Background: Anionic lipids compete for electrostatic interaction in membrane proteins.
Results: Despite competition, anionic lipids stabilize the integrin αIIbβ3 transmembrane complex.
Conclusion: Stabilizing anionic lipid-protein interactions exist and supersede destabilizing effects.
Significance: Anionic lipid-mediated stabilization of membrane proteins may be of a general nature.
Keywords: Biophysics, Integrin, Membrane Lipid, Membrane Protein, Molecular Dynamics, Nuclear Magnetic Resonance (NMR), Protein-Lipid Interaction
Abstract
Cationic membrane-proximal amino acids determine the topology of membrane proteins by interacting with anionic lipids that are restricted to the intracellular membrane leaflet. This mechanism implies that anionic lipids interfere with electrostatic interactions of membrane proteins. The integrin αIIbβ3 transmembrane (TM) complex is stabilized by a membrane-proximal αIIb(Arg995)-β3(Asp723) interaction; here, we examine the influence of anionic lipids on this complex. Anionic lipids compete for αIIb(Arg995) contacts with β3(Asp723) but paradoxically do not diminish the contribution of αIIb(Arg995)-β3(Asp723) to TM complex stability. Overall, anionic lipids in annular positions stabilize the αIIbβ3 TM complex by up to 0.50 ± 0.02 kcal/mol relative to zwitterionic lipids in a headgroup structure-dependent manner. Comparatively, integrin receptor activation requires TM complex destabilization of 1.5 ± 0.2 kcal/mol, revealing a sizeable influence of lipid composition on TM complex stability. We implicate changes in lipid headgroup accessibility to small molecules (physical membrane characteristics) and specific but dynamic protein-lipid contacts in this TM helix-helix stabilization. Thus, anionic lipids in ubiquitous annular positions can benefit the stability of membrane proteins while leaving membrane-proximal electrostatic interactions intact.
Introduction
Membrane proteins adopt pivotal physiological roles through mediating the exchange of matter, energy, and information between cells. Indispensable to the structural integrity of a membrane protein, membrane lipids engage membrane proteins in interactions that range from dynamic solvation to highly specific association, mediated by annular lipids and nonannular lipids, respectively (1–3). Protein-lipid interactions also play a critical role in defining the membrane topology of proteins. Membrane proteins are arranged such that positively charged residues localize to the intracellular membrane face (4). The structural basis of this phenomenon is the interaction of anionic lipids, which are concentrated to the intracellular membrane leaflet, with cationic protein residues (5, 6). This fundamental mechanism implies that anionic lipids also compete with electrostatic interactions between transmembrane (TM)3 helices within the intracellular lipid headgroup region. Anionic lipid competition with electrostatic interactions between TM helices would have profound consequences for the structure and stability of membrane proteins. To explore this scenario, we examine here the impact of anionic lipids on the thermodynamic stability of the integrin αIIbβ3 TM complex and the interaction of αIIbβ3 TM segments with anionic versus zwitterionic lipids.
The integrin αIIbβ3 TM complex exhibits an αIIb(Arg995)-β3(Asp723) electrostatic interaction within the cytosolic lipid headgroup region (see Fig. 1A) that is highly conserved among integrin receptors (7, 8). It stabilizes the TM complex, which suppresses the activation of the integrin αIIbβ3 cell adhesion receptor (7, 8). Integrin αIIbβ3 activation is a pivotal step in platelet aggregation and, thus, hemostasis and pathological thrombosis (9, 10). We employed biophysical, biochemical, and computational techniques to quantify the influence of anionic versus zwitterionic lipids on the stability of the integrin αIIbβ3 TM complex and the αIIb(Arg995)-β3(Asp723) interaction. Notwithstanding the competitive nature of anionic lipids reflected in preferential interactions with cationic αIIb and β3 residues including αIIb(Arg995), annular anionic lipids paradoxically stabilized the αIIbβ3 TM complex relative to electroneutral lipids.
FIGURE 1.
Integrin αIIbβ3 TM structure, sequence, and lipids studied. A, structure of the TM complex in phospholipid bicelles (Protein Data Bank entry 2k9j, model 17) (8). The side chains of most positively charged residues are depicted as sticks relative to approximate membrane borders (8). B, schematic depiction of a phospholipid bicelle, employed to reconstitute the TM complex. C, headgroup structures of POPC, POPG, and POPS. For reference, arginine guanidino and lysine amino moieties are depicted. The molecular volumes of choline, glycerol, and serine are 120.2, 87.1, and 90.6 Å3, respectively. D, amino acid sequences of the αIIb and β3 TM domains with charged residues highlighted. Membrane boundaries are indicated (8).
EXPERIMENTAL PROCEDURES
TM Peptide Preparation
Peptides encompassing human integrin αIIb(Ala958–Pro998) and β3(Pro685–Phe727), which incorporated β3(C687S), were produced as described (11, 12). Point mutations were introduced using QuikChange mutagenesis (Stratagene, Inc.) without having to modify subsequent peptide purification. Peptide concentrations were measured by UV spectroscopy using ϵ(αIIb)280 nm = 16,500 m−1 cm−1, ϵ(β3)280 nm = 5,500 m−1 cm−1, and ϵ(αIIbβ3)280 nm = 22,000 m−1 cm−1. Purified, freeze-dried peptides were dissolved in 67% CH3CN, 33% H2O for concentration measurements by UV spectroscopy.
KXY Measurements
The αIIb + β3 ⇄ αIIbβ3 equilibrium constant on the mole fraction scale (KXY) was quantified by NMR and ITC. For NMR measurements, fresh samples were prepared at the desired peptide concentrations for each titration point by mixing peptides in 67% CH3CN, 33% H2O. Subsequent to freeze-drying, peptides were taken up in 320 μl of 25 mm HEPES·NaOH, pH 7.4, 6% D2O, 0.02% (w/v) NaN3, 400 mm DHPC, and 120 mm of long chain lipids (see Table 1). HN-N correlation spectra were acquired on a Bruker Avance 700 spectrometer equipped with a cryoprobe at 28 °C, using the TROSY-HSQC pulse scheme (13) with acquisition times of 85.4 ms for 1HN and 71.7 ms for 15N. The volume of a monomeric, isotope-labeled β3 resonance at total peptide concentrations [β3] and [αIIb] was extracted (VM) and expressed as 1 − VM/VM,0, where VM,0 is the resonance volume in the absence of αIIb. For isotope labeled αIIb and unlabeled β3, analyte and titrant interchange. KXY was obtained by nonlinear curve fitting of 1 − VM/VM,0 as a function of unlabeled peptide concentration (see Fig. 2, A and B) as described in detail previously (14). ΔG° is obtained as −RT ln KXY, where R denotes the gas constant, and T denotes the absolute temperature.
TABLE 1.
