Background: Ssl1/p44 is a critical helicase stimulating factor for Rad3/XPD.
Results: Structure-based mutation analysis of the β4-α5 loop of Ssl1/p44 resulted in defects of Rad3/XPD stimulation and yeast cell lethality.
Conclusion: The β4-α5 loop of Ssl1/p44 is essential for the functional regulation of Rad3/XPD.
Significance: These findings provide insights into the interaction between Rad3/XPD and Ssl1/p44 in TFIIH complex.
Keywords: Crystal Structure, DNA Repair, Nucleotide Excision Repair, Transcription, Yeast Genetics, Rad3/XPD Helicase, TFIIH, Regulatory Domain of Ssl1/p44
Abstract
The Ssl1/p44 subunit is a core component of the yeast/mammalian general transcription factor TFIIH, which is involved in transcription and DNA repair. Ssl1/p44 binds to and stimulates the Rad3/XPD helicase activity of TFIIH. To understand the helicase stimulatory mechanism of Ssl1/p44, we determined the crystal structure of the N-terminal regulatory domain of Ssl1 from Saccharomyces cerevisiae. Ssl1 forms a von Willebrand factor A fold in which a central six-stranded β-sheet is sandwiched between three α helices on both sides. Structural and biochemical analyses of Ssl1/p44 revealed that the β4-α5 loop, which is frequently found at the interface between von Willebrand factor A family proteins and cellular counterparts, is critical for the stimulation of Rad3/XPD. Yeast genetics analyses showed that double mutation of Leu-239 and Ser-240 in the β4-α5 loop of Ssl1 leads to lethality of a yeast strain, demonstrating the importance of the Rad3-Ssl1 interactions to cell viability. Here, we provide a structural model for the Rad3/XPD-Ssl1/p44 complex and insights into how the binding of Ssl1/p44 contributes to the helicase activity of Rad3/XPD and cell viability.
Introduction
Initiation of eukaryotic transcription requires the assembly of RNA polymerase II and multiple general transcription factors, including TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, at a promoter region. TFIIH, which possess 10 subunits, exhibits at least four enzymatic activities, including ATPase, helicase, kinase, and ubiquitin ligase, which are responsible for several fundamental cellular functions of TFIIH, such as DNA repair, cell cycle regulation, and transcription (1, 2). Yeast/mammalian TFIIH comprises a core complex that is consisted of seven subunits (Rad3/XPD, Ssl2/XPB, Tfb1/p62, Tfb2/p52, Ssl1/p44, Tfb4/p34, and Tfb5/p8) and a Cdk-activating kinase (CAK)4 complex containing three subunits (Kin28/CDK7, Ccl1/Cylin H, and Tfb3/MAT1). Holo TFIIH comprising all 10 subunits is essential for transcription, whereas the core complex is required for DNA repair (2–4).
The four enzymatic activities of TFIIH are controlled by six of its subunits. The CAK subcomplex phosphorylates the C-terminal domain of the Rbp1 subunit of RNA polymerase II and controls its interaction with mRNA and histone-modifying enzymes (5–7). In addition, CAK phosphorylates several nuclear receptors, resulting in the transactivation of hormone-dependent genes in mammalian cells (2, 8). The XPB and XPD subunits possess ATP-dependent 3′ to 5′ and 5′ to 3′ helicase activities and unwind DNA around the promoter region to allow transcription by RNA polymerase II and the excision of damaged DNA via nucleotide excision repair (NER), respectively (9, 10). The XPD helicase and XPB ATPase activities are required for NER, and these activities are enhanced in the presence of the p44 and p52 subunits, respectively (11, 12). Finally, yeast Ssl1 subunit exhibits E3 ubiquitin ligase activity that is involved in the transcriptional response to DNA damage (13).
XPD helicase activity is important for the maintenance of genomic stability, and its mutation causes DNA repair defects and abnormal TFIIH assembly, resulting in the genetic disorders xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy, which cause premature aging and several types of cancer (14, 15). In a study of 50 individuals, 87% of xeroderma pigmentosum patients and 50% of trichothiodystrophy patients carried mutations in the C-terminal regions of XPD spanning residues 675–730 and 713–730, respectively (16). Biochemical analysis of human XPD-p44 and determination of the crystal structures of archaeal XPD homologs provided insights into how amino acid substitutions result in these disorders (10, 12, 17–20). Rad3/XPD comprises four domains, namely, RecA-like HD1, RecA-like HD2, 4FeS, and Arch. p44 is believed to interact with the C-terminal third (150 residues) of human XPD through its N-terminal domain (235 amino acids) and stimulates its helicase activity (10, 15). The C-terminal region of Thermoplasma acidophilum XPD is similar to that of eukaryotic XPD (17–20); however, it is ∼30 residues shorter than human XPD or yeast Rad3. Furthermore, an archaeal Ssl1/p44 homolog has not been identified.
The TFIIH p44 subunit consists of three domains, namely, the N-terminal (residues 1–235), zinc finger (residues 291–308), and ring finger (residues 345–385) domains (10, 21, 22). The N-terminal domain of p44 is responsible for regulation of the helicase activity of XPD through direct interaction with its C-terminal domain (10, 15).
Xeroderma pigmentosum- and trichothiodystrophy-causing mutations in the N- and C-terminal regions of XPD destabilize its interactions with MAT1 and p44, respectively, and interfere with both transcription and DNA repair (23, 24). The overall architectures of the yeast and human TFIIHs have been determined, and the subcomplex structures have been modeled into the overall electron microscopy (EM) density (3, 4). However, the structures of only a limited number of individual components are available, which hampers the modeling of complete TFIIH. It is unclear how Ssl1/p44 interacts with and stimulates Rad3/XPD helicase activity and how the disease-causing C-terminal mutations of Rad3/XPD are correlated with its interaction with Ssl1/p44. To address these questions, we have determined the crystal structure of the N-terminal regulatory domain of Saccharomyces cerevisiae Ssl1. The structure forms a nucleotide-binding fold with a central β-sheet flanked by two helical bundles. We show that the β4-α5 loop of Ssl1/p44 is important for the binding and stimulation of Rad3/XPD, and the interaction between Rad3/XPD and Ssl1/p44 is critical for cell viability. Based on the derived structure, mutation analysis, and genetics studies, we provide a model for the Rad3/XPD-Ssl1/p44 subcomplex.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of S. cerevisiae Ssl1
The cDNA encoding residues 1–350 of S. cerevisiae Ssl1 were inserted into the pET28a vector. Escherichia coli BL21 (DE3) cells containing the pET28a (Ssl1 1–350) vector were cultured in Luria broth. After induction with 0.4 mm isopropyl β-d-thiogalactopyranoside, the cells were incubated at 28 °C for 12 h. Cell pellets were resuspended in a buffer containing 20 mm Tris-Cl, pH 7.4, 300 mm NaCl, 7 mm β-mercaptoethanol, 5% glycerol) and lysed by sonication. The extract was centrifuged at 18,000 rpm for 1 h. The Ssl1 (1–350) protein was first purified by nickel-nitrilotriacetic acid affinity chromatography using the His tag at the N terminus of Ssl1 and then eluted with 500 mm imidazole in the same buffer. Fractions containing the Ssl1 (1–350) protein were subsequently purified using anion exchange (Mono Q) and gel filtration chromatography (Superdex 75). To aid crystallization of the Ssl1 N-terminal regulatory domain, the Ssl1 (1–350) fragment was subjected to limited proteolytic digestion by treatment with a 2000:1 ratio of subtilisin for 20 min at 10 °C. N-terminal sequencing and mass spectrometry analyses of the digested fragment revealed that the resulting Ssl1 fragment contained residues 108–328. The Ssl1(108–328) fragment was inserted into the pET28a vector, and the Ssl1 N-terminal regulatory domain was purified via nickel-nitrilotriacetic acid affinity chromatography. The protein was purified further using cation exchange (Mono S) and gel filtration chromatography. The selenomethionine-substituted Ssl1(108–328) protein was obtained from E. coli B834 (DE3) cells and purified using the same method as that used for native Ssl1. For biochemical assays, N-terminal histidine-tagged human TFIIH p44 proteins (wild type, T138R, or L174W/T175R) were purified using the same method for Ssl1 purification.
