agents (13, 21, 22). In line with these findings, we have also reported that this gene is up-regulated in a microarray analysis of glioblastoma cells treated with the metabolically active anticancer compound fenofibrate (4). Fenofibrate is a potent agonist of PPARα, which has exceptional anticancer properties, especially in tumors of neuroectodermal origin, including glioblastoma (5, 6, 23). To further analyze the effects of fenofibrate on this gene, we have exposed the human glioblastoma cell lines LN-229 and U87MG to 50 μm fenofibrate and monitored the expression of GDF15 at the 24 and 48 h time points. Note that up to 48 h, cells treated with fenofibrate look normal, and the toxic effect of fenofibrate is still reversible upon removal of the drug. However, the effects will become irreversible and the cells will die between 48 and 72 h (6). Following the treatment, total RNA was extracted and subjected to quantitative real-time PCR using GDF15-specific and GAPDH-specific primers. The results in Fig. 6A show a 60- and 35-fold up-regulation of GDF15 gene expression in the two cell lines. In agreement with these findings, a large increase in GDF15 protein content was detected by Western blot, and significant levels of secreted GDF15 were detected by ELISA at 48 h following fenofibrate treatment in LN-229 cells (Fig. 6, B and C, respectively). The 2-fold difference in the expression of GDF15 in the two cell lines was also reflected in the levels of miR-3189-3p because the amount of this microRNA in LN-229 was two times more than in U87MG cells (Fig. 6D). Expression of miR-3189-5p did not change in response to fenofibrate treatment in the cell lines tested. In addition to fenofibrate, we asked whether treatment of glioblastoma cells with common chemotherapeutic agents would trigger up-regulation of GDF15 and miR-3189-3p. Fig. 6E shows the results from the quantitative PCR performed to detect GDF15 mRNA and miR-3189-3p in LN-229 cells treated for 48 h with doxorubicin (18), paclitaxel (a mitotic inhibitor), and fenofibrate for 48 h. Relative expression compared with untreated cells indicates up-regulation of miR-3189-3p only in fenofibrate-treated cells, with a 4.8-fold-increase. GDF15 mRNA was also highly up-regulated by fenofibrate (more than 60-fold). This experiment confirms the specificity of the treatment and suggests an independent regulation of expression of GDF15 and miR-3189-3p.
FIGURE 6.
Fenofibrate treatment up-regulates GDF15 and miR-3189-3p expression in LN-229 and U87MG cells. A, real-time PCR to detect GDF15 mRNA expression at the indicated time points after fenofibrate treatment. Results are expressed as -fold change (2−ΔΔCt method) of the mRNA in fenofibrate-treated LN-229 cells compared with untreated (Unt) cells (p < 0.05). B, under the same experimental conditions, mature and precursor GDF15 proteins were detected by Western blots. GRB2 antibody was used to show equal loading of cellular lysates. C, ELISA to detect secreted mature (active) GDF15 protein in the culture medium obtained from LN-229 cells treated with FF and control (no FF). The data represent the change in GDF15 levels in medium from fenofibrate-treated cells compared with untreated cells. D, relative expression of miR-3189-3p and miR-3189–5p in LN-229 and U87MG cells treated for 48 h with fenofibrate. E, bar graph representing the relative expression of GDF15 and miR-3189-3p in LN-229 cells treated with doxorubicin (Doxo), paclitaxel (Paclit), and FF for 48 h. Note that, in D and E, a -fold change of <2 means unchanged levels compared with controls. The differences in expression levels depicted in A and C were statistically significant (p < 0.05), whereas, in D and E, only those with the asterisk (*) had a p < 0.05 compared with controls.
