Background: Pex3 is a docking factor for pexophagy receptors in yeast.
Results: A specific domain on Pex3 is responsible for interaction and activation of the pexophagy receptor Atg30 during pexophagy.
Conclusion: Pex3 regulates the initiation of pexophagy.
Significance: The peroxisomal ligand of the pexophagy receptor contributes to pexophagy signaling.
Keywords: Autophagy, Membrane Trafficking, Peroxisome, Protein-Protein Interaction, Receptor Regulation, Signaling, Pexophagy
Abstract
Pexophagy is a process that selectively degrades peroxisomes by autophagy. The Pichia pastoris pexophagy receptor Atg30 is recruited to peroxisomes under peroxisome proliferation conditions. During pexophagy, Atg30 undergoes phosphorylation, a prerequisite for its interactions with the autophagy scaffold protein Atg11 and the ubiquitin-like protein Atg8. Atg30 is subsequently shuttled to the vacuole along with the targeted peroxisome for degradation. Here, we defined the binding site for Atg30 on the peroxisomal membrane protein Pex3 and uncovered a role for Pex3 in the activation of Atg30 via phosphorylation and in the recruitment of Atg11 to the receptor protein complex. Pex3 is classically a docking protein for other proteins that affect peroxisome biogenesis, division, and segregation. We conclude that Pex3 has a role beyond simple docking of Atg30 and that its interaction with Atg30 regulates pexophagy in the yeast P. pastoris.
Introduction
Autophagy exists in yeast, plants, and mammalian cells as a homeostatic regulator for the destruction of superfluous or damaged cytosolic components or organelles, and its dysregulation has implications in many diseases (1, 2). Autophagy is referred to as a nonselective bulk degradation process (3, 4). However, many selective types of autophagy that lead to the elimination of specific organelles or protein aggregates have been described. All autophagy-related pathways share common components, which can be broken down into the following steps: phagophore assembly site (PAS)3 organization, elongation of the phagophore membrane around the cargo, vesicle formation to generate an autophagosome, fusion of the autophagosome with the vacuole (lysosome in mammals), and degradation of the cargo by vacuolar (lysosomal) hydrolases (5). Atg (autophagy-related) proteins involved in the formation of the autophagosome are considered to be the core autophagy machinery. Selective autophagy requires additional factors. Cargo selection is achieved by specific autophagy receptor proteins interacting with both the cargo and the autophagy machinery (6). Receptors have been described in yeast for the cytoplasm-to-vacuole targeting pathway (Atg19 and Atg34) and the selective autophagy of mitochondria (mitophagy; Atg32) and peroxisomes (pexophagy; Pichia pastoris Atg30 and Saccharomyces cerevisiae Atg36) (7–11). The organelle-selective autophagy receptors localize to their respective cargo surfaces and, upon phosphorylation by kinases (casein kinase 2 for Atg32 (12), Hrr25 for Atg19 (13, 14) and Atg36 (14), and an unknown kinase for Atg30), are then able to interact with the scaffold protein Atg11 and a ubiquitin-like component of the PAS and phagophore (Atg8) in a random-sequential manner (15). In addition, Atg30 interacts with other autophagy proteins at the receptor protein complex (RPC), such as the scaffold protein Atg17 and the acyl-CoA-binding protein Atg37, as well as two peroxisomal membrane proteins (PMPs), Pex3 and Pex14 (16, 17).
Peroxisomes are single membrane-bound organelles that perform essential cellular functions, such as β-oxidation of fatty acids and detoxification of reactive oxygen species. Peroxisome biogenesis factors or peroxins (encoded by PEX genes) are responsible for the biogenesis of the peroxisomal membrane and for the delivery of proteins carrying peroxisomal targeting signals (PTS1 and PTS2) to the organelle matrix (18). Biogenesis of the peroxisomal membrane requires Pex3 and Pex19 in all species and additionally requires Pex16 in mammals (19).