Thermodynamic parameters of αIIbβ3 TM association as a function of lipid environment
| Peptides | Lipida | KXY | ΔHo | ΤΔSo | ΔGo |
|---|---|---|---|---|---|
| kcal/mol | kcal/mol | kcal/mol | |||
| 2H/15N-αIIb + β3 | POPC | 1390 ± 30 | −4.33 ± 0.01 | ||
| 2H/15N-αIIb + β3 | 2 POPC:1 POPS | 2900 ± 100 | −4.77 ± 0.03 | ||
| αIIb + 2H/15N-β3 | POPC | 2070 ± 40 | −4.57 ± 0.01 | ||
| αIIb + 2H/15N-β3 | 2 POPC:1 POPS | 2900 ± 100 | −4.76 ± 0.02 | ||
| αIIb + 2H/15N-β3 | 1 POPC:2 POPS | 3400 ± 200 | −4.86 ± 0.03 | ||
| αIIb + 2H/15N-β3 | POPS | 3800 ± 100 | −4.94 ± 0.02 | ||
| αIIb + β3 | POPC | 3250 ± 60 | −16.0 ± 0.1 | −11.1 ± 0.1 | −4.84 ± 0.01 |
| αIIb + β3 | POPG | 5900 ± 200 | −18.9 ± 0.2 | −13.7 ± 0.2 | −5.20 ± 0.02 |
| αIIb + β3 | POPS | 7400 ± 300 | −19.4 ± 0.3 | −14.0 ± 0.3 | −5.34 ± 0.02 |
| αIIb + β3 17 mm l-O-Phosphoserine | POPC | 2630 ± 70 | −21.5 ± 0.3 | −16.8 ± 0.3 | −4.71 ± 0.01 |
| αIIb + β3 500 mm NaCl | POPC | 2220 ± 60 | −21.1 ± 0.3 | −16.5 ± 0.3 | −4.61 ± 0.01 |
| αIIb(R995A) + β3b | POPC | 250 ± 70 | −15 ± 4 | −12 ± 4 | −3.3 ± 0.2 |
| αIIb(R995A) + β3b | POPS | 300 ± 100 | −34 ± 16 | −31 ± 16 | −3.3 ± 0.3 |
| αIIb(K994A) + β3 | POPC | 3500 ± 300 | −10.5 ± 0.3 | −5.7 ± 0.3 | −4.88 ± 0.04 |
| αIIb(K994A) + β3 | POPS | 3600 ± 100 | −18.1 ± 0.3 | −13.2 ± 0.3 | −4.89 ± 0.02 |
a NMR measurements, for which no ΔH° and TΔS° values are available, were performed in 25 mm HEPES, pH 7.4, 0.02% NaN3 at 28 °C. ITC measurements were carried out in 25 mm NaH2PO4/Na2HPO4, pH 7.4, at 28 °C. For NMR and ITC, bicelles were formed from 400 mm DHPC, 120 mm of the indicated long chain lipids and 43 mm DHPC/17 mm of the indicated long chain lipid, respectively. The presence of 9 mm free, nonbicellar DHPC (21) and resulting effective q factors of 0.31 and 0.5 are noted.
b From competitive αIIb/αIIb(R995A) binding measurements (Fig. 4).
FIGURE 2.
Anionic lipid-mediated stabilization of the integrin αIIbβ3 TM complex. A and B, measurement of αIIb + β3 ⇄ αIIbβ3 equilibrium constant by NMR. The disappearance of the monomeric αIIb(Gly972) and β3(Gly702) resonances is plotted as 1 − VM/VM,0, where VM,0 denotes the resonance volume of pure monomer, and VM denotes the residual monomer volume at increasing concentrations of partnering peptide. C, free energy change, ΔG°, of αIIbβ3 association as a function of POPS fraction. Phospholipid bicelles consisting of 391 mm DHPC and 120 mm of the depicted long chain lipid (qeff = 0.31) were employed (Table 1). D, ITC measurements of αIIbβ3 TM association in phospholipid bicelles consisting of 34 mm DHPC and 17 mm of the depicted long chain lipid (qeff = 0.5) using a starting β3 concentration of 10 μm.
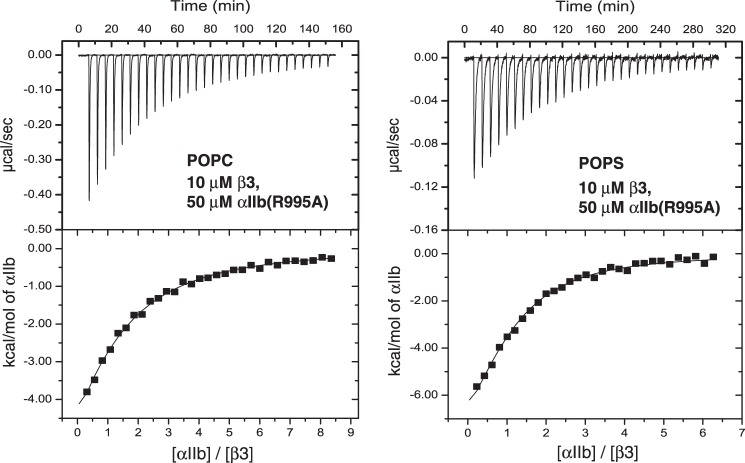
ITC measurements were carried on a Microcal VP-ITC calorimeter. 10 μm of β3 peptide in the 1.425-ml sample cell was titrated with αIIb peptide by injecting 9-μl aliquots over a period of 10 s each. Unless otherwise specified, measurements were carried out at 28 °C in 25 mm NaH2PO4/Na2HPO4, pH 7.4, 43 mm DHPC, and 17 mm of long chain lipid (see Table 1). Prior to data analysis, the measurements were corrected for the heat of dilutions of the αIIb and β3 peptides. The αIIbβ3 complex stoichiometry was fixed at 1:1 (14), and the reaction enthalpy (ΔH°) and KXY were calculated from the measured heat changes, δHi, as described previously (14). The entropy change, ΔS°, is obtained as (ΔH° − ΔG°)/T. For the relatively weak αIIb(R995A)β3 complex, KXY and ΔH° values were measured by competitive binding experiments (15, 16); specifically, the binding of αIIb to β3 was evaluated in the presence of competing αIIb(R995A).
We explicitly note that ITC experiments are often performed with the factor c = KXY × [peptide]0/[lipid] maintained above 1 when titrating to a 2:1 molar ratio of ligand-to-protein, which yields a sigmoidal titration curve (17). However, when titrating to binding saturation and when the complex stoichiometry is fixed, there is less restriction on c in terms of the accuracy of KXY (18, 19). To compare ΔG° values between different bicelle compositions, ideal solvent behavior of the hydrophobic phase (20) is required. Such behavior manifests in the independence of ΔG° from the employed peptide to lipid ratio (20) and requires relatively high peptide to lipid ratios for integrin αIIbβ3 in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bicelles (14). In POPC and POPG bicelles, we have experimentally verified ideal solvent behavior for the studied peptide to lipid range. Specifically, in 42 mm DHPC, 21 mm POPC; 34 mm DHPC, 17 mm POPC; and 26 mm DHPC, 13 mm POPC, ΔG° values were −4.76 ± 0.01, −4.84 ± 0.01, and −4.80 ± 0.01 kcal/mol, respectively. In 42 mm DHPC, 21 mm POPG; 34 mm DHPC, 17 mm POPG; and 26 mm DHPC, 13 mm POPG, ΔG° values were −5.24 ± 0.01, −5.20 ± 0.02, and −5.14 ± 0.02 kcal/mol, respectively. The free, nonbicellar DHPC concentration, estimated at 9 mm (21), was not counted toward the stated bicelle lipid concentration.