Crystallization and Data Collection
Crystals of the Ssl1 N-terminal regulatory domain (108–328) were grown at 18 °C by the hanging drop vapor diffusion method. The crystallization buffer contained 28% polyethylene glycol 3350 and 0.1 m Tris-HCl, pH 8.0, and the concentration of protein was 12 mg/ml. Diffraction data were collected at −170 °C using crystals flash-frozen in crystallization buffer containing 30% (w/v) glycerol. Diffraction data from selenomethionine-substituted Ssl1 core crystals were collected at 0.9791 Å on Beamline 4A using the Pohang Advanced Light Source. The Ssl1 regulatory domain crystals formed in the space group P3221 with the parameters a = 78.6 Å, b = 78.6 Å, c = 83.9 Å, α = 90, β = 90, and γ = 120, and contained one Ssl1 N-terminal regulatory domain protein in an asymmetric unit. Diffraction data integration, scaling, and merging were performed using the HKL2000 package (Table 1 and Ref. 25).
TABLE 1.
Statistics of data collection and refinement
The values in parentheses are for the highest shell. Rsym = ΣhΣi|Ih,i − Ih|IΣhΣlIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. r = Σ|Fobs − Fcalc|/ΣFobs, where Fobs = Fpi, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 10% of the reflections). Root mean square deviation in bond lengths and angles are the deviations from ideal values, and the root mean square deviation in B factors is calculated between bonded atoms.
| Ssl1 regulatory domain (residues 108–328) | |
|---|---|
| Diffraction data | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 78.6, 78.6, 83.9 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50-2.4 (2.48-2.4) |
| Measured reflections | 416,666 |
| Unique reflections | 20,596 |
| Rsym | 0.26 (0.48) |
| <I>/<σ> | 39.69 (8.21) |
| Completeness (%) | 100 (100) |
| Redundancy | 20.2 (20.0) |
| Phasing (SAD) Resolution range (Å) | 50–2.8 |
| Phasing (SAD) initial figure of merit | 0.33 |
| Refinement | |
| Resolution (Å) | 28.7-2.4 |
| No. of reflections | 12,054 |
| Rworking/Rfree | 0.195/0.214 |
| No. of atoms | |
| Protein | 1472 |
| Ligand/ion (SO4, glycerol) | 11 |
| Water | 81 |
| Root mean square deviation bond length (Å) | 0.008 |
| Root mean square deviation bond angles (°) | 1.17 |
| B-factors (Å2) | |
| Protein | 49.9 |
| Solvent | 49.4 |
| Ramachandran plot | |
| Favored/allowed/outlier (%) | 98.95/0.53/0.53 |
Structure Determination and Refinement
The structure of the Ssl1 N-terminal regulatory domain was determined by the single-wavelength anomalous scattering dispersion method. Five selenium sites were identified initially, and an electron density map was generated using the Phenix program (26). The electron density map generated at a resolution of 2.4 Å showed good quality, which allowed tracing of most of the chain. The complete structure was built through successive rounds of model building using COOT (27) and refinement (bulk solvent corrections, overall B-value refinement, and positional and individual B-value refinement) using Phenix program (26). The statistics are summarized in Table 1.
Expression of His-XPD, His Full-length p44, and His-p44 (1–252) or (252–395) Fragments
Sf9 cells were infected with baculoviruses expressing His-XPD, WT full-length His-p44, or the His-p44 1–252 or 252–395 fragment at the indicated multiplicity of infection (pfu/cell). In brief, the cells were collected 48 h after infection, washed once with 1× PBS containing 30% glycerol, resuspended in buffer A (20 mm Tris-HCl, pH 7.9, 20% glycerol, 150 mm NaCl, 0.1% Nonidet P40, and 1× protease inhibitor mixture), and then centrifuged at 14,000 × g for 30 min.
Yeast Viability Test
The S. cerevisiae PDY2 Matα leu2Δ1 ura3–52 trp1Δ63 his3Δ200 Ssl1::Kan (pSsl1/316) haploid strain was a gift from Dr. Charles Cole (Dartmouth University). We generated a mutant plasmid (pSsl1/pRS314) using site-directed mutagenesis. We transformed the PDY2 cells using the pSsl1/pRS314 mutant plasmid and selected transformants on Ura(−)/Trp(−) synthetic complete medium plates. The transformed strains were cultured in YPD broth until they reached an A600 nm of 1.0. The yeast cells were then serially diluted 5-fold and spotted onto Trp(−)/FOA medium.
Human XPD-p44 Binding Assay
G-Sepharose beads (5 μl) cross-linked with an anti-XPD antibody were washed with buffer B (20 mm Tris-HCl, pH 7.9, 20% glycerol, 150 mm KCl, 0.1% Nonidet P40, and 1× protease inhibitor mixture) and then incubated with a large excess of XPD cell crude extract at 4 °C for 4 h. After the incubation, the beads were washed four times with buffer A (20 mm Tris-HCl, pH 7.9, 20% glycerol, 1 m KCl, 0.1% Nonidet P40, and 1× protease inhibitor mixture) and twice with buffer B and then incubated with His-p44 WT or mutant cell crude extracts at 4 °C overnight. The beads were then washed four times with buffer C (20 mm Tris-HCl, pH 7.9, 20% glycerol, 400 mm KCl, 0.1% Nonidet P40, and 1× protease inhibitor mixture) and twice with buffer D (20 mm Tris-HCl, pH 7.9, 20% glycerol, 50 mm KCl, 0.01% Nonidet P40, and 1× protease inhibitor mixture). Following the washes, the beads were incubated in 1× Laemmli SDS buffer for electrophoresis at 95 °C for 30 min. The immobilized XPD-p44 complex was then analyzed by SDS-PAGE and Western blotting.