Pex3 has a dynamic role at the peroxisomal membrane, binding to many proteins and serving as a master regulator of peroxisome dynamics. Pex3 regulates peroxisome biogenesis by acting as a docking factor for Pex19, the protein that recognizes the membrane PTS on PMPs and facilitates their post-translational insertion into the peroxisomal membrane (20–23); as a tether for the peroxisome inheritance factor Inp1 (24, 25); as the receptor for Myo2 for peroxisome segregation in Yarrowia lipolytica (26); and as an anchor at the peroxisomal membrane for P. pastoris Atg30 and S. cerevisiae Atg36 (10, 16). Additionally, Pex3 has been implicated in pexophagy in other species, such as the methylotrophic yeast Hansenula polymorpha, in which removal of ubiquitinated Pex3 from the peroxisomal membrane induces pexophagy, and in mammals, in which Pex3 overexpression induces pexophagy through a ubiquitin-dependent mechanism (27, 28).
The crystal structure of human PEX3 has been determined with a peptide of PEX19 (29–32), elucidating the PEX3-PEX19 interaction region, which is likely similar in yeast based on a high degree of conservation of Pex3. Other highly conserved domains of PEX3 have also been described: the hydrophobic domain, which may play a role in PMP binding and peroxisome maturation, and a patch of acidic residues that do not have an assigned function (32). The Inp1-binding site on S. cerevisiae Pex3 was also determined by mutations affecting only inheritance while leaving biogenesis function intact (25). These data suggest that Pex3 has distinct regulatory domains that have been evolutionarily separated on the protein to maintain regulation of different processes.
Recently, pexophagy in higher eukaryotes has gained a lot of attention, and several breakthrough findings have emerged. A physiological association has been made between disease states and altered peroxisome turnover in primary endothelial cells, wherein impaired peroxisome homeostasis and function result from induced renal toxicity or sepsis (33). Artificial induction of pexophagy has been reported in mammalian cells by fusing ubiquitin moieties to PEX3 and PMP34 (34). NBR1 has been identified as a pexophagy receptor in mammalian cells, and SQSTM1/p62 was suggested to act as the pexophagy co-receptor (35). Interestingly, the peroxisome itself may be a signaling node for general autophagy regulation, as the peroxisome-localized (and peroxin-binding) tuberous sclerosis protein complex acts in the signaling cascade to suppress mTORC1 and induce autophagy in response to reactive oxygen species (36). The peroxisomal membrane could be the site of multiple signaling pathways.
The selective autophagy receptors in yeast (Atg19, Atg30, Atg32, and Atg36) depend on phosphoregulation for their interactions with components of the autophagy machinery, such as Atg8 and Atg11 (15). However, the upstream regulation of signaling for these receptors at the organelle membrane level is poorly understood. Both P. pastoris Atg30 and S. cerevisiae Atg36 bind Pex3, suggesting that Pex3 is a docking site at the peroxisomal membrane for both Atg30 and Atg36. Under peroxisome proliferation conditions, the synthesis of both Atg30 and Atg36 is up-regulated (15, 16). These proteins localize to the peroxisomal membrane and are then post-translationally modified under pexophagy conditions, which require at least glucose in P. pastoris and also nitrogen limitation in S. cerevisiae (10, 16). It is unclear whether the signal to degrade the peroxisome comes from the peroxisome itself. In view of the central role of Pex3 in coordinating many aspects of peroxisome dynamics, we wondered whether Pex3 might be a transducer protein that could inform the cell of nutrients or other signals regulating pexophagy. Understanding the role of Pex3 in pexophagy could provide further insight into the relationship between the targeted organelle or cargo and the selective autophagy machinery.
In this study, we report the identification of the Atg30-binding domain on P. pastoris Pex3. This domain is separate from other known protein-interacting domains implicated in peroxisome biogenesis. The Atg30-binding domain of Pex3 is not required to localize the pexophagy receptor to the peroxisome, but instead it affects the Atg30 phosphorylation status and the interaction of Atg30 with Atg11. Consequently, Atg11 is not targeted to the RPC, and the phagophore membrane (micropexophagic apparatus (MIPA)) is not formed. We conclude that Pex3 could serve a regulatory role in pexophagy rather than just as a docking factor as described previously.
EXPERIMENTAL PROCEDURES
Strains Used and Media
Yeast strains and plasmids used in this study are listed in Table 1. Media used to grow strains were as described previously (15, 16): synthetic dextrose medium lacking nitrogen (SD−N; 0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate and with 2% (w/v) glucose) and methanol medium (0.67% (w/v) yeast nitrogen base without amino acids and with 0.5% (v/v) methanol supplemented with the complete supplement mixture).
TABLE 1.