Quantification of Lipid-Protein Contacts and Mn2+EDDA2− Protection
To quantify NMR peak intensities of arginine ϵ-NH as a function of lipid type, 2H/15N-labeled freeze-dried peptides were dissolved in 320 μl of 25 mm HEPES·NaOH, pH 7.4, 6% D2O, 0.02% (w/v) NaN3, 200 mm DHPC, and 60 mm of long chain lipids (see Fig. 5A) yielding a final peptide concentration of 0.2 mm. HSQC-TROSY spectra were acquired at 28 °C and 700 MHz. The detection of arginine η-NH2 resonances was attempted by HSQC experiments (22). Solvent magnetization was maintained at +Iz throughout experiments whenever possible (23). Applying those same sample conditions, protection from paramagnetic Mn2+EDDA2− was evaluated at 35 °C. The ratio of H-N cross-peak intensities in the presence and absence of 1 mm Mn2+EDDA2−, I/I0, was calculated. To correct for small differences in sample conditions, I/I0 was uniformly scaled to 1 for the residues in the membrane center.
FIGURE 5.
Lipid headgroup-arginine side chain contacts. A, cumulative NMR signal intensities of arginine ϵ-HN resonances as function of bicelle long chain lipid composition and integrin subunit. Three arginines (residues 962, 995, and 997) are found in the αIIb TM segment, whereas the single Arg724 resides in the β3 TM segment (Fig. 1D). B, arginine ϵ-HN NMR cross-peak intensities of the depicted αIIb mutants reveal the relative contribution of each αIIb arginine to the cumulative signal intensity observed in A. Spectra were recorded at 28 °C and 700 MHz using TM peptides reconstituted at concentrations of 0.2 mm in 200 mm DHPC, 40 mm POPC, 20 mm POPS in 25 mm HEPES·NaOH, pH 7.4. C, arginine ϵ-HN contacts with PO4− of POPC and PO4−/COO− of POPS in MD simulations. For each lipid type, three 30-ns simulations starting from different initial starting coordinates were performed. D, signal intensity of αIIb(Arg995/ϵ-NH) in the absence and presence of β3 TM peptide. R962K/R997K-substituted αIIb peptide was reconstituted at a concentration of 0.2 mm in 200 mm DHPC, 40 mm POPC, 20 mm POPS in 25 mm HEPES·NaOH, pH 7.4. β3 peptide was added to a concentration of 0.3 mm. Spectra are shown at identical contour levels and were recorded at 28 °C and 700 MHz.
Assay of Integrin αIIbβ3 Receptor Activity in Lipid Nanodiscs
The receptor was purified from outdated human platelets based on a protocol modified from Ye et al. (24, 25). In brief, outdated platelets were centrifuged at 300 × g to remove erythrocytes and leukocytes, followed by centrifugation at 1800 × g to pellet platelets. The platelets were washed twice with Tris-buffered saline and membrane proteins extracted by incubating overnight in 20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 5 mm PMSF, 0.5 mm CaCl2, 10 μm leupeptin, 10 μm protease inhibitor E64 (Sigma), and 2.76 μm calpeptin buffer. Integrin αIIbβ3 was purified on a concanavalin A column and passed through a heparin column to remove thrombospondin 1. Subsequently, the receptor was purified by gel filtration chromatography, and the inactive fraction was isolated as the flowthrough of an immobilized KYGRGDS affinity matrix, as described by Steiner and co-workers (26). Purified integrins were stored in 20 mm Tris, pH 7.4, 150 mm NaCl, 0.1% Triton X-100, 1 mm MgCl2, and 1 mm CaCl2 buffer at −80 °C.
Integrin nanodiscs were assembled based on a protocol adapted from a previous report (27, 28). POPC, POPS, and POPG were solubilized in chloroform, mixed thoroughly, and dried onto a glass tube under a stream of nitrogen gas. The homogeneous lipid mixture was then solubilized in 100 mm cholate, 10 mm Tris, pH 7.4, and 100 mm NaCl, resulting in a lipid concentration of 50 mm. 72 μl of the lipid solution was then mixed with 200 μl of 200 μm membrane scaffold protein in H2O and 200 μl of 5 μm purified inactive integrin (see above). This resulted in a final lipids:membrane scaffold protein:protein ratio of 90:1:0.025 in a total volume of 472 μl. The integrin nanodiscs were assembled by removing the detergents with SM-2 biobeads overnight at room temperature. Finally, the assembled integrin nanodiscs were purified by gel filtration using a Superdex 200 column in 20 mm Tris, pH 7.4, 150 mm NaCl, 0.5 mm CaCl2 solution. Successful nanodisc assembly was verified by SDS-PAGE.
To assay the activation state of integrin αIIbβ3 obtained in nanodiscs as a function of lipid composition, the binding of activity state-dependent antibody PAC1 was quantified by ELISA as described previously (24). Briefly, ELISA plates were coated with 5 μg/ml AP3 antibody overnight at 4 °C and then blocked with BSA for 1 h at 37 °C. After washing the plate, integrin nanodiscs were added and incubated for 2 h at room temperature. The PAC1 antibody was used to detect active integrin receptors. PAC1 binding in the presence of activating antibody anti-LIBS6 was used as control for full activation. As negative control, PAC1 binding in the presence of 20 μm eptifibatide was evaluated. After 2 h of incubation with PAC1 antibody, the wells were washed again, and HRP-conjugated anti-mouse IgM was added for 1 h more of incubation. Subsequent to the final wash, luminescence of the added ECL reagent was read using a VICTOR2 plate reader. An activation index was calculated as (L − L0)/(Lmax − L0), where L denotes the luminescence intensity, L0 denotes the luminescence in the presence of 20 μm eptifibatide, and Lmax denotes the luminescence in the presence of anti-LIBS6 antibody.
Molecular Dynamics Simulations
Using the programs VMD 1.8.7 (29) and SOLVATE 1.0, αIIb(Glu961–Pro998) and β3(Pro688–Glu726) were immersed in lipid bilayers and solvated with TIP3P model water molecules (30). Na+ and Cl− ions were added to a concentration of 100 mm each and to neutralize the system. Monomeric αIIb and β3 TM and heterodimeric αIIbβ3 TM complex structures (Protein Data Bank codes 2k1a, 2rmz, and 2k9j) constituted starting structures. The energy-minimized average αIIbβ3 TM complex structure without αIIb(Arg995)-β3(Asp723) salt bridge was calculated as described previously (8) in the absence of an explicit αIIb(Arg995)-β3(Asp723) distance restraint (31). Initial membrane embedding oriented the monomeric αIIb helix axis (Ile966–Lys989) along the bilayer normal and centered it to the bilayer center. The αIIbβ3 TM complex was aligned on this αIIb orientation, and the embedding of the monomeric β3 helix (Ile693–Ile721) replicated its embedding in the αIIbβ3 TM complex. To assess the dependence of protein-lipid contacts on starting conditions, two additional starting orientations were studied for each monomeric αIIb and β3 TM segment. These starting orientations were related to the original orientations by the following transformations: αIIb rotated relative to the z axis (membrane normal) by 15° and translated relative to the y axis (membrane surface) by −2 Å. Alternatively, αIIb translated relative to the x axis (membrane surface) by 2 Å and relative to the z axis by −2 Å. β3 rotated relative to the z axis by 10° and translated relative to the x axis by 2 Å. Alternatively, β3 translated relative to the x axis by 2 Å and relative to the z axis by −2 Å.
