Background: 1,8-Cineole, a commercially important monoterpene, was identified as a fungal product.
Results: The 1,8-cineole synthase was identified from a Hypoxylon fungal genome, and mutagenesis revealed a critical asparagine residue.
Conclusion: The fungal 1,8-cineole synthase uses a mechanism similar to the plant version.
Significance: This is the first identified fungal monoterpene synthase and may facilitate future terpene synthase identification and production.
Keywords: Enzyme Mechanism, Fungi, Mutagenesis, Phylogenetics, Terpenoid
Abstract
Terpenes are an important and diverse class of secondary metabolites widely produced by fungi. Volatile compound screening of a fungal endophyte collection revealed a number of isolates in the family Xylariaceae, producing a series of terpene molecules, including 1,8-cineole. This compound is a commercially important component of eucalyptus oil used in pharmaceutical applications and has been explored as a potential biofuel additive. The genes that produce terpene molecules, such as 1,8-cineole, have been little explored in fungi, providing an opportunity to explore the biosynthetic origin of these compounds. Through genome sequencing of cineole-producing isolate E7406B, we were able to identify 11 new terpene synthase genes. Expressing a subset of these genes in Escherichia coli allowed identification of the hyp3 gene, responsible for 1,8-cineole biosynthesis, the first monoterpene synthase discovered in fungi. In a striking example of convergent evolution, mutational analysis of this terpene synthase revealed an active site asparagine critical for water capture and specificity during cineole synthesis, the same mechanism used in an unrelated plant homologue. These studies have provided insight into the evolutionary relationship of fungal terpene synthases to those in plants and bacteria and further established fungi as a relatively untapped source of this important and diverse class of compounds.
Introduction
Fungi have long been studied for the ability to produce secondary metabolites with useful properties (1). Many of the most well studied molecules are bioactive natural products that may have antimicrobial or other properties useful in medicine or agriculture (2, 3). More recently, there has been an increased focus on biological compounds that may serve as biofuels, commodity chemicals, or their precursors (4, 5). These molecules are often volatile organic compounds (VOCs)2 that mimic the properties of compounds found in petroleum mixtures.
Terpenoids represent the largest class of secondary metabolites, with over 40,000 known structures, and are produced as VOCs from many fungal species (6). This large amount of chemical diversity is generated by the terpene synthase enzymes (TPS) that produce these molecules (7, 8). These enzymes take a prenyl diphosphate precursor and, through a variety of cyclizations and rearrangements of a carbocation intermediate, produce the final terpene scaffold. The prenyl diphosphate precursors can have variable chain lengths in five-carbon intervals, which adds to the diversity of this compound class. For example, monoterpenes (C10) derive from geranyl pyrophosphate (GPP), sesquiterpenes (C15) from farnesyl pyrophosphate (FPP), and diterpenes (C20) from geranylgeranyl pyrophosphate. The terpene backbone can be further modified, often through oxidation by P450 enzymes, to create a huge array of natural products.
The diversity of terpene products is due in large part to the plasticity of terpene synthase enzyme active sites. The TPS enzymes can be either specific or promiscuous, synthesizing anywhere from a single product to more than 50 different compounds. The identity and number of compounds produced by a TPS enzyme is determined in the active site, where the substrate is activated by a metal binding motif (DDXX(D/E)) to form a carbocation intermediate and guided through a variety of cyclization mechanisms. Structurally, all TPS enzymes share a common terpene synthase fold, which is conserved in bacteria, fungi, and plants; however, outside of the DDXX(D/E) motif, there is only limited sequence similarity between plant and microbial enzymes (9, 10). Evidence from plants suggests that terpene synthases have evolved rapidly and that sequence similarity is dominated by species relationships rather than the terpene product (8, 11, 12). The “evolvability” and plasticity of these enzymes have recently allowed for directed evolution and rational engineering of TPS activity (13, 14). Site-directed mutagenesis was used by Yoshikuni et al. (15) to identify plasticity residues in a promiscuous γ-humulene synthase and convert it into a variety of more specific synthases. Furthermore, Kampranis et al. (16) were able to convert a 1,8-cineole synthase from the sage plant Salvia fruticosa to produce non-oxygenated monoterpenes sabinene or limonene as well as sesquiterpenes by mutation of a single asparagine residue. Such plasticity has also been observed in a fungal trichodiene synthase, demonstrating that this is a conserved feature of TPS enzymes (17).
Many terpenoids are bioactive and may have medicinal properties, such as anticancer (taxanes), antimicrobial, or antimalarial activity (artemisinin) (18–21). These compounds, particularly the mono- and sesquiterpenes, are often used as fragrance or flavoring compounds. More recently, a number of terpene hydrocarbons have been studied as potential biofuels. Farnesane and bisabolane, fully reduced forms of the sesquiterpenes farnesene and bisabolene, have both been produced as alternative diesel fuels (22). These molecules have an energy density similar to that of diesel fuel and much greater than that of ethanol, currently the most widely used biofuel. In addition, these terpene derived fuels are immiscible in water and compatible with current engines, which are further advantages over ethanol. Monoterpenes have also been studied as biofuel targets. Pinene can be chemically dimerized for use as a potential jet fuel, whereas limonene has been studied for use as a biofuel and a precursor for commodity chemicals (23–25). Both of these compounds are traditionally derived from plant biomass, but alternative biosynthetic production platforms have been explored.
Because of the interest in terpenes for a variety of uses, the pathways that produce these molecules have been engineered for increased production in Sacharomyces cerevisiae and Escherichia coli. In E. coli, this involved expression of components of the yeast mevalonate pathway in addition to the endogenous DXP pathway in the bacterium. This pathway was originally developed for the overproduction of amorpha-4,11-diene, a sesquiterpene precursor of the antimalarial drug artemisinin (26–28). Additional optimization and expression of a bisabolene synthase allowed the pathway to be adapted for production of the biofuel precursor bisabolene at a yield of 900 mg/liter (22). Recently, this pathway was adapted for expression of the monoterpene limonene by replacing the FPP synthase gene ispA with a GPP synthase from Mentha spicata and expression of a limonene synthase from Abies grandis (23). In each of the described cases, a plant TPS gene was used from terpene production, which is unsurprising because the plant TPS enzymes are better characterized than microbial versions. Because this step of the pathway has been identified as a bottleneck, there remains a need for identification of additional and potentially more efficient terpene synthases.
Fungi are another source of terpenes, and the TPS genes that produce them are underexplored compared with plants. Current biodiversity estimates suggest that there exist ∼400,000 plant species compared with between 3 and 5 million fungal species, which represents a tremendous amount of biodiversity that remains largely untapped (29). Only a handful of fungal terpene synthases have been characterized, including aristolochene synthase, trichodiene synthase, presilphiperfolan-8β-ol synthase, and the sesquiterpene synthases of Basidiomycota Coprinus cinereus, Armillaria gallica, and Omphalotus olearius (30–36). No fungal monoterpene synthases have been identified to date.
Endophytic fungi are a diverse group of microbes that reside within plant tissues. They have been explored for the production of VOCs, and many of them produce an abundance of terpenes (37). Recently, three endophytic fungi in the family Xylariaceae, identified as Hypoxylon sp. (isolate CI-4) and related anamorph Nodulisporium sp. (isolates EC12 and Ni25–2A), were discovered to produce a series of VOCs that included 1,8-cineole (38–40). Also known as eucalyptol, 1,8-cineole is an oxygenated monoterpene that is the main component of the essential oil of Eucalyptus globulus. Cineole is widely used in a number of applications, including medicinally in cough suppressants and mouth wash, and as a flavoring and fragrance additive. It has also been explored as a biofuel with potential as a gasoline additive that may prevent separation in ethanol and petroleum fuel blends as well as a precursor for commodity chemicals, such as p-cresol and benzylic esters (41–44).
Here we have identified eight new fungal isolates in the family Xylariaceae that produce 1,8-cineole and a variety of other potentially interesting compounds. Through genome sequencing, heterologous gene expression in E. coli, and in vitro enzymology, we identify the fungal cineole synthase. A mutational analysis of the enzyme active site reveals a remarkable similarity in the mechanism of the otherwise divergent fungal and plant cineole synthases for incorporating the oxygen moiety during terpene cyclization and excluding sesquiterpene precursor substrates. Finally, we test the yield of cineole production in an overexpression strain to demonstrate the viability of fungal synthases for terpene production.
EXPERIMENTAL PROCEDURES
Fungal Isolation and Culturing
Plant stem material was collected from various locations in Ecuador as well as Kubah National Park in Sarawak, Malaysia. Stems were stored in clean plastic bags and transported back to Yale University, where they were surface-sterilized and plated on isolation medium comprised of 1× potato dextrose agar (PDA) (24 g/liter potato dextrose), 0.1× PDA (2.4 g/liter potato dextrose) (EMD Millipore), or water agar (all 15 g/liter agar). Several fungi were isolated in the presence of 3-day-old Muscodor albus, which was used to enrich for VOC-producing isolates, as described previously (45, 46). Isolates observed growing from the stems were transferred to a new 1× PDA plate. For VOC analysis, agar plugs were inoculated into sealed vials with both 1× PDA and potato dextrose broth, as described previously (37). Vials were sampled after growth at 23 °C for 4, 8, and 12 days.
GC/MS Analysis of Fungal Volatile Compounds
Volatile compounds in the headspace of the fungal cultures were sampled by solid phase microextraction (SPME) with a 50/30-μm divinylbenzene/carboxen/polydimethylsiloxane StableFlex SPME fiber (Supelco). Compounds were injected into the gas chromatograph (Agilent 7890A) with a ZB-624 column (30 meters × 0.25 mm ID × 1.40-μm film thickness; Phenomenex) paired with a time-of-flight mass spectrometer (GCT Premier, Waters). Electron ionization spectral data were analyzed with the MassLynx Software Suite (Waters), and initial peak identification was carried out by comparison of the electron impact spectra with the Wiley Registry of Mass Spectral Data (8th Ed.) and the National Institute of Standards and Technology (NIST) database. Whenever possible, the compound identification was verified by comparison of retention time and mass spectra with authentic standards (Sigma-Aldrich).
Phylogenetic Analysis of Fungal Isolates
The internal transcribed spacer (ITS) rDNA of the cineole-producing fungi was amplified by the colony-PCR method described by Van Zeijl et al. (47) using trichoderma lysing enzyme (Sigma) in place of Novozyme 234. The primers used for amplification were ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATGC), and the resulting PCR products were sequenced by the W. M. Keck Foundation (48). A phylogenetic tree was constructed comparing the ITS sequence of cineole-producing isolates from this study with previous cineole producers (CI-4, Ni25–2A, and EC12) and taxa reported in several recent molecular phylogenetic studies of genera in the family Xylariaceae (Table 1) (49, 50). Sequences were aligned using MUSCLE version 3.8.31, and the phylogenetic tree was constructed using Bayesian (MrBayes version 3.2.0) and maximum likelihood methods (RAxML version 7.2.8) (51, 52).
TABLE 1.