Helicase Assay
G-Sepharose beads bound to human XPD (150 ng) and the 2-fold excess of WT or mutant p44 complex were generated by the procedure described for the XPD-p44 binding assay. Helicase assays were performed as described previously (10). Briefly, reactions were incubated for 45 min at 37 °C in the presence of 4 mm MgCl2, 4 mm ATP, 50 mm KCl, 50 μg/ml BSA, and 1–3 ng of radioactive DNA substrate and were stopped by the addition of EDTA and SDS. The samples were loaded onto a 14% nondenaturing acrylamide gel, which was dried and analyzed by autoradiography. The DNA substrate for helicase assays was prepared by mixing oligonucleotides corresponding to fragment 6219–6253 of the single-stranded M13mp18(+) DNA with single-stranded M13mp18(−) DNA.
RESULTS
Overall Structure of the N-terminal Domain of Ssl1
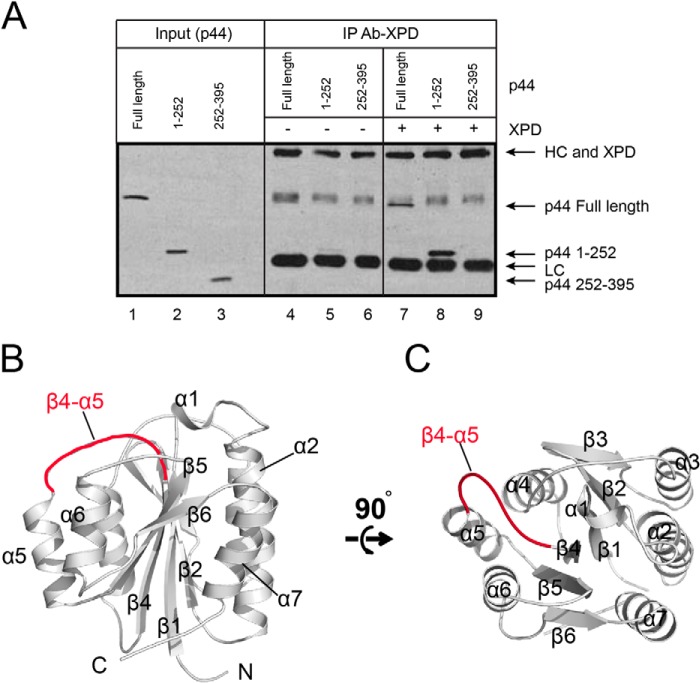
Our initial attempt to crystallize full-length yeast Ssl1 (or human p44) or a fragment containing residues 1–350 was unsuccessful, presumably because of the presence of highly flexible regions. To identify the stable and compact domain of Ssl1, we performed a limited digestion analysis of Ssl1 (1–350) and identified a compact fragment comprising residues 108–328, with which we successfully obtained high diffraction quality crystals. In conjunction with previous results, our analysis showed that the human p44 fragment containing residues 1–252 (corresponding to residues 50–322 of Ssl1) is sufficient to interact with XPD (Fig. 1A, lane 8), whereas the C-terminal fragment (residues 252–395) does not bind to XPD (Fig. 1A, lane 9). Furthermore, residues 1–235 of p44 participate in stimulating the helicase activity of XPD (10); thus we named this region the “regulatory domain.” The structure of the Ssl1 regulatory domain was determined using the selenium single-wavelength anomalous dispersion scattering method at a resolution of 2.4 Å (Table 1). The final structure of Ssl1 contained residues 119–310, of which the N-terminal 11 residues and C-terminal 18 residues were disordered.
FIGURE 1.
The overall structure of the Ssl1 regulatory domain. A, residues 1–252 of the human p44 fragment (residues 50–322 of ScRad3), but not the C-terminal fragment comprising residues 252–395, interact with human XPD. Sf9 cells infected with XPD baculoviruses were lysed and incubated with either full-length p44, p44 (residues 1–252), or p44 (residues 252–395) Sf9 cell extracts. The Ab-XPD immunoprecipitated (IP) complexes were resolved by SDS-PAGE and immunoblotted. Lanes 1–3 represent full-length p44 (full-length), p44 (residues 1–252), and p44 (residues 252–395), respectively; lanes 4–6 represent the negative control, in which the Ab-XPD were incubated with various p44 proteins only. Lanes 7–9 represent the immunoprecipitated complexes containing Ab-XPD, which were incubated with XPD, and subsequently with p44 proteins. The positions of XPD, p44 full-length, p44 (residues 1–252) fragment, p44 (residues 252–395) fragment, immunoglobulin heavy chain (HC), and light chain (LC) are indicated in the right panel. B and C, in Ssl1, a central β-sheet is surrounded by the seven amphipathic helices. The β4-α5 loop is indicated as a red line.
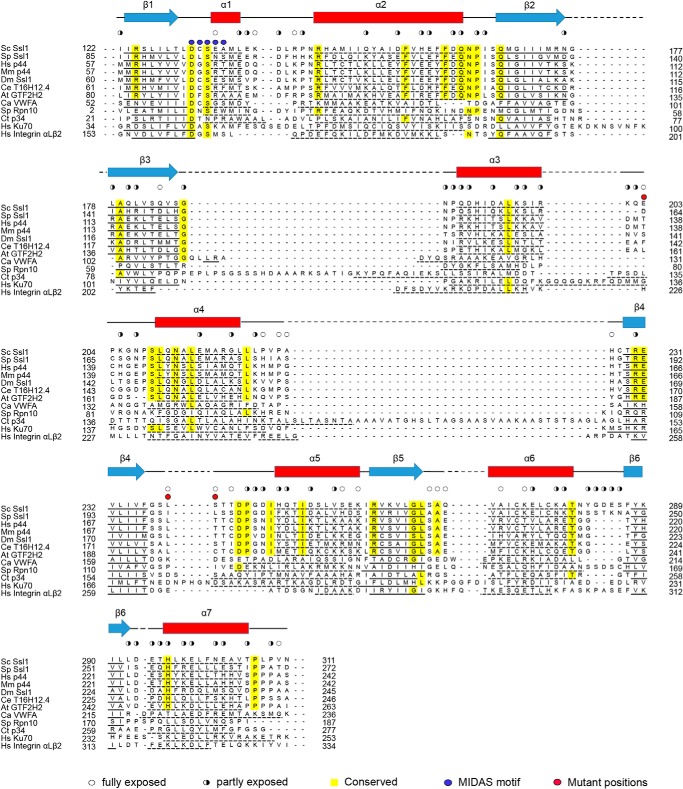
The overall shape of the Ssl1 regulatory domain is a cuboid with six faces (Fig. 1, B and C). Ssl1 folds into an α/β structure in which a six-stranded sheet (in the order 6, 5, 4, 1, 2, and 3) forms a central core flanked by three amphipathic helices on each side. In addition, a short helix is located at the top of the sheet. With the exception of the β3 strand, all other strands are oriented in a parallel manner. The domain is 40 Å high, 30 Å wide across the β-sheet, and 40 Å broad. The top and bottom of the structure are both comprised of loops that connect the sheet to the helices (Fig. 1B). Because the central sheet is surrounded by helices and loops, fully or partially exposed residues are located primarily in the N-terminal α1-α2 loop, the β3-α3, α3-α4, and β4-α5 loops, the α5 and α6 helices, and the C-terminal end (Figs. 1, B and C, and 2). Among these exposed residues, highly conserved residues are located in the N-terminal region; in the α2-β2, β3-α3, and β4-α5 loops; and at the end of the α7 helix (Fig. 2).