Dimer/monomer simulations in POPC typically consisted of 115/98 lipids, 6661/5851 water molecules, 10/9 Na+ atoms, and 15/13 Cl− ions. Analogous simulations in POPS typically contained 115/98 lipids, 7089/6145 water molecules, 125/107 Na+ atoms, and 15/13 Cl− ions. All-atom molecular dynamics (MD) simulations were carried out using CHARMM22 and CHARMM27 force fields for proteins and lipids, respectively, in the context of the program NAMD 2.9 (32–34). A uniform integration time step of 1 fs and periodic boundary conditions were employed. Electrostatic interactions were calculated using the particle mesh Ewald algorithm with a grid size of <1 Å (35). Only water molecules were treated rigidly using the SETTLE algorithm (36). The melting of lipid tails with all other atoms fixed was followed by minimization and equilibration with protein constrained and equilibration with protein released, each for a period of 0.5 ns. During simulations of 30 or 100 ns duration, the area per lipid was kept constant, and the cell dimension was variable. Simulations were carried out at 301.2 K and at constant pressure of 1 atm. Constant temperature was maintained using Langevin dynamics (37), with a damping coefficient of 1.0 ps−1. Constant pressure was enforced using a Nosé-Hoover Langevin piston with a period of 200 fs and a time constant of 50 fs.
RESULTS AND DISCUSSION
Anionic Lipids Stabilize the αIIbβ3 TM Complex Relative to Electroneutral Lipids
To explore the influence of anionic lipids on αIIbβ3 complex stability, we quantified the αIIb + β3 ⇄ αIIbβ3 equilibrium constant on the mole fraction scale, termed KXY, as a function of lipid type. Phospholipid bicelles, which consist of a long chain lipid bilayer disc that is stabilized by a rim of short chain lipids (Fig. 1B), served as proven membrane mimics for integrin αIIbβ3 (12, 38). The symmetrical lipid bilayer of bicelles places anionic lipids in both membrane leaflets. Within the headgroup region of extracellular lipids, αIIbβ3 residues are dynamically unstructured without defined intersubunit contacts (Fig. 1, A and D). Consequently, contributions to αIIbβ3 TM stability from specific interactions with extracellular lipid headgroups are unlikely. As long chain lipids, zwitterionic POPC, anionic POPG, and anionic POPS were tested in combination with the short chain lipid DHPC. The long chain lipids differ only in their headgroup structure and charge (Fig. 1C), which have no significant effect on the backbone conformation of either the αIIb or β3 TM segments (11, 12). Moreover, the bicelles served as a thermodynamically ideal solvent for αIIbβ3 at the employed peptide to lipid ratio (see “Experimental Procedures”). Determination of KXY was facilitated by the documented absence of αIIb or β3 homodimerization (14, 39). To provide independent measurements of KXY and the associated change in free energy, termed ΔG°, complex formation was evaluated by both NMR spectroscopy and ITC. ITC additionally provides the enthalpy and entropy changes upon αIIbβ3 TM complex formation, denoted by ΔH° and ΔS°, respectively.
In NMR experiments, integrin αIIbβ3 monomers and heterodimer are observable independently at 28 °C (8, 39). This exchange behavior permits the direct extraction of the fraction of αIIbβ3 dimer by quantifying the decline of peak volumes of monomer residues. We labeled either αIIb or β3 with 2H/15N isotopes and quantified the peak volumes of labeled monomer as a function of concentration of unlabeled partner peptide. Nonlinear curve fitting of the obtained αIIbβ3 complex fractions yielded KXY (Fig. 2, A and B, and Table 1). The results were similar for isotope labeling of either αIIb or β3 and, perhaps surprisingly, revealed a POPS-mediated αIIbβ3 complex stabilization. For instance, at a POPC:POPS ratio of 2:1, which approximates the ratio of anionic lipids in the intracellular membrane leaflet (40), the ΤΜ complex is stabilized by ≥0.19 ± 0.02 kcal/mol compared with POPC only. At POPC/POPS = 2:1, POPS is at least in 50-fold excess of αIIb. However, TM complex stabilization still increased at higher POPS ratios (Fig. 2C and Table 1), showing that αIIbβ3 was unable to attract anionic lipids with high affinity. This inability demonstrates that annular lipids were responsible for the observed stabilizing effect.
ITC measurements detected the heat changes upon titrating β3 with αIIb to binding saturation and yielded KXY and ΔH° from nonlinear curve fitting (Fig. 2D). We note that nonsigmoidal titration curves were chosen to ensure ideal solvent behavior of the hydrophobic phase (see “Experimental Procedures”). To control for the size of the long chain lipid bilayer area (Fig. 1B) and for any lipid headgroup-dependent difference in short and long chain lipid mixing (41), larger bicelles were employed in ITC than in NMR experiments (qeff = 0.5 versus 0.31), which merely offset ΔG° to lower values (Table 1) because of a more favorable binding entropy (14). For pure POPS, complex stabilization relative to POPC was 0.50 ± 0.02 kcal/mol, and the corresponding value for pure POPG was 0.36 ± 0.02 kcal/mol (Table 1). Relative to ΔG° of −4.84 ± 0.01 kcal/mol in POPC (Table 1), lipid composition modulates significantly TM complex stability. In sum, NMR and ITC measurements show that annular anionic lipids stabilize the integrin αIIbβ3 TM complex in a lipid headgroup structure-dependent manner.
Influence of Lipid Composition on Integrin Receptor Activity
Before we provide a structural view on ΔG°POPS < ΔG°POPC, we note that the platelet membrane that harbors integrin αIIbβ3 undergoes large scale changes in anionic lipid membrane distribution. The activation of the blood clotting enzyme thrombin requires the emergence of PS lipids in the outer membrane leaflet of platelets (42). Its failure results in the bleeding disorder known as Scott syndrome (43), in which the phospholipid scramblase TMEM16F is compromised (44). Anionic lipid scrambling will reduce the concentration of these lipids in the intracellular membrane leaflet, which may affect αIIbβ3 TM complex stability and thus the adhesive state of the receptor. The substrate binding abilities of two integrins have been examined in liposomes of different, symmetric lipid compositions (45, 46). The affinity of integrin αVβ3 to bind vitronectin is larger in phosphatidylethanolamine/phosphatidylcholine (PE/PC)-based liposomes than liposomes containing solely PC (46). PE/PC-based lipids are both net neutral. A further increase in substrate binding is detected when incorporating PS and phosphatidylinositol-based lipids and cholesterol (46), which affect many different membrane characteristics. For integrin αIIbβ3, it is reported that the anionic lipid phosphatidic acid increases fibrinogen binding (45). Accordingly, membrane lipid characteristics may influence integrin activity.
To deepen our understanding of integrin αIIbβ3 activity as a function of membrane lipid composition, we examined the adhesive state of integrin αIIbβ3 in lipid nanodiscs that incorporated POPC, POPG, or POPS. For this purpose, integrin αIIbβ3 was purified from human platelets, reconstituted in the nanodiscs, and assayed using conformation-specific antibodies. If the TM structure in bicelles and nanodiscs match, the free energy difference between receptors in POPC and POPS nanodiscs is 0.50 ± 0.02 kcal/mol. Except for a slightly higher receptor activity in POPG, no significant differences in integrin activity were detected between the different lipid compositions (Fig. 3). To assess the veracity of this result, we examined the effect of the alanine substitution of αIIb(Arg995) on TM complex stability in POPC bicelles by ITC. Disturbing the αIIb(Arg995)-β3(Asp723) interaction in mammalian cell membranes leads to integrin activation (7), and a ΔG° loss of 1.5 ± 0.2 kcal/mol was detected for αIIb(R995A) (Table 1 and Fig. 4). The loss of 0.50 ± 0.02 kcal/mol upon replacing all POPS with POPC is indeed too small to trigger integrin activation. Notwithstanding, the threshold of integrin activation must shift with anionic lipid content, and the physiological scrambling of anionic lipid upon platelet activation has the capacity to facilitate integrin activation.