Yeast strains and plasmids used in this study
| Name | Genotype and plasmid used for construction | Source/Ref. |
|---|---|---|
| PPY12h | arg4 his4 | Ref. 39 |
| Sew1 | PPY12h Δpex3::ARG4 | Ref. 40 |
| Sjcf936 | PPY12h Δatg30::ZEOR | Ref. 41 |
| Sjcf959 | PPY12h Δatg30::KANR | Ref. 15 |
| Stn98 | Sjcf936 his4::pTW51 (PAOX1-GFP-PTS1, HIS4) | This study |
| Stn636 | PPY12h PAOX1::pPICz-BFP-PTS1 (PAOX1-BFP-PTS1, ZEOR), arg4::pJCF291 (PATG11-GFP-ATG11, ARG4) | Ref. 17 |
| Stn640 | Sjcf959 PAOX1::pPICz-BFP-PTS1 (PAOX1-BFP-PTS1, ZEOR), arg4::pJCF291 (PATG11-GFP-ATG11, ARG4) | Ref. 17 |
| Srrm197 | PPY12h Δypt7::KANR | Ref. 43 |
| Ssl12 | Sew1 PAOX1::pSJ7 (PAOX1-GFP-PTS1, ZEOR) | This study |
| Ssl13 | Ssl12 his4::pSJ40 (PPEX3-PEX3, HIS4) | This study |
| Ssl20 | Ssl12 his4::pSJ56 (PPEX3-PEX3L320P, HIS4) | This study |
| Ssfb20 | Ssl12 his4::pSFB127 (PPEX3-PEX3I152V,L320P,N325D, HIS4) | This study |
| Ssfb24 | Ssl12 his4::pSFB139 (PPEX3-PEX3N325D, HIS4) | This study |
| Ssfb28 | Ssl12 his4::pSFB145 (PPEX3-PEX3Q218H, HIS4) | This study |
| Ssfb40 | Ssl12 his4::pIB1 (HIS4) | This study |
| Ssfb52 | Sew1 his4::pSJ40 (PPEX3-PEX3, HIS4), his4::pJCF419 (PATG8-GFP-ATG8, HIS4, HYGROR), his4::pJCF401 (PGAP-BFP-PTS1, HIS4, KANR) | This study |
| Ssfb58 | Ssl12 his4::pSFB165 (PPEX3-PEX3L320P,N325D, HIS4) | This study |
| Ssfb64 | Ssl12 his4::pSFB164 (PPEX3-PEX3I152V, HIS4) | This study |
| Ssfb82 | Sew1 his4::pSFB165 (PPEX3-PEX3L320P,N325D, HIS4), his4::pJCF419 (PATG8-GFP-ATG8, HIS4, HYGROR), his4::pJCF401 (PGAP-BFP-PTS1, HIS4, KANR) | This study |
| Ssfb83 | Sew1 his4::pSJ40 (PPEX3-PEX3, HIS4), ATG30::pJCF105 (PATG30-ATG30-YFP, ZEOR), his4::pJCF401 (PGAP-BFP-PTS1, HIS4, KANR) | This study |
| Ssfb86 | Sew1 ATG30::pJCF105 (PATG30-ATG30-YFP, ZEOR), his4::pJCF401 (PGAP-BFP-PTS1, HIS4, KANR) | This study |
| Ssfb87 | Sew1 his4::pSFB165 (PPEX3-L320P,N325D, HIS4), ATG30::pJCF105 (PATG30-ATG30-YFP, ZEOR), his4::pJCF401 (PGAP-BFP-PTS1, HIS4, KANR) | This study |
| Ssfb431 | Srrm197 PEX3::pSFB202 (PPEX3-PEX3L320P,N325D, HYGROR), arg4::pJCF340 (PATG11-FLAG-ATG11, ARG4) | This study |
| Ssfb432 | Srrm197 PEX3::pSFB203 (PPEX3-PEX3, HYGROR), arg4::pJCF340 (PATG11-FLAG-ATG11, ARG4) | This study |
| Ssfb435 | Srrm197 PEX3::pSFB202 (PPEX3-PEX3L320P,N325D, HYGROR), arg4::pJCF340 (PATG11-FLAG-ATG11, ARG4), ATG30::pJCF75 (PATG30-ATG30-TAP, ZEOR) | This study |
| Ssfb436 | Srrm197 PEX3::pSFB203 (PPEX3-PEX3, HYGROR), arg4::pJCF340 (PATG11-FLAG-ATG11, ARG4), ATG30::pJCF75 (PATG30-ATG30-TAP, ZEOR) | This study |
| Y2H Gold | S. cerevisiae MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-Gal1TATA-His3, GAL2UAS-Gal2TATA-Ade2, URA3::MEL1UAS-Mel1TATA, AUR1-C MEL1 | Clontech |
| Ssfb461 | Y2H Gold trp1::pKSN65 (GAL4BD-PEX3, TRP1), leu2::pJCF226 (GAL4AD-ATG30, LEU2) | This study |