FIGURE 2.
Sequence alignment of seven Ssl1/p44 homologs and five different VWA proteins. Shown are Ssl1 from S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Homo sapiens (Hs), Mus musculus (Mm), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), and Arabidopsis thaliana (At), S. pombe (Sp) Rpn10, Chaetomium thermophilum (Ct) Tfb4/p34, Catenulispora acidiphila (Ca) VWFA, H. sapiens Ku70, and H. sapiens integrin αLβ2. α helices and β strands are shown as red rectangles and blue arrows, respectively. The secondary structures of the proteins are underlined. The conserved residues are indicated by yellow boxes, and fully and partly exposed residues are indicated by open and half-filled circles, respectively. Metal ion-dependent adhesion site motif residues are indicated by blue dots. The positions of the mutations (Leu-174/Ser-175 and Glu-203) are indicated as red dots. Promals program (34) was used for multiple sequence alignments.
Ssl1 Structural Homologs and Their Interactions with Partners
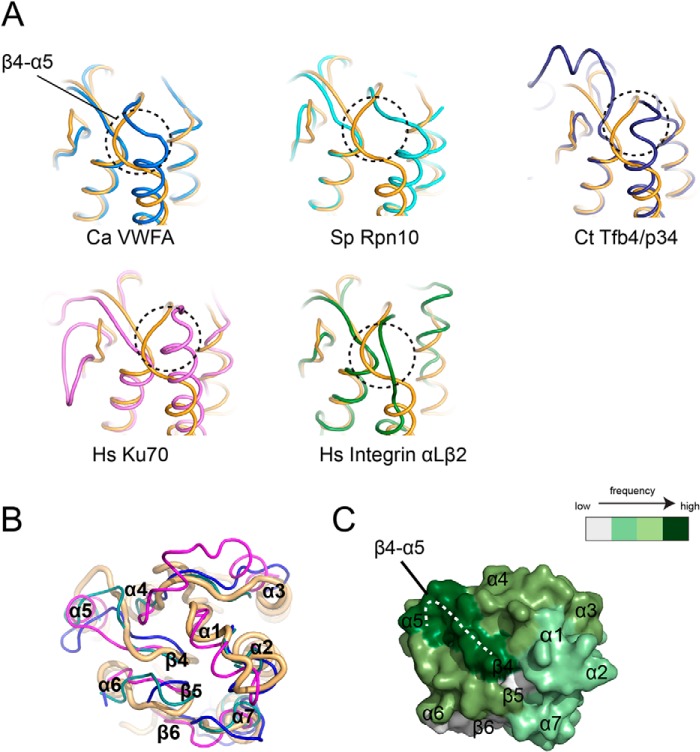
A database search using DALI revealed that Ssl1 resembles several members of the von Willebrand factor A (VWA) family proteins, including von Willebrand factor type A (PDB code 4FX5), 26S proteasome regulatory subunit RPN10 (PDB code 2X5N), Ku70 (PDB code 1JEY), TFIIH Tfb4/p34 subunit (PDB code 4PN7), and integrin αLβ2 (PDB code 1T0P), with root mean square deviations of 2.0–2.6 Å for 172–178 Cα atoms (Refs. 28–33 and Fig. 3A). Although the secondary structures of Ssl1 are similar to those of other VWA family members, the lengths of some regions differ; for example, the lengths of the α1, α3, and α4 helices of Ssl1 are notably different from those of the corresponding helices of the TFIIH Tfb4/p34 subunit (Fig. 2). In addition, the loops that connect each secondary structure show marked differences (Fig. 3A). The largest structural differences between the Ssl1 and VWA proteins occur in the β3-α3, α4-β4, β4-α5, and β5-α6 loops, which exhibit up to a 10 Å difference. The sequences of Ssl1 and the VWA proteins are poorly conserved (Fig. 2); no noticeable conservation was observed in a multiple sequence alignment. The length of the loop corresponding to the β3-α3 loop varies from 2–25 amino acid residues and the length of the α4-β4 loop varies from 7 to 35 residues. From the aspects of the length, loop β4-α5 and β5-α6 showed the smallest differences (5–8 and 4–7 residues, respectively). Unlike many VWA family members, Ssl1 has a five-residue insertion (α1 helix) between β1 and α2, which is similar to the equivalent β1-α2 region of RPN10 (Fig. 3B). Furthermore, the metal ion-dependent adhesion site (DXSXS motif) that is found in a number of VWA proteins is replaced by a DCSEA motif in β1-α1 in Ssl1 (Fig. 2).
FIGURE 3.
Structural comparisons of Ssl1 and VWA family proteins. A, from left, Ssl1 (orange) was superimposed onto C. acidiphila VWFA (Protein Data Bank 4FX5, RMSD 2.0 Å, blue), S. pombe Rpn10 (Protein Data Bank code 2X5N, RMSD 2.5 Å, cyan), C. thermophilum Tfb4/p34 (Protein Data Bank code 4PN7, RMSD 2.2 Å, deep blue), H. sapiens Ku70 (Protein Data Bank code 1JEY, RMSD 2.6 Å, pink), or H. sapiens integrin αLβ2 (Protein Data Bank code 1T0P, RMSD 2.6 Å, green). The β4-α5 loop is indicated by dotted black circles. The structures are shown in the same orientation. B, structural superposition of Ssl1 (light orange) and VWA family proteins. The variable regions of VWFA (Protein Data Bank code 4FX5), RPN10 (Protein Data Bank code 2X5N), and Tfb4/p34 (Protein Data Bank code 4PN7) are shown in magenta, blue, and teal, respectively. C, surface representation of the Ssl1 regulatory domain. The frequencies with which specific regions of VWA family members interact with their counterparts are indicated by dark green, green, and pale green, which represent high, moderate, and low frequencies. The binding frequencies were calculated from the interface analyses of VWA family protein structures that are complexed with their binding partners (30, 33, 35–38). The β4-α5 loop is indicated as a white dotted line.
To predict the surface of Ssl1 that interacts with Rad3, we first analyzed the interfaces between VWA domain-containing proteins and their binding partners based on nine available structures. The most frequently used binding surface at the VWA domain is the region that corresponds to the β4-α5 loop in Ssl1 (white dotted line in Fig. 3C). In five VWA protein structures, a loop equivalent to the β4-α5 loop of Ssl1 participates in the recognition of partners (dark green in Fig. 3C); these proteins include anthrax toxin-receptor (PDB code 1T6B), VWFA1 domain-GpIbα (PDB code 1M10), integrin αLβ2-ICAM3 (PDB code 1T0P), integrin β3-integrin αIIb (PDB code 3NIG), and Sec23/24-Sar1 (PDB code 1M2O) (30, 33, 35–38). Four VWA protein structures utilize a loop corresponding to the α3-α4 loop and α4 and α6 helices of Ssl1 for interactions with cellular counterparts (Fig. 3C).