Summary of organisms used in the Xylariaceae phylogenetic tree
| Taxon | Strain ID | ITS accession no. | NCBI taxon ID |
|---|---|---|---|
| Annulohypoxylon atroroseum | SUT009 | DQ223733.1 | 42309 |
| A. annulatum | H3M | AJ390395 | 327058 |
| A. cohaerens | F-160,842 | AY909025 | 326618 |
| A. minutellum | H9A | AJ390399 | 326621 |
| A. multiforme | F160,843 | AY909003 | 326624 |
| A. nitens | ST2313 | DQ223751.1 | 326626 |
| A. stygium | ntu44 | FJ205451.1 | 326628 |
| A. stygium | 105F | FJ008986.1 | 326628 |
| A. stygium | SUT243 | DQ223761.1 | 326628 |
| Biscogniauxia atropunctata | B5A | AJ390411 | 109358 |
| B. atropunctata | ATCC38987 | AF201705 | 109358 |
| B. bartholomaei | ATCC38992 | AF201719 | 109359 |
| B. marginata | B74A | AJ390417 | 110534 |
| B. mediterranea | B71C | AJ390413 | 97377 |
| B. nummularia | B72C | AJ390415 | 97378 |
| B. repanda | B75A | AJ390418 | 110535 |
| Daldinia cf. fissa | P. D. Rabenborg (C) | AF176981 | 103427 |
| D. loculata | HJ 107 | AF176969 | 103429 |
| D. petriniae | HJ103 | AF176975 | 103430 |
| Hypoxylon begae | 215 JDR | JN660820.1 | 326653 |
| H. cinnabarinum | H7R | AJ390398 | 110541 |
| H. fragiforme | SGLAf55 | EU715619.1 | 63214 |
| H. fragiforme | H12C | AJ390401.1 | 327061 |
| H. fragiforme | ATCC36662 | AF201709 | 63214 |
| H. fragiforme | H12C | AJ390401 | 327061 |
| H. fuscum | CBS 113048 | AY616699.1 | 109372 |
| H. fuscum | H16A | AJ390405 | 109372 |
| H. haematostroma | SUT292 | DQ223739.1 | 152305 |
| H. intermedium | H4A | AJ390396 | 110540 |
| H. investiens | CBS 118185 | FJ185308.1 | 326663 |
| H. monticulosum | SUT116 | DQ223749.1 | 326669 |
| H. monticulosum | H076 | FM209439.1 | 326669 |
| H. ochraceum | H17R | AJ390406 | 110544 |
| H. papillatum | ATCC58729 | AF201710 | 58729 |
| H. perforatum | H18R | AJ390407.1 | 110542 |
| H. perforatum | H18R | AJ390407 | 110542 |
| H. rickii | H19R | AJ390408 | 110545 |
| H. rubiginosum | SUT221 | DQ223759.1 | 110542 |
| Nemania aenea var. aureolutea | ATCC 60819 | AF201704 | 109375 |
| N. bipapillata | CL8 | AY541610 | 110536 |
| N. chestersii | N23A | AJ390430 | 110537 |
| N. serpens | N20A | AJ390431 | 109381 |
| N. serpens | N112C | AJ390436 | 109381 |
| Xylaria allantoidea | F-165,173 | AY909005 | 323539 |
| X. arbuscula | WARMI | AY183369 | 114810 |
| X. digitata | CBS 161.22 | AY909006 | 323540 |
| X. globosa | PR7 | AY909007 | 323541 |
| X. hypoxylon | CBS 868.72 | AY909011 | 37992 |
| X. liquidambaris | ATCC 42766 | AY909021 | 323546 |
| X. longipes | CBS 148.73 | AY909013 | 114818 |
| X. multiplex | PR111 | AY909019 | 323545 |
| EC12 | EC12 | JQ424940 | 1001832 |
| CI-4 | CI-4 | GU797135 | 755336 |
| Ni25–2A | Ni25–2A | JQ968613 | 1210998 |
| B10820A | YU.101024 | KJ412996 | 1488202 |
| E6826D | YU.101016 | KJ412992 | 1488199 |
| E13415F | YU.101019 | KJ412990 | 1488201 |
| E13732D | YU.101018 | KJ412994 | 1488200 |
| E7015B | YU.101017 | KJ412993 | 1488204 |
| E13615I | YU.101020 | KJ412997 | 1488205 |
| E7406B | YU.101015 | KJ412991 | 1488203 |
| E14012A | YU.101021 | KJ412995 | 1488206 |
Genome Sequencing of E7406B and Terpene Synthase Gene Identification
Genomic DNA from isolate E7406B was obtained from a 7-day-old PDA culture using a Plant DNeasy kit (Qiagen) as described previously (53). This DNA was used for 2 × 100 paired-end sequencing on an Illumina Hiseq 2000 of both a 180-bp fragment library and a 3-kb mate-pair library. The sequencing data were assembled using ALLPATHS-LG into a genome of 45.3 Mbp (54). The assembly yielded 504 scaffolds with an N50 of 855 kbp and genome coverage of 450%. Genome data were submitted to GenBankTM (Bioproject ID PRJNA244689). Gene models were defined using the GeneMark-ES self-training algorithm, identifying 11,551 protein-coding genes.
TPS genes were identified using Pfam based on similarity to the terpene synthase C-terminal domain family (PF03936). Putative terpene synthase gene identities were also checked using BLAST for similarity to known terpene synthase genes. A total of 13 putative TPS genes were identified, and several criteria were used to determine candidates for expression. Alignment of the TPS protein sequences with MUSCLE version 3.8.31 was performed to confirm the presence of the known catalytic motifs DDXX(D/E) and NSE/DTE. Two of the enzymes did not contain the required catalytic residues and were excluded from further analysis, whereas the remaining 11 enzymes were included in the construction of a phylogenetic tree along with protein sequences of terpene synthases with known products from plants, fungi, and bacterial species (Table 2). TPS protein sequences were again aligned using MUSCLE version 3.8.31, and the phylogenetic tree was constructed using RAxML version 7.2.8.
TABLE 2.
Summary of TPS enzymes used in the phylogenetic tree
| TPS gene | Accession no. | Organism |
|---|---|---|
| (−)-(4S)-Limonene synthase | AAB70907.1 | Abies grandis |
| Pinene synthase | AAB71085.1 | A. grandis |
| β-Phellandrene synthase | AAF61453.1 | A. grandis |
| d-Selinene synthase | AAC05727.1 | A. grandis |
| γ-Humulene synthase | AAC05728.1 | A. grandis |
| Taxadiene synthase | AAC49310.1 | Taxus brevifolia |
| (−)-Limonene synthase | AAS47694.1 | Picea abies |
| (−)-α/β-Pinene synthase | AAS47692.1 | P. abies |
| Myrcene synthase | AAS47696.1 | P. abies |
| (E,E)-α-Farnesene synthase | AAS47697.1 | P. abies |
| Longifolene synthase | AAS47695.1 | P. abies |
| (E)-α-bisabolene synthase | AAS47689.1 | P. abies |
| AtTPS-CIN2 | P0DI77.1 | Arabidopsis thaliana |
| Cineole synthase | ABH07677.1 | Salvia fruticosa |
| TPS8 cineole synthase | G1JUH5 | Solanum lycopersicum |
| d-Cadinene synthase A | Q43714.1 | Gossypium arboreum |
| d-Cadinene synthase | P93665.1 | Gossypium hirsutum |
| 5-epi-Aristolochene synthase | Q40577.3 | Nicotiana tabacum |
| (+)-Bornyl diphosphate synthase | AAC26017.1 | Salvia officinalis |
| (+)-Sabinene synthase | AAC26018.1 | S. officinalis |
| Limonene synthase | AAD50304.1 | Mentha longifolia |
| Germacrene C synthase | AAC39432.1 | S. lycopersicum |
| (E)-B-farnesene synthase | AAB95209.1 | Mentha x piperita |
| 2-Methylisoborneol synthase | YP_001105919.1 | Saccharopolyspora erythraea NRRL 2338 |
| Germacradienol synthase | NP_823339.1 | Streptomyces avermitilis MA-4680 |
| epi-Isozizaene synthase | NP_824208.1 | S. avermitilis MA-4680 |
| Pentalenene synthase | NP_824174.1 | S. avermitilis MA-4680 |
| Cineole synthase | ZP_05003209.1 | Streptomyces clavuligerus ATCC 27064 |
| Cadinene synthase | ZP_05004823.1 | S. clavuligerus ATCC 27064 |
| Muurolene synthase | ZP_05006242.1 | S. clavuligerus ATCC 27064 |
| Linalool synthase | ZP_06776363.1 | S. clavuligerus ATCC 27064 |
| Germacrendiol synthase | NP_630182.1 | Streptomyces coelicolor A3(2) |
| 2-Methylisoborneol synthase | NP_733742.1 | S. coelicolor A3(2) |
| Selinene synthase | YP_001867159.1 | Nostoc punctiforme PCC 73102 |
| Pentalenene synthase | Q55012.4 | Streptomyces exfoliatus |
| Aristolochene synthase | AAA33694 | Penicillium roqueforti |
| Tri5 trichodiene synthase | P27679 | Fusarium sambucinum |
| Tri5 trichodiene synthase | AAN05035 | Fusarium sporotrichioides |
| Longiborneol synthase | ACY69978 | Gibberella zeae |
| Aristolochene synthase | AAF13264.1 | Aspergillus terreus |
| BOT2 presilphiperfolan-8-β-ol synthase | Q6WP50.1 | Botryotinia fuckeliana |
| Fusicoccadiene synthase | EEQ31327.1 | Arthroderma otae CBS 113480 |
| Omp1 α-muurolene synthase | Wawrzyn et al. (35) | Omphalotus olearius |
| Omp2 terpene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp3 germacrene A synthase | Wawrzyn et al. (35) | O. olearius |
| Omp4 δ-cadinene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp5a γ-cadinene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp5b γ-cadinene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp6 δ-6 protoilludene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp7 δ-6 protoilludene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp8 terpene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp9 α-barbatene synthase | Wawrzyn et al. (35) | O. olearius |
| Omp10 trans-dauca-4(11),8-diene synthase | Wawrzyn et al. (35) | O. olearius |
| Cop1 germacrene A synthase | XP_001832573.2 | Coprinopsis cinerea okayama7 (#130) |
| Cop2 germacrene A synthase | XP_001836556.1 | C. cinerea okayama7 (#130) |
| Cop3 α-muurolene synthase | XP_001832925.1 | C. cinerea okayama7 (#130) |
| Cop4 δ-cadinene synthase | XP_001836356.1 | C. cinerea okayama7 (#130) |
| Cop5 terpene synthase | XP_001834007.1 | C. cinerea okayama7 (#130) |
| Cop6 α-cuprenene | XP_001832549.1 | C. cinerea okayama7 (#130) |
| Hyp1 | KJ433269 | Hypoxylon sp. |
| Hyp2 | KJ433270 | Hypoxylon sp. |
| Hyp3 | KJ433271 | Hypoxylon sp. |
| Hyp4 | KJ433272 | Hypoxylon sp. |
| Hyp5 | KJ433273 | Hypoxylon sp. |
| Hyp6 | KJ433274 | Hypoxylon sp. |
| Hyp7 | KJ433275 | Hypoxylon sp. |
| Hyp8 | KJ433276 | Hypoxylon sp. |
| Hyp9 | KJ433277 | Hypoxylon sp. |
| Hyp10 | KJ433278 | Hypoxylon sp. |
| Hyp11 | KJ433279 | Hypoxylon sp. |
Cloning and Expression of Terpene Synthase Genes in E. coli
Candidate TPS genes were synthesized by GENEWIZ and codon-optimized for expression in E. coli (supplemental sequences). These genes were then subcloned into vector pTrc99 between restriction sites NcoI and XmaI and transformed into E. coli DH5α (22). The bacteria were also co-transformed with pJBEI-2999, which contains the mevalonate pathway optimized for FPP production (22).
Overnight cultures of E. coli with terpene synthase plasmids were grown in LB broth (Sigma) and diluted 1:100 in 50 ml of production medium (Teknova, EZ-Rich with 1% glucose) as described previously (22). Cultures were induced with 500 μm isopropyl 1-thio-β-d-galactopyranoside at OD 0.6, and 5 ml was removed for growth in sealed GC/MS vials. Dodecane, which has been shown to trap volatile terpenes and relieve product inhibition while not inhibiting E. coli growth, was overlaid on the shaker flask sample at 10% (v/v) as reported (55). After 48 h of growth, the GC/MS vials were sampled by SPME, and the dodecane layer of shaker flask samples was extracted and diluted at 1:10 and 1:100 in ethyl acetate for direct injection into the GC/MS. Both sampling methods were used because SPME is the more sensitive method for detecting minor compounds, whereas direct injection is less biased toward more volatile compounds, allowing for the determination of relative compound amounts. Production peaks were matched to authentic standards when available for identification, as done for the fungal samples. In addition, retention indices were measured on a DB5-MS column (Agilent) by comparison with an alkane mix (Fluka), and production peaks were compared with known compounds in the MassFinder terpene library and the NIST database. Peak areas in the direct injection samples were also measured to give a relative amount of each product.