| Ssfb462 | Y2H Gold trp1::pSFB171 (GAL4BD-PEX3L320P,N325D, TRP1), leu2::pJCF226 (GAL4AD-ATG30, LEU2) | This study |
| Ssfb463 | Y2H Gold trp1::pKSN65 (GAL4BD-PEX3, TRP1), leu2::pGAD-GH (GAL4AD, LEU2) | This study |
| Ssfb464 | Y2H Gold trp1::pGBKT7 (GAL4BD, TRP1), leu2::pJCF226 (GAL4AD-ATG30, LEU2) | This study |
Yeast Two-hybrid Analysis
The GAL4-based yeast two-hybrid Matchmaker system (Clontech) was used. Full-length P. pastoris PEX3 was inserted into pGBKT7 (Gal4 DNA-binding domain), and subsequently, a QuikChange XL site-directed mutagenesis Kit (Stratagene) was used to produce the pex3 mutant plasmids. All Gal4 DNA-binding domain plasmids were cotransformed into the yeast two-hybrid Gold strain (Clontech) in a pairwise manner with full-length P. pastoris ATG30 (inserted into pGADGH, Gal4 activation domain), as shown in Fig. 3A. All strains were first selected on SD medium without Leu and Trp and then streaked in duplicate on SD medium without His, Leu, and Trp and with 2.5 mm 3-amino-1,2,4-triazole.
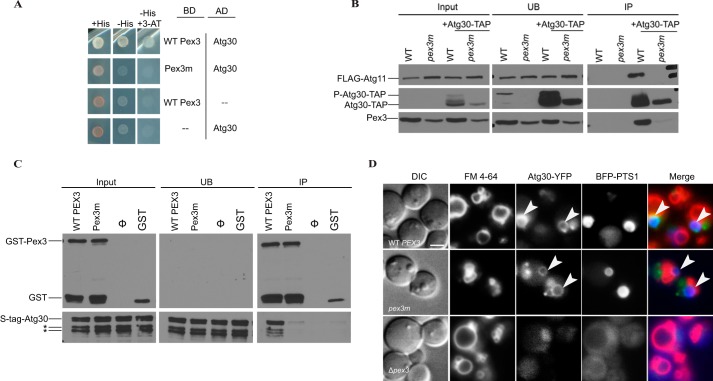
FIGURE 3.
Pex3-Atg30 interaction is not required to localize Atg30 to the peroxisome. A, yeast two-hybrid analysis for interaction between Atg30 and WT Pex3 or Pex3m. Atg30 was fused to the Gal4 activation domain (AD), and Pex3 was fused to the Gal4 DNA-binding domain (BD). Cell growth in the absence of histidine was used as a measure of protein-protein interaction. Dashes denote empty plasmids used as controls: pGBKT7 (Gal4 DNA-binding domain) and pGADGH (Gal4 activation domain). B, Δypt7 cells with endogenous PEX3 replaced with WT PEX3-HYGROR or pex3m-HYGROR were transformed with FLAG-Atg11 and Atg30-TAP (denoted as +Atg30-TAP). Immunoprecipitation (IP) was performed using IgG-agarose beads under pexophagy conditions (SD−N). Lysates of cells taken at absorbances of 0.8, 29.2, and 0.8 were loaded as input, immunoprecipitate, and unbound fractions (UB; long exposure), respectively. Immunoprecipitated proteins were visualized with anti-FLAG, anti-calmodulin-binding peptide, and anti-P. pastoris Pex3. P-Atg30 denotes the phosphorylated species. C, His-Atg30 bound to GST-Pex3ΔN, but not to GST-Pex3mΔN or GST, in vitro. There was equivalent loading in the input, unbound fraction, and immunoprecipitate (pulldown) lanes. Ø contained His-Atg30, but no GST species. Immunoblotting was performed with anti-S·Tag (to detect Atg30) and anti-GST antibodies. D, Δpex3 cells were transformed with Atg30-YFP, BFP-PTS1, and plasmids containing either WT PEX3 (upper panels) or pex3m (middle panels) and subjected to peroxisome proliferation and subsequent pexophagy conditions (SD−N, 1 h), followed by the localization of Atg30 to peroxisomal membranes (white arrows). FM 4-64 was used to label the vacuolar membrane. Δpex3 cells did not form peroxisomes, and Atg30 was cytosolic (lower panels). Scale bar = 2 μm.
