Background: Adenylylation/AMPylation by Fic proteins alters cellular signaling. HYPE, the sole human Fic protein, is an adenylyltransferase.
Results: BiP is identified as a substrate for HYPE. HYPE adenylylates BiP and is required for UPR induction.
Conclusion: HYPE regulates ER homeostasis.
Significance: Adenylylation/AMPylation is a new mode of UPR regulation. This is the first demonstration of a physiological role for human HYPE.
Keywords: AMP, Apoptosis, Post-translational Modification, Signaling, Unfolded Protein Response (UPR), AMPylation, BiP, Filamentation Induced by cAMP, HYPE/FicD, Adenylylation
Abstract
The maintenance of endoplasmic reticulum (ER) homeostasis is a critical aspect of determining cell fate and requires a properly functioning unfolded protein response (UPR). We have discovered a previously unknown role of a post-translational modification termed adenylylation/AMPylation in regulating signal transduction events during UPR induction. A family of enzymes, defined by the presence of a Fic (filamentation induced by cAMP) domain, catalyzes this adenylylation reaction. The human genome encodes a single Fic protein, called HYPE (Huntingtin yeast interacting protein E), with adenylyltransferase activity but unknown physiological target(s). Here, we demonstrate that HYPE localizes to the lumen of the endoplasmic reticulum via its hydrophobic N terminus and adenylylates the ER molecular chaperone, BiP, at Ser-365 and Thr-366. BiP functions as a sentinel for protein misfolding and maintains ER homeostasis. We found that adenylylation enhances BiP's ATPase activity, which is required for refolding misfolded proteins while coping with ER stress. Accordingly, HYPE expression levels increase upon stress. Furthermore, siRNA-mediated knockdown of HYPE prevents the induction of an unfolded protein response. Thus, we identify HYPE as a new UPR regulator and provide the first functional data for Fic-mediated adenylylation in mammalian signaling.
Introduction
Protein folding and maturation take place in the endoplasmic reticulum (ER),2 where proteins enter as unfolded polypeptides and are either folded into mature proteins or misfolded into aggregates for degradation. When there is an imbalance between the rate of unfolded proteins entering the ER and the rate of protein folding by ER chaperones, ER stress is induced. Under such a condition, the ER activates an unfolded protein response (UPR), mediated by the following three ER transmembrane receptors: inositol-requiring protein-1 (IRE1), activating transcription factor-6 (ATF6). and protein kinase RNA-like ER kinase (PERK) (1, 2). The UPR signaling cascade works at three different levels. First, it lowers protein synthesis and translocation into the ER, helping the cell to transiently adapt to the stress. Second, it up-regulates the transcription of ER chaperones and other protein folding machinery to increase the rate of protein folding. Third, when these responses cannot establish homeostasis, programmed cell death is triggered.
Here, we show that the human Fic protein, HYPE (Huntingtin yeast interacting protein E), is a player in the UPR cascade. HYPE belongs to the Fic (filamentation induced by cAMP) family of enzymes that uses cellular sources of nucleotides to carry out various post-translational modifications (3–5). Predominant among these post-translational modifications is the covalent addition of AMP to target substrates such as mammalian Rho GTPases (4–8). Fic proteins are now known to utilize ATP, UTP, GTP, and CTP and their derivatives to adenylylate (AMPylate), uridinylate (UMPylate), phosphocholinate (PCylate), as well as phosphorylate protein targets in vitro and/or in vivo (3–14). Fic proteins are defined by an HXFX(D/E)(G/A)N(G/K)RXXR motif, with the invariant His being required for enzymatic activity (3, 7). Their activity is often regulated by an inhibitory helix, defined by a conserved (S/T)XXXE(G/N) sequence, that occupies the Fic active site to prevent nucleotide docking (15). Removal of the helix from the active site by mutating the conserved Glu relieves inhibition.
The human genome encodes a single Fic protein, called HYPE/FicD (5). HYPE was identified in a yeast two-hybrid screen where it interacted with the polyQ region of huntingtin, mutations within which cause Huntington disease (16). HYPE is highly conserved among eukaryotes, with human HYPE displaying high amino acid sequence similarity to its eukaryotic orthologs as follows: mouse (Mus musculus), 89.5%; zebrafish (Danio rerio), 76%; fruit fly (Drosophila melanogaster), 55%; and worm (Caenorhabditis elegans), 45.4%.
Using Rho GTPases, RhoA, Rac1, and Cdc42, as artificial substrates in an in vitro adenylylation assay, we previously showed that HYPE preferentially uses ATP as a nucleotide source and functions as an adenylyltransferase (4). However, compared with bacterial Fic proteins like Histophilus somni IbpA that adenylylate Rho GTPases, enzymatic activity associated with WT HYPE was extremely weak (4, 5). Accordingly, overexpression of HYPE in HeLa cells failed to induce a cytotoxic phenotype typical of that seen with IbpA (5). We reasoned that HYPE's weak enzymatic activity against Rho GTPases suggests that Rho GTPases may not be its physiological targets and/or that HYPE's active site was blocked by its inhibitory helix. Accordingly, a mutation of HYPE's Glu-234 to Gly in the context of HYPE's Fic domain alone (aa 187–437) was shown to relieve this inhibition by removing the inhibitory helix from the Fic active site (15).
Here, we have characterized HYPE's enzymatic activity in vitro and in tissue culture cells, and we show that uncontrolled HYPE activity induces apoptosis. Furthermore, we demonstrate that HYPE localizes to the lumen of the endoplasmic reticulum and adenylylates an ER resident chaperone, BiP, at Ser-365 and Thr-366. BiP is critical for protein quality control and maintaining ER homeostasis (1, 2, 17). Our findings indicate that HYPE expression is up-regulated upon induction of UPR, and knockdown of HYPE prevents activation of UPR. We postulate that HYPE regulates UPR progression by adenylylating BiP. Our study is the first functional demonstration of the physiological significance of Fic-mediated adenylylation in mammalian cell signaling.
EXPERIMENTAL PROCEDURES
Cloning and Site-directed Mutagenesis
HYPE clones were obtained by PCR using human HYPE cDNA (from Origene) as template. Accession numbers for HYPE and its orthologs are as follows: Homo sapiens HYPE/FicD, NP_009007.2; M. musculus HYPE, NP_001010825.2; D. rerio HYPE, B1350952.1; D. melanogaster HYPE, NP_609026.1; and C. elegans HYPE, NP_502036.1. Rho GTPase clones were obtained from the Missouri S&T cDNA Resource Center and re-cloned as described previously (5). For protein production, HYPE wild-type gene encoding amino acids 46–458 and GRP78 (BiP) wild-type gene encoding amino acids 19–637 were cloned as an N-terminal His6-SUMO fusion in pSMT3 (Addgene). For expression in mammalian cells, full-length HYPE wild-type gene was cloned with a C-terminal FLAG tag into pCDNA3.1 (Addgene) or with a C-terminal GFP tag into pEGFPN1 (Clontech). Mutations of HYPE's residues E234G, E234G/H363A, T76A/S77A, S79A/T80A, T183A, S79A/T80A/T183A, N275Q, N446Q, and N275Q/N446Q and BiP's residues S365A/T366A were constructed by site-directed mutagenesis using primers listed in Table 1.
TABLE 1.
List of primers used for site-directed mutagenesis of HYPE and BiP
| Primer | Sequence |
|---|---|
| HYPE E234G forward | 5′ GTGGCCATCGGCGGCAACACCCTCACCCTCTC 3′ |
| HYPE E234G reverse | 5′ GTGTTGCCGCCGATGGCCACTGTGTGGTAG 3′ |
| HYPE S79AT80A forward | 5′ CACCAAGTGCACCAGCCCGGCCGCGGAGCTC 3′ |
| HYPE S79AT80A reverse | 5′ GAGCTCCGCGGCCGGGCTGGTGTGATGCTGA 3′ |
| HYPE T183A forward | 5′CCGCGATCGGGCACTGCCTCTTGTG 3′ |
| HYPE T183A reverse | 5′ CAAGAGGCAGTGCCCGATCGCGGTT 3′ |
| HYPE N275Q forward | 5′ ATGAAGTACATCCAGACGACTCTGGTT 3′ |
| HYPE N275Q reverse | 5′ AACCAGAGTCGTCTGGATGTACTTCAT 3′ |
| HYPE N446Q forward | 5′ GAAGCCCAACCCCAGCACTCTGGGTTC 3′ |
| HYPE N446Q reverse | 5′ GAACCCAGAGTGCTGGGGTTGGGCTTC 3′ |
| BiP S365AT366A forward | 5′ TGTTGGTGGCGCGGCTCGAATTCCAAAG 3′ |
| BiP S365AT366A reverse | 5′ TTGGAATTCGAGCCGCGCCACCAACAAG 3′ |
Protein Expression and Purification
GST fusion, His6-SUMO fusion, and MBP-His-TEV fusion proteins were expressed in Escherichia coli BL21-DE3-RILP (Stratagene) in LB medium containing 50 μg/ml kanamycin (pSMT3) or 100 μg/ml ampicillin (pET-GSTx or pSJ8) to a density of A600 = 0.6. Protein expression was induced for 12–16 h at 18 °C with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were lysed in lysis buffer (50 mm HEPES, pH 8.0, 250 mm NaCl, 5 mm imidazole) containing 1 mm PMSF protease inhibitor and purified using a cobalt resin. Resin was washed with wash buffer (50 mm HEPES, pH 8.0, 250 mm NaCl, 40 mm imidazole). Next, His6-SUMO-tagged wild-type HYPEΔN45, HYPEΔN45-E234G, HYPEΔN45-E234G/H363A, and their corresponding autoadenylylation site mutants (T76A/S77A, S79A/T80A, T183A, and S79A/T80A/T183A) were eluted from the resin using elution buffer (50 mm HEPES, pH 8.0, 250 mm NaCl, 350 mm imidazole). Wild-type HYPEΔ102 (used for ATPase assays) as well as WT BiP (aa 19–637) and its adenylylation site mutant BiP-S365A/T366A were also purified as described above. The protein mixture was desalted into buffer (100 mm Tris-HCl, pH 8.8, 50 mm NaCl) using PD-10 desalting columns (GE Healthcare) and re-applied to a cobalt column, and flow-through containing HYPE proteins was further purified by ion exchange chromatography using 100 mm Tris-HCl, pH 8.8, with a salt gradient from 10 mm to 1 m NaCl. Fractions containing HYPE were further purified by size exclusion chromatography in buffer containing 10 mm Tris-HCl, pH 8.8, 100 mm NaCl. Protein concentrations were measured using the Bradford method. Purity was determined by SDS-PAGE. Proteins were stored at −80 °C.
In Vitro Adenylylation Assays
0.1 μg of His6-SUMO-tagged wild-type or mutant HYPE (aa 46–458) protein was incubated either alone or with 8 μg of WT-BiP (aa 19–637) or BiP-S365A/T366A mutant in an adenylylation reaction containing 5 mm HEPES, pH 7.5, 1 mm manganese chloride tetrahydrate, 0.5 mm EDTA, and 0.01 mCi [α-32P]ATP for 15 min. Where mentioned, manganese was replaced with 1 mm calcium chloride, 1 mm cobalt chloride, 1 mm zinc sulfate, or 1 mm magnesium chloride in the buffer. Reactions were stopped with SDS-PAGE loading buffer. Where mentioned, 20 mm EDTA was added to the adenylylation reaction to chelate metal ions. Reaction products were separated on 10 or 4–20% polyacrylamide gels and visualized by autoradiography. Bands were quantified using Typhoon phosphorimager and ImageJ software.