Structural Modeling and Mutational Analysis of Hyp3 Enzyme
The protein sequence of Hyp3 was analyzed for subcellular localization signaling motifs using the programs WoLF PSORT and PSORTII online tools (56, 57). The enzyme structure was modeled using the structure of the most closely related TPS enzyme based on phylogeny, Aspergillus terreus aristolochene synthase (Protein Data Bank entry 2E4O) (58), using the Modeler software (59). We also aligned this modeled structure with the FPP substrate-bound aristolochene synthase (Protein Data Bank entry 4KUX) and generated the figure using PyMOL version 1.7.2.1.
PCR primers matching to the hyp3 gene in the pTrc99 vector were used to alter the codon at position Asn-136 to produce valine (N136V), isoleucine (N136I), serine (N136S), and alanine (N136A) mutants. These mutants were expressed in E. coli DH5α with pJBEI-2999 as described for other TPS genes and were sampled by SPME after a 2-h induction to ensure that production peaks did not saturate the mass spectrometry detector. GC/MS conditions were the same as described previously for fungal volatile compounds.
Hyp3 Protein Purification and Enzyme Assays
The codon-optimized hyp3 gene was PCR-amplified from the pTrc99 construct using primers designed for Gibson assembly (forward primer, 5′-CAGCAGCGGCCATATCGAAGGTCGTCATATGGCCCGCCCGATTAC; reverse primer, 5′-GCTTTGTTAGCAGCCGGATCCTCGAGCATACTATTAGATACCACGTAAGCC). The hyp3 gene was cloned using Gibson assembly into a pET16b vector following the His tag (Gibson Assembly Kit, New England Biolabs). This Hyp3 expression vector was transformed into E. coli BL21, and cells were grown at 37 °C in Terrific Broth with ampicillin (100 μg/ml) to an OD of 0.6–0.8. When the OD was reached, the cells were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and transferred to a 30 °C incubator for 4 h. Cells were lysed using a microfluidizer and clarified by centrifugation. Hyp3 was purified from the clarified lysate using nickel-nitrilotriacetic acid resin (Qiagen) in wash buffer (20 mm Tris, pH 7.5, 10 mm MgCl2, 200 mm NaCl, 1 mm 2-mercaptoethanol, 10 mm imidazole) and eluted with elution buffer (20 mm Tris, pH 7.5, 10 mm MgCl2, 1 mm 2-mercaptoethanol, 300 mm imidazole). For storage, the purified protein was dialyzed into buffer A (20 mm Tris, pH 7.5, 10 mm MgCl2, 1 mm 2-mercaptoethanol, 10% glycerol).
GC/MS enzyme activity assays were carried out by incubating a 500-μl reaction overnight at 30 °C in a 5-ml scintillation vial sealed with Parafilm. The reaction contained 100 μm GPP and 10 μm Hyp3 enzyme in GC/MS assay buffer (buffer A with no glycerol). The reaction vials were sampled by SPME and analyzed by GC/MS as described for the bacterial cultures.
Steady-state kinetics of fungal cineole synthase was assayed in a plate reader (Synergy 4, BioTek) by measuring pyrophosphate (PPi) release via conversion to ATP in an ATP-dependent luminescence assay (PPiLight kit, Lonza). Final concentrations of GPP (Sigma-Aldrich) and (E,E)-FPP (Sigma-Aldrich) were varied between 0.25 and 5 μm. Sample containing no substrate was used as a blank, and known concentrations of PPi were used to obtain the time-dependent calibration curves. Reaction mixtures (total volume 75 μl) contained 15 μl of PPiLight solution reconstituted according to the supplier's instructions, 10 nm cineole synthase, and assay buffer (buffer A with 50 mm HEPES rather than Tris). Incubation time was 90 min at 30 °C. The activity was determined as the difference in luminescence/min of the samples with and without the substrate. Concentrations of PPi released during each assay were calculated from the linear fit equations for each minute of recorded luminescence from the known concentration of PPi (varied between 0.025 and 10 μm). The scatter plots of initial rate versus substrate concentration were fitted to the equation,
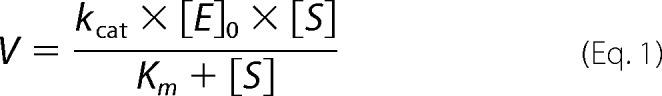 |
from which Km and kcat values were determined. KaleidaGraph version 4.1.3 was used for data analysis.
Quantification of 1,8-Cineole Production
For cineole quantification, shaker flasks (50 ml) of the fungus in PDA were grown for 4 days, and E. coli DH5α harboring hyp3, with and without pJBEI-2999, were grown for 2 days in EZ-Rich production medium. A 10% dodecane layer was overlaid on each of the samples, and an aliquot was removed and sampled after the allotted growth time as described above, with the inclusion of an internal camphor standard. The amount of cineole in each sample was calculated as compared with a standard cineole curve with a camphor internal standard included.
RESULTS
VOC Analysis of Fungal Isolates
Fungi isolated after collection trips to tropical forests in Ecuador and the Malaysian state of Sarawak were subjected to GC/MS analysis of the volatile compounds produced in the culture headspace. Cultures were grown in sealed vials on potato dextrose broth medium and sampled after 4, 8, or 20 days of growth. We discovered eight unique isolates producing the monoterpene 1,8-cineole based on comparison with the MS fragmentation pattern in the NIST database. These isolates were identified as E7406B, E6826D, E7015B, E13732D, E13415F, E13615I, and E14012A from locations in Ecuador and B10820A from Kubah National Park in Sarawak. These isolates, along with the originally identified 1,8-cineole producer strain CI4 isolated in the Canary Islands, were then retested on both solid PDA and liquid potato dextrose broth to confirm the production of 1,8-cineole and further catalogue the spectrum of VOCs that they produce. The major VOCs produced by these organisms are presented in Table 3.
TABLE 3.
Major volatiles produced by fungal isolates
| Formula | Compound ID | CI4 | E7406B | E6826D | E7015B | E13732D | E13415F | E13615I | E14012A | B10820A |
|---|---|---|---|---|---|---|---|---|---|---|
| Alcohols etc. | ||||||||||
| C4H10O | 2-Methyl propanol | × | × | × | × | × | × | |||
| C5H12O | 3-Methyl butanola | × | × | × | × | × | × | × | × | × |
| C5H12O | 2-Methyl butanola | × | × | × | × | × | × | × | × | × |
| C7H14O2 | 2-Methylbutyl acetate | × | × | × | × | × | ||||
| Monoterpenoids | ||||||||||
| C10H16 | α-Pinenea | × | × | × | × | × | ||||
| C10H16 | α-Phellandrenea | × | × | × | × | |||||
| C10H16 | α-Terpinenea | × | × | × | × | × | ||||
| C10H16 | Limonenea | × | × | × | × | × | × | × | × | |
| C10H14 | p-Cymenea | × | × | × | × | × | × | × | ||
| C10H16 | Phellandrene | × | × | × | × | × | × | × | × | |
| C10H18O | 1,8-Cineolea | × | × | × | × | × | × | × | × | × |
| C10H16 | γ-Terpinenea | × | × | × | × | × | ||||
| C10H16 | Terpinolenea | × | × | × | × | × | × | |||
| Sesquiterpenoids | ||||||||||
| C15H24 | Unidentified sesquiterpenes | ×9 | ×24 | ×32 | ×12 | ×9 | ×16 | ×10 | ×14 | ×18 |
| Polyenes | ||||||||||
| C5H8 | Pentadiene | × | × | |||||||
| C7H10 | Heptatriene | × | × | × | × | × | × | |||
| C7H10 | Unknown | × | × | × | × | × | ||||
| C9H12 | Nonatetraenea,b | × | × | × | × | × | × | × | ||
| C9H12 | Nonatetraene isomer | × | × | × | × | × | × | |||
| C9H12 | Nonatetraene isomer | × | × | × | × | × | × | |||
| C9H12 | Nonatetraene isomer | × | × | × | × | × | × | |||
| C9H12 | Nonatetraene isomer | × | × | × | × | |||||
a Verified by authentic GC/MS standard.
b Verified by NMR.
The identity of 1,8-cineole was confirmed by comparison with an authentic standard with a match of both the retention time and fragmentation pattern to the fungal product. We also observed a series of other monoterpene compounds produced in a majority of the isolates (Table 3). Authentic standards matched the identity of limonene, p-cymene, γ-terpinene, α-pinene, α-phellandrene, α-terpinene, and terpinolene. Other monoterpenes were identified in the spectrum of several of the isolates based on the appearance of a 136 mass peak consistent with compounds with the formula C10H16 and comparison with the NIST database, which gave a tentative identification of these compounds as α-pinene, α-phellandrene, α-terpinene, and terpinolene.
The cineole producing strains also produced an expansive series of sesquiterpenes. The number observed ranged from 9 compounds in CI4 and E13732D up to 32 in isolate E6826D. The sesquiterpenes were identified by a characteristic 204 Da peak in the mass spectral data and comparison with the NIST database. These compounds eluted in the range where sesquiterpene standards were observed, but with only a single exception, commercially available compounds did not match the product peaks. We were only able to match the compound δ-cadinene in seven of our isolates to the known compound from the extracts of Atlas Cedar and Cade Juniper. We were unable to confirm the identity of the remaining sesquiterpene compounds.
A series of alcohols and acetate derivatives, which are common fungal metabolites, were also among the compounds in the VOC profile of our isolates. The observed compounds included 3-methyl butanol and 2-methyl butanol, produced by all of the strains tested, the related derivatives 3-methyl and 2-methyl butyl acetate in six strains, and the shorter chain 2-methyl propanol in six of the strains. Other compounds commonly observed in the tested organisms included dimethylfuran, benzene ethanol, benzene, and styrene.
Many of the isolates tested produced an extensive series of volatile polyene compounds with the formulas C7H10 and C9H12. Compounds with these formulas were previously reported as products of isolate CI-4. The previously identified C7H10 compound methyl-1,4-cyclohexadiene was not produced in our cultures based on comparison with an authentic standard, indicating that we may be observing a different subset of these compounds. The largest of the polyene compound peaks was a C9H12 compound that matched the retention time and fragmentation pattern of a compound identified as (3E,5E,7E)-nona-1,3,5,7-tetraene by NMR in a different fungus (82). This is also consistent with the NIST database match for this compound peak. Four smaller peaks eluted immediately after the nonatetraene peak and are probably isomers of this compound, having identical electron impact fragmentation patterns. The major C7H10 compound produced by our fungi was identified in the database as 1,3,5-heptatriene, which is probably related to the nonatetraene peak.