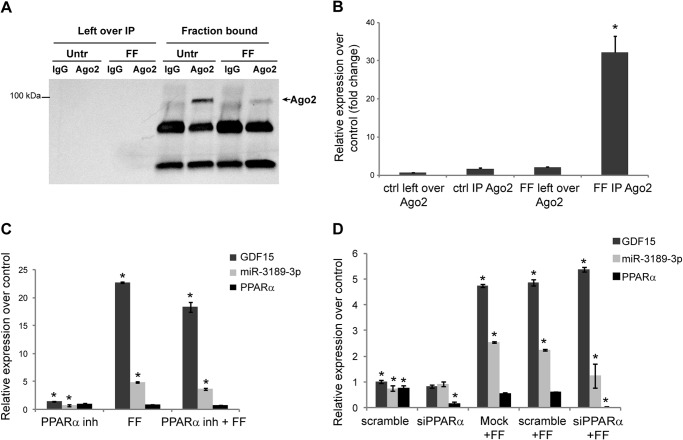
Fenofibrate exerts its proapoptotic action in cancer cells through PPARα-dependent and -independent mechanisms (5, 6, 23), and, therefore, the contribution of an increased expression of miR-3189-3p to fenofibrate-mediated tumor cell toxicity could be difficult to determine. Indeed, treatment of LN-229 cells with anti-miR-3189-3p did not rescue cells from fenofibrate-induced cell death (data not shown). However, in the presence of fenofibrate miR-3189-3p is functionally associated with Ago2 complexes, as determined by RNA-IP (Fig. 7). Although endogenous levels of Ago2 were undetectable in LN-229 cells by Western blot, it was immunoprecipitated efficiently under all conditions tested (Fig. 7A). When compared with untreated cells, a nearly 35-fold overexpression of miR-3189-3p linked to Ago2 immunocomplexes was measured in extracts from FF-treated cells (Fig. 7B). Finally, we asked whether the fenofibrate-mediated increase in GDF15 and miR-3189-3p was a PPARα-dependent event. PPARα activity was inhibited at the level of receptor by a specific inhibitor, GW 9662 (6, 23) or through its down-regulation via siRNAs. Total RNA was extracted 48 h after the treatment/transfection, and levels of GDF15 and miR-3189-3p were determined by quantitative PCRs. The results shown in Fig. 7, C and D, indicate no significant impact of either compound on the levels of GDF15 and miR-3189-3p because their expression triggered by fenofibrate was comparable with the controls. Although miR-3189-3p expression showed a reproducible, moderate reduction in both experiments, this difference was not statistically significant. Efficacy of the siRNA to down-regulate PPARα was determined by RT-PCR and is included in Fig. 7D.
FIGURE 7.
MiR-3189-3p is up-regulated and incorporated into the RNA-induced silencing complex in cells treated with fenofibrate. A, Western blot to detect Ago2 after immunoprecipitation of lysates obtained from untreated (Untr) and fenofibrate-treated cells. Left over IP represents the fraction of lysates obtained after overnight incubation with Ago2 antibody or the control isotype IgG and is used as a negative control. B, real-time PCR to detect miR-3189-3p. Results are expressed as -fold change of the microRNAs in fenofibrate-treated cells compared with untreated cells. The enrichment of microRNAs or mRNAs in the RNA-induced silencing complex was calculated according to the formula 2−(CtAgo2 − CtIgG) and normalized over RNU6B. ctrl, control. C and D, relative expression of GDF15 and miR-3189-3p under the indicated conditions (treatments and transfections) compared with controls. *, p < 0.05 compared with the respective controls.
DISCUSSION
MiR-3189 has been identified previously as a possible mirtron expressed in melanoma (24), and an inhibitory effect of miR-3189–5p on TGFβR2 has been hypothesized (25), but a function for miR-3189-3p or miR-3189–5p has not been validated experimentally. Mirtrons are microRNAs encoded within introns, and their biogenesis follows a non-canonical, Drosha/DGCR8-independent pathway that relies on mRNA splicing and on RNA lariats debranching enzymes (reviewed in Ref. 26). Differently from canonical pre-microRNA stem loops, microRNAs generated from the 3′ (-3p) of the mirtron hairpin appear to be more stable than those generated from the 5′ (-5p) (27). This may explain why, in our experiments, even when miR-3189–5p expression was induced slightly and variably by mitogenic stimuli (10% FBS), this microRNA was not detected in the Ago2-immunoprecipitated complex (data not shown).