Proteomic Analysis
Protein bands were excised from the gel, chopped into small (∼1 mm) pieces, destained with 25 mm NH4HCO3 in 50% (v/v) acetonitrile, and digested as described previously (18). Resulting peptides were loaded into a capillary C18 column (15 cm × 75 μm, Agilent, Santa Clara, CA) coupled to a nano-HPLC system (1100 series, Agilent) equilibrated with 5% (v/v) mobile phase B (mobile phase A, 0.1 formic acid in water; mobile phase B, 0.1% (v/v) formic acid in acetonitrile), and eluted with the following gradient: 5% B for 5 min, 5–40% B in 50 min, 40–90% B in 8 min, and hold at 90% B for 6 min. The flow rate was set at 300 nl/min. Eluting peptides were analyzed online by electrospray ionization on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Full-MS scans were collected at 30,000 mass resolution at 400 m/z in a range of 300–2000 m/z, and the eight most intense ions were selected for collision-induced dissociation in the linear trap (35% normalized collision energy), before being dynamically excluded for 1 min.
Data were processed and searched against the human SwissProt database (22,723 sequences, downloaded on July 9, 2013) using the MaxQuant package (version 1.4.0.8) in two rounds (19). In the first round, peptides were searched using cysteine carbamidomethylation (+57.0215 Da) and 20 ppm and 0.5 Da for precursor and fragment mass tolerances, respectively. After recalibration, peptides were re-searched using the following parameters: 1) trypsin digestion in both peptide termini because of missed cleavage sites; 2) methionine oxidation (+15.9949 Da) and adenylylation (+329.0525 Da) of serine, threonine, and tyrosine as variable modifications; 3) cysteine carbamidomethylation as fixed modification; and 4) 4.5 ppm and 0.5 Da for precursor and fragment mass tolerances, respectively. The data were filtered with a false discovery rate of ≤0.01 in both peptide and protein levels.
Mammalian Cell Culture Assays
Human epithelium HEK293A, HEK293T, and HeLa cells and human breast cancer MCF7 cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 5% (v/v) fetal bovine serum (Life Technologies) and 100 μg/ml penicillin/streptomycin (Sigma) at 37 °C with 5% CO2. FuGENE HD (Promega) was used for transfections according to the manufacturer's directions. Cells were transfected for 10 h or 30 h, as indicated for each experiment. For detection of apoptosis, cells were incubated with DMSO for 3 h or 25 μm Z-VAD-fmk (R&D Systems) or DMSO for 2 h, prior to treatment with 1 μm staurosporine (Sigma). Samples were analyzed either by microscopy or by Western blot analysis following a sample preparation.
For induction of unfolded protein response, HEK293T cells were treated with 1 μg/ml thapsigargin (Sigma) or 2.5 μg/ml tunicamycin (Calbiochem) for specific time points. Cells were harvested, washed, and prepared for RNA isolation.
For siRNA-mediated knockdown of HYPE, HEK293T cells were transfected with 10 μm FicD siRNA (Life Technologies, s22000 and s21999) for 48 h, using FuGENE HD (Promega) according to the manufacturer's directions. Cells were harvested, washed, and prepared for RNA isolation.
Immunofluorescence, Fluorescence Protease Protection (FPP) Assay, and Tissue Fractionation
For immunofluorescence, cells were washed with 1× PBS, fixed with 2% (w/v) paraformaldehyde, blocked with 2.5% (v/v) FBS in PBS for 30 min, and permeabilized with 0.1% (v/v) Triton X-100 in PBS for 5 min. Primary antibody to HYPE (Abcam), BiP (Cell Signaling), GM130 (BD Biosystems), EEA1 (Cell Signaling), calreticulin (Abcam), or Mitotracker Red (Life Technologies) was incubated for 1 h and washed with PBS; the secondary antibody was incubated for 1 h, washed with PBS, and visualized by confocal microscopy.
Fluorescence protease protection assay was done as described previously (20). Mitoneet-GFP and BiP-GFP were used as mitochondrial and ER lumen-specific controls (21).
ER-enriched fractions were prepared from fresh rat livers as described previously (22). The postnuclear supernatant was spun at 15,000 × g to pellet the ER/microsome-enriched fraction, which was further separated over Histogenz density gradients. Equal amounts of protein from each fraction were separated by SDS-PAGE and analyzed by Western blotting. The ER fraction was then treated with trypsin as described previously (23). Primary antibodies against HYPE (Abcam), BiP (Cell Signaling), calnexin (Cell Signaling), and mitochondrial complex I (Mitosciences) were used to detect these proteins in the tissue fractions.
For microscopy, HEK293A cells transfected with HYPE or HYPE mutants were observed by fluorescence microscopy (Zeiss Axiovert 200 M and AMG EVOS). EGFP was excited at 488 nm; fluorescence was monitored between 510 and 520 nm. Cell morphology was observed using differential interference contrast (DIC) microscopy (Zeiss Axiovert 200 M). Fluorescence and DIC images were overlaid using the Zeiss software.
Immunoprecipitation and Western Blotting
For immunoprecipitation, HEK293T and MCF7 cells were lysed with modified RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% (v/v) Nonidet P40, containing 1.0 mm Pefabloc (Roche Applied Science), 1.0 mm benzamidine hydrochloride, 1 μm leupeptin, 1 μm E64). The homogenates were pre-cleared with 25 μl of protein A/G beads (Pierce), centrifuged at 14,000 × g at 4 °C for 10 min, and the supernatant was incubated with anti-HYPE antibody (Abcam) or anti-BiP antibody (Cell Signaling) for 2–4 h at 4 °C with gentle rotation followed by incubation with 25 μl of protein A/G beads (Pierce) for 30 min at 4 °C with gentle rotation. Beads were washed three times with ice-cold modified RIPA buffer, resuspended in 20 μl of 1× SDS loading buffer, and denatured at 95 °C for 5–7 min.
For Western blot analysis, HEK293A and HeLa cells were lysed in cell lysis buffer (50 mm HEPES, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 100 mm NaF, 1 mm DTT, 100 μm PMSF, 10% glycerol, 1% (v/v) Triton X-100, pH 7.4) for 15 min. The lysates were centrifuged at 18,000 × g for 15 min at 4 °C, and the supernatant was mixed with SDS-PAGE loading buffer and denatured at 95 °C for 6 min. Protein samples were resolved on an SDS-polyacrylamide gel, transferred to nitrocellulose membrane, and blocked with 5% bovine serum albumin (Sigma). Blots were probed with primary antibodies to cleaved poly(ADP-ribose) polymerase (cPARP; Abcam), FLAG-M2 (Sigma), GFP (Clontech), HYPE (Abcam), actin (Sigma), GAPDH (Amicon), AMPylated threonine (Millipore), AMPylated tyrosine (Millipore), or BiP (Cell Signaling). Secondary antibodies conjugated to fluorescent dyes or horseradish peroxidase were used for detection using the Odyssey detection system (LI-COR Biosciences) or film.
Endoglycosidase H Treatment
HEK293A cells were collected, resuspended in cell lysis buffer (50 mm HEPES, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 100 mm NaF, 1 mm DTT, 100 μm PMSF, 10% (v/v) glycerol, 1% (v/v) Triton X-100, pH 7.4), and kept on ice for 15 min. The supernatant was mixed with anti-FLAG M2-agarose (Sigma) and incubated at 4 °C with gentle shaking for 1 h. The precipitate was washed three times and heated in Glycoprotein denaturing buffer (New England Biolabs) at 100 °C for 10 min. The samples were then either left untreated or treated with endoglycosidase H (New England Biolabs) for 1 h at 37 °C as per the manufacturer's protocol. The products were analyzed by Western blot.
RNA Isolation, RT, and qRT-PCR
Total cell RNA was extracted with the RNeasy mini kit (Qiagen). 5 μg of RNA were treated with DNase (New England Biolabs) and reverse-transcribed with random primers using the Superscript III First Strand Synthesis System (Life Technologies). 2 μl of a 1:10 dilution of cDNA were used for quantitative PCR with specific primers and SYBR Green PCR master mix 2 (Applied Biosystems) for 40 cycles in a CFX96 real time PCR system (Bio-Rad), according to the manufacturer's instructions, HYPE, GRP78, CHOP, and Xbp1 Ct was normalized to GAPDH Ct for each condition. Primers used for analysis of various genes are listed in Table 2. UPR induction was done as indicated.
TABLE 2.
List of primers used for qRT-PCR analysis of HYPE and UPR-specific and UPR-independent control genes
| Primer | Sequence |
|---|---|
| HYPE forward | 5′ TGAAGCCATTGGTATGAGCA 3′ |
| HYPE reverse | 5′ GCGGTTGGGGTTCTGAC 3′ |
| GRP78/BiP forward | 5′ GGCCAAATTTGAAGAGCTGA 3′ |
| GRP78/BiP reverse | 5′ GCTCCTTGCCATTGAAGAAC 3′ |
| XBP-1(s) forward | 5′ CCGCAGCAGGTGCAGG 3′ |
| XBP-1(s) reverse | 5′ GAGTCAATACCGCCAGAATCCA 3′ |
| Total Xbp1 forward | 5′ GGCATCCTGGCTTGCCTCCA 3′ |
| Total Xbp1 reverse | 5′ GCCCCCTCAGCAGGTGTTCC 3′ |
| CHOP forward | 5′ GCACCTCCCAGAGCCCTCACTCTCC 3′ |
| CHOP reverse | 5′ GTCTACTCCAAGCCTTCCCCCTGCG 3′ |
| GAPDH forward | 5′ AGAAGGCTGGGGCTCATTTG 3′ |
| GAPDH reverse | 5′ AGGGGCCATCCACAGTCTTC 3′ |
| Actin forward | 5′ TCTACAATGAGCTGCGTGTG 3′ |
| Actin reverse | 5′ GGTGAGGATCTTCATGAGGT 3′ |
RNase Binding Assay
Bovine pancreatic RNase A (Sigma) was chemically coupled to CNBr-activated Sepharose (GE Healthcare) according to the manufacturer's protocol at 0.2 μmol per ml of resin volume. When mentioned, 30 nmol of RNase A matrix was denatured in 1.3 ml of 6 m guanidine hydrochloride, 0.1 m DTT, 10 mm Tris, pH 8.6, at room temperature overnight. Before use, the denaturant and DTT were washed away, and free thiols were blocked with 50 mm N-ethylmaleimide. 10 μg of His-tagged wild-type BiP or His-SUMO-tagged BiP S365A/T366A was incubated with 1 μg of wild-type, E234G, or E234G/H363A HYPE in an adenylylation reaction containing 5 mm HEPES, pH 7.5, 1 mm manganese chloride tetrahydrate, 0.5 mm EDTA, and 1 μm ATP for 15 min. The proteins were then incubated with 10 μl of either the native or denatured RNase A resin in a total volume of 500 μl of Buffer K at room temperature, with rotation for 1 h. The resins were washed with cold Buffer K, resuspended in SDS loading buffer, separated by SDS-PAGE, and probed for the respective proteins by Western blot.
ATPase Assay
ATPase assay was conducted using the phosphate assay kit (ab65622, Abcam). This assay measures the abundance of free inorganic phosphate (Pi) released from ATP hydrolysis reaction, which forms a chromogenic complex with malachite green and ammonium molybdate that gives an absorption peak at 650 nm. Sample was prepared using 1 μg of wild-type or E234G or E234G/H363A HYPEΔ102 and 10 μg of unmodified or adenylylated BiP with 1 mm ATP in a 50-μl volume and incubated for 30 min. The sample was diluted to a final volume of 200 μl in water; 30 μl of phosphate reagent was added and incubated for 30 min. The absorbance was read at 650 nm using Tecan. Phosphate concentration was calculated from a standard curve and represented as a bar chart.
Statistical Analysis
Data were analyzed using t test and two-way ANOVA (for Fig. 6) with Tukey's post hoc correction. Specifically, we conducted a two-way ANOVA with “siRNA treatment” and “stress (UPR) treatment” as the fixed main effects, the “siRNA treatment × stress treatment” as the interaction parameter, and the “number of dead cells” as the response variable. Our data met the assumptions of ANOVA. Separate models were used for the 48- and 72-h trials, because these were run as separate experiments. Because each two-way ANOVA indicated that the main effects and the interaction were all highly significant, we then conducted post hoc comparisons between control siRNA under UPR and siRNA1 under UPR; control siRNA under UPR and siRNA3 under UPR; control siRNA under no UPR and siRNA1 under no UPR; control siRNA under no UPR; and siRNA3 under no UPR. The p values that were used to evaluate statistical significance were adjusted for multiple comparisons using Tukey's correction.