Phylogenetic Analysis of Cineole-producing Fungi
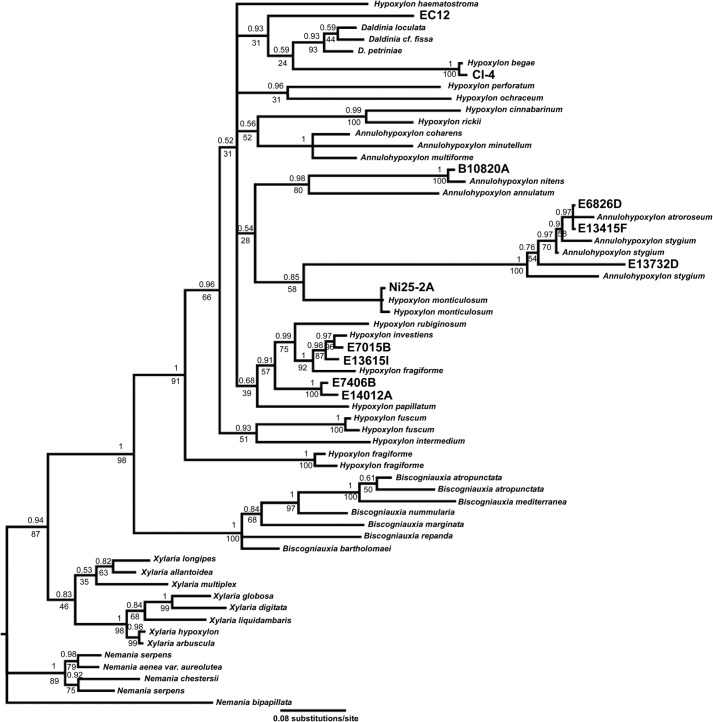
The isolates presented here had a number of VOCs in common and were typified by the production of 1,8-cineole. We reasoned that these fungi may also be phylogenetically related although they derived from sources around the world. To test this idea, we sequenced the internal transcribed spacers of our isolates and compared them with the published cineole-producing strains CI-4, Ni25–2A, and EC12. These cineole-producing strains all grouped to a clade comprising fungi in the genera Hypoxylon, Annulohypoxylon, and Daldinia within the family Xylariaceae (Fig. 1). This clade is clearly resolved from other genera, Nemania, Xylaria, and Biscogniauxia, by both maximum likelihood and Bayesian phylogenetic approaches. The genera containing cineole producers are closely related, and although their phylogenetic relationship was not resolved by ITS sequencing alone in our tree, consistent with previous publications, we can observe to which of these groups our isolates are most closely related (49).
FIGURE 1.
Phylogenetics of cineole-producing fungi in the family Xylariaceae. Phylogram of cineole-producing fungal isolates (boldface type) in the context of members of the family Xylariaceae, genera Nemania, Xylaria, Biscogniauxia, Hypoxylon, Annulohypoxylon, and Daldinia. ITS sequences were analyzed by both Bayesian and maximum likelihood methods. Nodes are labeled with Bayesian posterior probabilities (top) and maximum likelihood bootstrap values (bottom).
We were largely able to recapitulate the phylogenetic relationships of published strains CI-4, EC12, and Ni25–2A, with EC12 appearing closely related to species of Daldinia, Ni25–2A close to Hypoxylon monticulosum, and CI-4 close to Hypoxylon begae (38–40). Strains E6826D, E13415F, and E13732D grouped in a clade containing species of Annulohypoxylon, whereas a second Annulohypoxylon clade contained B10820A. Isolates E7015B, E13615I, E7406B, and E14012A all appeared closely related to each other and group in a clade containing strains of Hypoxylon fragiforme, Hypoxylon investiens, and Hypoxylon rubiginosum. These data suggest that the ability to produce cineole is widespread in these genera of Xylariaceae.
Identification and Phylogenetic Analysis of Terpene Synthases from Isolate E7406B
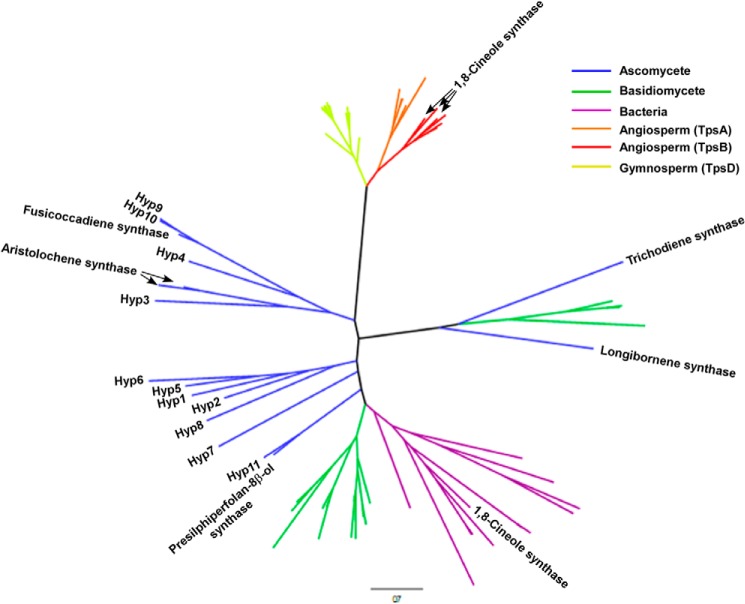
We wanted to identify the terpene synthase gene responsible for 1,8-cineole production in our fungi. To accomplish this, we sequenced the genome of the isolate E7406B. Assembly of the draft genome and gene prediction using ALLPATHS-LG and GeneMark-ES identified 11,551 protein-coding genes. Thirteen of those were identified as terpene synthase genes using BLAST and Pfam matches, but two were excluded from further analysis because they lack the conserved DDXX(D/E) motif required for catalysis. Phylogenetic analysis of the remaining terpene synthases, termed Hyp1–11, revealed that they do not form a monophyletic clade but are unsurprisingly most closely related to other ascomycete terpene synthases (Fig. 2). In particular, Hyp9 and Hyp10 appear closely related to a fusicoccadiene synthase, whereas Hyp11 appears related to a TPS producing presilphiperfolan-8-β-ol, a botrydial precursor (33). The plant enzymes in the tree recapitulated the known TPS subfamilies Tpsa, Tpsb, and Tpsd. Three known plant 1,8-cineole synthases group among the other angiosperm monoterpene synthases (Tpsb), whereas the recently identified 1,8-cineole synthase from the bacterium Streptomyces clavuligerus groups with other bacterial TPS enzymes. None of the TPS enzymes from E7406B appear closely related to these known 1,8-cineole synthases. Because fungal monoterpene synthases have yet to be identified, we reasoned that the 1,8-cineole synthase from Xylariaceae fungi may be divergent from the previously identified ascomycete sesquiterpene synthases. We went on to select TPS genes for heterologous expression to identify their products, prioritizing those that were divergent from known TPS genes.
FIGURE 2.
Phylogenetic tree of plant, fungal, and bacterial terpene synthases. Gene products of the ascomycete terpene synthases are labeled along with Hyp1–11 from E7406B. Known 1,8-cineole synthases from bacteria and angiosperm species are also labeled.
Identification of Fungal 1,8-Cineole Synthase Hyp3 and Other TPS Genes
The eight selected genes, hyp1–8, were expressed in E. coli DH5α harboring the pJBEI-2999 plasmid for high production of terpene precursors through the mevalonate pathway. The volatile products were measured by GC/MS analysis of both the culture headspace by SPME in sealed vials and direct injection of a dodecane overlay in 50-ml shaker flasks. This analysis revealed that hyp3 is the gene responsible for 1,8-cineole production in our fungal isolate. E. coli expressing Hyp3 produced a major monoterpene product matching the retention time and mass spectrum of a 1,8-cineole standard. Hyp3 appeared specific for monoterpenes, producing 1,8-cineole at about 90% of the product peak area, along with a minor peak of the unoxygenated monoterpene limonene (Table 4). No sesquiterpene products were observed from this enzyme. This result is especially interesting considering that pJBEI-2999 was optimized for production of farnesyl pyrophosphate, the precursor to sesquiterpene biosynthesis. None of the other TPS genes tested exhibited monoterpene specificity.
TABLE 4.
Terpene products of Hyp1–8 in E. coli DH5α with pJBEI-2999
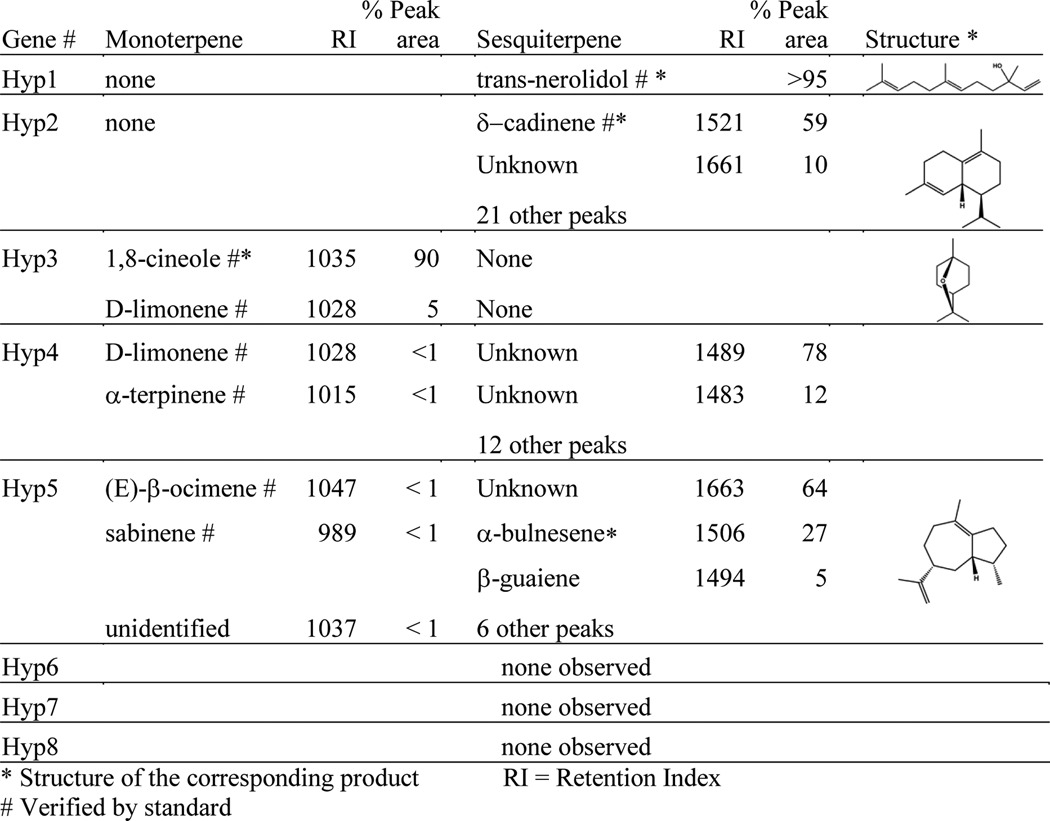
E. coli expressing pJBEI-2999 with four of the remaining genes produced sesquiterpenes as a majority of their terpene products (Table 4). hyp1 appeared to be specific for the oxygenated, uncyclized sesquiterpene trans-nerolidol, as identified by comparison with an authentic nerolidol standard. The other three sesquiterpene synthases appeared to be more promiscuous enzymes, producing a variety of products. The major product of Hyp2 was δ-cadinene, identified by similarity to the known compound from plant extracts, with 22 other minor products, most of which the NIST database identified as containing a naphthalene-like bicyclic ring structure. Products of the other two sesquiterpene synthases, Hyp4 and Hyp5, could not be definitively identified by authentic GC/MS standards. Hyp4 produced unknown compounds tentatively identified in the NIST database as β-caryophyllene and germacrene A, whereas Hyp5 produced compounds with an apparent azulene-type bicyclic ring structure, the major components identified as α-bulnesene, β-guaiene, and an unknown oxygenated derivative based on comparison of the mass spectra and retention indices. Both of these genes also produced minor monoterpene products at less than 5% of the total peak area. Three of the genes, hyp6–8, produced no observed products, despite containing what appeared to be the required catalytic domains. There are a number of reasons this might occur, including misidentified splice sites in the original gene model, low expression of the enzymes in E. coli, toxicity of their products, or production of compounds that are not observable with our column and GC/MS conditions, including some diterpenes or highly oxygenated sesquiterpenes. Because our goal was to identify the 1,8-cineole synthase, we did not explore these non-producing genes further.