Overexpression of miR-3189-3p in LN-229 and U87MG cells resulted in morphological changes accompanied by attenuation of cell proliferation and migration (Fig. 1). Indeed, the anti-proliferative effect miR-3189-3p prevented the generation of stable cell lines expressing this microRNA (data not shown). MicroRNA gene target prediction databases indicate the splicing factor SF3B2 and the Rho guanine nucleotide exchange factor p63RhoGEF as the top putative targets. Given their validated role in proliferation and migration (see below), we sought to investigate their role in the biological effects of miR-3189-3p. Overexpression of miR-3189-3p in LN-229 and U87MG cells resulted in down-regulation of SF3B2 and p63RhoGEF mRNAs (Fig. 2A) and proteins (Fig. 2B). Although the down-regulation of SF3B2 and p63RhoGEF mRNAs was comparable in the two cell lines, it was stronger at the level of proteins in LN-229 cells. This may suggest a different mechanism of posttranscriptional regulation for these proteins in the two types of cells tested. Nevertheless, expression of miR-3189-3p had a strong biological effect on both LN-229 and U87MG cells, impairing their migration and growth. These effects have been shown to be mediated through down-regulation of p63RhoGEF and SF3B2, respectively. With respect to p63RhoGEF, previous reports have also shown a role for this protein in cell migration (28, 29). Specifically, Hayashi et al. (20) demonstrated that the expression of p63RhoGEF is essential for lamellipodial polarization during serum-induced chemotaxis. Our findings are in agreement with a role for p63RhoGEF in cell motility because siRNA against p63RhoGEF impaired migration of LN-229 cells (Fig. 3D). Our results showing the inhibitory effect of miR-3189-3p on p63RhoGEF-overexpressing cells (Fig. 3C) demonstrate that inhibition of cell migration by miR-3189-3p is only partially due to the down-regulation of p63RhoGEF. Such a result may not be surprising because other members of the RhoA family of guanine nucleotide exchange factors, such as SLIT-ROBO Rho GTPase-activating protein 2 (SRGAP2) and Rho guanine nucleotide exchange factor (GEF) 12 (ARCHGEF12) are other putative targets of miR-3189-3p and may equally contribute to the impaired cell migration by miR-3189-3p. Likewise, the striking change in cellular morphology upon expression of miR-3189-3p might be the result of this microRNA targeting multiple genes involved in cytoskeletal remodeling.
It has been reported that the expression of the transcription factor E2F-1 is dependent on the presence of SF3B2 in the cell (19). Furthermore, E2F-1 has been shown to be a master regulator of cell cycle progression (30, 31). Therefore, it is not surprising that SF3B2 down-regulation by miR-3189-3p in glioblastoma delayed cell growth. In our studies, this effect has been shown to be dependent on SF3B2 because overexpression of this gene in the presence of miR-3189-3p restored the proliferative capacity of LN-229 cells (Fig. 3A). The antiproliferative activity of miR-3189-3p was also demonstrated in vivo in nude mice bearing subcutaneous LN-229/mCherry (Fig. 4, A and B) or U87MG/luciferase tumors (data not shown). Even more impressive were the results obtained with intracranial injection of U87MG/luciferase cells treated previously with vehicle or miR-3189-3p (Figs 4, C and E). Further in vivo experiments should clarify whether miR-3189-3p, besides preventing tumor growth, could also have a therapeutic effect by reducing tumor size.
When we analyzed the genomic sequence of miR-3189-3p and found that it was located in the intron of GDF15, we thought that this microRNA could be coexpressed and could have been involved in the activity of GDF15. In fact, our data indicate that these two molecules are regulated differentially in various experimental and clinical settings. Our analysis of clinical samples indicates that GDF15 is undetectable in control brain tissues and up-regulated in gliomas, whereas miR-3189-3p is present in normal brain tissue and down-regulated in gliomas (Figs. 5B and 1A, respectively). Experimentally, we serum-starved LN-229 cells and then stimulated them with 10% FBS or EGF (50 ng/ml) for various times and, surprisingly, found that FBS treatment down-regulated GDF15 and up-regulated miR-3189-3p and miR-3189–5p, whereas EGF up-regulated GDF15 and did not significantly change the expression of either microRNAs (data not shown). Further work is needed to clarify the posttranscriptional events that lead to the differential expression of those coencoded molecules.