FIGURE 6.
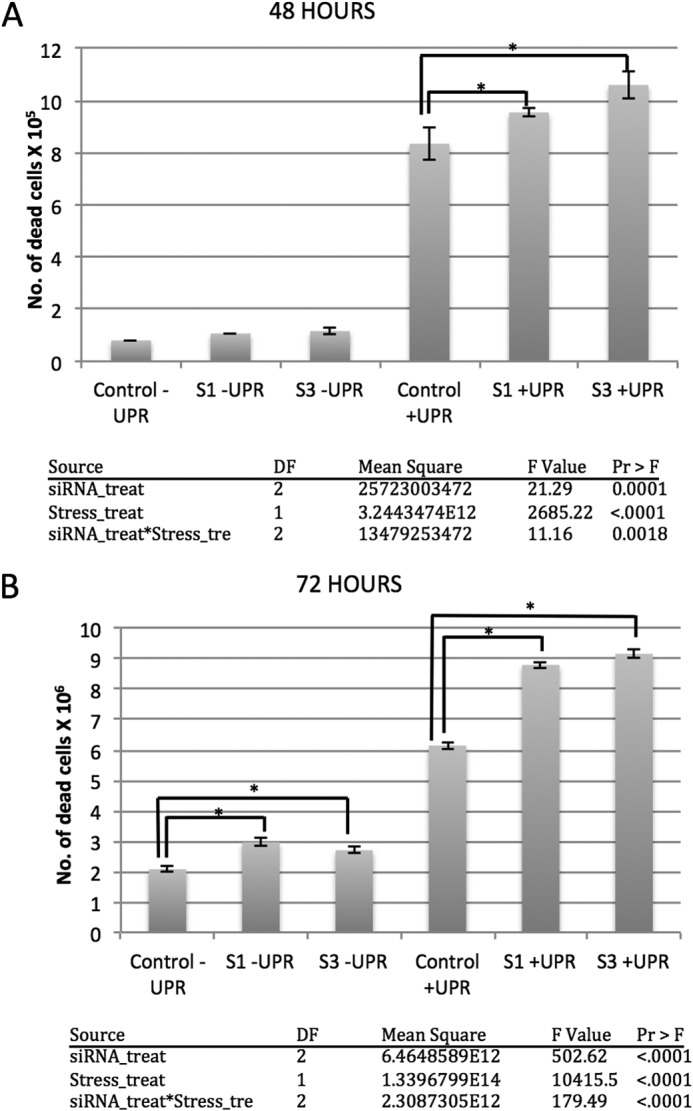
HYPE is needed for cell survival during ER stress. HEK293T cells knocked down for HYPE via two different siRNA treatments (S1 and S3) were compared with untreated cells for their ability to survive upon UPR induction using 1 μg/ml thapsigargin for 48 h (A) or 72 h (B). Scrambled siRNA was used as a control. As expected, sustained UPR induction (+UPR) induced cell death in both control and HYPE siRNA treated samples compared with the unstressed cells (−UPR). Cell death, however, was significantly more pronounced in cells treated with siRNA targeting HYPE. Tables respective for the 48- and 72-h panels indicate details of a two-way ANOVA statistical analysis, with “siRNA treatment” (siRNA_treat) and “stress (UPR) treatment” (stress_treat) as the fixed main effects, the “siRNA treatment X stress treatment” (siRNA_treat*stress-tre) as the interaction parameter, and the “number of dead cells” as the response variable. * on the histograms denotes p < 0.0001.
RESULTS
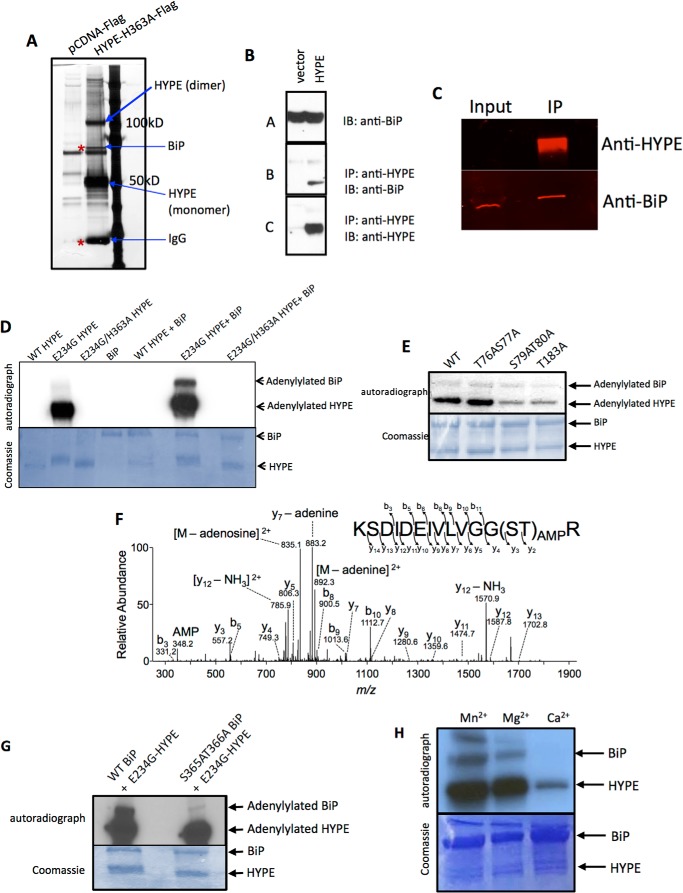
HYPEΔN45-E234G Is a Highly Active Adenylyltransferase That Autoadenylylates at Thr-183, Ser-79, and Thr-80 in Vitro
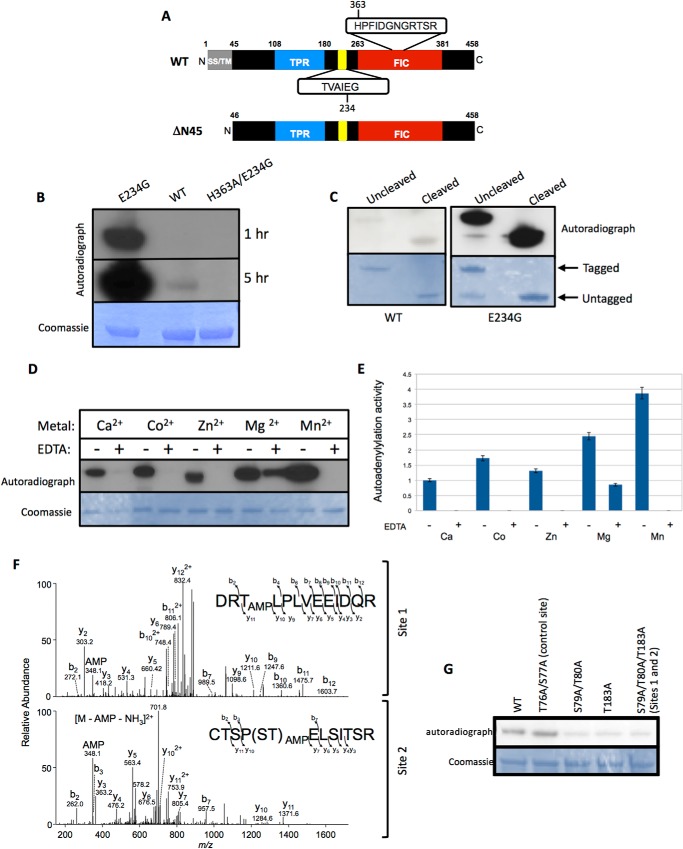
HYPE is a 52-kDa protein that is expressed in all tissues, albeit at very low levels (5). SMART (Simple Molecular Architecture Research Tool) analysis of the domain architecture of HYPE predicts an N-terminal signal peptide and/or a hydrophobic stretch of amino acids that could specify a transmembrane sequence (Fig. 1A). This is followed by a tetratricopeptide repeat (TPR) domain that is typically implicated in protein-protein interactions and a Fic domain at its C terminus (Fig. 1A). In addition, HYPE contains an intrinsic inhibitory helix between its TPR and Fic domains (Fig. 1A). All previous reports on HYPE activity have been with its Fic domain only. To test the enzymatic activity of HYPE in the context of both its Fic and TPR domains, we mutated Glu-234 in its inhibitory helix to Gly using site-directed mutagenesis. Next, His6-SUMO-tagged wild-type (WT; aa 46–458) protein, its constitutively active HYPEΔN45-E234G mutant, and the corresponding catalytically inactive HYPEΔN45-E234G/H363A protein were bacterially expressed and purified. Amino acids 1–45 of HYPE's hydrophobic N terminus were truncated to facilitate solubility. The enzymatic activity of the purified proteins was tested by assessing the ability of each protein to autoadenylylate in an in vitro adenylylation assay using [α-32P]ATP as a nucleotide source. As shown in Fig. 1B, HYPEΔN45-E234G displayed significantly higher autoadenylylation activity compared with HYPEΔN45-WT. In contrast, HYPEΔN45-E234G's catalytically inactive mutant (HYPEΔN45-E234G/H363A) failed to display any autoadenylylation activity. The presence of the His6-SUMO tag did not affect the adenylyltransferase activity of HYPE, as the autoadenylylation levels between tagged (uncleaved) versus untagged (cleaved) versions of HYPEΔN45-WT or HYPEΔN45-E234G were similar (Fig. 1C).
FIGURE 1.
HYPE-E234G is a highly active adenylyltransferase that autoadenylylates Thr-183, Ser-79, and Thr-80, residues that lie outside the Fic domain. A, schematic representation of full-length (WT) HYPE indicating its hydrophobic transmembrane (TM) domain with a potential signal sequence (SS, gray box), followed by a tetratricopeptide domain (TPR, blue box) and its Fic domain (red box). HYPE's conserved Fic motif of sequence HPFIDGNGRTSR and its inhibitory domain (yellow) of sequence TVAIEG are indicated. Bottom panel, scheme of N-terminally truncated HYPE (ΔN45). The hydrophobic first 45 amino acids of HYPE were truncated to facilitate solubility of the purified protein. B, autoadenylylation activity of HYPEΔN45-E234G is more robust than wild-type HYPEΔN45, whereas HYPEΔN45-H363A/E234G is catalytically dead. Bacterially expressed His6-Sumo-HYPEΔN45-WT, His6-Sumo-HYPEΔN45-E234G, or His-Sumo-HYPEΔN45-H363A/E234G was incubated in an in vitro adenylylation assay using [α-32P]ATP. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). Autoradiographs obtained at 1- and 5-h exposure times are shown, because the weak autoadenylylation activity of WT HYPE is evident only at the later time point. C, autoadenylylation activity of untagged and tagged HYPEΔN45 wild-type and HYPEΔN45-E234G were similar. Bacterially expressed His6-Sumo-HYPEΔN45-WT or His6-Sumo-HYPEΔN45-E234G was incubated in an in vitro adenylylation assay using [α-32P]ATP. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). Where indicated the His6-Sumo tag was cleaved off of HYPE. D, autoadenylylation activity of HYPEΔN45-E234G with various divalent cations as cofactors. HYPEΔN45-E234G was incubated with [α-32P]ATP in buffers containing 1 mm Ca2+, Co2+, Zn2+, Mg2+, or Mn2+, in the presence or absence of EDTA. HYPE displays maximum activity in the presence of Mn2+. This activity is completely eliminated when Mn2+ is chelated by the addition of 20 mm EDTA. E, quantitative analysis of HYPE's autoadenylylation levels plotted as fold activity. F, tandem mass spectra (MS/MS) fragmentation patterns of peptides from autoadenylylated HYPEΔN45-WT indicate the presence of AMP on residues Ser-79, Thr-80, and Thr-183. Thr-183 is labeled as Site 1, whereas Ser-79 and Thr-80 are together labeled as Site 2. G, in vitro adenylylation assay with purified WT and autoadenylylation site mutants of HYPE, validating Ser-79, Thr-80, and Thr-183 as autoadenylylation sites. Thr-76 and Ser-77 (labeled as control site) are residues within the peptide containing AMP modifications on Site 2 and serve as a negative control.