Hyp3 Enzyme Kinetics Demonstrate a Strong Specificity for GPP Substrate
We were intrigued by the apparent specificity as well as potential biotechnological applications of the fungal 1,8-cineole synthase. To further explore this enzyme, we tested it in in vitro activity assays. The purified enzyme was incubated overnight with GPP substrate, and products were analyzed by GC/MS. This assay confirmed that 1,8-cineole was the major monoterpene product, with no other products observed.
In order to more definitively measure the substrate specificity of Hyp3, steady-state kinetic data with GPP and FPP were compared. Enzymatic activity of the fungal 1,8-cineol synthase was measured with purified enzyme using a luminescence-based pyrophosphate (PPi) detection assay because both GPP and FPP release PPi prior to cyclization step. Fig. 3 clearly shows more specific activity with GPP compared with FPP, as expected for a monoterpene synthase, giving a kcat value for GPP of 17.7 ± 2.0 min−1 and an apparent Km value of 2.5 ± 0.6 μm. Measurable activity with the FPP substrate was not observed. This kcat value compares favorably with those of 1,8-cineole synthases from organisms in other kingdoms; for example, those from the sage plant S. fruticosa and streptomycete Streptomyces lividans were measured at 3.2 and 4.7 min−1, respectively (16, 60). Substrate affinity, as measured by Km, varied widely between these enzymes, with S. fruticosa cineole synthase having about 25-fold weaker affinity and S. lividans about 15-fold tighter affinity. We can also compare activity of the fungal Hyp3 monoterpene synthase to the more phylogenetically related fungal sesquiterpene synthases. The kcat/Km of 7.0 min−1 μm−1 for Hyp3 1,8-cineole synthase was similar to those of the fungal aristolochene synthases, although those enzymes had better affinity for FPP (0.13 μm for A. terreus and 0.6 for Penicillium roqueforti) and a 7–17-fold slower kcat than Hyp3 had for GPP substrate catalysis (61).
FIGURE 3.
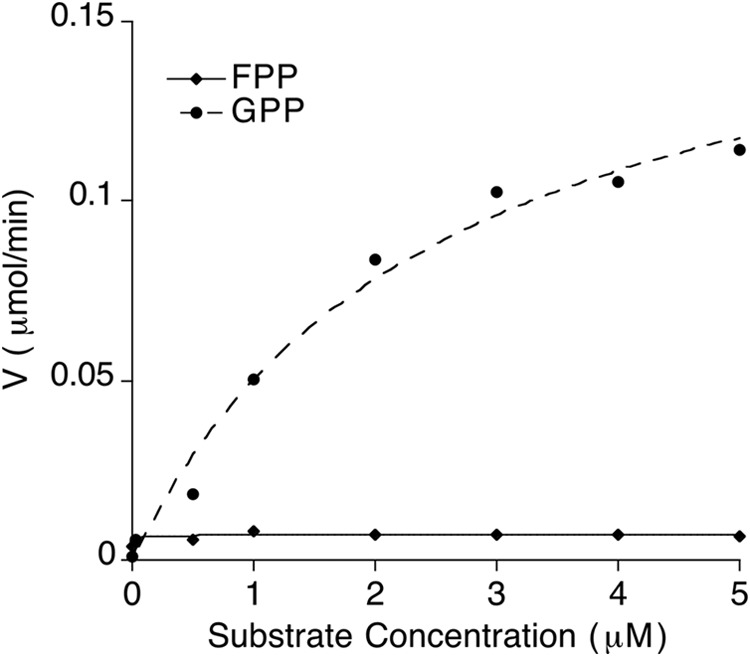
Steady-state kinetic analysis of the fungal 1,8-cineol synthase. Shown is a comparison of the Hyp3-catalyzed reactions with GPP (dashed line) and FPP (solid line) substrates.
Hyp3 Enzyme Sequence and Structural Modeling Analysis
In plants, monoterpene synthase enzymes contain a signal peptide for plastid localization, whereas the sesquiterpene synthases are largely cytosolic. A sequence alignment of the Hyp3 enzyme with the related aristolochene synthases showed that Hyp3 contains a slightly expanded N-terminal sequence, 9 amino acids more than P. roqueforti aristolochene synthase, but does not contain an obvious signaling sequence. We used several tools for subcellular localization prediction, including WoLF PSORT and PSORT II, to identify any analogous localization signals on the fungal Hyp3 monoterpene synthase. Both of these programs predict that Hyp3 is a cytosolic enzyme (61% cytosolic by PSORT II).
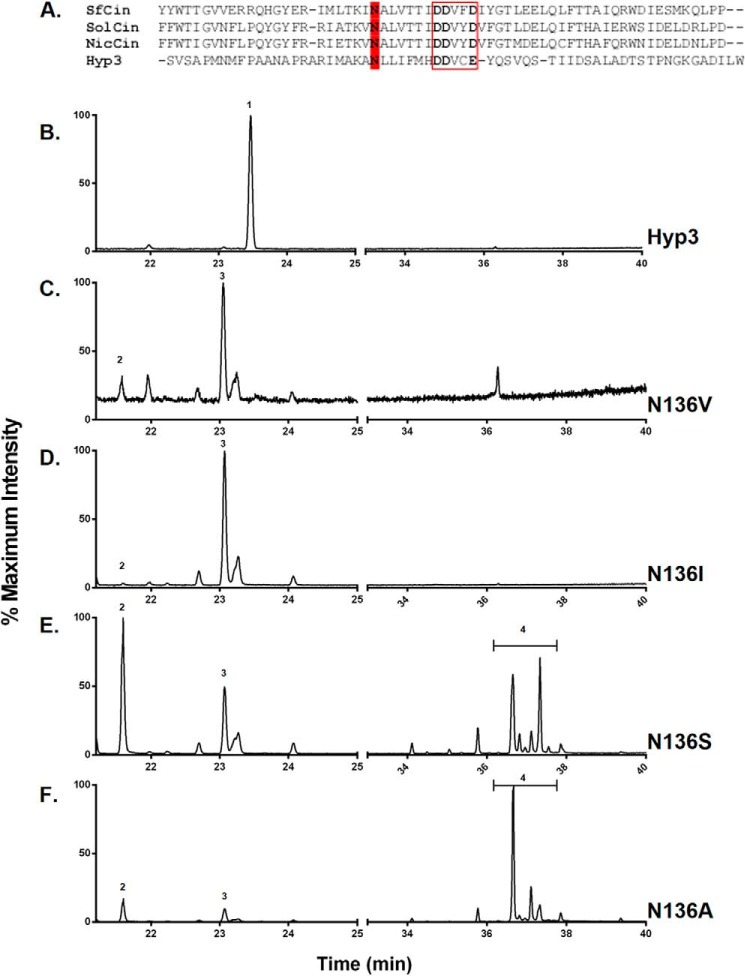
We also created a sequence alignment and structural model of Hyp3 compared with the Aspergillus terreus aristolochene synthase, the most closely related terpene synthase in the phylogenetic analysis (Fig. 4A) (58). These models reveal several features in the active site pocket that may influence enzyme specificity and 1,8-cineole catalysis. In substrate-bound structures of A. terreus aristolochene synthase, the FPP substrate C12 and C13 carbons are positioned near the NSE motif-containing α-helix (62). Near this position, Leu-209 in the aristolochene synthase is replaced with the bulkier aromatic Phe-267 in Hyp3, which may interfere with the FPP substrate (Fig. 4B). Several other aromatic amino acids, such as Phe-233 and Phe-237, are found near the Hyp3 active site, replacing smaller hydrophobic residues, and, although they are not predicted to be oriented near the substrate, they may serve to decrease the overall size of the active site pocket. We were most intrigued by an asparagine residue, Asn-136, found in Hyp3 near the DDXX(D/E) active site motif but not observed in the aristolochene synthases (Leu-77 in A. terreus aristolochene synthase) or other Hyp enzymes. In the 1,8-cineole synthase from S. fruticosa (Sf-CinS1), it was determined that a conserved asparagine residue near the active site was responsible for activating water for attack on the carbocation intermediate (16). The Hyp3 structural model and an alignment of plant cineole synthases with our fungal Hyp3 revealed that Asn-136 resides in the same location adjacent to the active site DDXX(D/E) motif (Fig. 5A).
FIGURE 4.

Structural model of Hyp3 based on the A. terreus aristolochene synthase crystal structure. A, structural model of Hyp3 featuring the DDXX(D/E) motif (red) (N/D)XXSXXXE motif (orange), and Asn-136 and Phe-267 (cyan with side chain). B, aligned model of Hyp3 (blue) with substrate-bound aristolochene synthase (yellow). The FPP substrate of the aristolochene synthase is shown colored by element.
FIGURE 5.
Mutational analysis of Hyp3 Asn-136. A, alignment of Hyp3 sequence near the conserved DDXX(D/E) motif (red box) with plant cineole synthases from Solanum lycopersicum (SolCin), Nicotiana suaveolens (NicCin), and S. fruticosa (SfCin). The asparagine (N) critical for plant synthase function is highlighted in red near the catalytic motif. Shown is a total ion chromatogram of terpene products of Hyp3 (B) and mutant enzymes N136V (C), N136I (D), N136S (E), and N136A (F) in E. coli with pJBEI-2999. The x axis is split to show both monoterpene and sesquiterpene regions of the spectrum. The indicated compounds are 1,8-cineole (1), myrcene (2), limonene (3), and farnesene isomers (4).
Hyp3 Amino Acid Asn-136 Is Critical for Cineole Oxygen Incorporation and Monoterpene Specificity
We hypothesized that the fungal synthase may use the same mechanism as the plant version to produce 1,8-cineole. To test this hypothesis, we made a series of mutations at the Asn-136 position in Hyp3 and tested them in our E. coli expression assay. We found that in each of our mutations (N136V, N136I, N136S, and N136A), removing the asparagine eliminated cineole production by the enzyme. In each case, the enzyme was not inactive but rather produced unoxygenated monoterpenes. The major product of N136V and N136I was limonene, which can be made from the same carbocation intermediate as cineole (Fig. 5, B–D). When the side chain at this position is shortened to a serine or alanine, the major monoterpene product becomes the uncyclized myrcene, indicating that the enzyme can no longer catalyze cyclization of the GPP precursor (Fig. 5, E and F).
Similar to the plant cineole synthase, this asparagine is also responsible for the specificity of Hyp3 for the monoterpene GPP precursor. Hyp3 normally does not produce sesquiterpene products (Fig. 5B), but when Asn-136 is shortened to serine, sesquiterpene products begin to appear. In the N136A mutant, sesquiterpene compounds, which match to farnesene isomers, form the majority of the terpene products from the enzyme.
Expression of Hyp3 in E. coli Overexpression Strains Increases 1,8-Cineole Titers
Engineered production of terpenes in E. coli has been intensely explored recently as an alternative to the natural sources, usually derived from plants. High production titers would be required for microbial production of 1,8-cineole to be a cost-effective alternative to current eucalyptus oil sources. As a first step to test the legitimacy of microbial cineole production, we attempted to quantify cineole production from both fungus E7406B and heterologous expression in E. coli. The dodecane layer from a 4-day-old E7406B shake flask culture and 2-day-old E. coli culture was extracted, and the amount of cineole produced was quantified in the GC/MS by comparison with an authentic cineole standard curve.
The fungal isolate produced 0.54 ± 0.18 mg/liter cineole after 4 days of growth at 30 °C (Fig. 6). When a codon optimized version of the fungal cineole synthase was transferred into E. coli DH5α, it initially produced a lower yield than the fungus. The amount produced in E. coli was near our limit of detection in our direct injection assay, and we only observed the cineole product in one of the three replicates at an amount of 0.09 mg/liter. This low amount of production is consistent with previous reports of the monoterpene production capacity of E. coli, which is limited by low production of the geranyl pyrophosphate precursor (63). When expressed in an E. coli strain harboring the mevalonate pathway plasmid pJBEI-2999, the amount of cineole produced increased significantly to 21 ± 11 mg/liter. The overexpression strain increased cineole yield at least 230-fold over the E. coli alone and about 38-fold over the production level in the fungus.