FIGURE 3.
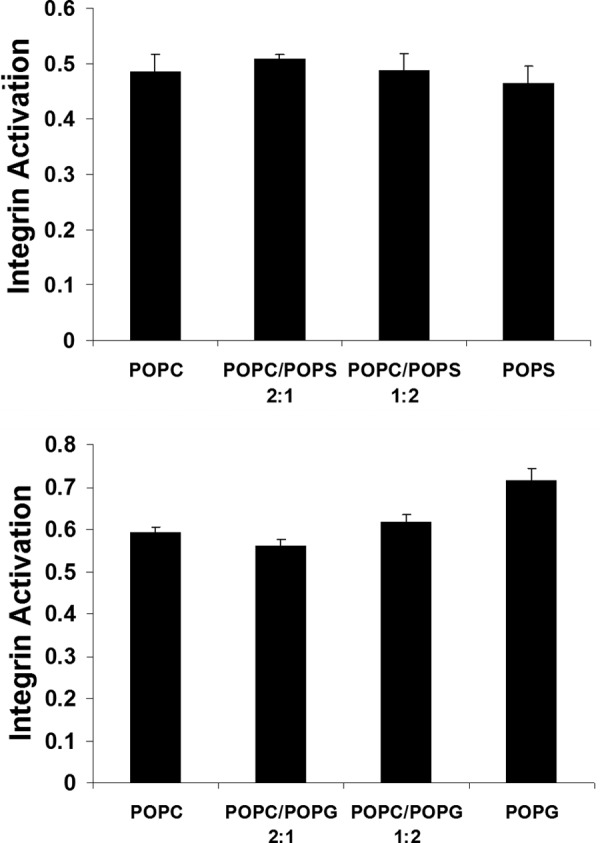
Integrin αIIbβ3 activity as a function of nanodisc lipid composition. The ratio of active adhesive receptor, as evaluated by Pac1 antibody binding, was quantified by the activation index (L − L0)/(Lmax − L0), where L denotes the degree of Pac1 binding, L0 is the degree of Pac1 binding in the presence of the small molecule inhibitor, eptifibatide, and Lmax denotes the degree of Pac1 binding in the presence of integrin-activating antibody anti-LIBS6.
FIGURE 4.
Competitive ITC measurement of αIIb(R995A)β3 TM complex association. The association of the wild-type αIIbβ3 TM complex was measured in the presence of competing αIIb(R995A) peptide. In the sample cell, starting β3 and αIIb(R995A) concentrations of 10 and 50 μm, respectively, were used. Measurements were carried out in phospholipid bicelles consisting of 34 mm DHPC and 17 mm of the depicted long chain lipid (qeff = 0.5). The experimental uncertainties for KXY and ΔH° of wild-type αIIbβ3 propagate to the measured αIIb(R995A)β3 TM values (16).
Anionic Lipids Establish Headgroup Structure-dependent Contacts with αIIb and β3
The stabilization of the αIIbβ3 TM complex by anionic lipids raises the question of whether anionic lipids were at all competing for the αIIb(Arg995)-β3(Asp723) interaction. Unlike the PC headgroup, PS and PG headgroups form intra- and interlipid hydrogen bonds and engage their Na+ counterion in lipid-bridging interactions (47, 48). We therefore evaluated whether αIIb(Arg995) established contacts with lipid headgroups. Electrostatic interactions, notably hydrogen bonding, prolong the lifetime of labile 1H nuclei such as the ϵ-NH and η-NH2 nuclei of arginine (Fig. 1C). Lipid headgroup contacts to these nuclei may therefore increase their peak intensities in NMR spectra. 180° flips around the Nϵ-Cζ partial double bond are often sufficient to prohibit the NMR detection of η-NH2 nuclei (49), and we were indeed unable to observe these nuclei at pH 7.4 and 28 °C. However, the ϵ-NH nuclei of the three arginines of αIIb (residues 962, 995, and 997; Fig. 1D) and the one arginine of β3 (residue 724) were detectable. Initially, we compared the cumulative ϵ-NH signal intensities of αIIb and β3 arginines as a function of bicelle lipid composition. Relative to POPC, ϵ-NH nuclei were stabilized by POPS and POPG (Fig. 5A). Moreover, POPS surpassed POPG in stabilizing ϵ-NH. The electrostatic character of this stabilization was confirmed by observing a reduction in signal intensity in the presence of 120 and 240 mm NaCl (Fig. 5A). Thus, the zwitterionic nature of POPC disfavored PO4− electrostatic interactions external to the lipid. In contrast, this functional group was available in POPG and POPS, which additionally offers a COO− moiety that further aided electrostatic ϵ-NH-lipid contacts. The electrostatic interaction of lipids with arginine side chains therefore confirmed the competitive nature of anionic lipids and paralleled the order of lipid-mediated αIIbβ3 TM complex stabilization.
Arg995 of αIIb is Positioned for Electrostatic Contacts with Lipid Headgroups
The ϵ-NH resonances of the three arginines of αIIb overlapped. To observe individual resonances, Arg to Lys substitutions were carried out. Individual arginine resonance intensities were quite different and decreased in the order of 962 > 997 > 995 (Fig. 5B). The ϵ-NH resonance of αIIb(Arg995) was barely detectable. Among the three arginines, Arg995 is closest to the membrane core (Fig. 1D), and perhaps it was able to evade headgroup contacts either through its positioning or through shielding by surrounding cationic residues (Fig. 1, A and D). To seek an atomistic understanding of αIIb(Arg995)-lipid contacts, all atom MD simulations were carried out for αIIb and β3, which were immersed in a bilayer of either POPC or POPS. To consider bias from the starting configuration, three 30-ns simulations with different initial αIIb/β3 bilayer immersions were performed for each lipid type. For all arginines, lipid contacts were observed and, in accordance with solvent exchange (Fig. 5A), the incidence of electrostatic ϵ-NH contacts to lipid headgroups was on average higher in POPS than in POPC bilayers (Fig. 5C), an observation that is likely of a general nature (50). Remarkably, αIIb(Arg995/ϵ-NH) produced the highest incidence of lipid headgroup contacts in both POPC and POPS bilayers relative to the other arginines (Fig. 5C). Simultaneously, contacts to water were scarcest for αIIb(Arg995/ϵ-NH) (data not shown). As a result, it appears that Arg995 is suitably positioned to interact with lipid headgroups. As a consequence, the difficulty of observing the αIIb(Arg995/ϵ-NH) resonance by NMR is not indicative of its inability to establish lipid contacts. Its low peak intensity could arise from a high intrinsic hydrogen exchange rate in the particular chemical environment of Arg995, which would imply that anionic lipids only marginally stabilize its lifetime. Alternatively, if the ϵ-NH nucleus exchanges between different chemical environments on an unfavorable time scale relative to the NMR chemical shift time scale, its resonance would severely broaden. Interestingly, the addition of β3 peptide did only slightly increase the low signal intensity of αIIb(Arg995/ϵ-NH) (Fig. 5D). In light of the strong effects of αIIb(Arg995) mutations on TM complex stability (Table 1), the low αIIb(Arg995/ϵ-NH) signal intensity suggests that the αIIb(Arg995)-β3(Asp723) interaction is dynamic. In conclusion, it is unlikely that αIIb(Arg995) is able to evade lipid headgroup contacts, which means anionic lipids will compete for αIIb(Arg995) with β3(Asp723).