Cytotoxic agents such as etoposide and doxorubicin have been shown to increase GDF15 expression (7). However, in our experimental setting, among doxorubicin, paclitaxel, and fenofibrate, only the latter was able to increase both GDF15 and miR-3189-3p expression (Fig. 6E). It is likely that the discrepancy of our results with previous work is due to the type of cells utilized. Indeed, fenofibrate treatment triggered two times more GDF15 and miR-3189-3p expression in LN-229 than in U87MG cells, suggesting a variability in the responses within the same type of tumor cells. Furthermore, we observed that, at least in LN-229 cells, although fenofibrate treatment always induced expression of GDF15, such an expression ranged between 5- and 6-fold to 60-fold (compare Figs. 6, A and E, and 7, C and D) within 48 h treatment, whereas the range of miR-3189-3p expression was consistently between 2.5 and 5. Again, this discrepancy may indicate that expression of GDF15 and miR-189-3p is regulated independently and that expression of GDF15 likely follows a Gaussian curve with the peak in a dynamic range. A detailed time course monitoring of GDF15 levels up to 72 h (or until cells detach and die) after fenofibrate treatment should clarify its dynamic range of expression.
Overexpression of GDF15 protein or treatment of LN-229 cells with its soluble version did not elicit any morphological or biological effects in LN-229 cells in vitro (data not shown). Similarly, anti-miR-3189-3p did not protect fenofibrate-treated cells from apoptosis, indicating that up-regulation of miR-3189-3p is not required for fenofibrate-mediated cell death. Given the broad range of anticancer effects triggered by fenofibrate (5, 6, 22, 23, 32, 33) and the activity of a single microRNA, it is not surprising that miR-3189-3p is not contributing to the massive cell death mediated by fenofibrate. The microRNA itself seems to have a cytostatic rather than cytotoxic effect on the cells (Fig. 1). Treatment of glioblastoma cells with fenofibrate results in massive cell death that occurs between 48 and 72 h post-treatment and involves PPARα-dependent and -independent mechanisms (5, 6, 22, 23, 32, 33). Up to 48 h post-treatment cells appear normal in morphology, although they are still suffering from mitochondrial dysfunction and metabolic crisis (6). Increased expression of GDF15 and/or miR-3189-3p appears to be a PPARα-independent event (Fig. 7, C and D) and could be the result of mitochondrial stress and, similar as induction of autophagy (6), it may reflect an attempt by the cells to counteract the toxic effects of fenofibrate. However, the fact that miR-3189-3p is highly incorporated into the Ago2 complex (Fig. 7) indicates that it is functionally active in this context. One could hypothesize that the effect of miR-3189-3p on its targets may augment a parallel, irreversible action (perhaps transcriptional) on the same targets exerted by fenofibrate through microRNA-independent mechanisms. Although further investigation may help to clarify those points, the tumor suppressor activity in vitro and in vivo of an overexpressed miR-3189-3p remains an important result from this study that could be exploited further as a potential therapy for this devastating cancer.
Acknowledgments
We thank Constance Porretta (LSUHSC Comprehensive Alcohol Research Center) for technical assistance with flow cytometry acquisition and analysis.
This work was supported, in whole or in part, by NIGMS/National Institutes of Health Grant 1U54 GM104940, which funds the Louisiana Clinical and Translational Science Center, and by National Institutes of Health Grants P20 GM103501 (to F.P. and K.R.) and RO1 CA095518 (to K.R.). This work was also supported by the Stanley S Scott Cancer Research Center, LSUHSC, Louisiana Cancer Research Center.