FIGURE 6.
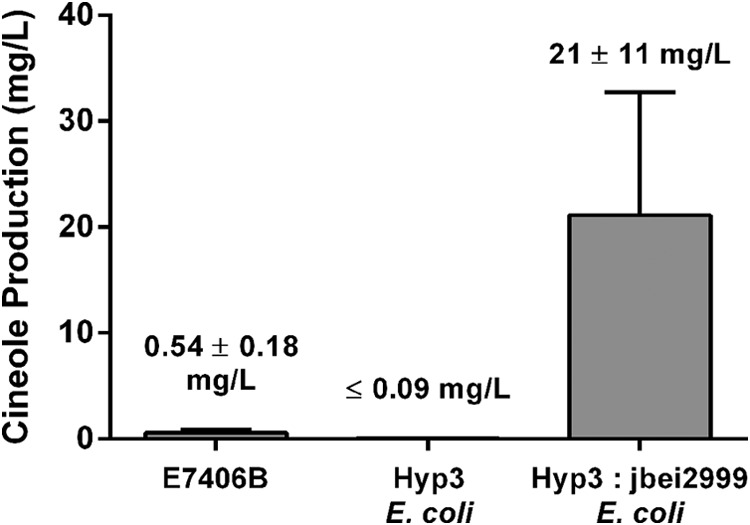
Quantification of 1,8-cineole products in producing microbes. Cineole was quantified by comparison with a standard curve of authentic cineole standard and normalized to an internal camphor standard. Error bars, S.D. of three independent culture growths. Compound was only observed in one replicate of E. coli DH5α with the hyp3 gene.
DISCUSSION
We were intrigued by the capacity of a number of fungal isolates to produce an interesting series of VOCs, including terpenes, in particular 1,8-cineole. Endophytic fungi such as these are known to produce an extensive array of terpene molecules, but the enzymes that produce them have been understudied compared with their plant counterparts. To begin to fill this gap, we set out to identify the gene responsible for 1,8-cineole biosynthesis in a collection of endophytic fungi within the genera Hypoxylon, Annulohypoxylon, and Daldinia. We were able to identify this gene as hyp3 from the genome of isolate E7406B. This enzyme uses a similar mechanism to produce 1,8-cineole as the plant enzyme, in an apparent example of convergent evolution.
In addition to cineole, these fungi produced a wide variety of volatile compounds. An extensive series of monoterpenoids was observed in each of the isolates except for E7015B, which only produced 1,8-cineole. Several of the commonly produced monoterpenes were identified by authentic standards as d-limonene, p-cymene, α-pinene, α-phellandrene, α- and γ-terpinene, and terpinolene. Each of the isolates also produced a series of sesquiterpenes, varying in number from 9 to 32 different species, which were more difficult to identify by GC/MS. The diversity of these molecules produced by these Xylariaceae fungal isolates highlights the large capacity for terpenoid biosynthesis in these fungi.
Only a handful of terpene synthase genes have been identified in ascomycete fungi, and in an effort to identify new terpene synthases, including the gene for 1,8-cineole production, we chose to express TPS genes from isolate E7406B that appeared divergent from known synthases. The four sesquiterpene synthase genes identified, hyp1, hyp2, hyp4, and hyp5, all produced sesquiterpenes with different structures which probably result from different cyclization reactions catalyzed by these enzymes (64). The Hyp1 enzyme produced only the uncyclized trans-nerolidol, which is generated by water capture of the nerolidyl cation (65). Hyp2 probably catalyzes a 1,10-ring closure followed by 1,6-ring closure from both farnesyl and nerolidyl cations to produce the bicyclic, cadalane skeleton compounds δ-cadinene, α-muurolene, and other minor peaks (66, 67). Products of the Hyp4 and Hyp5 enzymes were not positively identified by standards, but products of Hyp5 were identified in the NIST database as guaienes or the related bulnesene derivatives, which are thought to form by 1,10-closure to form germacrene A, followed by 6,2-closure to form the azulenic skeleton (68). The E. coli harboring hyp4 and hyp5 with pJBEI-2999 also produced minor monoterpene peaks, but because these strains are optimized for the FPP precursor, they should be biased against monoterpene production. More accurate determination of their specificity would require further in vitro assays with both GPP and FPP precursors.
The TPS Hyp3 was identified as the cineole synthase in the E7406B fungal genome. 1,8-cineole represented the major product of this terpene synthase at about 90% of the observed peak area, with the remainder primarily composed of d-limonene. Unlike the other TPS genes tested, it exclusively produced monoterpenes, making it, to our knowledge, the first identified fungal monoterpene synthase. The specificity of this enzyme was confirmed in the in vitro enzymatic assay, showing no activity with the FPP substrate, and a kcat/Km with the GPP substrate that compares favorably with other 1,8-cineole synthases from plants. This monoterpene specificity is intriguing in light of the fact that the overexpression plasmid pJBEI-2999 contains the ispA gene, which produces the FPP sesquiterpene precursor, and that E. coli lack a dedicated GPP synthase, which should bias production against monoterpenes. The Hyp3 enzyme is apparently able to bind any GPP substrate not converted to FPP and exclude the 15-carbon substrate.
Hyp3 from the E7406B genome is the first cineole synthase identified from fungi, but plant and bacterial cineole synthases have been characterized structurally and biochemically (16, 60, 69–71). Analysis of cineole synthases in the sage plants S. officinalis and S. fruticosa (Sf-cin) identified a conserved asparagine (Asn-338) that is critical for activating the water molecule for attack on the α-terpinyl cation, resulting in cineole synthesis. Terpene synthases from plants and fungi are known to diverge greatly at the sequence level, but the overall terpene synthase fold is well conserved in structures of these enzymes. Because of this, we thought it was interesting to note that an asparagine, Asn-136, was present in our cineole synthase in the same position as Asn-338 of Sf-cin relative to the DDXX(D/E) motif in the active site (Fig. 5A) (16). We observed that, like in the plant enzyme, mutation of this asparagine abolished cineole production. Both the valine and isoleucine mutants (N136V and N136I) in the Hyp3 enzyme resulted in production of limonene, which results from proton elimination from the α-terpinyl cation rather than carbocation capture by water in the wild type (Figs. 5 (B–D) and 7). The serine mutant (N136S) in our enzyme was also unable to activate water for nucleophilic attack. In fact, both the serine and alanine mutant (N136A) were unable to catalyze cyclization of the GPP substrate, resulting in production of myrcene (Figs. 5 (E and F) and 7). These results place the attacking water molecule in the same position within the fungal enzyme as the observed water pocket in the plant version.
FIGURE 7.
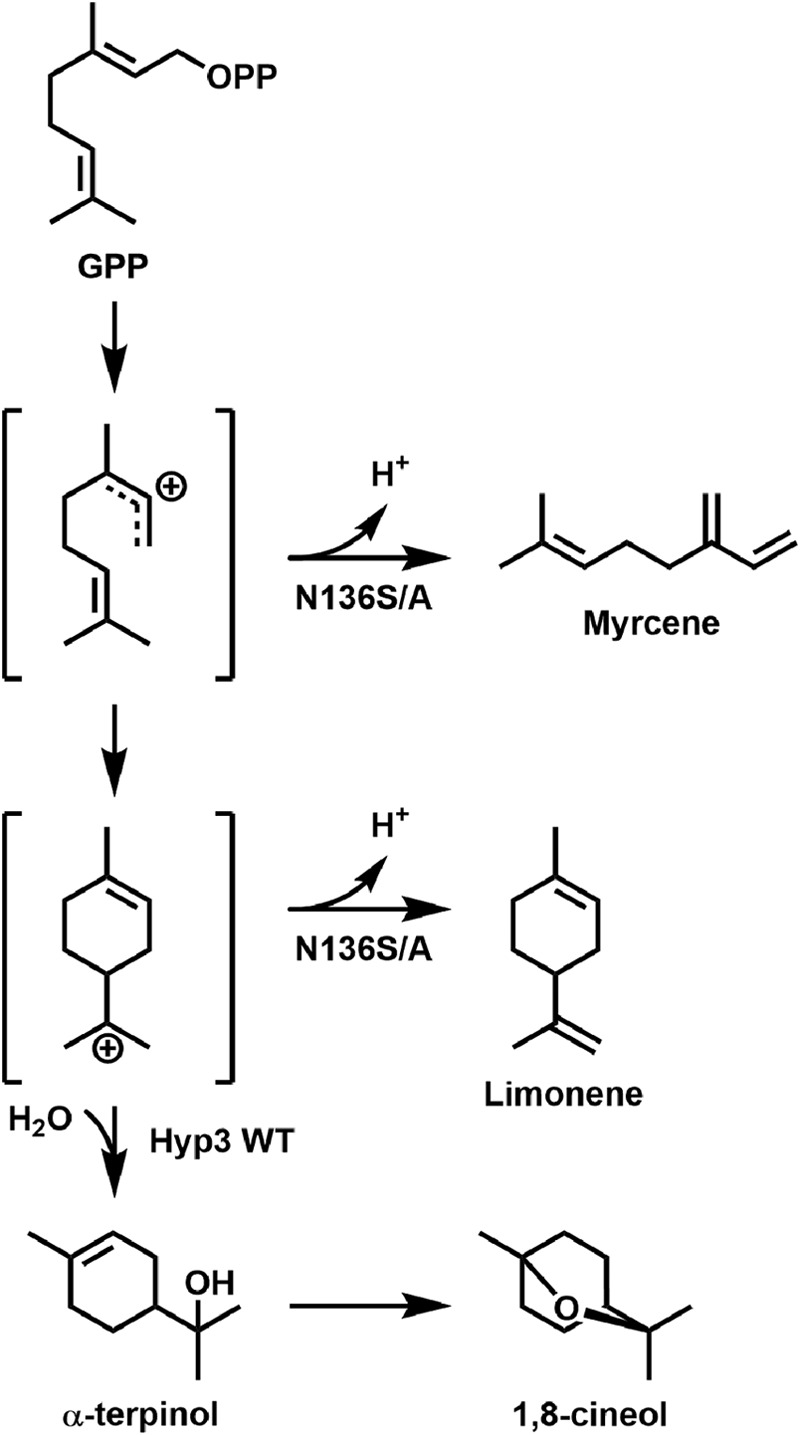
Schematic of 1,8-cineole product formation by Hyp3 and mutants. Derived from the proposed mechanism of the S. fruticosa cineole synthase (16). Reaction pathways catalyzed by the wild type, N136I/V, or N136S/A enzymes are indicated.
In addition, the serine and alanine mutants lost monoterpene specificity and are able to use the FPP substrate for sesquiterpene production. In the case of N136A, the major products are farnesene isomers, which are the sesquiterpene equivalent of the myrcene product. This indicates that, as in the plant cineole synthase, the bulky asparagine side chain prevents the 15-carbon substrate from fitting into the active site pocket. In the related aristolochene synthase enzymes, the residue at this position is a leucine, which is similarly sized as residues that do not support FPP utilization in Hyp3. Clearly, other residues that shape the active site pocket are also critical for monoterpene specificity in Hyp3, potentially including aromatic residues that may be positioned near the substrate (Phe-167) or active site pocket (Phe-233 and Phe-237). That we are able to convert Hyp3 from a 1,8-cineole synthase to a limonene, myrcene, or farnesene synthase by mutating a single amino acid is yet another example of the plasticity of the active sites in terpene synthase enzymes. Rational conversion of terpene synthases has been achieved previously in a number of studies. Plasticity residues controlling the product profiles of several sesquiterpene synthases were identified through contact mapping and site-directed mutagenesis (13, 15). In the analogous class I active site of diterpene synthases, small changes (a single amino acid in some cases) were sufficient to switch the enzyme product specificity (72–74). The ability to convert terpene synthases with a minimal number of mutations to produce different products is thought to be one reason why they seem to diverge so quickly to produce the variety of terpene products seen in nature (15, 75).