Mutational Analysis of p44 in Vitro
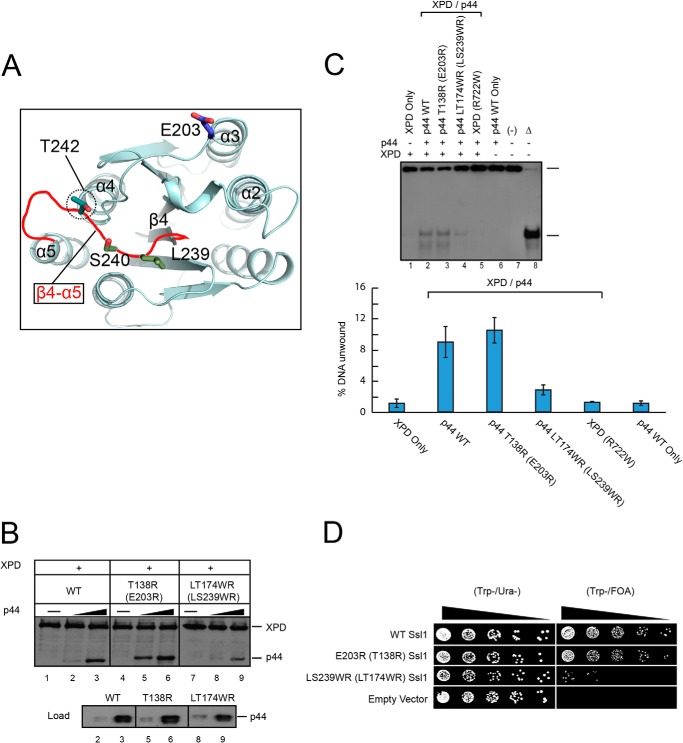
Previous biochemical and genetics analyses provide an important clue for the regions essential for the function of Ssl1. First, the C-terminal region of XPD (residues 616–730) interacts with the N-terminal region of p44 (residues 1–252) (15). Second, in a yeast genetics analysis, the T242I Ssl1 mutant (corresponding to Cys-177 of human p44) exhibited reduced helicase activity and caused a defect in recombination between short repeats, as well as translational defects (39). Thr-242 is exposed in the β4-α5 loop and does not make contact with any neighboring residues. We hypothesized that the β4-α5 loop could be involved in binding to ScRad3 (Figs. 3B and 4A). Furthermore, the region equivalent to the β4-α5 loop in VWA proteins is one of the most commonly used segments involved in interactions with partner proteins (30, 33, 35–38). Based on these data, we predicted that the β4-α5 loop of Ssl1 may participate in binding to XPD.
FIGURE 4.
Protein interaction, helicase activity assays, and yeast viability tests of XPD and mutant p44 proteins. A, the positions of the following residues that were mutated in this study are indicated: Glu-203 (Thr-138 of p44, blue line) and Leu-239/Ser-240 (Leu-174/Thr-175 of p44, dark green lines) at the top side of Ssl1. The β4-α5 loop is indicated as red lines. The Thr-242 residue is indicated by a dotted circle. The top figure is in the same orientation as that of Fig. 1C. B, binding of WT, T138R, or L174W/T175R p44 proteins to XPD. Equivalent residues of Ssl1 are marked in parentheses. The positions of XPD and p44 are indicated at the right side of the panel. C, the stimulation of XPD helicase activity by WT and mutant p44 proteins. Lane 2, WT; lane 3, T138R; lane 4, L174W/T175R. The positions of the substrate and product are shown at the right side of the panel. Lanes 1 and 6 show results for WT-XPD and WT-p44 only, respectively. Lane 5 contains XPD mutant (R722W) and p44. Lanes 7 (−) and 8 (Δ) show the results for nonboiled and boiled substrate, respectively. The helicase activities were quantified using ImageJ software and are shown as a bar graph (bottom panel). D, 5-fold serial dilutions of yeast strains containing the pRS316_WT Ssl1 plasmid and the pRS314_mutant or WT Ssl1 plasmid were spotted onto Trp−/Ura− and Trp−/FOA plates and photographed after growth for 2 days at 30 °C in the dark. The positions of the Ssl1 mutants are indicated at the left side of the panel. The strain that harbored the pRS314 vector only was spotted at the empty vector line.
To validate our prediction, we performed mutational analyses of the solvent-exposed residues of Ssl1 and examined the Rad3 binding and helicase stimulating activities of the mutant proteins (Fig. 4, B and C). Because we could not isolate the full-length yeast Rad3 in a soluble form, we used human p44, a homolog of Ssl1, for the XPD binding and helicase stimulation assays (“Experimental Procedures”).
Leu-239 and Ser-240 are solvent-exposed residues in the β4-α5 loop of yeast Ssl1; therefore, we mutated the equivalent Leu-174 and Thr-175 residues in human p44 to Trp and Arg, respectively (L174W/T175R). In contrast to wild type p44, L174W/T175R p44 exhibited significantly reduced binding to XPD (Fig. 4B, lanes 3 and 9). The α3-α4 loop is also frequently used to interact with partners by VWA family proteins, which is closely placed to the β4-α5 loop (closest Cα-Cα distance between two loops is 6.3 Å). We mutated the solvent-exposed Thr-138 of the α3-α4 loop, to Arg (T138R; Glu-203 in the α3-α4 loop of Ssl1). The T138R mutant showed similar or slightly increased XPD binding activity relative to WT p44 (Fig. 4B, lanes 3 and 6).
Next, we examined the XPD helicase-stimulating activity by the p44 mutants. Full-length p44 alone and a XPD mutant that failed to associate with p44 (R722W) did not exhibit any helicase activity, which established the critical relationship of p44 binding to XPD and helicase stimulating activity (10, 15). The XPD helicase-stimulating activity of the L174W/T175R p44 was significantly lower than that of WT p44, which is consistent with its decreased XPD binding (Fig. 4C, lanes 2 and 4 and histogram). By contrast, T138R p44 retained XPD helicase stimulating activity similar to that of WT p44 (∼110% helicase activity compared with WT p44; Fig. 4, B, lanes 3 and 6, and C, lanes 2 and 3). We conclude that Leu-174 and Thr-175 in the β4-α5 loop play a critical role for p44 in XPD binding and helicase stimulation.