The Stability of the αIIb(Arg995)-β3(Asp723) Interaction Is Similar in POPC and POPS
To further show electrostatic competition for αIIb(Arg995)-β3(Asp723) by the headgroup of POPS, we examined the effects of the compound O-phospho-l-serine (OPS), which corresponds to the POPS headgroup (Fig. 1C). For equimolar amounts of POPC and OPS, the TM complex was destabilized by ΔΔG°OPS = ΔG°POPC/OPS − ΔG°POPC = 0.13 ± 0.01 kcal/mol (Table 1). As a reference, for 500 mm NaCl the corresponding ΔΔG°NaCl was 0.23 ± 0.01 kcal/mol (Table 1). Electrostatic competition is thus apparent. However, as noted earlier, the αIIb(R995A) substitution destabilized the TM complex by 1.5 ± 0.2 kcal/mol in POPC bicelles (Table 1). The relatively weak competition observed suggests that OPS and NaCl concentrations near αIIb(Arg995) were reduced by solute concentration gradients in lipid headgroup regions. The headgroup of POPS should accordingly be more destabilizing to αIIb(Arg995)-β3(Asp723) than OPS. If so, αIIb(Arg995)-β3(Asp723) will contribute less to overall TM complex stability in POPS than in POPC, and ΔΔG°αIIb(R995A) of 1.5 ± 0.2 kcal/mol in POPC will diminish in POPS. Paradoxically, αIIb(R995A) destabilized the TM complex by 2.0 ± 0.3 kcal/mol in POPS (Table 1 and Fig. 4), demonstrating that the stability of the αIIb(Arg995)-β3(Asp723) interaction in POPS was not diminished relative to POPC. For reference, the stability of the mutant αIIb(R995A)β3 TM complex is similar to the TM complex stability of the fibroblast growth factor receptor 3 at −2.8 ± 0.1 kcal/mol (51). By what means αIIb(Arg995)-β3(Asp723) was able to compensate for the competition for αIIb(Arg995) from POPS?
Anionic Lipids Induce a Ternary αIIb(Arg995)·POPS·β3 Complex
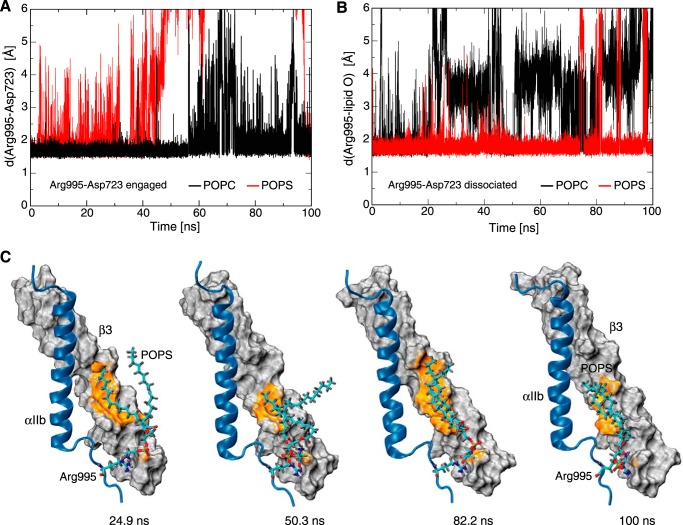
To gain insight into lipid-protein interactions in the context of the αIIbβ3 TM complex, αIIb(Arg995)-lipid and αIIb(Arg995)-β3(Asp723) contacts were compared in MD simulations of 100-ns duration. The suitability of MD simulations for studying the αIIbβ3 TM complex is well established (52). The TM complex was examined in the presence of POPC and POPS bilayers starting with engaged and disengaged αIIb(Arg995)-β3(Asp723) contact, respectively. When starting with an engaged salt bridge, it dissociated intermittently in both lipid environments (Fig. 6A). However, αIIb(Arg995)-β3(Asp723) distance fluctuations in POPS were significantly larger in amplitude and duration than in POPC, which is indicative of the competitive nature of POPS lipids. When starting with the salt bridge dissociated, lipid interactions of αIIb(Arg995) were substantially more stable in POPS as compared with POPC (Fig. 6B). Interestingly, the lipid that is interacting with αIIb(Arg995) formed dynamic but persistent contacts with the β3 TM helix (Fig. 6C and supplemental Movie S1). Relative to the lifetime of the αIIbβ3 TM complex of 0.5 s (14), the MD simulations afford only a brief glimpse into protein-lipid interactions. However, our observations agree with dynamic αIIb(Arg995)-β3(Asp723) interaction (Fig. 5D) and reflect the different lifetimes of Arg(ϵ-NH)-lipid headgroup contacts in POPC and POPS (Fig. 5A). To explain the similar stability of αIIb(Arg995)-β3(Asp723) in POPC and POPS, we present the following hypothesis.
FIGURE 6.
Relationship between αIIb(Arg995), β3(Asp723), and lipid molecules in molecular dynamics simulations of the αIIbβ3 TM complex. A, stability of the αIIb(Arg995)-β3(Asp723) salt bridge in POPC and POPS bilayers, respectively, with αIIb(Arg995)-β3(Asp723) initially engaged. Minimal Arg995(ϵ/η-H)-Asp723(δ-O) distances are plotted. B, stability of αIIb(Arg995)-lipid contacts in POPC and POPS bilayers, respectively, with αIIb(Arg995)-β3(Asp723) initially dissociated. Minimal distances of Arg995(ϵ/η-H) to the closest lipid oxygen atom are plotted. C, illustration of POPS lipid-mediated contacts between αIIb and β3 TM subunits as a function of simulation time (MD simulation with αIIb(Arg995)-β3(Asp723) initially dissociated). POPS and αIIb(Arg995) are shown in ball and stick representation. The surface of the β3 TM helix is shown in gray; β3 atoms within a distance of 3.5 Å to any POPS atom are highlighted in orange.
POPC is neutral with respect to αIIb(Arg995)-β3(Asp723), but the salt bridge is still not engaged at all times in POPC. On the other hand, POPS is able to break αIIb(Arg995)-β3(Asp723) frequently and engage αIIb(Arg995), resulting in a ternary αIIb(Arg995)·POPS·β3 complex (Fig. 6C and supplemental Movies S1-S2). The concurrence of anionic lipid headgroup-αIIb and lipid tail-β3 contacts in this complex offsets the perturbation of αIIb(Arg995)-β3(Asp723) by anionic lipids. The dual contacts of αIIb(Arg995) with β3(Asp723) and POPS would render the αIIb(Arg995)-β3(Asp723) salt bridge asymmetric. That is, the substitution of αIIb(Arg995) in the presence of anionic lipids should destabilize the TM complex more than the corresponding substitution of β3(Asp723). In cellular assays, the extent of receptor activation has been quantified for αIIb(Arg995) and β3(Asp723) substitutions, and, in instances where significant differences were observed, αIIb(Arg995) substitutions were indeed more activating (7, 53).