We also assessed HYPE-E234G's adenylyltransferase activity in the presence of various divalent cations. HYPE was maximally active in a buffer containing Mn2+ (Fig. 1D and “Experimental Procedures”), displaying nearly a 2-fold increase in activity over the Mg2+-containing buffer and a 4-fold increase over the Ca2+-containing buffer (Fig. 1E). All Fic proteins characterized so far have been shown to use Mg2+ in vitro. The need for divalent cations for HYPE's adenylyltransferase activity was confirmed by chelating the autoadenylylation reactions with EDTA, which significantly reduced autoadenylylation (Fig. 1, D and E).
Mass spectrometric analysis of autoadenylylated HYPEΔN45-WT and HYPEΔN45-E234G HYPE revealed Thr-183, Ser-79, and Thr-80 as common sites of adenylylation between the two proteins (Fig. 1F). To validate this observation, we made Ala substitutions in the context of HYPEΔN45-WT at each of the sites of modification and tested the HYPEΔN45-T183A (Site 1) mutant and HYPEΔN45-S79A/T80A (Site 2) double mutants for their ability to autoadenylylate. As a control, we tested additional mutations in the Site 2 peptide at Ser and Thr residues that were not found to be adenylylated (HYPEΔN45-T76A/S77A double mutant; control site). The samples were analyzed by autoradiography, and the signal for each sample was quantified. The autoadenylylation signal for HYPEΔN45-WT was set as 1 arbitrary unit (AU). As expected, the HYPEΔN45-T76A/S77A control was autoadenylylated at levels similar to HYPEΔN45-WT (Fig. 1G; AU 1.19 for HYPEΔN45-T76A/S77A). Furthermore, the combined mutation in Ser-79 and Thr-80 abrogated autoadenylylation at levels similar to the mutation at Thr-183 (Fig. 1G; AU 0.36 for HYPEΔN45-S79A/T80A and AU 0.39 for HYPEΔN45-T183A). These data indicate that, in vitro, HYPE is autoadenylylated at Sites 1 and 2 at similar levels. We therefore constructed and tested a HYPEΔN45-S79A/T80A/T183A triple mutant, which contained Ala substitutions at both Sites 1 and 2. Interestingly, the HYPEΔN45-S79A/T80A/T183A triple mutant also failed to fully eliminate autoadenylylation (AU 0.32), suggesting that, similar to what is seen with other post-translational modifications such as phosphorylation and ubiquitination, additional sites can be autoadenylylated when HYPE's preferred sites of modification are missing (24).
Constitutive Activity of HYPE Induces Caspase-dependent Apoptosis
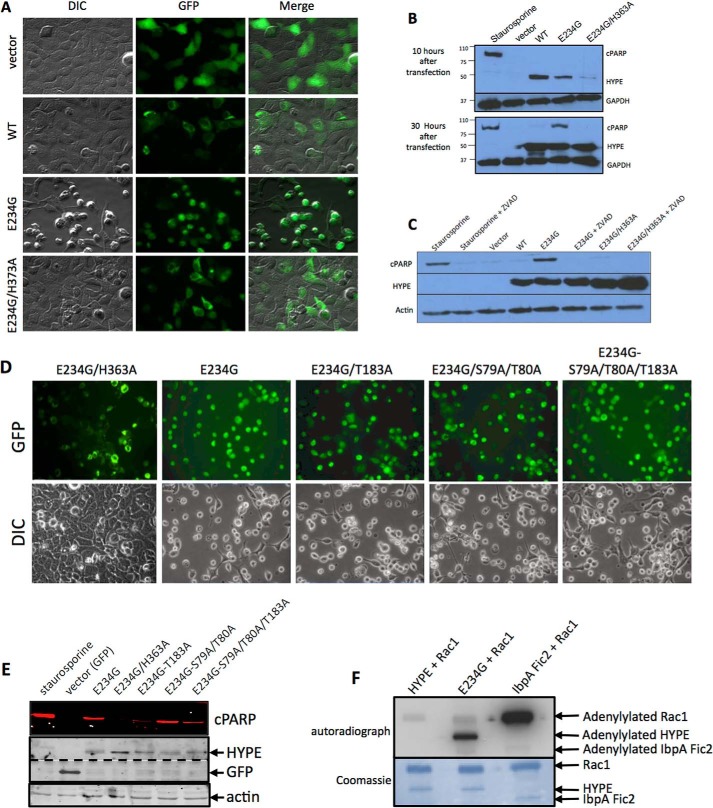
To test the cellular effect of HYPE activation, we transfected human cell lines, HEK293A and HeLa, with C-terminally GFP-tagged full-length WT HYPE (HYPEFL-WT) or its E234G (HYPEFL-E234G) or E234G/H363A (HYPEFL-E234G/H363A) mutants expressed from a mammalian expression vector. Results with HEK293A are shown. Cells were transfected for 30 h and observed by microscopy. Expression of WT HYPE resulted in a normal cellular morphology similar to the control GFP-transfected cells, in agreement with our previously published results (Fig. 2A) (5). Furthermore, HYPE displayed perinuclear localization. In contrast, cells transfected with HYPEFL-E234G displayed a cell rounding phenotype, reminiscent of the cytotoxic phenotype associated with bacterial Fic proteins such as IbpA (Fig. 2A) (5). This cell rounding phenotype was abrogated when cells were transfected with the catalytically inactive HYPEFL-E234G/H363A mutant, indicating that this phenotype was due to HYPE's enzymatic activity. Because WT HYPE does not induce a cell rounding phenotype, we reasoned that this is because WT-HYPE is intrinsically inhibited by its inhibitory helix and that simply overexpression is not sufficient to relieve this inhibition. Thus, the physiological signal involved in activating HYPE remains to be determined. Furthermore, these data clearly show that HYPE's activity needs to be tightly regulated within the cell, as constitutive expression of active HYPE is detrimental to the cell.
FIGURE 2.
Constitutively active HYPE induces caspase-dependent apoptosis. A, pEGFPN1 vector, WT, E234G, or E234G/H363A HYPE were transfected into HEK293A cells. Fluorescence (GFP) and cell morphology (DIC) were observed under the microscope 30 h after transfection. Only cells expressing E234G-HYPE displayed a cell rounding/cytotoxic phenotype. B, Western blot analysis of HEK293A cells transfected with FLAG-tagged WT-, E234G-, or E234G/H363A-HYPE for 10 and 30 h. Lysates were assessed for apoptosis by probing for cleaved PARP and the loading control GAPDH. Staurosporine-treated cells served as a positive control for apoptosis. HYPE-induced apoptosis is seen at 30 h when HYPE protein levels are maximal. C, Western blot analysis of HEK293A cells transfected as in B for 30 h in the presence or absence of the Z-VAD-fmk pan-caspase inhibitor and probed for cPARP, HYPE, and actin (loading control). D, autoadenylylation is not required for HYPE-mediated cytotoxicity. HEK293A cells were transfected with the pEGFP-N1 vector or GFP-tagged HYPEFL-E234G or its autoadenylylation site mutants, T183A, S79A/T80A or S79A/T80A/T183A for 30 h, and visualized by fluorescence microscopy. E, cell lysates were assessed for apoptosis by probing for cPARP. Staurosporine-treated cells served as a positive control for apoptosis. The level of HYPE and GFP expression was determined by probing with antibody to GFP. Bands corresponding to HYPE and GFP are indicated. The dotted line indicates different exposure times, because expression of GFP alone was far greater than the expression of GFP-tagged HYPE. Equal loading of cell lysates was determined by probing for actin. HYPEFL-E234G/T183A and HYPEFL-E234G/T183A/S79A/T80A induced lower levels of cPARP than HYPEFL-E234G. All HYPE mutants induced cPARP cleavage and the corresponding cellular morphology. F, cytoplasmic Rho GTPases are not HYPE's physiological targets. Bacterially expressed GST-Rac1 was incubated with HYPEΔN45-WT or HYPEΔN45-E234G in an in vitro adenylylation assay using [α-32P]ATP. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). The positions of His6-Sumo-HYPE and the GST-Rac1 on the gel are indicated by arrows. HYPEΔN45-E234G does not significantly enhance adenylylation of Rac1 when compared with HYPEΔN45-WT. Robust adenylylation of Rac1 by IbpA-Fic2 serves as a positive control.
To determine the mechanism of HYPE-mediated cell rounding, we assessed HYPE transfected cells for necrosis, apoptosis, and autophagy. Although we found no evidence for autophagy and necrosis (data not shown), we found that expression of HYPEFL-E234G induced apoptosis, as measured by PARP cleavage (Fig. 2B). PARP is cleaved and inactivated by caspases upon induction of programmed cell death, and it is used as an indicator of apoptosis. Compared with the 10-h time point, HYPE-mediated apoptosis was apparent only at 30 h post-transfection (Fig. 2B). Cells expressing HYPEFL-E234G displayed cPARP similar to the positive control, i.e. cells treated with the kinase inhibitor staurosporine. HYPEFL-WT induced extremely low levels of PARP cleavage, although the HYPEFL-E234G/H363A catalytically inactive mutant did not induce PARP cleavage. Furthermore, treatment of cells with the Z-VAD-fmk pan-caspase inhibitor prevented PARP cleavage in HYPEFL-E234G-transfected cells, indicating that constitutive activation of HYPE induces apoptosis in a caspase-dependent manner (Fig. 2C). Similar results were obtained with HeLa cells (data not shown).
Autoadenylylation appears to play a limited role in apoptosis and HYPE's function in cells. Specifically, we tested HYPE's autoadenylylation Site 1 and Site 2 mutants for their ability to induce cytotoxicity in mammalian cells. Each of the Site 1 (T183A), Site 2 (S79A/T80A), and Site 1/Site 2 (S79A/T80A/T183A) mutants was generated in a HYPEFL-E234G background and tested for their ability to induce apoptosis, as assayed by microscopy and PARP cleavage (Fig. 2, D and E). The level of cPARP detected by Western analysis was quantified as a ratio of cPARP/HYPE expression for each corresponding sample. PARP cleavage by HYPEFL-E234G was set at AU = 1. As expected, HYPEFL-E234G induced apoptosis when expressed in HEK293A cells for 30 h, although HYPEFL-E234G/H363A did not (AU = 0.05). Surprisingly, based on cellular morphology, all the autoadenylylation site mutants induced a cell rounding phenotype similar to HYPEFL-E234G (Fig. 2D). Western blot analysis confirmed that all the autoadenylylation mutants displayed PARP cleavage at 30 h (Fig. 2E), although their levels of cPARP varied (T183A = 0.41; S79A/T80A = 1.65; S79A/T80A/T183A triple mutant = 1.44). Although T183A-HYPE displayed somewhat lower cPARP levels, this difference was not evident for the triple mutant.