- PPARα
- peroxisome proliferator-activated receptor α
- RIP
- RNA-binding protein immunoprecipitation
- IP
- immunoprecipitation
- FF
- fenofibrate
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1. Nakada M., Nakada S., Demuth T., Tran N. L., Hoelzinger D. B., Berens M. E. (2007) Molecular targets of glioma invasion. Cell. Mol. Life Sci. 64, 458–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagasawa D. T., Chow F., Yew A., Kim W., Cremer N., Yang I. (2012) Temozolomide and other potential agents for the treatment of glioblastoma multiforme. Neurosurg. Clin. N. Am. 23, 307–322, ix [DOI] [PubMed] [Google Scholar]
- 3. Hegi M. E., Diserens A. C., Gorlia T., Hamou M. F., de Tribolet N., Weller M., Kros J. M., Hainfellner J. A., Mason W., Mariani L., Bromberg J. E., Hau P., Mirimanoff R. O., Cairncross J. G., Janzer R. C., Stupp R. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 [DOI] [PubMed] [Google Scholar]
- 4. Jeansonne D., Pacifici M., Lassak A., Reiss K., Russo G., Zabaleta J., Peruzzi F. (2013) Differential effects of MicroRNAs on glioblastoma growth and migration. Genes 4, 46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drukala J., Urbanska K., Wilk A., Grabacka M., Wybieralska E., Del Valle L., Madeja Z., Reiss K. (2010) ROS accumulation and IGF-IR inhibition contribute to fenofibrate/PPARα-mediated inhibition of glioma cell motility in vitro. Mol. Cancer 9, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilk A., Wyczechowska D., Zapata A., Dean M., Mullinax J., Marrero L., Parsons C., Peruzzi F., Culicchia F., Ochoa A., Grabacka M., Reiss K. (2014) Molecular mechanisms of fenofibrate-induced metabolic catastrophe and glioblastoma cell death. Mol. Cell. Biol. 35, 182–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mimeault M., Batra S. K. (2010) Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J. Cell. Physiol. 224, 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Baek S. J., Eling T. E. (2013) The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem. Pharmacol. 85, 597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eling T. E., Baek S. J., Shim M., Lee C. H. (2006) NSAID activated gene (NAG-1), a modulator of tumorigenesis. J. Biochem. Mol. Biol. 39, 649–655 [DOI] [PubMed] [Google Scholar]
- 10. Kim J. M., Kosak J. P., Kim J. K., Kissling G., Germolec D. R., Zeldin D. C., Bradbury J. A., Baek S. J., Eling T. E. (2013) NAG-1/GDF15 transgenic mouse has less white adipose tissue and a reduced inflammatory response. Mediators Inflamm. 2013, 641851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li A., Walling J., Ahn S., Kotliarov Y., Su Q., Quezado M., Oberholtzer J. C., Park J., Zenklusen J. C., Fine H. A. (2009) Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 69, 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiu S. C., Wang M. J., Yang H. H., Chen S. P., Huang S. Y., Chen Y. L., Lin S. Z., Harn H. J., Pang C. Y. (2011) Activation of NAG-1 via JNK signaling revealed an isochaihulactone-triggered cell death in human LNCaP prostate cancer cells. BMC Cancer 11, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshioka H., Kamitani H., Watanabe T., Eling T. E. (2008) Nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) expression is increased by the histone deacetylase inhibitor trichostatin A. J. Biol. Chem. 283, 33129–33137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eletto D., Russo G., Passiatore G., Del Valle L., Giordano A., Khalili K., Gualco E., Peruzzi F. (2008) Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J. Cell. Physiol. 216, 764–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pacifici M., Delbue S., Ferrante P., Jeansonne D., Kadri F., Nelson S., Velasco-Gonzalez C., Zabaleta J., Peruzzi F. (2013) Cerebrospinal fluid miRNA profile in HIV-encephalitis. J. Cell. Physiol. 