In Vivo Analysis of Ssl1/p44 Mutants
To examine the biological significance of interactions between Ssl1/p44 and Rad3/XPD, we created yeast strains containing mutations of the equivalent residues of the p44 mutants tested above and examined their viabilities in budding yeast (Fig. 4D). Because Ssl1 is essential for cell viability, we tested the effects of these mutations on cell viability using the plasmid shuffling method (40, 41). A pRS316 plasmid containing mutant Ssl1 was transformed into a yeast cell that lacked the chromosomal Ssl1 gene but expressed WT Ssl1 from pRS314. The WT Ssl1 pRS314 plasmid was shuffled out by growth on SC(Trp-)/FOA medium. We generated yeast strains harboring plasmids expressing the E203R (T138 of p44) or L239W/S240R (L174/T175 of p44) Ssl1 mutants. The strain harboring the L239W/S240R mutant shows significantly reduced viability on FOA medium. By contrast, the E203R mutation had no negative effect on cell viability (Fig. 4D). Collectively, these in vivo analyses confirmed the significance of the interaction between Rad3 and the β4-α5 loop of Ssl1 to cell viability.
Structural Prediction of the Interface between Rad3 and Ssl1
To understand how Ssl1/p44 interacts with Rad3/XPD and stimulates its helicase activity, we built a model onto the EM density of budding yeast TFIIH (ScTFIIH) using high resolution structures of T. acidophilum XPD homolog (TaXPD; PDBID:2VSF) and Ssl1 (3, 42). We used the TaXPD structure because it contains the longest C-terminal region among the available XPD homologs and is most similar to yeast Rad3. Nevertheless, this model lacks a region correspond to residues 727–778 of ScRad3. We initially fitted the structure of TaXPD to two well defined segments of ScTFIIH cryo-EM density using a chimera program (Ref. 42 and Fig. 5A). The correlation coefficient of the fitting trials of TaXPD structures into the EM density was 0.92. Subsequently, we performed rigid body fitting of Ssl1 into the electron density in the presence of TaXPD, such that the β4-α5 loop was directed toward Rad3 (TaXPD) (Fig. 5B). The overall structure fit into the Ssl1 density reasonably well; however, a small empty space was present between Rad3 (TaXPD) and Ssl1, and we presume that the C-terminal region (residues 727–778) of ScRad3 may be placed in this region. This model can explain the previous data that the C-terminal region of ScRad3 interacts with the N-terminal segment of Ssl1 (43). In previous cross-linking analysis, only Lys-113 of Ssl1 was cross-linked with Lys-731 of Rad3 in the Rad3-Ssl1 interface (43). This could be due to the following: (i) the interface between Rad3 and Ssl1 may be too close and/or (ii) no lysine residues are present in the β4-α5 loop and neighboring region of Ssl1, which limit the high resolution cross-linking analysis (performed by bis-sulfosuccinimidyl suberate) in this interface. We note that, because of the lack of structure of the C-terminal region of Rad3, the model presented here provides only limited information (Fig. 5, A and B).
FIGURE 5.
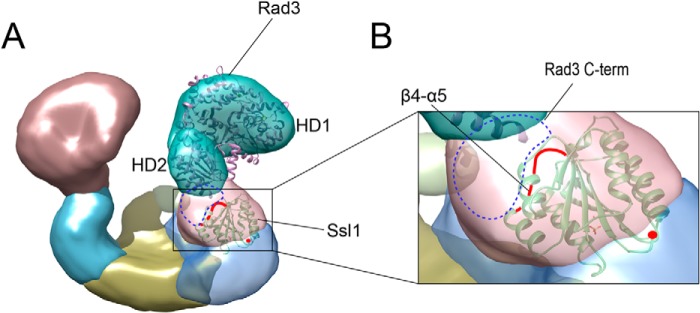
The predicted position of Ssl1 in the TFIIH complex. A, the x-ray structures of T. acidophilum XPD (Protein Data Bank code 2VSF) and Ssl1 were fitted into the EM density of the ScTFIIH complex. The predicted locations of the Rad3 C-terminal is depicted as blue dotted circles. The β4-α5 loop is shown as a red line. The Ssl1 N-terminal end is indicated by a red dot. B, a close-up view of the interface between Rad3 and Ssl1 in the complex model.
DISCUSSION
The Ssl1/p44 subunit is critical for maintaining the architecture of the TFIIH transcription/DNA repair factor and stimulation of the Rad3/XPD helicase, which are required for accurate transcription and NER, respectively (2, 12, 44). In the present work, we showed that the N-terminal regulatory domain of Ssl1 forms a VWA fold, identified a key interaction between Rad3/XPD and Ssl1/p44, and provided a model for the Rad3/XPD-Ssl1/p44 complex through structural, biochemical, and genetic analyses.
The VWA domain interacts with a diverse range of cellular proteins involved in cell adhesion and signal transduction (38). Structural analyses have shown that VWA family members exhibit noticeable structural diversity with weak conservation. Although VWA proteins recognize cellular proteins through various motifs, the region corresponding to the β4-α5 loop is most frequently involved in protein recognition (38). Our analyses suggest that the β4-α5 loop of Ssl1/p44 are involved in binding to Rad3/XPD, its helicase stimulating activity, and the cell viability. Although it is located at the same face as the β4-α5 loop, mutation of the α3-α4 loop of p44 did not influence XPD binding or helicase stimulating activity, which suggests that the binding to Rad3/XPD may not require an extensive surface of Ssl1/p44.
In a previous study, mutation of the conserved residues of p44, including Asp-66, Glu-166, Asp-178, and Gly-200, abolished the helicase activity of XPD, suggesting that these residues are involved in binding to XPD (12). However, these residues are either partly or completely buried in the structure of Ssl1 (Fig. 6, A–D).
FIGURE 6.
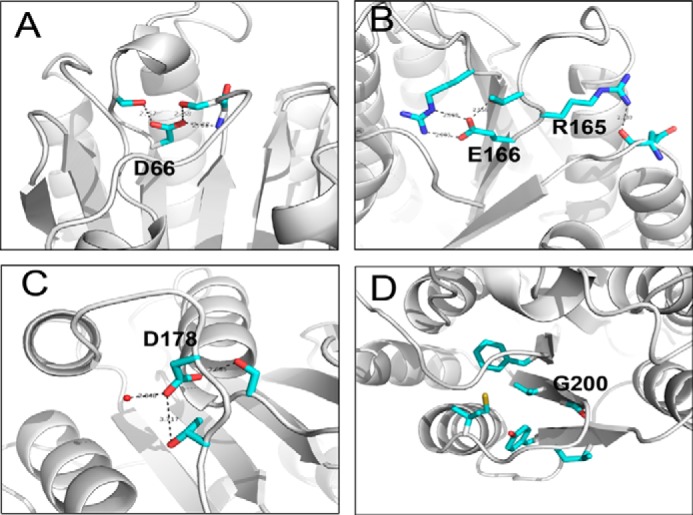
Close-up views of the p44 mutants. A–D, previously studied p44 mutant residues Asp-66, Arg-165, Glu-166, Asp-178, and Gly-200 are shown in stick representation and colored in cyan (12). Water molecules are shown as red dots, and hydrogen bonds are indicated by black dotted lines.