Rho GTPases Are Not the Physiological Targets for HYPE
We have previously shown that bacterial Fic proteins, like IbpA from H. somni, induce cytoskeletal collapse in host cells by adenylylating and inhibiting mammalian Rho GTPases (5, 14). The apoptosis phenotype that we observed in Fig. 2A was reminiscent of the cell rounding phenotype associated with adenylylation of Rho GTPases, like RhoA, Rac1, and Cdc42, by IbpA. Also, Rho GTPases like Rac1 have previously been used as artificial substrates for assessing HYPE's adenylyltransferase activity, with most success when tested with just HYPE's Fic domain (4, 5, 15). We therefore assessed the ability of the activated HYPEΔN45-E234G mutant to adenylylate Rho GTPases in the context of its TPR (putative substrate binding) domain. For this, we incubated GST-tagged Rac1 with His6-SUMO-tagged HYPEΔN45-WT and HYPEΔN45-E234G in an in vitro adenylylation reaction using [α-32P]ATP and a Mn2+ containing buffer (see under “Experimental Procedures”). His6-SUMO-tagged IbpA-Fic2 (25) was used as a positive control and incubated with Rac1 in a similar reaction except Mg2+ was substituted for Mn2+, which is optimal for IbpA activity. As shown in Fig. 2F, Rac1 adenylylation is not significantly increased even upon incubation with HYPEΔN45-E234G, although IbpA-Fic2 robustly modifies Rac1. Increased autoadenylylation of HYPEΔN45-E234G confirms that it is active. Thus, Rho GTPases are unlikely to be physiological targets for HYPE. Accordingly, we failed to detect adenylylated Rac1 or Cdc42 when cells were cotransfected with GFP-tagged full-length HYPE-E234G and FLAG-tagged Rac1 or Cdc42, and the Rho GTPase pulled out with anti-FLAG M2 affinity agarose was analyzed by Western blot analysis using anti-AMPylated Thr and anti-AMPylated Tyr antibodies (data not shown). Thus, HYPE-mediated apoptosis is unlikely to be a result of HYPE adenylylating cytoplasmic Rho GTPases.
HYPE Localizes to the Lumen of the ER
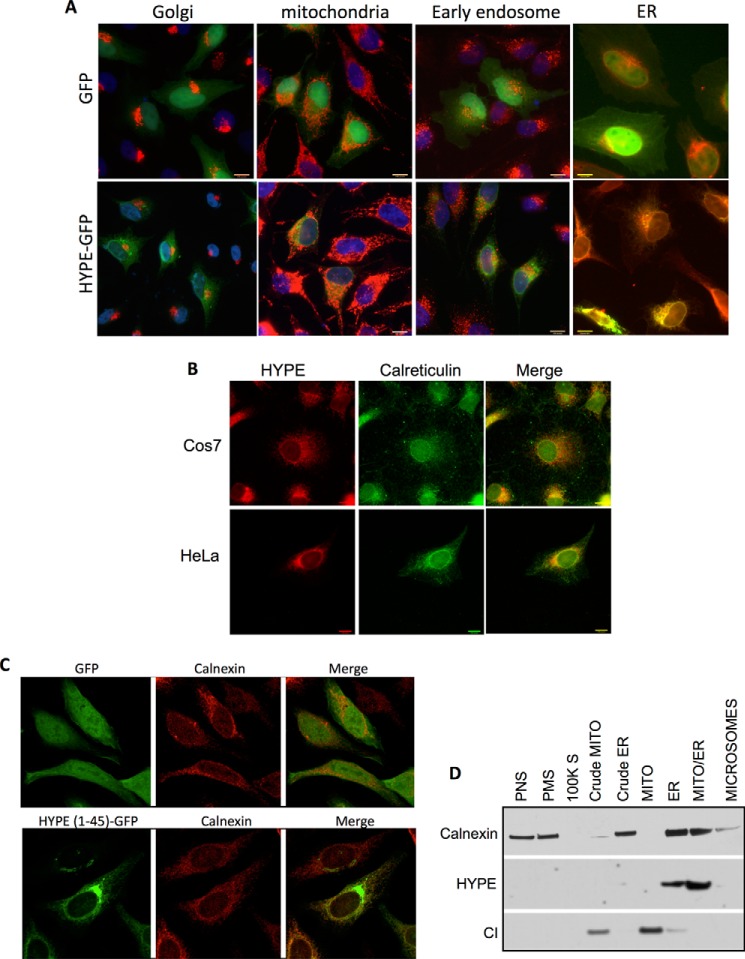
To identify HYPE's substrates, we sought to determine HYPE's subcellular localization, which would help in narrowing down potential substrates. C-terminally GFP-tagged HYPE was expressed in HeLa cells via transfection, and the immunofluorescence was visualized by microscopy. These samples were also stained for various organellar markers like Mitotracker Red for mitochondria, LAMP1 for the lysosome, GM130 for the Golgi, EEA1 for endosomes, and calreticulin for the ER (Fig. 3A). HYPE-GFP colocalized with calreticulin, indicating that HYPE localizes to the ER. No colocalization was observed with any of the other organellar markers. We further confirmed the ER localization of endogenous HYPE by fixing several mammalian cell lines, including COS7 and HeLa cells, staining for HYPE with α-HYPE antibody, and visualizing the immunofluorescence by microscopy. Immunofluorescence assays confirmed that endogenous HYPE also colocalizes with the ER marker calreticulin, thus confirming that HYPE localizes to the ER (Fig. 3B).
FIGURE 3.
HYPE localizes to the ER. A, immunofluorescence of HYPE-GFP (green) and organellar markers (red) in HeLa cells showing colocalization of HYPE with ER marker calreticulin but not with Golgi marker GM130, mitochondrial marker Mitotracker Red, or early endosomal marker EEA1. B, staining of endogenous HYPE and ER marker calreticulin in HeLa and COS7 cells showing localization of HYPE in the ER. C, GFP fused N-terminally with hydrophobic N terminus (aa 1–45) of HYPE is targeted to the ER, as indicated by colocalization of GFP with the ER marker calnexin (lower panel). Untagged GFP, used as negative control, localizes to both the cytoplasm and nucleus of the cell (upper panel). D, rat liver tissue fractionation showing HYPE is present in ER-enriched tissue fractions (ER and mito-ER) but not in other cellular fractions as follows: post-nuclear spin (PNS), plasma membrane (PMS), 100,000 spin, mitochondria, or microsomes. Calnexin is a marker for ER. Complex I (CI) is a marker for mitochondria.
Because HYPE's N terminus is hydrophobic, we hypothesized that it may harbor an ER-targeting signal. We therefore fused HYPE's aa 1–45 to GFP, transfected HeLa cells with this fusion construct, and assessed GFP localization by immunofluorescence microscopy. Unlike the GFP vector control, where GFP localizes to both the cytoplasm and nucleus, the HYPE(1–45)-GFP fusion protein targeted to the ER as determined by colocalization with calnexin (Fig. 3C, compare upper panels with the two lower panels). Thus, the ER localization signal sequence for HYPE is located in its first 45 amino acids (Fig. 3C). We next extended these analyses by testing cellular fractions of rat liver tissue to confirm that endogenous HYPE localizes to the ER. Specifically, a freshly harvested rat liver was homogenized and subjected to a series of gradient centrifugations to isolate subcellular fractions, as described previously (21, 23). Each fraction was analyzed by Western blot using antibody to HYPE and other organellar control antibodies. Endogenous HYPE consistently colocalized with subcellular fractions that were enriched for the ER (ER and Mito-ER, Fig. 3D).
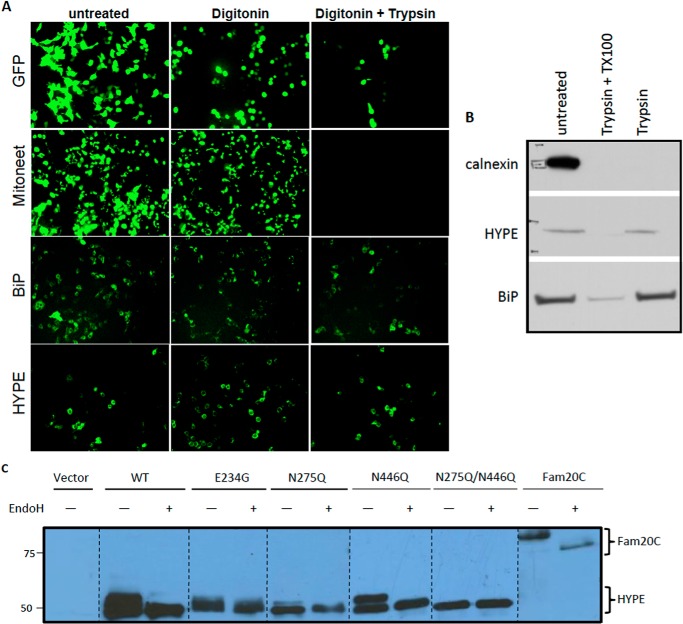
HYPE is predicted to be a single pass transmembrane protein. Therefore, we sought to determine the topology of HYPE in the ER. For this, we transfected HeLa cells with HYPE C-terminally tagged with GFP and assessed its orientation in the ER membrane using an FPP assay (Fig. 4). The FPP assay uses the restricted proteolytic digestibility of GFP-tagged membrane proteins to reveal their intramembrane orientation. In the FPP assay, the cholesterol-binding drug digitonin is used to selectively permeabilize the plasma membrane, which allows cytosolic contents to diffuse across the plasma membrane, whereas intracellular organelles are retained. A subsequent proteinase K treatment then destroys the fluorescence associated with any GFP molecule attached to a protein of interest facing the cytosol but not with an organellar protein that is membrane-associated or lumenal. HeLa cells were transfected with a vector expressing GFP alone (control for cytoplasmic protein) or GFP-tagged proteins like Mitoneet (a mitochondrial protein that faces the cytoplasm), BiP (an ER lumenal protein), or HYPE (Fig. 4A). FPP analysis of these samples shows that HYPE's expression/fluorescence patterns mimic the ER lumen protein, BiP, indicating that HYPE likely faces the ER lumen (Fig. 4A). Furthermore, trypsin treatment of the ER tissue fractions revealed that HYPE could be digested only after pretreatment with detergent in a manner similar to the ER lumen protein BiP, confirming that HYPE is a type II ER membrane protein (Fig. 4B).
FIGURE 4.
HYPE localizes to the ER lumen and is glycosylated predominantly at Asn-275. A, localization of HYPE in the lumen was determined by FPP analysis. GFP-tagged HYPE or controls (GFP only; GFP-tagged Mitoneet, a mitochondrial protein that faces the cytoplasm; and GFP-tagged BiP, an ER protein that resides in the lumen) were transfected into HeLa cells, and the fluorescence was observed following treatment with digitonin or digitonin + trypsin. HYPE's fluorescence pattern matches that of the known ER lumen protein, BiP. B, ER fraction of rat liver tissue treated with trypsin in the presence and absence of detergent shows that HYPE is not susceptible to proteolytic cleavage in the absence of detergent, indicating luminal topology of HYPE. BiP (ER lumen protein) and calnexin (ER membrane protein facing cytosol) are used as controls. C, FLAG-tagged WT, E234G, and glycosylation site mutants of HYPE (N275Q, N446Q, and N275Q/N446Q-HYPE) were transfected into HEK293A cells. The expressed protein was pulled down using anti-FLAG M2 agarose beads and either left untreated or treated with endoglycosidase H. Samples were then analyzed by Western blotting with antibody to the FLAG epitope. WT, E234G, and N446Q HYPE are heavily glycosylated, whereas N275Q-HYPE displays much less glycosylation. The N275Q/N446Q-HYPE double mutant displays no glycosylation, indicating that although both sites are glycosylated, the bulk of the glycosylation occurs on Asn-275. Fam20C, a known glycosylated protein, was used as a positive control.
HYPE Is N-Glycosylated Predominantly at Residue Asn-275
Based on HYPE's orientation in the ER lumen, we predicted that HYPE would be N-glycosylated. To test this, we transfected HEK293A cells with FLAG-tagged WT or E234G-HYPE. The human secreted kinase Fam20C was used as a positive control for glycosylation, although the vector alone served as a negative control (26). At 30 h following transfection, cell lysates were bound to M2-agarose beads to isolate expressed FLAG-tagged proteins. Recovered proteins were then separated by SDS-PAGE and analyzed by Western blot by probing with anti-M2 FLAG antibody. As indicated in Fig. 4C, both WT- and E234G-HYPE, as well as the Fam20C positive control, displayed a higher mobility shift that could be eliminated by treating the sample with endoglycosidase H, an endoglycosidase that cleaves sugar moieties from N-glycosylated proteins.