Peroxisome Biogenesis and Pexophagy Assays
Peroxisome biogenesis was induced by incubation of cells in methanol medium at a starting A600 of 0.2. For pexophagy assay, cells were grown from 8 to 16 h in methanol medium and then transferred to SD−N at an A600 of 2 to induce pexophagy. One-ml samples were prepared using TCA precipitation and analyzed by Western blotting.
Fluorescence Microscopy
Pexophagy assays were performed as described above with the addition of FM 4-64 to the medium. Cells were visualized at 0 and 3 h in SD−N. For Atg30 localization and Atg8 and Atg11 studies, cells were grown as described above to induce peroxisomes and then transferred to SD−N for 0.5–1 h at an A600 of 2. GFP-Atg8 was used to label the PAS/MIPA. Quantification was performed by taking 10 Z-stack images 0.2 μm apart and deconvoluting images using Zeiss software. Microscopy parameters such as exposure time and gain were constant for all samples pictured per figure.
Co-immunoprecipitation
The Δypt7 strains expressing Atg30-TAP (calmodulin-binding peptide followed by tandem protein A) and FLAG-Atg11 (with endogenous PEX3 replaced by PEX3(HYGROR) or pex3m(HYGROR)) were used in all experiments as described previously (15, 16). Peroxisome proliferation was induced with methanol medium (8 h), and pexophagy was induced by transfer to SD−N. The relative levels of Pex3, Atg30-TAP, and FLAG-Atg11 were calculated by densitometry using NIH ImageJ (version 1.48t).
Protein Modeling
Protein modeling was done using 3D-JIGSAW software (Cancer Research UK) and human PEX3 (Protein Data Bank ID 3MK4 (30)). A Protein Data Bank format of P. pastoris Pex3 was generated and then visualized using PyMOL software.
Protein Purification
GST-Pex3ΔN was mutated to obtain GST-Pex3mΔN. The GST-Pex3ΔN and GST-Pex3mΔN proteins were purified as described (17). ATG30 from plasmid pJCF226 (16) was digested with restriction enzymes EcoRV and SalI, and the full-length ATG30 gene was ligated into pET-32b (Novagen) after digestion with the SmaI and SalI enzymes. The resulting fusion protein (His-Atg30) contained a His tag, followed by the S·Tag, at the N terminus of Atg30. This plasmid, pSFB342, was transformed into Escherichia coli BL21(DE3) cells. His-Atg30 expression was induced by adding 0.3 mm isopropyl β-d-thiogalactopyranoside for 3 h at 30 °C, and the fusion protein was purified using nickel-nitrilotriacetic acid-agarose (Qiagen). Cells were resuspended in lysis buffer (50 mm Tris-Cl (pH 7.5), 150 mm NaCl, 20 mm imidazole, and 0.05% Nonidet P-40) with lysozyme and protease inhibitors and disrupted by sonication. The 14,000 × g supernatant was incubated with the nickel-nitrilotriacetic acid-agarose for 1 h at 4 °C. The beads were washed twice with wash buffer (50 mm Tris-Cl (pH 7.5), 500 mm NaCl, 20 mm imidazole, and 0.05% Nonidet P-40), and the bound protein was collected using elution buffer (lysis buffer containing 100 mm imidazole). Pooled fractions were subjected to buffer exchange and desalted using Amicon filters with a 30-kDa molecular mass cutoff (Millipore).