Structural View of Anionic Lipid-mediated αIIbβ3 TM Complex Stabilization
The net stabilization of the αIIbβ3 TM complex by anionic lipids raises the question of its structural basis. Next to lipid contacts involving αIIb(Arg995), we consider differences in physical membrane characteristics and specific TM helix-lipid contacts to be relevant to differences in overall αIIbβ3 TM complex stability. Electroneutral and anionic lipids give rise to differences in physical membrane characteristics (47, 48) that may directly modulate TM helix-helix interactions (1, 54). Here, we examined whether differences in POPC and POPG headgroup characteristics (Fig. 1C) lead to differences in the protection of protein backbone HN nuclei from the net neutral paramagnetic agent Mn2+EDDA2− (12). For the αIIb and β3 TM segments, two to four additional residues were protected by POPC compared with POPG at both the intra- and extracellular membrane faces (Fig. 7, A and B). This observation indicates that small molecules can penetrate the headgroup region of PG more deeply than of PC by a range of 3–6 Å. Accordingly the concentration gradients of solvent and ions in the PC and PG/PS headgroup region will differ. We anticipate PS to resemble PG based on their similar headgroup volumes (Fig. 1C) and molecular interactions (47, 48). Independent of differences in Mn2+EDDA2− protection, we noted that the “charge content” of the lipid headgroup region stemming from lipid charges, charged protein residues, and small ions affected the magnitude of enthalpy changes (ΔH°) that accompanied αIIbβ3 TM complex association (Table 1). Thus, substantial differences in the structural context of integrin αIIbβ3 TM segments in zwitterionic and anionic lipids entail the capacity of contributing to αIIbβ3 TM complex stabilization by annular anionic lipids. For instance, it is conceivable that the hydrophobic effect in the lipid headgroup region increased in POPG/POPS relative to POPC.
FIGURE 7.
Lipid immersion of αIIb, β3, and αIIb(K994A) TM segments in POPC and POPG bicelles. Protection of backbone HN nuclei from the paramagnetic Mn2+EDDA2− agent in the aqueous phase as quantified by the ratio of HN NMR resonances intensity in the presence and absence of 1 mm Mn2+EDDA2−, I/I0. A–B, comparison of POPC and POPG protection for αIIb and β3, respectively; C–D, comparison of αIIb(K994A) and αIIb protection in POPC and POPG, respectively.
TM complex stability increased in POPS compared with POPG lipids (Table 1) in correlation with an increase of the lifetime of lipid-protein contacts in POPS relative to POPG (Fig. 5A). This apparent correlation, which relates to the wider charge distribution of the PS than the PG headgroup (Fig. 1C), suggests that specific lipid-protein contacts may contribute to anionic lipid-mediated complex stabilization. In the structured region of the TM complex, four cationic residues are found (Fig. 1, A and D). Next to αIIb(Arg995) and the first positively charged residues on the intracellular side, αIIb(Lys989) and β3(Lys716), αIIb(Lys994) is noteworthy for its high conservation among human integrin α subunits at 89% (8). The Ala substitution of Lys994 altered the protection profile of the αIIb TM segment from paramagnetic Mn2+EDDA2− (Fig. 7, C and D). In both POPG and POPC, increased HN exposures of especially Gly991 and Ala994 were detectable relative to the wild-type peptide. These residues are structurally adjacent, and it is evident that lipids rearranged around residue 994 in support of altered lipid-protein contacts. If an electrostatic nature of such contacts were important to TM complex-stabilizing lipid-protein contacts, αIIb(K994A) would increase ΔG° in POPS and not POPC. In POPC, αIIb(K994A) did not change ΔG° relative to wild type, but in POPS ΔG° was increased by 0.45 ± 0.03 kcal/mol (Table 1). Thus, αIIb(Lys994)-lipid contacts appear to be important for anionic lipid-mediated TM complex stabilization. The stabilizing effect of αIIb(Lys994)-anionic lipid interactions may arise from a topographical stabilization of the αIIb helix in the membrane (14) and/or by providing an “optimal” structural environment at residue 994.
Conclusions
The evolution of membrane proteins in a bilayer, where anionic lipids are concentrated in the inner leaflet, has resulted in the usage of electrostatic contacts between anionic lipids and cationic protein residues to establish membrane protein topology (4–6). Although the headgroup of anionic lipids did compete for electrostatic interactions within the integrin αIIbβ3 TM complex, the net stability of its key αIIb(Arg995)-β3(Asp723) electrostatic interaction was not compromised, and an overall TM complex stabilization was induced. The observed complex stabilization of up to 0.5 kcal/mol is a noteworthy contribution to the threshold of integrin αIIbβ3 receptor activation, which is reached at 1.5 ± 0.2 kcal/mol in POPC. Our work provides thermodynamic insight into principles of membrane protein-lipid interactions that is of general interest to the study of membrane proteins. For instance, compared with the affinity of other receptor TM complexes (14), the complex stability of integrin αIIbβ3 at −4.84 ± 0.01 kcal/mol is relatively high, indicating the potential of lipids to significantly modulate the strength of physiological TM helix-helix interactions (58). We emphasize that integrin αIIbβ3 does not possess a high affinity anionic lipid-binding site, which means that solely annular lipids were responsible for the observed effects. Membrane proteins often use nonannular lipids, which conceptually resemble co-factors of soluble proteins, to stabilize their conformation (1–3). Moreover, high affinity binding sites for annular anionic lipids have been observed in membrane proteins (55–57). However, by stabilizing the TM helix-helix association of integrin αIIbβ3 in ubiquitous low affinity annular positions, our results point to a general role of anionic lipids in enhancing membrane protein stability.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL31950, HL078784, and HL117807 (to M. H. G.).

This article contains supplemental Movies S1 and S2.
- TM
- transmembrane
- DHPC
- 1,2-dihexanoly-sn-glycero-3-phosphocholine
- MD
- molecular dynamics
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- ITC
- isothermal titration calorimetry
- HSQC
- heteronuclear single quantum coherence
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- OPS
- O-phospho-l-serine.