Phylogenetic analysis of these terpene synthases suggests that Hyp3 of E7406B is not closely related to the plant or bacterial version of the enzyme but is more closely related to fungal aristolochene synthases. This would argue against a horizontal gene transfer between the endophytic fungus and any plant it inhabited, including the Clarisia racemosa tree from which it was isolated. Thus, the 1,8-cineole synthase mechanism using an asparagine to activate water in the active site has apparently evolved independently in plants and fungi, an intriguing example of convergent evolution that has been observed for other TPS enzymes and secondary metabolic enzymes in general (76, 77).
ITS-based sequence analysis of the cineole-producing isolates reported here, in addition to three previously reported isolates (CI-4, EC-12, and Ni25–2A), placed all of the cineole producers in a clade within the family Xylariaceae, comprising the genera Daldinia, Hypoxylon, and Annulohypoxylon. The phylogenetic relationship between these groups was difficult to resolve based on ITS sequence alone, consistent with previous reports; however, they were clearly distinguished from the related genera of Biscogniauxia, Xylaria, and Nemania (49). These results reveal that the capacity to produce 1,8-cineole is common and widespread in these genera of fungi. Most of the identified Daldinia, Hypoxylon, and Annulohypoxylon isolates have not been screened for volatile compound production, but it is possible that a large fraction of them have cineole production capacity. Considering the close phylogenetic relationship between these cineole producers and the fact that cineole has not been commonly observed in other genera, it seems unlikely that the cineole synthase has arisen completely independently in each of the 11 cineole-producing isolates. Rather, this suggests that the gene for cineole synthesis or a closely related gene that will convert to produce cineole with minimal mutations evolved in a common ancestor of these isolates. Identification of the 1,8-cineole synthase from additional members of this clade will be required to more thoroughly study its evolution.
There has been interest recently in categorizing terpene synthases in the hope that identifying TPS enzyme phylogenetic relationships will help guide genome mining efforts and predict their terpene product. This has proven difficult in plants, where TPS sequences are dominated by species relationships rather than product, but the addition of a large number of sequenced fungal and bacterial genomes has reignited these efforts (78). Recent work on terpene synthases from Basidiomycota has attempted to categorize them based on the type of ring closure catalyzed by the enzymes (34, 35, 79). Most ascomycete terpene synthases in our phylogeny, including those from E7406B, appear distinct from the basidiomycete enzymes as well as plants and bacteria. Efforts to categorize TPS enzymes from Ascomycota in a similar fashion will require the identification of enzymes and products from additional ascomycete genomes.
1,8-Cineole is the main component of eucalyptus oil and is a commodity chemical used in non-prescription pharmaceuticals, fragrances, and flavorings, and it is known to have natural insecticidal and insect-repellant properties (80, 81). It has been explored as a potential precursor for synthesis of p-cresol and benzylic esters, which themselves are commodity chemicals used in the production of pharmaceuticals, dyes, fertilizers, and agrochemicals (43). Cineole has also been proposed as a biofuel additive in ethanol-gasoline fuel blends (44). On its own, cineole has a heat of combustion of 36 MJ/liter, comparable with fossil fuels (32–39 MJ/liter) and much higher than ethanol (21 MJ/liter). It has an octane rating of 100 and can be used as an additive to raise the octane of gasoline mixtures, giving an octane rating of 95 in a mixture of 8 parts cineole to 1 part gasoline. One hindrance to cineole as a biofuel may come from its high melting point (1.5 °C) which would be unsuitable for cold weather use on its own. Other monoterpenes, such as limonene (−96.7 °C freezing point), have good cold weather properties and could be suitable biofuel additives in mixtures with 1,8-cineole.
For use as a commodity chemical, microbial derived 1,8-cineole would need to be cost-competitive with plant extract sources. Eucalyptus oil production has been estimated at 3000 metric tons annually, so efficient yield from any microbial source would be critical. The cineole yield from a 4-day-old fungal culture was 0.54 mg/liter. By identifying the cineole synthase gene and transferring it to an E. coli strain harboring a plasmid for terpene precursor production through the mevalonate pathway, we were able to increase the yield nearly 40-fold to 21 mg/liter. The pJBEI-2999 plasmid was optimized for production of the sesquiterpene biofuel target bisabolene, and, because it is designed to produce the FPP substrate, it is probably not optimal for production of monoterpenes, such as 1,8-cineole. Thus, only small changes, including introduction of a GPP synthase rather than the ispA FPP synthase, may increase cineole yield substantially. A recent effort to produce limonene in E. coli was able to achieve a yield of 400 mg/liter, indicating that substantial increases in monoterpene yield are possible (23). Furthermore, when terpene pathways are optimized, the activity of the terpene cyclase often becomes a bottleneck. Because plant homologues have been used in these efforts to date, screening the tremendous diversity of fungal terpene synthases may reveal enzymes with high activity. Future efforts to correlate the huge variety of fungal terpenes with the genes that produce them may allow production of new terpene-derived drugs, commodity chemicals, or biofuels.
Supplementary Material
Acknowledgments
We thank Gary Strobel for providing strain CI-4, Percy Vargas Nunez for help with host plant identification, and Lauren Eyler and Shana Berwick for fungal isolation. For collection in Sarawak, we thank Dr. Charlie Yeo, Dr. Rita Manurung, Noreha Mahidi, Jugah anak Taqi, Ajuwin ak Lain, Michael Lim Yee Liang, and Affidah Binti Sabran of the Sarawak Biodiversity Center. In Ecuador, this research was performed with a collecting and research permit provided to S. A. S. by the Ministerio del Ambiente of Ecuador and, in Sarawak, by the Sarawak Biodiversity Council of the Government of Sarawak. For genome sequencing assistance, we thank Shrikant Mane and John Overton of the Yale Center for Genome Analysis as well as Nicholas J. Carriero and Robert D. Bjornson of the Yale University Biomedical High Performance Computing Center.
This work was supported by Office of Assistant Secretary of Defense for Research and Engineering NSSEFF Grant N00244-09-1-0070 and a Howard Hughes Medical Institute Professorship (to S. A. S.).

This article contains supplemental sequences.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JYCQ00000000.
- VOC
- volatile organic compound
- TPS
- terpene synthase
- GPP
- geranyl pyrophosphate
- FPP
- farnesyl pyrophosphate
- PDA
- potato dextrose agar
- SPME
- solid phase microextraction
- ITS
- internal transcribed spacer.
REFERENCES
- 1. Turner W. B., Aldridge D. C. (1983) Fungal Metabolites, Academic Press, New York [Google Scholar]
- 2. Strobel G., Daisy B., Castillo U., Harper J. (2004) Natural products from endophytic microorganisms. J. Nat. Prod. 67, 257–268 [DOI] [PubMed] [Google Scholar]
- 3. Schulz B., Boyle C., Draeger S., Römmert A.-K., Krohn K. (2002) Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol. Res. 106, 996–1004 [Google Scholar]
- 4. Peralta-Yahya P. P., Keasling J. D. (2010) Advanced biofuel production in microbes. Biotechnol. J. 5, 147–162 [DOI] [PubMed] [Google Scholar]
- 5. Fortman J. L., Chhabra S., Mukhopadhyay A., Chou H., Lee T. S., Steen E., Keasling J. D. (2008) Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 26, 375–381 [DOI] [PubMed] [Google Scholar]
- 6. Christianson D. W. (2008) Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Degenhardt J., Köllner T. G., Gershenzon J. (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70, 1621–1637 [DOI] [PubMed] [Google Scholar]
- 8. Bohlmann J., Meyer-Gauen G., Croteau R. (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. U.S.A. 95, 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lesburg C. A., Zhai G., Cane D. E., Christianson D. W. (1997) Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science 277, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 10. Starks C. M., Back K., Chappell J., Noel J. P. (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820 [DOI] [PubMed] [Google Scholar]
- 11. Aubourg S., Lecharny A., Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745 [DOI] [PubMed] [Google Scholar]
- 12. Sharkey T. D., Yeh S., Wiberley A. E., Falbel T. G., Gong D., Fernandez D. E. (2005) Evolution of the isoprene biosynthetic pathway in kudzu. Plant Physiol. 137, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenhagen B. T., O'Maille P. E., Noel J. P., Chappell J. (2006) Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc. Natl. Acad. Sci. U.S.A. 103, 9826–9831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lauchli R., Rabe K. S., Kalbarczyk K. Z., Tata A., Heel T., Kitto R. Z., Arnold F. H. (2013) High-throughput screening for terpene-synthase-cyclization activity and directed evolution of a terpene synthase. Angew. Chem. Int. Ed. 52, 5571–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshikuni Y., Ferrin T. E., Keasling J. D. (2006) Designed divergent evolution of enzyme function. Nature 440, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 16. Kampranis S. C., Ioannidis D., Purvis A., Mahrez W., Ninga E., Katerelos N. A., Anssour S., Dunwell J. M., Degenhardt J., Makris A. M., Goodenough P. W., Johnson C. B. (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vedula L. S., Jiang J., Zakharian T., Cane D. E., Christianson D. W. (2008) Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+ B motif,. Arch. Biochem. Biophys. 469, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trombetta D., Castelli F., Sarpietro M. G., Venuti V., Cristani M., Daniele C., Saija A., Mazzanti G., Bisignano G. (2005) Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 49, 2474–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inoue Y., Shiraishi A., Hada T., Hirose K., Hamashima H., Shimada J. (2004) The antibacterial effects of terpene alcohols on Staphylococcus aureus and their mode of action. FEMS Microbiol. Lett. 237, 325–331 [DOI] [PubMed] [Google Scholar]
- 20. Haynes R. K. (2001) Artemisinin and derivatives: the future for malaria treatment? Curr. Opin. Infect. Dis. 14, 719–726 [DOI] [PubMed] [Google Scholar]
- 21. Jennewein S., Croteau R. (2001) Taxol: biosynthesis, molecular genetics, and biotechnological applications. Appl. Microbiol. Biotechnol. 57, 13–19 [DOI] [PubMed] [Google Scholar]
- 22. Peralta-Yahya P. P., Ouellet M., Chan R., Mukhopadhyay A., Keasling J. D., Lee T. S. (2011) Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alonso-Gutierrez J., Chan R., Batth T. S., Adams P. D., Keasling J. D., Petzold C. J., Lee T. S. (2013) Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 19, 33–41 [DOI] [PubMed] [Google Scholar]
- 24. Harvey B. G., Wright M. E., Quintana R. L. (2010) High-density renewable fuels based on the selective dimerization of pinenes. Energy Fuels 24, 267–273 [Google Scholar]
- 25. Yang J., Nie Q., Ren M., Feng H., Jiang X., Zheng Y., Liu M., Zhang H., Xian M. (2013) Metabolic engineering of Escherichia coli for the biosynthesis of α-pinene. Biotechnol. Biofuels 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dietrich J. A., Yoshikuni Y., Fisher K. J., Woolard F. X., Ockey D., McPhee D. J., Renninger N. S., Chang M. C. Y., Baker D., Keasling J. D. (2009) A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450BM3. ACS Chem. Biol. 4, 261–267 [DOI] [PubMed] [Google Scholar]