In Vitro Binding
Purified recombinant proteins GST-Pex3ΔN, GST-Pex3mΔN, and His-Atg30 were quantified by Coomassie Brilliant Blue staining following SDS-PAGE and by comparing the intensities of the protein bands against a BSA standard. For the binding assay, we mixed GST-Pex3ΔN, GST-Pex3mΔN, or GST (23 μm) with His-Atg30 (81 μm) in 200 μl of binding buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 0.05% (v/v) Nonidet P-40) for 1 h at 4 °C. The His-Atg30/GST-Pex3ΔN, His-Atg30/GST-Pex3mΔN, and His-Atg30/GST reaction mixtures were used along with Atg30 alone as an additional control. For each binding assay, 50 μl of the reaction volume was taken as input, and 150 μl was incubated 1 h at 4 °C with 50 μl of prewashed glutathione-Sepharose 4B (GE Healthcare) to bind the GST and GST-tagged Pex3 species. The beads were washed three times with binding buffer at room temperature. Elution of bound proteins was done with 150 μl of 2× SDS loading buffer, and the samples were boiled for 5 min. Equivalent samples were subjected to SDS-PAGE and analyzed by immunoblotting with anti-S·Tag or anti-GST antibodies (Novagen).
RESULTS
A pex3 Mutant with Pexophagy Defects
P. pastoris Pex3 has a known interaction with Atg30 and has been suggested as a docking factor for the pexophagy receptor on the peroxisomal membrane (16). To determine the nature of the Pex3-Atg30 interaction, we used pex3 mutant strains generated from a previous laboratory screen. We rescreened the pex3 mutants for the following attributes: they expressed the full-length Pex3 protein, grew on peroxisome proliferation medium, imported PTS1 proteins, and did not degrade GFP-PTS1 by pexophagy. Two mutants were selected: pex3L320P and pex3I152V,Q218H,N325D (Fig. 1, A and B). In an effort to confine the important residues of pex3I152V,Q218H,N325D, single mutants pex3I152V, pex3Q218H, and pex3N325D were also tested. Only pex3N325D had a pexophagy phenotype (Fig. 1, A and B). Leu-320 and Asn-325 are in close proximity, and when these two residues were combined (pex3L320P,N325D), the pexophagy phenotype was exaggerated (Fig. 1, A and B).
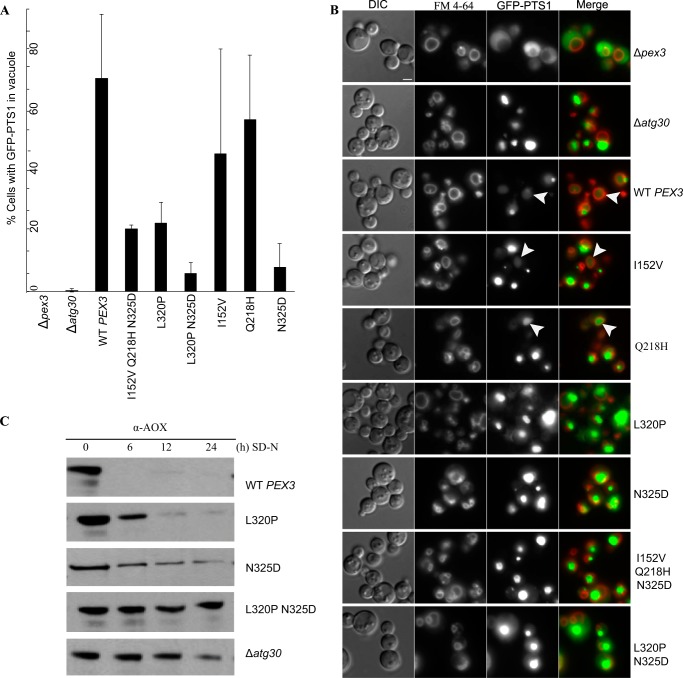
FIGURE 1.
Pex3 mutants with pexophagy defect. A, the Δpex3 strain expressing a peroxisome reporter (GFP-PTS1) was transformed with plasmids expressing either wild-type PEX3 or mutant pex3 ORFs. Strains Δpex3 and Δatg30 were used as controls. The percentage of cells that contained GFP fluorescence in the vacuole after 3 h in SD−N was estimated (mean % ± S.E. of three experiments). Quantification was done on at least 300 cells of each strain. B, representative images in A showing cells under pexophagy conditions. FM 4-64 was used to stain the vacuolar membrane. Arrowheads indicate GFP fluorescence in the vacuole. Scale bar = 2 μm. DIC, differential interference contrast. C, biochemical pexophagy assay. Cell lysates were prepared as described under “Experimental Procedures” and resolved by SDS-PAGE. Western blotting was performed with antibodies against P. pastoris AOX.