Recently, the structure of Tfb4/p34, another component of TFIIH, was reported (31). Notably, Tfb4/p34 also forms a VWA fold. Despite the structural similarity between Tfb4/p34 and Ssl1, the lengths of several helices and loops are clearly different. In addition, the loop of Tfb4/p34 equivalent to the β4-α5 loop of Ssl1 exhibits significant difference, suggesting that this region may be a unique structural feature that allows Ssl1/p44 to interact with Rad3/XPD.
Which parts of Rad3/XPD interact with Ssl1/p44? Four disease-causing mutations at Asp-673, Gly-675, Asp-681, and Arg-683 of XPD are important for its interaction with p44, which suggests that the HD2 domain is important for the binding to p44 (16). However, it is unlikely that these residues from the HD2 domain are directly involved in binding to p44, because they play structural roles in maintaining the integrity of the HD2 domain (17–20). In addition, residues Gly-713, Arg-722, and residues 716–730 of XPD are also involved in binding to p44 (16). We note that the C-terminal 30 residues (residues 749–778) of yeast Rad3 are disordered in the structural prediction presented here; this region may undergo a structural transition to become ordered upon binding to Ssl1. Because a 30-residue peptide (residues 749–778) from the C-terminal tail of yeast Rad3 did not bind to Ssl1,5 we speculate that a coordinated action of HD2 and the C-terminal tail of Rad3 may be required for the stable assembly of Rad3 and Ssl1.
The Rad3/XPD-Ssl1/p44 interface is crucial for maintaining the architecture and the functional role of TFIIH in both DNA repair and transcription. First, by interacting with XPD, p44 is able to regulate its helicase activity in DNA repair required for opening DNA lesions induced by UV irradiation and chemical adducts caused by agents such as cisplatin (10, 11). Mutation of the Arg-658, Gly-713, and Arg-722 residues located in the C-terminal end of human XPD disrupt the XPD-p44 interaction and consequently suppress the NER function of TFIIH (10, 15). In a previous study, a strain expressing the G265R mutant of Ssl1 exhibited defective NER and conditionally defective transcription and cell viability (40–41, 45). Gly-265 is located at the β5 strand, next to the β4-α5 loop (Fig. 1, B and C); thus mutation of this residue to Arg likely perturbs the local structure in this region, supporting our data (Fig. 1, B and C). Second, p44 also allows the anchoring of the CAK subcomplex that interacts with XPD through the MAT1 subunit to the core TFIIH (23). Disruption of the XPD-p44 interaction also perturbs the anchoring of the CAK kinase to the TFIIH core and subsequently its ability to phosphorylate the activators (such as the retinoic acid receptor, estrogen receptor, and/or peroxisome proliferator-activated receptors) that would result in a defect of the transactivation process (2). This event ultimately leads to the developmental and neurological defects observed in some xeroderma pigmentosum patients (14). A weak or absent interaction between Ssl1/p44 and Rad3/XPD may lead to the incorrect assembly of TFIIH, resulting in decreased transcriptional activation, and thereby limits cell viability. In human XPD, the K75Q polymorphism located at this region induces a number of cancers, including esophageal, gastric, colorectal, breast, and bladder cancer (46–50), highlighting the importance of the interaction of XPD and p44 through this region.
In conclusion, we determined the structure of the N-terminal regulatory domain of Ssl1 and provided insights into the mechanism of stimulation of Rad3/XPD by Ssl1/p44 using multidisciplinary approaches. The structural and biological data presented here should contribute to understanding the architecture of TFIIH and its function in DNA repair and transcription.
Acknowledgments
We are grateful to Angele Koh, Cathy Braun, Jie-Ae Kim, and Isabelle Kolb for invaluable technical help. We thank Dr. Charles Cole for providing yeast strains lacking Ssl1. We also thank Dr. Caroline Kisker for helpful comments.
This work was supported by National R&D Program for Cancer Control, Ministry for Health and Welfare Grant 1020280; National Research Foundation of Korea Grants 2012R1A2A1A01004028, 2013M3A6A4044580, and 2010-0019706 funded by the Korea government (Ministry of Education, Science and Technology; MEST); the Pohang University of Science and Technology rising star program; the Ministry of Education BK21 Program (to Y. C.); and a European Research Council advanced grant (to J. M. E.). This work was also supported by grants from the European Research Council Advanced Scientists; Agence Nationale de la Recherche Grants ANR-08-MIEN-022-03, ANR-10-BLANC-1231-02, and FRETNET; and Association de la Recherche contre le Cancer Grant SL220100601335.
The atomic coordinates and structure factors (code 4WFQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
J. S. Kim, C. Saint-André, H. S. Lim, C.-S. Hwang, J. M. Egly, and Y. Cho, unpublished data.
- CAK
- Cdk-activating kinase
- VWA or VWFA
- von Willebrand factor A
- NER
- nucleotide excision repair
- EM
- electron microscopy
- FOA
- 5-fluoroorotic acid
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation.