Computational programs predict Asn-275 and Asn-446 as potential sites of glycosylation on HYPE. We therefore mutated both these sites to a Gln, which should eliminate glycosylation at these sites without altering the protein structure. FLAG-tagged N275Q- and N446Q-HYPE, as well as an N275Q/N446Q-HYPE double mutant, were transfected into HeLa cells and assessed for glycosylation as described above. Although the mutation of Asn-275 significantly reduced the level of glycosylation, total glycosylation of HYPE could be eliminated only upon mutating both Asn-275 and Asn-446 (Fig. 4C). These data further confirm HYPE's location in the ER lumen and show that HYPE is glycosylated predominantly at Asn-275. Also, the data in Figs. 2F, 3, and 4 taken together support our findings that, given HYPE's topology within the lumen of the ER, cytoplasmic Rho GTPases, RhoA, Rac1, and Cdc42 are not HYPE's physiological targets.
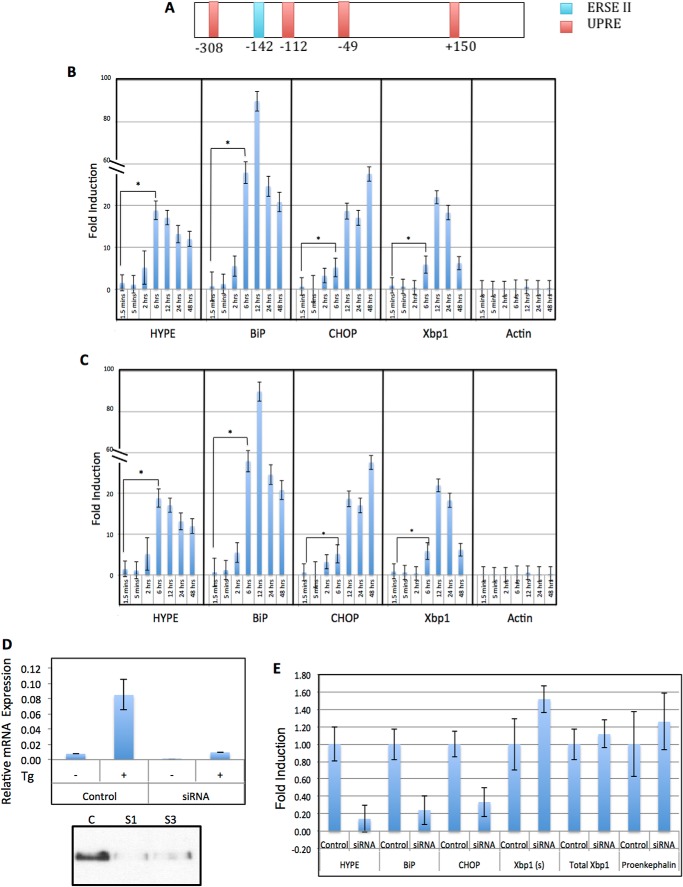
HYPE Expression Is Up-regulated in Response to Activation of a UPR
The human UPR pathway consists of three branches regulated by ER transmembrane receptor proteins IRE1, ATF6, and PERK. The UPR integrates these three pathways to activate an ER stress response involving both translational and transcriptional mechanisms that function to relieve ER stress. However, upon sustained stress, the UPR pathway activates a pro-apoptotic signaling cascade. The transcriptional induction of UPR target genes is mediated by cis-acting promoter elements like the UPR element and the ER stress-response elements (I and II). Analysis of HYPE's promoter revealed the presence of potential UPR element at +150, −49, −112, −308, and ER stress-response element II at −142 (Fig. 5A). HYPE's ER localization and its stress-responsive promoter elements led us to ask whether HYPE was involved in UPR. To assess the effect of UPR induction on HYPE expression, we induced UPR in mammalian cells using the ER calcium channel blocker thapsigargin (Tg) and the N-linked glycosylation blocker tunicamycin (Tn), which are drugs known to induce a UPR stress response. RNA was isolated from lysates of untreated and Tg- or Tn-treated samples and assessed by quantitative RT-PCR for expression of HYPE and other UPR target genes like BiP, CHOP, XBP1, and negative controls, like actin or proencephalin. As expected from transcriptomics data, HYPE levels were very low in untreated cells. However, HYPE expression was significantly up-regulated at 6 h post-induction and remained high until 24 h post-induction by Tg (Fig. 5B) and Tn (Fig. 5C).
FIGURE 5.
HYPE plays an important role in the unfolded protein response pathway. A, schematic of HYPE promoter containing UPR element sites at +150, −49, −112, −308, and ER stress-response element II site at −142. B, HYPE gene expression is up-regulated in response to UPR induction by 1 μg/ml thapsigargin; C, 2.5 μg/ml tunicamycin. qRT-PCR analysis was done for HYPE, UPR genes (BiP, CHOP, Xbp1(s); positive control), and actin (negative control). All values are normalized to GAPDH internal control. Each histogram from left to right corresponds to 1.5 and 5 min and 2, 6, 12, 24, and 48 h post-induction. D, upper panel, RNAi knockdown of HYPE using two siRNA constructs, followed by UPR induction using 1 μg/ml Tg analyzed by qRT-PCR (same as B) shows down-regulation of HYPE in the HYPE siRNA cells but not in the scrambled control. Lower panel, control (C; scrambled siRNA) or HYPE-specific siRNA (S1 and S3)-treated samples were immunoprecipitated for HYPE using a mouse polyclonal antibody to HYPE and analyzed by Western blot using a rabbit polyclonal antibody to HYPE. HYPE protein could not be detected in siRNA-treated samples but was detected in the control sample. E, qRT-PCR analysis of siRNA-mediated HYPE knockdown samples indicates decreased BiP and CHOP expression in the absence of HYPE, whereas Xbp1 remains unaffected. Actin and proenkephalin are used as negative controls.
HYPE Is Required for UPR Induction via ATF6 and PERK
The UPR-induced expression of HYPE led us to ask whether HYPE might play a role in the UPR pathway. We therefore knocked down HYPE in HEK293T cells using siRNA specific to HYPE. Scrambled siRNA was used as a control. As shown in Fig. 5D, although siRNA treatment did reduce HYPE expression levels, this decrease was difficult to quantify because HYPE is known to be expressed at very low levels in untreated cells. Therefore, to assess the effectiveness of our siRNA treatment, we induced UPR in control and siRNA-treated samples with Tg for 6 h, and then measured HYPE expression by qRT-PCR. Fig. 5D shows that unlike control samples, which showed the expected up-regulation of HYPE expression upon UPR induction, cells treated with siRNA targeting HYPE failed to induce HYPE expression even upon Tg treatment, indicating efficient silencing of HYPE expression. We further confirmed the effectiveness of the siRNA-mediated knockdown of HYPE by immunoprecipitating HYPE from cells treated with control or HYPE-specific siRNA and by checking protein levels of HYPE by Western analysis. Again, siRNA treatment eliminated HYPE at the protein level, whereas the control treatment did not (Fig. 5D, lower panel). In these siRNA-mediated HYPE knockdown cells, we next induced UPR by treating with Tg for 6 h and assayed UPR progression by qRT-PCR analysis of UPR effector genes, BiP, CHOP, spliced XBP1 (Xbp1(s)), and total XBP1 (Fig. 5E). Xbp1(s) up-regulation serves as a readout for IRE1 activation, and BiP up-regulation is a readout primarily of ATF6 activation, whereas CHOP is up-regulated via the activation of PERK.
We observed that in the absence of HYPE, UPR progression failed to up-regulate canonical UPR target genes like BiP and CHOP, although, interestingly, levels of XBP1(s) compared with total XBP1 remained unaltered between the control and HYPE siRNA samples (Fig. 5E). These data provide strong evidence that HYPE positively regulates UPR, with a stronger influence on the ATF6 and PERK branches of the UPR. We are currently mapping out the details of the exact UPR branches affected by HYPE activity.
HYPE Is Needed for Cell Survival under UPR Induction
Our findings presented in Fig. 5 indicate that HYPE positively regulates UPR progression. As mentioned earlier, UPR activation entails an early response that involves up-regulation of chaperones such as BiP to increase folding of misfolded proteins. However, upon sustained stress, UPR triggers activation of pro-apoptotic pathways. Accordingly, the inability of cells to mount a normal UPR makes them more susceptible to stress resulting in increased cell death. Because the absence of HYPE prevents UPR activation, we hypothesized that knockdown of HYPE would reduce cell survival in response to ER stress. To test this, HEK293T cells were transfected in duplicate with siRNA to HYPE or with the scrambled siRNA control. Tg was added at 1 h post-transfection, and both siRNA and UPR treatments were allowed to progress for up to 48 or 72 h. One set of duplicate samples was assessed via qRT-PCR to confirm knockdown of HYPE and induction of UPR using BiP and CHOP as readouts, as described for Fig. 5 (data not shown). Cell survival was determined as a measure of the total number of dead cells that stained positive for trypan blue in control or HYPE siRNA-treated samples at 48 and 72 h. As predicted, a significantly greater number of dead cells were observed in samples treated with HYPE siRNA at both 48 and 72 h of UPR induction, with a more pronounced increase at 72 h (Fig. 6). These data indicate that the inability to manifest a normal UPR in the absence of HYPE affects cell survival, and the data provide overwhelming evidence that HYPE plays a critical role in regulating UPR.
HYPE Adenylylates the ER Chaperone BiP
To gain insight into the mechanism by which HYPE regulates UPR, we sought to identify protein interactors and potential substrates for HYPE. Using MCF7 breast cancer cells stably expressing C-terminally FLAG-tagged H363A-HYPE as a substrate trap, we conducted pulldown assays using anti-FLAG M2-agarose followed by mass spectrometry and identified the ER molecular chaperone BiP (binding immunoglobulin protein) as an interacting partner for HYPE (Fig. 7A). Specifically, analysis of indicated bands on the silver-stained gel shown in Fig. 7A revealed bands corresponding to BiP (∼78 kDa), IgG (∼25 kDa; known to bind to BiP), HYPE monomer (∼50 kDa), and HYPE dimer (∼100 kDa). HYPE's interaction with BiP was confirmed by Western blot analysis of the pulled down samples using antibody to HYPE and BiP (Fig. 7B). BiP is a 78-kDa heat shock protein that, like HYPE, also resides in the ER lumen. It has a canonical Hsp70 architecture consisting of an N-terminal ATPase domain and a C-terminal substrate-binding domain (27). By binding to misfolded proteins generated in response to cellular stresses, BiP maintains ER homeostasis and determines cell fate in response to ER stress. To ensure that the HYPE-BiP interaction was not an artifact of overexpressing HYPE, we repeated immunoprecipitation of BiP from untreated HEK293T cells and assessed the eluted interacting partners by Western blot analysis using antibody against HYPE (Fig. 7C). Because HYPE expression levels are very low in cells, only BiP could be detected in our input samples. However, a significant amount of HYPE could be clearly detected in the immunoprecipitated eluate, confirming that endogenous HYPE interacts with BiP.
FIGURE 7.
HYPE adenylylates BiP at Ser-365 and Thr-366. A, silver-stained gel indicating proteins immunoprecipitated with FLAG-H363A-HYPE and identified by mass spectrometry analysis. Bands marked with arrows were identified as BiP, HYPE (monomer and dimer), and IgG, respectively. B, bead-based pulldown of FLAG-tagged HYPE, showing input of the pCDNA-FLAG vector control and cells overexpressing HYPE-FLAG, probed with α-BiP antibody to detect total levels of BiP in the cells (A), immunoprecipitation (IP) with α-HYPE antibody, and Western blot (immunoblot; IB) probed with α-BiP antibody to show amount of BiP pulled down with HYPE (B) and immunoprecipitation with α-HYPE antibody and Western blot probed with α-HYPE antibody to show total amount of HYPE pulled down (C). C, immunoprecipitation of endogenous BiP showing input probed with α-BiP and α-HYPE antibodies to detect total levels of BiP and HYPE in the cells, and immunoprecipitated (IP) beads probed with α-BiP and α-HYPE antibody to detect total amount of HYPE pulled down with BiP. D, in vitro adenylylation assay with purified WT, E234G, and E234G/H363A HYPE and WT BiP showing adenylylation of BiP by HYPE. E, autoadenylylation of HYPE is not required for adenylylation of BiP. WT BiP was incubated with recombinant WT HYPE or its Site 1 (Thr-183) and Site 2 (S79A/T80A) autoadenylylation site mutants, as well as the control T76A/S77A mutant, in an in vitro adenylylation reaction using [α-32P]ATP. The S79A/T80A and Thr-183 HYPE autoadenylylation site mutants are still able to modify BiP, albeit at levels weaker than WT HYPE, indicating that autoadenylylation is not a prerequisite for BiP adenylylation. F, tandem mass spectra (MS/MS) fragmentation patterns of peptides from adenylylated BiP indicating sites of modification of BiP to be Ser-365 and Thr-366. G, in vitro adenylylation assay with purified wild-type and S365A/T366A BiP with E234G HYPE showing loss of modification due to loss of adenylylation sites on BiP, confirming the LC-MS/MS data. H, in vitro adenylylation of BiP by HYPE in the presence of 1 mm Mg2+, Mn2+, or Ca2+. HYPE displays robust modification of BiP in the presence of Mn2+ and Mg2+ but not Ca2+.