228, 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rom S., Rom I., Passiatore G., Pacifici M., Radhakrishnan S., Del Valle L., Piña-Oviedo S., Khalili K., Eletto D., Peruzzi F. (2010) CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J. 24, 2292–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curtale G., Mirolo M., Renzi T. A., Rossato M., Bazzoni F., Locati M. (2013) Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc. Natl. Acad. Sci. U.S.A. 110, 11499–11504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marrero L., Wyczechowska D., Musto A. E., Wilk A., Vashistha H., Zapata A., Walker C., Velasco-Gonzalez C., Parsons C., Wieland S., Levitt D., Reiss K., Prakash O. (2014) Therapeutic efficacy of aldoxorubicin in an intracranial xenograft mouse model of human glioblastoma. Neoplasia 16, 874–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orr S. J., Boutz D. R., Wang R., Chronis C., Lea N. C., Thayaparan T., Hamilton E., Milewicz H., Blanc E., Mufti G. J., Marcotte E. M., Thomas N. S. (2012) Proteomic and protein interaction network analysis of human T lymphocytes during cell-cycle entry. Mol. Syst. Biol. 8, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayashi A., Hiatari R., Tsuji T., Ohashi K., Mizuno K. (2013) p63RhoGEF-mediated formation of a single polarized lamellipodium is required for chemotactic migration in breast carcinoma cells. FEBS Lett. 587, 698–705 [DOI] [PubMed] [Google Scholar]
- 21. Shimizu S., Kadowaki M., Yoshioka H., Kambe A., Watanabe T., Kinyamu H. K., Eling T. E. (2013) Proteasome inhibitor MG132 induces NAG-1/GDF15 expression through the p38 MAPK pathway in glioblastoma cells. Biochem. Biophys. Res. Commun. 430, 1277–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Araki H., Tamada Y., Imoto S., Dunmore B., Sanders D., Humphrey S., Nagasaki M., Doi A., Nakanishi Y., Yasuda K., Tomiyasu Y., Tashiro K., Print C., Charnock-Jones D. S., Kuhara S., Miyano S. (2009) Analysis of PPARα-dependent and PPARα-independent transcript regulation following fenofibrate treatment of human endothelial cells. Angiogenesis 12, 221–229 [DOI] [PubMed] [Google Scholar]
- 23. Wilk A., Urbanska K., Grabacka M., Mullinax J., Marcinkiewicz C., Impastato D., Estrada J. J., Reiss K. (2012) Fenofibrate-induced nuclear translocation of FoxO3A triggers Bim-mediated apoptosis in glioblastoma cells in vitro. Cell Cycle 11, 2660–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stark M. S., Tyagi S., Nancarrow D. J., Boyle G. M., Cook A. L., Whiteman D. C., Parsons P. G., Schmidt C., Sturm R. A., Hayward N. K. (2010) Characterization of the melanoma miRNAome by deep sequencing. PloS ONE 5, e9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sivadas V. P., George N. A., Kattoor J., Kannan S. (2013) Novel mutations and expression alterations in SMAD3/TGFBR2 genes in oral carcinoma correlate with poor prognosis. Genes Chromosomes Cancer 52, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 26. Lai E. C. (2003) microRNAs: runts of the genome assert themselves. Curr. Biol. 13, R925–936 [DOI] [PubMed] [Google Scholar]
- 27. Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C. (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swenson-Fields K. I., Sandquist J. C., Rossol-Allison J., Blat I. C., Wennerberg K., Burridge K., Means A. R. (2008) MLK3 limits activated Gαq signaling to Rho by binding to p63RhoGEF. Mol. Cell 32, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang X., Jin R., Qu G., Wang X., Li Z., Yuan Z., Zhao C., Siwko S., Shi T., Wang P., Xiao J., Liu M., Luo J. (2013) GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73, 6206–6218 [DOI] [PubMed] [Google Scholar]
- 30. DeGregori J., Kowalik T., Nevins J. R. (1995) Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell Biol. 15, 4215–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsumura I., Tanaka H., Kanakura Y. (2003) E2F1 and c-Myc in cell growth and death. Cell Cycle 2, 333–338 [PubMed] [Google Scholar]
- 32. Urbanska K., Pannizzo P., Grabacka M., Croul S., Del Valle L., Khalili K., Reiss K. (2008) Activation of PPARα inhibits IGF-I-mediated growth and survival responses in medulloblastoma cell lines. Int. J. Cancer 123, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Panigrahy D., Kaipainen A., Huang S., Butterfield C. E., Barnés C. M., Fannon M., Laforme A. M., Chaponis D. M., Folkman J., Kieran M. W. (2008) PPARα agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc. Natl. Acad. Sci. U.S.A. 105, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]