REFERENCES
- 1. Kim T. K., Ebright R. H., Reinberg D. (2000) Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 [DOI] [PubMed] [Google Scholar]
- 2. Compe E., Egly J. M. (2012) TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354 [DOI] [PubMed] [Google Scholar]
- 3. Gibbons B. J., Brignole E. J., Azubel M., Murakami K., Voss N. R., Bushnell D. A., Asturias F. J., Kornberg R. D. (2012) Subunit architecture of general transcription factor TFIIH. Proc. Natl. Acad. Sci. U.S.A. 109, 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schultz P., Fribourg S., Poterszman A., Mallouh V., Moras D., Egly J. M. (2000) Molecular structure of human TFIIH. Cell 102, 599–607 [DOI] [PubMed] [Google Scholar]
- 5. Serizawa H., Conaway J. W., Conaway R. C. (1993) Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature 363, 371–374 [DOI] [PubMed] [Google Scholar]
- 6. Cho E. J., Takagi T., Moore C. R., Buratowski S. (1997) mRNA capping enzyme is recruted to the transcritption complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11, 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komarnitsky P., Cho E. J., Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tirode F., Busso D., Coin F., Egly J. M. (1999) Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and Cdk7. Mol. Cell. 3, 87–95 [DOI] [PubMed] [Google Scholar]
- 9. Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16, 6559–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coin F., Marinoni J. C., Rodolfo C., Fribourg S., Pedrini A. M., Egly J. M. (1998) Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20, 184–188 [DOI] [PubMed] [Google Scholar]
- 11. Coin F., Oksenych V., Egly J. M. (2007) Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell. 26, 245–256 [DOI] [PubMed] [Google Scholar]
- 12. Seroz T., Perez C., Bergmann E., Bradsher J., Egly J. M. (2000) p44/SSL1, the regulatory subunit of the XPD/RAD3 helicase, plays a crucial role in the transcriptional activity of TFIIH. J. Biol. Chem. 275, 33260–33266 [DOI] [PubMed] [Google Scholar]
- 13. Takagi Y., Masuda C. A., Chang W. H., Komori H., Wang D., Hunter T., Joazeiro C. A., Kornberg R. D. (2005) Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol. Cell. 18, 237–243 [DOI] [PubMed] [Google Scholar]
- 14. Lehmann A. R. (2001) The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15, 15–23 [DOI] [PubMed] [Google Scholar]
- 15. Dubaele S., Proietti De Santis L., Bienstock R. J., Keriel A., Stefanini M., Van Houten B., Egly J. M. (2003) Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell. 11, 1635–1646 [DOI] [PubMed] [Google Scholar]
- 16. Cleaver J. E., Thompson L. H., Richardson A. S., States J. C. (1999) A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 14, 9–22 [DOI] [PubMed] [Google Scholar]
- 17. Wolski S. C., Kuper J., Hänzelmann P., Truglio J. J., Croteau D. L., Van Houten B., Kisker C. (2008) Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 6, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuper J., Wolski S. C., Michels G., Kisker C. (2012) Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 31, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan L., Fuss J. O., Cheng Q. J., Arvai A. S., Hammel M., Roberts V. A., Cooper P. K., Tainer J. A. (2008) XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H., Rudolf J., Johnson K. A., McMahon S. A., Oke M., Carter L., McRobbie A. M., Brown S. E., Naismith J. H., White M. F. (2008) Structure of the DNA repair helicase XPD. Cell 133, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tremeau-Bravard A., Perez C., Egly J. M. (2001) A role of the C-terminal part of p44 in the promoter escape activity of transcription factor IIH. J. Biol. Chem. 276, 27693–27697 [DOI] [PubMed] [Google Scholar]
- 22. Fribourg S., Kellenberger E., Rogniaux H., Poterszman A., Van Dorsselaer A., Thierry J. C., Egly J. M., Moras D., Kieffer B. (2000) Structural characterization of the cysteine-rich domain of TFIIH p44 subunit. J. Biol. Chem. 275, 31963–31971 [DOI] [PubMed] [Google Scholar]
- 23. Sandrock B., Egly J. M. (2001) A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J. Biol. Chem. 276, 35328–35333 [DOI] [PubMed] [Google Scholar]
- 24. Taylor E. M., Broughton B. C., Botta E., Stefanini M., Sarasin A., Jaspers N. G., Fawcett H., Harcourt S. A., Arlett C. F., Lehmann A. R. (1997) Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl. Acad. Sci. U.S.A. 94, 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walker J. R., Corpina R. A., Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 30. Huizinga E. G., Tsuji S., Romijn R. A., Schiphorst M. E., de Groot P. G., Sixma J. J., Gros P. (2002) Structures of glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science 297, 1176–1179 [DOI] [PubMed] [Google Scholar]
- 31. Riedinger C., Boehringer J., Trempe J. F., Lowe E. D., Brown N. R., Gehring K., Noble M. E., Gordon C., Endicott J. A. (2010) Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J. Biol. Chem. 285, 33992–34003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmitt D. R., Kuper J., Elias A., Kisker C. (2014) The structure of the TFIIH p34 subunit reveals a von Willebrand factor A like fold. PLoS One 9, e102389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song G., Yang Y., Liu J. H., Casasnovas J. M., Shimaoka M., Springer T. A., Wang J. H. (2005) An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin αLβ2. Proc. Natl. Acad. Sci. U.S.A. 102, 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pei J., Grishin N. V. (2007) PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23, 802–808 [DOI] [PubMed] [Google Scholar]
- 35. Zhu J., Zhu J., Negri A., Provasi D., Filizola M., Coller B. S., Springer T. A. (2010) Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood 116, 5050–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bi X., Corpina R. A., Goldberg J. (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277 [DOI] [PubMed] [Google Scholar]
- 37. Santelli E., Bankston L. A., Leppla S. H., Liddington R. C. (2004) Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature 430, 905–908 [DOI] [PubMed] [Google Scholar]
- 38. Springer T. A. (2006) Complement and the multifaceted functions of VWA and integrin I domains. Structure 14, 1611–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maines S., Negritto M. C., Wu X., Manthey G. M., Bailis A. M. (1998) Novel mutations in the RAD3 and SSL1 genes perturb genome stability by stimulating recombination between short repeats in Saccharomyces cerevisiae. Genetics 150, 963–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoon H., Miller S. P., Pabich E. K., Donahue T. F. (1992) SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 6, 2463–2477 [DOI] [PubMed] [Google Scholar]
- 41. Wang Z., Buratowski S., Svejstrup J. Q., Feaver W. J., Wu X., Kornberg R. D., Donahue T. F., Friedberg E. C. (1995) The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol. Cell. Biol. 15, 2288–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C. C., Pettersen E. F., Goddard T. D., Meng E. C., Sali A., Ferrin T. E. (2012) UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 179, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murakami K., Elmlund H., Kalisman N., Bushnell D. A., Adams C. M., Azubel M., Elmlund D., Levi-Kalisman Y., Liu X., Gibbons B. J., Levitt M., Kornberg R. D. (2013) Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342, 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bardwell L., Bardwell A. J., Feaver W. J., Svejstrup J. Q., Kornberg R. D., Friedberg E. C. (1994) Yeast RAD3 protein binds directly to both SSL2 and SSL1 proteins: Implications for the structure and function of transcription/repair factor b. Proc. Natl. Acad. Sci. U.S.A. 91, 3926–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsui P., DePaulo J., Buratowski S. (1995) An interaction between the Tfb1 and Ssl1 subunits of yeast TFIIH correlates with DNA repair activity. Nucleic Acids Res. 23, 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ding D. P., Ma W. L., He X. F., Zhang Y. (2012) XPD Lys751Gln polymorphism and esophageal cancer susceptibility: a meta-analysis of case-control studies. Mol. Biol. Rep. 39, 2533–2540 [DOI] [PubMed] [Google Scholar]
- 47. Xue H., Lu Y., Lin B., Chen J., Tang F., Huang G. (2012) The effect of XPD/ERCC2 polymorphisms on gastric cancer risk among different ethnicities: a systematic review and meta-analysis. PLoS One 7, e43431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Ding D., Wang X., Zhu Z., Huang M., He X. (2011) Lack of association between XPD Lys751Gln and Asp312Asn polymorphisms and colorectal cancer risk: a meta-analysis of case-control studies. Int. J. Colorectal Dis. 26, 1257–1264 [DOI] [PubMed] [Google Scholar]
- 49. Pabalan N., Francisco-Pabalan O., Sung L., Jarjanazi H., Ozcelik H. (2010) Meta-analysis of two ERCC2 (XPD) polymorphisms, Asp312Asn and Lys751Gln, in breast cancer. Breast Cancer Res. Treat 124, 531–541 [DOI] [PubMed] [Google Scholar]
- 50. Wang M., Gu D., Zhang Z., Zhou J. (2009) XPD polymorphisms, cigarette smoking, and bladder cancer risk: a meta-analysis. J. Toxicol. Environ. Health A. 72, 698–705 [DOI] [PubMed] [Google Scholar]