REFERENCES
- 1. Marsh D. (2008) Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575 [DOI] [PubMed] [Google Scholar]
- 2. Palsdottir H., Hunte C. (2004) Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- 3. Lee A. G. (2011) Biological membranes: the importance of molecular detail. Trends Biochem. Sci. 36, 493–500 [DOI] [PubMed] [Google Scholar]
- 4. von Heijne G. (1989) Control of topology and mode of assembly of a polytopic membrane-protein by positively charged residues. Nature 341, 456–458 [DOI] [PubMed] [Google Scholar]
- 5. van Klompenburg W., Nilsson I., von Heijne G., de Kruijff B. (1997) Anionic phospholipids are determinants of membrane protein topology. EMBO J. 16, 4261–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogdanov M., Xie J., Dowhan W. (2009) Lipid-protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J. Biol. Chem. 284, 9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. (1996) Breaking the integrin hinge: a defined structural constraint regulates integrin signaling. J. Biol. Chem. 271, 6571–6574 [DOI] [PubMed] [Google Scholar]
- 8. Lau T.-L., Kim C., Ginsberg M. H., Ulmer T. S. (2009) The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hynes R. O. (2002) Integrins: Bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 10. Bennett J. S., Berger B. W., Billings P. C. (2009) The structure and function of platelet integrins. J. Thromb. Haemost. 7, 200–205 [DOI] [PubMed] [Google Scholar]
- 11. Lau T.-L., Dua V., Ulmer T. S. (2008) Structure of the integrin αIIb transmembrane segment. J. Biol. Chem. 283, 16162–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau T.-L., Partridge A. W., Ginsberg M. H., Ulmer T. S. (2008) Structure of the integrin β3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 47, 4008–4016 [DOI] [PubMed] [Google Scholar]
- 13. Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Situ A. J., Schmidt T., Mazumder P., Ulmer T. S. (2014) Characterization of membrane protein interactions by isothermal titration calorimetry. J. Mol. Biol. 426, 3670–3680 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y. L., Zhang Z. Y. (1998) Low-affinity binding determined by titration calorimetry using a high-affinity coupling ligand: a thermodynamic study of ligand binding to protein tyrosine phosphatase 1B. Anal. Biochem. 261, 139–148 [DOI] [PubMed] [Google Scholar]
- 16. Sigurskjold B. W. (2000) Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 277, 260–266 [DOI] [PubMed] [Google Scholar]
- 17. Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 18. Turnbull W. B., Daranas A. H. (2003) On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 [DOI] [PubMed] [Google Scholar]
- 19. Tellinghuisen J. (2008) Isothermal titration calorimetry at very low c. Anal. Biochem. 373, 395–397 [DOI] [PubMed] [Google Scholar]
- 20. Fleming K. G. (2002) Standardizing the free energy change of transmembrane helix-helix interactions. J. Mol. Biol. 323, 563–571 [DOI] [PubMed] [Google Scholar]
- 21. Chou J. J., Baber J. L., Bax A. (2004) Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR 29, 299–308 [DOI] [PubMed] [Google Scholar]
- 22. Mori S., Abeygunawardana C., Johnson M. O., van Zijl P. C. (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B 108, 94–98 [DOI] [PubMed] [Google Scholar]
- 23. Ulmer T. S., Campbell I. D., Boyd J. (2004) Amide proton relaxation measurements employing a highly deuterated protein. J. Magn. Reson. 166, 190–201 [DOI] [PubMed] [Google Scholar]
- 24. Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., Ginsberg M. H. (2010) Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye F., Liu J., Winkler H., Taylor K. A. (2008) Integrin αIIbβ3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J. Mol. Biol. 378, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kouns W. C., Hadvary P., Haering P., Steiner B. (1992) Conformational modulation of purified glycoprotein (Gp)-IIb-IIIa allows proteolytic generation of active fragments from either active or inactive GpIIb-IIIa. J. Biol. Chem. 267, 18844–18851 [PubMed] [Google Scholar]
- 27. Nath A., Atkins W. M., Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 28. Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 29. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 30. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 31. Ulmer T. S. (2010) Structural basis of transmembrane domain interactions in integrin signaling. Cell Adh. Migr. 4, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gumbart J., Wang Y., Aksimentiev A., Tajkhorshid E., Schulten K. (2005) Molecular dynamics simulations of proteins in lipid bilayers. Curr. Opin. Struct. Biol. 15, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 34. Feller S. E., MacKerell A. D. (2000) An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B 104, 7510–7515 [Google Scholar]
- 35. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 36. Miyamoto S., Kollman P. A. (1992) Settle: an analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 13, 952–962 [Google Scholar]
- 37. Brunger A., Brooks C. L., Karplus M. (1984) Stochastic boundary-conditions for molecular-dynamics simulations of St2 water. Chem. Phys. Lett. 105, 495–500 [Google Scholar]
- 38. Suk J. E., Situ A. J., Ulmer T. S. (2012) Construction of covalent membrane protein complexes and high-throughput selection of membrane mimics. J. Am. Chem. Soc. 134, 9030–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim C., Lau T.-L., Ulmer T. S., Ginsberg M. H. (2009) Interactions of platelet integrin αIIb and β3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood 113, 4747–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. García-Guerra R., García-Domínguez J. A., González-Rodríguez J. (1996) A new look at the lipid composition of the plasma membrane of human blood platelets relative to the GPIIb/IIIa (integrin αIIbβ3) content. Platelets 7, 195–205 [DOI] [PubMed] [Google Scholar]
- 41. Triba M. N., Warschawski D. E., Devaux P. F. (2005) Reinvestigation by phosphorus NMR of lipid distribution in bicelles. Biophys. J. 88, 1887–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zwaal R. F., Comfurius P., van Deenen L. L. (1977) Membrane asymmetry and blood-coagulation. Nature 268, 358–360 [DOI] [PubMed] [Google Scholar]
- 43. Zwaal R. F., Comfurius P., Bevers E. M. (2004) Scott syndrome, a bleeding disorder caused by defective scrambling of membrane phospholipids. Biochim. Biophys. Acta 1636, 119–128 [DOI] [PubMed] [Google Scholar]
- 44. Suzuki J., Umeda M., Sims P. J., Nagata S. (2010) Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 [DOI] [PubMed] [Google Scholar]
- 45. Smyth S. S., Hillery C. A., Parise L. V. (1992) Fibrinogen binding to purified platelet glycoprotein-IIb-IIIa (Integrin-αIIb-β3) is modulated by lipids. J. Biol. Chem. 267, 15568–15577 [PubMed] [Google Scholar]
- 46. Conforti G., Zanetti A., Pasquali-Ronchetti I., Quaglino D., Jr., Neyroz P., Dejana E. (1990) Modulation of vitronectin receptor-binding by membrane lipid-composition. J. Biol. Chem. 265, 4011–4019 [PubMed] [Google Scholar]
- 47. Zhao W., Róg T., Gurtovenko A. A., Vattulainen I., Karttunen M. (2007) Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidylglycerol lipid bilayers with Na+ counterions. Biophys. J. 92, 1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petrache H. I., Tristram-Nagle S., Gawrisch K., Harries D., Parsegian V. A., Nagle J. F. (2004) Structure and fluctuations of charged phosphatidylserine bilayers in the absence of salt. Biophys. J. 86, 1574–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henry G. D., Sykes B. D. (1995) Determination of the rotational-dynamics and pH-dependence of the hydrogen-exchange rates of the arginine guanidino group using NMR-spectroscopy. J. Biomol. NMR 6, 59–66 [DOI] [PubMed] [Google Scholar]
- 50. Koldso H., Sansom M. S. (2013) Local lipid reorganization by a transmembrane protein domain. J. Phys. Chem. Lett. 3, 3498–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li E., You M., Hristova K. (2006) FGFR3 dimer stabilization due to a single amino acid pathogenic mutation. J. Mol. Biol. 356, 600–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kalli A. C., Hall B. A., Campbell I. D., Sansom M. S. (2011) A helix heterodimer in a lipid bilayer: prediction of the structure of an integrin transmembrane domain via multiscale simulations. Structure 19, 1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhu J., Luo B. H., Barth P., Schonbrun J., Baker D., Springer T. A. (2009) The structure of a receptor with two associating transmembrane domains on the cell surface: integrin αIIbβ3. Mol. Cell 34, 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee A. G. (2004) How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 [DOI] [PubMed] [Google Scholar]
- 55. Powl A. M., East J. M., Lee A. G. (2005) Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: hot spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry 44, 5873–5883 [DOI] [PubMed] [Google Scholar]
- 56. Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 Angstrom resolution. Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brini M., Di Leva F., Ortega C. K., Domi T., Ottolini D., Leonardi E., Tosatto S. C., Carafoli E. (2010) Deletions and mutations in the acidic lipid-binding region of the plasma membrane Ca2+ pump: a study on different splicing variants of isoform 2. J. Biol. Chem. 285, 30779–30791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arkhipov A., Shan Y. B., Das R., Endres N. F., Eastwood M. P., Wemmer D. E., Kuriyan J., Shaw D. E. (2013) Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.