Given that our data thus far indicate that HYPE interacts with BiP and regulates UPR, we asked whether HYPE adenylylated BiP. We bacterially expressed and purified N-terminally truncated (lacking BiP's hydrophobic signal peptide) His6-SUMO-BiP (aa 19–637), and we incubated it with WT-HYPE, constitutively active E234G-HYPE, or catalytically inactive E234G/H363A-HYPE in an in vitro adenylylation assay. E234G-HYPE robustly adenylylated BiP, whereas WT HYPE had weak adenylyltransferase activity toward BiP (Fig. 7, D and E). E234G/H363A-HYPE did not adenylylate BiP (Fig. 7D). The autoradiograph in Fig. 7E was quantitated, and the level of adenylylation signal on BiP when incubated with WT-HYPE was set as an AU = 1. As shown in Fig. 7E, HYPE autoadenylylation also does not appear to be a prerequisite for BiP adenylylation, as both the HYPE autoadenylylation mutants, S79A/T80A-HYPE and T183A-HYPE, modified BiP, albeit at levels lower than WT- or T76A/S77A-HYPE (S79A/T80A-HYPE, AU = 0.696; T183A-HYPE, AU = 0.282; T76A/S77A-HYPE, AU = 0.997). LC-MS/MS analysis confirmed that HYPE adenylylated BiP and identified AMP moieties on Ser-365 and Thr-366 of BiP (Fig. 7F). Both these sites lie within BiP's ATPase domain. Site-directed mutagenesis of BiP at Ser-365 and Thr-366 confirmed these residues as the sites of adenylylation, as S365A/T366A-BiP when incubated with E234G-HYPE in an in vitro adenylylation reaction failed to be radiolabeled (Fig. 7G). Thus, BiP is a bona fide target of HYPE-mediated adenylylation.
Drosophila Fic, dFicD, was recently shown to adenylylate BiP preferentially using Ca2+ as a divalent cation (28). We thus compared HYPE's ability to adenylylate BiP in the presence of Ca2+, Mg2+, and Mn2+ cations. Unlike dFicD, HYPE activity against BiP was highest in the presence of Mn2+ and Mg2+ but not Ca2+ (Fig. 7H). Specifically, BiP was robustly modified in the presence of both Mg2+ and Mn2+, with slightly higher activity with Mn2+, whereas HYPE failed to adenylylate BiP in the presence of Ca2+ (Fig. 7H).
BiP is known to be ADP-ribosylated by an unknown enzyme at residues Arg-470 and Arg-492 of its substrate binding domain. We then tested whether HYPE could utilize nicotinamide adenine dinucleotide (NAD) as a nucleotide source for ADP-ribosylating BiP. WT BiP was incubated with E234G-HYPE in a reaction buffer containing 32P-labeled NAD as a nucleotide source. However, HYPE failed to ADP-ribosylate BiP (data not shown). Thus, BiP undergoes different post-translational modifications as follows: ADP-ribosylation at its substrate binding domain and adenylylation at its ATPase domain.
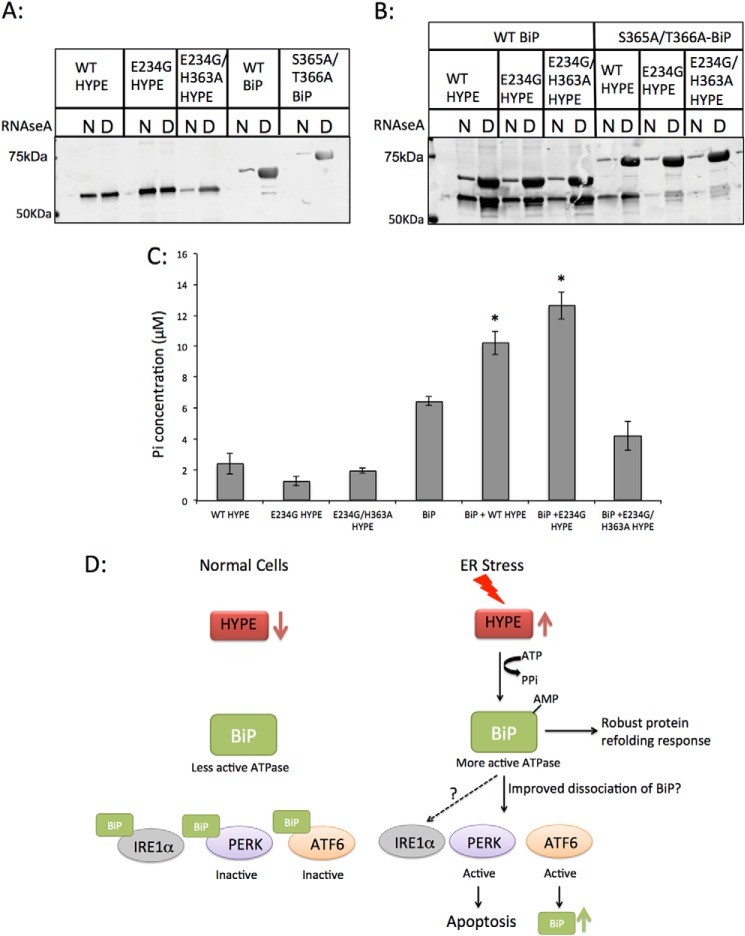
HYPE-mediated Adenylylation of BiP Does Not Affect BiP's Binding to Misfolded Proteins
Our data thus far establish that BiP is a bona fide endogenous substrate of HYPE and led us to hypothesize that HYPE may regulate UPR by post-translationally modifying BiP. We therefore assessed the consequence of adenylylation on BiP activity and function. As mentioned earlier, BiP is an Hsp70 chaperone that uses its ATPase activity to bind and refold misfolded proteins during UPR. Using purified wild-type or S365A/T366A-BiP (aa 19–637) with purified denatured or native RNase A, we found that adenylylation of BiP does not affect its binding to denatured RNase A (Fig. 8). Specifically, we incubated native or denatured RNase A with WT or S365A/T366A-BiP that was either adenylylated by HYPE or left untreated and compared the ability of adenylylated versus unmodified BiP to bind denatured RNase A. Fig. 8B shows that adenylylated BiP bound to denatured RNase A at levels similar to nonadenylylated BiP. Furthermore, S365A/T366A-BiP retained the ability to bind misfolded proteins, as seen from its binding to denatured RNase A (Fig. 8, A and B). Hence, adenylylation of BiP does not affect its function of binding misfolded proteins. Interestingly, HYPE itself was observed to bind misfolded proteins in an activity independent manner, because WT-, E234G-, and E234G/H363A-HYPE bound to RNase A (Fig. 8A). Furthermore, this interaction was enhanced in the presence of WT BiP (Fig. 8B).
FIGURE 8.
Adenylylation of BiP does not affect BiP's ability to bind misfolded proteins, but it does enhance BiP's ATPase activity. A, purified recombinant WT HYPE or its E234G and E234G/H363A mutants and WT BiP or its S365A/T366A mutant were incubated with native (N) or denatured (D) RNase A bound to Sepharose beads. The proteins were then eluted from beads, separated on SDS-PAGE, and probed for HYPE and BiP. HYPE binds similarly to native and denatured RNase A, although both WT and S365A/T366A BiP bind robustly to denatured RNase. B, unmodified or adenylylated WT and S365A/T366A BiP incubated with native or denatured RNase A show that irrespective of its modification status, BiP robustly binds to denatured RNase A, indicating that BiP's binding to misfolded proteins is not affected by HYPE-mediated adenylylation. C, unmodified or adenylylated WT BiP was assessed for ATPase activity using colorimetric detection of inorganic phosphate released by ATP hydrolysis. BiP modified by WT or E234G HYPE shows significantly higher ATPase activity than unmodified BiP. BiP incubated with E234G/H363A HYPE shows ATPase activity similar to BiP alone, suggesting that HYPE-mediated adenylylation of BiP is responsible for enhancing its ATPase activity. D, model for UPR regulation involving HYPE-mediated adenylylation of BiP. Upon ER stress, HYPE is up-regulated and adenylylates BiP. Adenylylation enhances BiP's ATPase activity, thus enabling it to more efficiently refold misfolded proteins. Adenylylation also enhances BiP's disassociation from PERK and ATF6, thus efficiently activating these pathways. Although our data thus far indicate that IRE1α is not affected by HYPE activity, more work is needed to confirm this *, p < 0.005.
HYPE-mediated Adenylylation Enhances BiP's ATPase Activity
The adenylylation sites for BiP (Ser-365 and Thr-366) lie in its ATPase domain. Therefore, we assessed the consequence of BiP adenylylation on its ATPase activity. WT BiP was incubated with WT-, E234G-, or E234G/H363A-HYPE in an in vitro adenylylation reaction. Following adenylylation, the ATPase activity of modified BiP was compared with unmodified BiP as measured colorimetrically by the release of Pi following ATP hydrolysis (see “Experimental Procedures”). Proteins used in this assay were first cleaved to remove their purification tags and then purified further by ion exchange and size exclusion to ensure no background ATPase activity was observed from residual bacterial chaperone contaminants from the protein purification process (see “Experimental Procedures”). As shown in Fig. 8C, ATPase activity of BiP was significantly higher when incubated with WT-HYPE and even higher when incubated with E234G-HYPE. Incubation with E234G/H363A-HYPE did not significantly alter BiP's ATPase activity, nor did any of the HYPE proteins display significant ATPase activity (Fig. 8C). These data indicate that adenylylation enhances BiP's ATPase activity and fits with all our data indicating a positive role for HYPE in UPR activation and progression.
DISCUSSION
We have identified a previously unknown role for Fic-mediated adenylylation in regulating the UPR pathway. Our data show that HYPE is up-regulated upon UPR induction (Fig. 5, B and C). Furthermore, our siRNA-mediated knockdown of HYPE reveals that HYPE is required for normal induction and progression of UPR likely via the ATF6 and PERK pathways (Fig. 5E). Consequently, cells knocked down for HYPE fail to cope with ER stress and show decreased survival (Fig. 6). Additionally, sustained expression of HYPE activity induces caspase-mediated apoptosis (Fig. 2), in keeping with the programmed cell death seen in response to sustained UPR activation.
We ruled out cytoplasmic Rho GTPases, which have been used as in vitro substrates for HYPE, as physiological targets for HYPE by showing that activation of HYPE in the context of both its TPR and Fic domains does not enhance modification of Rac1 (Fig. 2F). Additionally, we show that HYPE is targeted via its N terminus to the lumen of the endoplasmic reticulum (Figs. 3 and 4), providing additional proof that the physiological target of HYPE must also localize to the ER lumen. By this same logic, histone H3, which has previously been used as an in vitro substrate for HYPE, is also not likely to be a physiologically relevant target (29). In agreement with HYPE's ER localization, we show for the first time that HYPE is glycosylated predominantly at Asn-275 (Fig. 4C). Furthermore, we identify BiP, a sentinel chaperone in the activation of UPR, as a substrate for HYPE and identify Ser-365 and Thr-366 as sites of adenylylation on BiP (Fig. 7).