- 27. Martin V. J. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802 [DOI] [PubMed] [Google Scholar]
- 28. Redding-Johanson A. M., Batth T. S., Chan R., Krupa R., Szmidt H. L., Adams P. D., Keasling J. D., Lee T. S., Mukhopadhyay A., Petzold C. J. (2011) Targeted proteomics for metabolic pathway optimization: application to terpene production. Metab. Eng. 13, 194–203 [DOI] [PubMed] [Google Scholar]
- 29. Blackwell M. (2011) The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438 [DOI] [PubMed] [Google Scholar]
- 30. Cane D. E., Shim J. H., Xue Q., Fitzsimons B. C., Hohn T. M. (1995) Trichodiene synthase: identification of active site residues by site-directed mutagenesis. Biochemistry 34, 2480–2488 [DOI] [PubMed] [Google Scholar]
- 31. Hohn T. M., Plattner R. D. (1989) Purification and characterization of the sesquiterpene cyclase aristolochene synthase from Penicillium roqueforti. Arch. Biochem. Biophys. 272, 137–143 [DOI] [PubMed] [Google Scholar]
- 32. Hohn T. M., Beremand P. D. (1989) Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79, 131–138 [DOI] [PubMed] [Google Scholar]
- 33. Pinedo C., Wang C.-M., Pradier J.-M., Dalmais B., Choquer M., Le Pêcheur P., Morgant G., Collado I. G., Cane D. E., Viaud M. (2008) Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem. Biol. 3, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agger S., Lopez-Gallego F., Schmidt-Dannert C. (2009) Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 72, 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wawrzyn G. T., Quin M. B., Choudhary S., López-Gallego F., Schmidt-Dannert C. (2012) Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem. Biol. 19, 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Engels B., Heinig U., Grothe T., Stadler M., Jennewein S. (2011) Cloning and Characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. J. Biol. Chem. 286, 6871–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griffin M. A., Spakowicz D. J., Gianoulis T. A., Strobel S. A. (2010) Volatile organic compound production by organisms in the genus Ascocoryne and a re-evaluation of myco-diesel production by NRRL 50072. Microbiology 156, 3814–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riyaz-Ul-Hassan S., Strobel G., Geary B., Sears J. (2013) An endophytic Nodulisporium sp. from Central America producing volatile organic compounds with both biological and fuel potential. J. Microbiol. Biotechnol. 23, 29–35 [DOI] [PubMed] [Google Scholar]
- 39. Tess Mends M., Yu E. (2012) An Endophytic Nodulisporium sp. producing volatile organic compounds having bioactivity and fuel potential. J. Pet. Environ. Biotechnol. 10.4172/2157-7463.1000117 [DOI] [Google Scholar]
- 40. Tomsheck A. R., Strobel G. A., Booth E., Geary B., Spakowicz D., Knighton B., Floerchinger C., Sears J., Liarzi O., Ezra D. (2010) Hypoxylon sp., an endophyte of Persea indica, producing 1,8-cineole and other bioactive volatiles with fuel potential. Microb. Ecol. 60, 903–914 [DOI] [PubMed] [Google Scholar]
- 41. Silvestre A. J. D., Válega M., Cavaleiro J. A. S. (2000) Chemical transformation of 1,8-cineole: synthesis of seudenone, an insect pheromone. Ind. Crops Prod. 10.1016/S0926-6690(99)00067-9 [DOI] [Google Scholar]
- 42. Silvestre A. J. D., Cavaleiro J. A. S., Feio S. S., Roseiro J. C., Delmond B., Filliatre C. (1999) Synthesis of some new benzylic ethers from 1,8-cineole with antimicrobial activity. Monatshefte Für Chem. Chem. Mon. 130, 589–595 [Google Scholar]
- 43. Leita B. A., Gray P., O'Shea M., Burke N., Chiang K., Trimm D. (2011) The conversion of 1,8-cineole sourced from renewable eucalyptus oil to p-cymene over a palladium doped γ-Al2O3 catalyst. Catal. Today 10.1016/j.cattod.2011.05.037 [DOI] [Google Scholar]
- 44. Barton A. F. M., Tjandra J. (1989) Eucalyptus oil as a cosolvent in water-ethanol-gasoline mixtures. Fuel 10.1016/0016-2361(89)90004-5 [DOI] [Google Scholar]
- 45. Strobel G. A., Knighton B., Kluck K., Ren Y., Livinghouse T., Griffin M., Spakowicz D., Sears J. (2008) The production of myco-diesel hydrocarbons and their derivatives by the endophytic fungus Gliocladium roseum (NRRL 50072). Microbiology 154, 3319–3328 [DOI] [PubMed] [Google Scholar]
- 46. Ezra D., Hess W. M., Strobel G. A. (2004) New endophytic isolates of Muscodor albus, a volatile-antibiotic-producing fungus. Microbiology 150, 4023–4031 [DOI] [PubMed] [Google Scholar]
- 47. Van Zeijl C. M. J., van de Kamp E. H. M., Punt P. J., Selten G. C. M., Hauer B., van Gorcom R. F. M., van den Hondel C. A. M. J. J. (1997) An improved colony-PCR method for filamentous fungi for amplification of PCR-fragments of several kilobases. J. Biotechnol. 59, 221–224 [DOI] [PubMed] [Google Scholar]
- 48. White T. J., Bruns T., Lee S., Taylor J. (1990) in PCR Protocols: A Guide to Methods and Application, pp. 315–322, Academic Press, New York [Google Scholar]
- 49. Suwannasai N., Martín M. P., Phosri C., Sihanonth P., Whalley A. J. S., Spouge J. L. (2013) Fungi in Thailand: A Case Study of the Efficacy of an ITS Barcode for Automatically Identifying Species within the Annulohypoxylon and Hypoxylon Genera. PLoS One 8, e54529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hsieh H.-M., Ju Y.-M., Rogers J. D. (2005) Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 97, 844–865 [DOI] [PubMed] [Google Scholar]
- 51. Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 52. Ronquist F., Huelsenbeck J. P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 53. Gianoulis T. A., Griffin M. A., Spakowicz D. J., Dunican B. F., Alpha C. J., Sboner A., Sismour A. M., Kodira C., Egholm M., Church G. M., Gerstein M. B., Strobel S. A. (2012) Genomic analysis of the hydrocarbon-producing, cellulolytic, endophytic fungus Ascocoryne sarcoides. PLoS Genet. 8, e1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gnerre S., Maccallum I., Przybylski D., Ribeiro F. J., Burton J. N., Walker B. J., Sharpe T., Hall G., Shea T. P., Sykes S., Berlin A. M., Aird D., Costello M., Daza R., Williams L., Nicol R., Gnirke A., Nusbaum C., Lander E. S., Jaffe D. B. (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 108, 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Newman J. D., Marshall J., Chang M., Nowroozi F., Paradise E., Pitera D., Newman K. L., Keasling J. D. (2006) High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol. Bioeng. 95, 684–691 [DOI] [PubMed] [Google Scholar]
- 56. Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horton P., Nakai K. (1997) Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 147–152 [PubMed] [Google Scholar]
- 58. Shishova E. Y., Di Costanzo L., Cane D. E., Christianson D. W. (2007) X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry 46, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fiser A., Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 60. Nakano C., Kim H.-K., Ohnishi Y. (2011) Identification of the first bacterial monoterpene cyclase, a 1,8-cineole synthase, that catalyzes the direct conversion of geranyl diphosphate. Chembiochem 12, 1988–1991 [DOI] [PubMed] [Google Scholar]
- 61. Felicetti B., Cane D. E. (2004) Aristolochene synthase: mechanistic analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 126, 7212–7221 [DOI] [PubMed] [Google Scholar]
- 62. Chen M., Al-lami N., Janvier M., D'Antonio E. L., Faraldos J. A., Cane D. E., Allemann R. K., Christianson D. W. (2013) Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase. Biochemistry 52, 5441–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carter O. A., Peters R. J., Croteau R. (2003) Monoterpene biosynthesis pathway construction in Escherichia coli. Phytochemistry 64, 425–433 [DOI] [PubMed] [Google Scholar]
- 64. Davis E. M., Croteau R. (2000) Cyclization Enzymes in the Biosynthesis of Monoterpenes, Sesquiterpenes, and Diterpenes. Top. Curr. Chem. 10.1007/3-540-48146-X_2 [DOI] [Google Scholar]
- 65. Degenhardt J., Gershenzon J. (2000) Demonstration and characterization of (E)-nerolidol synthase from maize: a herbivore-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta 210, 815–822 [DOI] [PubMed] [Google Scholar]
- 66. Benedict C. R., Alchanati I., Harvey P. J., Liu J., Stipanovic R. D., Bell A. A. (1995) The enzymatic formation of δ-cadinene from farnesyl diphosphate in extracts of cotton. Phytochemistry 10.1016/0031-9422(95)00066-G [DOI] [Google Scholar]
- 67. Chen X.-Y., Chen Y., Heinstein P., Davisson V. J. (1995) Cloning, expression, and characterization of (+)-δ-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch. Biochem. Biophys. 324, 255–266 [DOI] [PubMed] [Google Scholar]
- 68. Kumeta Y., Ito M. (2010) Characterization of δ-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 154, 1998–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fähnrich A., Krause K., Piechulla B. (2011) Product variability of the “cineole cassette” monoterpene synthases of related Nicotiana species. Mol. Plant. 4, 965–984 [DOI] [PubMed] [Google Scholar]
- 70. Fähnrich A., Brosemann A., Teske L., Neumann M., Piechulla B. (2012) Synthesis of “cineole cassette” monoterpenes in Nicotiana section alatae: gene isolation, expression, functional characterization and phylogenetic analysis. Plant Mol. Biol. 79, 537–553 [DOI] [PubMed] [Google Scholar]
- 71. Croteau R., Alonso W. R., Koepp A. E., Johnson M. A. (1994) Biosynthesis of monoterpenes: partial purification, characterization, and mechanism of action of 1,8-cineole synthase. Arch. Biochem. Biophys. 309, 184–192 [DOI] [PubMed] [Google Scholar]
- 72. Xu M., Wilderman P. R., Peters R. J. (2007) Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. U.S.A. 104, 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wilderman P. R., Peters R. J. (2007) A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J. Am. Chem. Soc. 129, 15736–15737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Keeling C. I., Weisshaar S., Lin R. P. C., Bohlmann J. (2008) Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc. Natl. Acad. Sci. U.S.A. 105, 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Katoh S., Hyatt D., Croteau R. (2004) Altering product outcome in Abies grandis (−)-limonene synthase and (−)-limonene/(−)-α-pinene synthase by domain swapping and directed mutagenesis. Arch. Biochem. Biophys. 425, 65–76 [DOI] [PubMed] [Google Scholar]
- 76. Köllner T. G., Held M., Lenk C., Hiltpold I., Turlings T. C. J., Gershenzon J., Degenhardt J. (2008) A maize (E)-β-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most american maize varieties. Plant Cell 20, 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pichersky E., Gang D. R. (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 5, 439–445 [DOI] [PubMed] [Google Scholar]
- 78. Cane D. E., Ikeda H. (2012) Exploration and mining of the bacterial terpenome. Acc. Chem. Res. 45, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wawrzyn G. T., Bloch S. E., Schmidt-Dannert C. (2012) Discovery and characterization of terpenoid biosynthetic pathways of fungi. Methods Enzymol. 515, 83–105 [DOI] [PubMed] [Google Scholar]
- 80. Wang J. L., Li Y., Lei C. L. (2009) Evaluation of monoterpenes for the control of Tribolium castaneum (Herbst) and Sitophilus zeamaise Motschulsky. Nat. Prod. Res. 23, 1080–1088 [DOI] [PubMed] [Google Scholar]
- 81. Klocke J. A., Darlington M. V., Balandrin M. F. (1987) 1,8-Cineole (Eucalyptol), a mosquito feeding and ovipositional repellent from volatile oil of Hemizonia fitchii (Asteraceae). J. Chem. Ecol. 13, 2131–2141 [DOI] [PubMed] [Google Scholar]
- 82. Shaw J. J., Spakowicz D. J., Dalal R. S., Davis J. H., Lehr N. A., Dunican B. F., Orellana E. A., Narváez-Trujillo A., Strobel S. A. (2015) Biosynthesis and genomic analysis of medium-chain hydrocarbon production by the endophytic fungal isolate Nigrograna mackinnonii E5202H. Appl. Microbiol. Biotechnol. 10.1007/s00253-014-6206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.