We propose that HYPE regulates UPR via its interactions with BiP. Accordingly, HYPE-mediated adenylylation of BiP enhances its ATPase activity (Fig. 8C), which is required for refolding misfolded proteins to cope with ER stress (30). These data suggest a positive role for HYPE in the UPR pathway, in keeping with our qRT-PCR data showing up-regulation of HYPE upon ER stress and the role of HYPE in cell survival under sustained ER stress.
During the course of submitting this manuscript, a report describing the Drosophila homolog of HYPE, dFicD, was published (28). In agreement with our findings, dFicD is also up-regulated in response to UPR induction and adenylylates BiP at Thr-366. However, there are some differences in the experimental strategies and results obtained between our study and the report on dFicD. For instance, although both HYPE and dFicD use divalent cations for their activity, unlike what is reported for dFicD (28), we failed to see an increase in BiP adenylylation by HYPE in the presence of Ca2+. Instead, Mn2+ and Mg2+ served as optimal cations, at physiologically relevant concentrations, for BiP adenylylation (Fig. 7H). The ability to use both Mg2+ and Mn2+ may give HYPE an advantage over other Mg2+-dependent enzymes; a similar property is seen for other proteins of the secretory pathways, such as the Fam20C kinase (26). Next, using the ATPase domain of BiP with or without its linker region as an adenylylation target and to measure ATPase activity, Ham et al. suggest that adenylylation disrupts the ATPase activity of BiP (28). This observation is surprising, because both HYPE and BiP are up-regulated during UPR and inhibiting BiP function would be detrimental to coping with stress. A more likely explanation is that the ATPase assays with Drosophila Fic were not done with full-length BiP, and the inhibitory effect of adenylylation was inferred indirectly. In contrast, using proteins purified and separated by ion exchange and size exclusion, to ensure purity and lack of ATPase activity from contaminating bacterial chaperones from the purification process, we show that adenylylation enhances the ATPase activity of BiP (Fig. 8C), which fits with our observation that HYPE positively regulates UPR.
Our studies further reveal that adenylylation of BiP does not alter its ability to bind to misfolded proteins (Fig. 8, A and B). Interestingly, however, we find that HYPE itself is able to bind to misfolded proteins, a function that is independent of its adenylyltransferase activity (Fig. 8A). Furthermore, its binding to misfolded proteins is enhanced in the presence of BiP (Fig. 8B). These findings provide useful insight to speculate that HYPE may form a complex with BiP and misfolded proteins under UPR stress.
During UPR activation, misfolded proteins are first bound by TPR domain containing proteins like Hsp40 before handing them off to BiP (31). This mechanism stalls unnecessary activation of UPR cascades, offering the cell a chance to recover from ER stress. These co-chaperones also regulate interaction between Hsp70 and Hsp90 via their TPR domains and coordinate the cycle of binding of misfolded proteins and release of folded polypeptides from the chaperone machinery (30, 32). Based on our structural analysis of IbpA Fic in complex with its Cdc42 substrate, domains outside the Fic enzymatic core are likely to be important for substrate recognition (25). To this end, HYPE contains a TPR domain, which may be important for its protein-protein interactions. It is possible that HYPE interacts with misfolded proteins in complex with BiP and other co-chaperones, and adenylylation may be important for stabilizing this interaction.
We are currently mapping exactly which branches of UPR are directly affected by HYPE's activity. Our data so far show a more dramatic effect on PERK and ATF6. Why HYPE appears to affect predominantly the PERK and ATF6 pathways remains to be determined. One possibility is that HYPE may have targets other than BiP. This idea is supported by the fact that even though HYPE adenylylates BiP, the presence of BiP enhances HYPE's autoadenylylation levels (Fig. 7D). It is therefore possible that in addition to being adenylylated by HYPE, BiP may also function as a cofactor for HYPE. We are currently assessing other ER-localized members of the UPR pathway as adenylylation targets of HYPE.
Recently, the ER protein PARP16 was shown to regulate PERK and Ire1α-mediated UPR activation by ADP-ribosylating these proteins and improving their disassociation from BiP (33). One possible explanation for HYPE's effect on the PERK and ATF6 pathways, but not Ire1α pathway, is that adenylylation of BiP specifically alters its ability to interact with PERK and ATF6. We hypothesize that adenylylation promotes BiP's disassociation from PERK and ATF6, thus activating these pathways (Fig. 8D). This also explains why sustained activation of HYPE, such as with expression of E234G-HYPE, induces apoptosis, which is controlled by PERK activation. The observed increase in BiP's ATPase activity upon adenylylation would also promote induction of this pathway.
The physiological signal that activates HYPE remains a critical question. BiP could be adenylylated in vitro only when incubated with E234G-HYPE but not WT-HYPE (Fig. 7, D and E). This observation suggests that simply an interaction between WT-HYPE and its substrate is not sufficient to relieve HYPE's intrinsic inhibition. Thus, substrate binding is not the signal for the enzymatic activation of HYPE. Autoadenylylation also does not appear to be a signal for regulating HYPE activity, as autoadenylylation mutants were minimally affected in their ability to induce apoptosis and adenylylate BiP (Figs. 2D and 7E).
Our study is the first characterization of the human Fic protein, and the first functional demonstration of the physiological significance of this enigmatic protein in eukaryotic signaling. Our knockdown studies and determination of the consequence of BiP modification further provide mechanistic insight into the role of Fic-mediated adenylylation in mammalian signal transduction.
Several lines of evidence support the activation of UPR during disease, for which BiP's ability to monitor and recover from these stress perturbations is critical. As a protein that modifies and activates BiP under stress conditions, HYPE poses as an attractive candidate for disease therapeutics.
Acknowledgments
We thank Drs. Stephen Konieczny and David Hess for advice, reagents, and helpful comments for our UPR studies. We are very grateful to Dr. Nancy Emery for guidance with statistical analysis. We also thank Sandy Wiley for technical advice regarding tissue fractionation. Fam20C/pEGFP and wild-type HYPEΔ102/pSMT3 were kind gifts from Drs. Vincent Tagliabracci and Junyu Xiao. Finally, we are grateful to members of the Mattoo laboratory and the Dr. Jack Dixon laboratory for their helpful discussions.
This work was supported in part by American Cancer Society Institutional Research Grant ACS IRG 58-006-53, Indiana Clinical and Translational Research Grant CTSI 106564, a Purdue Research Foundation Summer Faculty Grant, Showalter Research Trust Award 207655 (to S. M.), and a Yeunkyung Woo Achieve Excellence Travel Grant (to A. S.).
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- PERK
- PKR-like ER kinase
- aa
- amino acid
- PARP
- poly(ADP-ribose) polymerase
- cPARP
- cleaved PARP
- ANOVA
- analysis of variance
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- FPP
- fluorescence protease protection
- DIC
- differential interference contrast
- qRT
- quantitative RT
- TPR
- tetratricopeptide repeat
- AU
- arbitrary unit
- Tg
- thapsigargin
- Tn
- tunicamycin.
REFERENCES
- 1. Bernales S., Papa F. R., Walter P. (2006) Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 [DOI] [PubMed] [Google Scholar]
- 2. Wiseman R. L., Haynes C. M., Ron D. (2010) SnapShot: the unfolded protein response. Cell 140, 590–590 [DOI] [PubMed] [Google Scholar]
- 3. Cruz J. W., Woychik N. A. (2014) Teaching Fido New ModiFICation Tricks. PLoS Pathog. 10, e1004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mattoo S., Durrant E., Chen M. J., Xiao J., Lazar C. S., Manning G., Dixon J. E., Worby C. A. (2011) Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J. Biol. Chem. 286, 32834–32842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., Dixon J. E. (2009) The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cruz J. W., Rothenbacher F. P., Maehigashi T., Lane W. S., Dunham C. M., Woychik N. A. (2014) Doc toxin is a kinase that inactivates elongation factor Tu. J. Biol. Chem. 289, 7788–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia-Pino A., Zenkin N., Loris R. (2014) The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem. Sci. 39, 121–129 [DOI] [PubMed] [Google Scholar]
- 8. Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 [DOI] [PubMed] [Google Scholar]
- 9. Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N. A., Loris R., Zenkin N. (2013) The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J. M. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118 [DOI] [PubMed] [Google Scholar]
- 11. Mukherjee S., Liu X., Arasaki K., McDonough J., Galán J. E., Roy C. R. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Palanivelu D. V., Goepfert A., Meury M., Guye P., Dehio C., Schirmer T. (2011) Fic domain catalyzed adenylylation: insight provided by the structural analysis of the type IV secretion system effector BepA. Protein Sci. 20, 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan Y., Arnold R. J., Luo Z. Q. (2011) Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U.S.A. 108, 21212–21217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zekarias B., Mattoo S., Worby C., Lehmann J., Rosenbusch R. F., Corbeil L. B. (2010) Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect. Immun. 78, 1850–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Engel P., Goepfert A., Stanger F. V., Harms A., Schmidt A., Schirmer T., Dehio C. (2012) Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482, 107–110 [DOI] [PubMed] [Google Scholar]
- 16. Faber P. W., Barnes G. T., Srinidhi J., Chen J., Gusella J. F., MacDonald M. E. (1998) Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 7, 1463–1474 [DOI] [PubMed] [Google Scholar]
- 17. Schröder M., Kaufman R. J. (2005) The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 18. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.–range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Lorenz H., Hailey D. W., Lippincott-Schwartz J. (2008) Addressing membrane protein topology using the fluorescence protease protection (FPP) assay. Methods Mol. Biol. 440, 227–233 [DOI] [PubMed] [Google Scholar]
- 21. Wiley S. E., Rardin M. J., Dixon J. E. (2009) Chapter 13 Localization and function of the 2Fe-2S outer mitochondrial membrane protein mitoNEET. Methods Enzymol. 456, 233–246 [DOI] [PubMed] [Google Scholar]
- 22. Wiley S. E., Andreyev A. Y., Divakaruni A. S., Karisch R., Perkins G., Wall E. A., van der Geer P., Chen Y. F., Tsai T. F., Simon M. I., Neel B. G., Dixon J. E., Murphy A. N. (2013) Wolfram Syndrome protein, Miner1, regulates sulphydryl redox status, the unfolded protein response, and Ca2+ homeostasis. EMBO Mol. Med. 5, 904–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zelenski N. G., Rawson R. B., Brown M. S., Goldstein J. L. (1999) Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 21973–21980 [DOI] [PubMed] [Google Scholar]
- 24. Miranda M., Dionne K. R., Sorkina T., Sorkin A. (2007) Three ubiquitin conjugation sites in the amino terminus of the dopamine transporter mediate protein kinase C-dependent endocytosis of the transporter. Mol. Biol. Cell 18, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao J., Worby C. A., Mattoo S., Sankaran B., Dixon J. E. (2010) Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tagliabracci V. S., Engel J. L., Wen J., Wiley S. E., Worby C. A., Kinch L. N., Xiao J., Grishin N. V., Dixon J. E. (2012) Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tavaria M., Gabriele T., Kola I., Anderson R. L. (1996) A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones 1, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ham H., Woolery A. R., Tracy C., Stenesen D., Krämer H., Orth K. (2014) Unfolded protein response-regulated dFic reversibly AMPylates BiP during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059–36069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewallen D. M., Sreelatha A., Dharmarajan V., Madoux F., Chase P., Griffin P. R., Orth K., Hodder P., Thompson P. R. (2014) Inhibiting AMPylation: a novel screen to identify the first small molecule inhibitors of protein AMPylation. ACS Chem. Biol. 9, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris J. A., Dorner A. J., Edwards C. A., Hendershot L. M., Kaufman R. J. (1997) Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 272, 4327–4334 [DOI] [PubMed] [Google Scholar]
- 31. Fan C. Y., Lee S., Cyr D. M. (2003) Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Höhfeld J., Minami Y., Hartl F. U. (1995) Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 83, 589–598 [DOI] [PubMed] [Google Scholar]
- 33. Jwa M., Chang P. (2012) PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat. Cell Biol. 14, 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]