In another test of pexophagy, we examined the ability of the pex3L320P,N325D mutant, hereafter referred to as pex3m, to affect peroxisome degradation as measured by the loss of the peroxisomal matrix protein AOX after the shift of cells to pexophagy-inducing conditions. Partial degradation of AOX was seen after 24 h compared with 0-h levels in pex3L320P and pex3N325D, whereas Δatg30 and pex3m showed a complete block of pexophagy, with similar AOX levels at 0 and 24 h (Fig. 1C).
The pex3m cells had a severe block in pexophagy (Fig. 1), but not in peroxisome biogenesis, as evidenced by the presence of peroxisomes, by normal peroxisomal import of PTS1-containing proteins (Fig. 1B), and by their growth in methanol medium, which requires peroxisomal metabolism (Fig. 2A). Pex3 is an essential protein for peroxisomes biogenesis, so Δpex3 cells did not contain peroxisomes, could not import GFP-PTS1 (Fig. 1B), and could not grow in methanol medium (Fig. 2A), as expected, whereas the pex3m allele partially complemented Δpex3 cells.
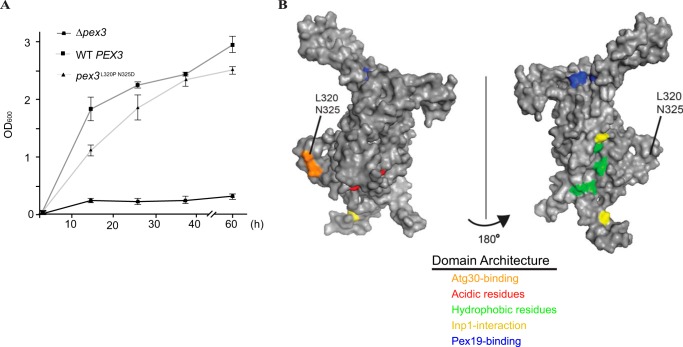
FIGURE 2.
Identification of a novel pexophagy-specific domain. A, strains Δpex3 and Δpex3 complemented with plasmids containing either WT PEX3 or pex3m were evaluated for proper peroxisome proliferation via growth in methanol medium. Mean absorbance ± S.E. from three individual experiments is presented. B, hypothetical predicted structure of P. pastoris Pex3 modeled in PyMOL based on human PEX3. The surface of the protein is visualized in gray, and protein-binding domains are shown in colors as indicated.
We used the crystal structure of the cytosolic domain of human PEX3 (29, 30) to create a hypothetical model of P. pastoris Pex3 (Fig. 2B). Leu-320 and Asn-325 are found in a structural loop that is distinct from any defined protein-interacting domains of yeast Pex3 thus far (25, 32).
Pex3 Mutants Defective in Binding Atg30
To elucidate whether Leu-320 and Asn-325 are the crux of the Pex3-Atg30 interaction domain, we analyzed Pex3m by yeast two-hybrid analysis for interaction with full-length Atg30. The WT protein maintained interaction with Atg30, whereas Pex3m no longer interacted with Atg30, suggesting the importance of these residues of Pex3 in Pex3-Atg30 interaction (Fig. 3A). Therefore, mutation of the key residues in this structural loop might affect pexophagy through the disruption of Atg30 interaction.
In vivo interaction and localization of Atg30 with Pex3m were performed to test whether or not Pex3 acts as the docking factor for Atg30, as well as to test the importance of the Leu-320- and Asn-325-containing structural loop in Pex3 as the potential domain mediating Pex3-Atg30 interaction. Verification of the yeast two-hybrid data that Pex3m interaction with Atg30 is impaired (Fig. 3A) was obtained by immunoprecipitation of the Pex3 and Pex3m proteins in P. pastoris cells defective in autophagosome-vacuole fusion (Δypt7) to accumulate Atg30 and Pex3 on cytosolic pexophagosomes (Fig. 3B). As determined by densitometry, Atg30 levels were always ∼2 times lower in pex3m than in WT PEX3. This ratio was considered during interpretation of our immunoprecipitation experiments. Furthermore, the Atg30-interacting proteins (Pex3 and Atg11) were not a limiting factor because their levels were comparable in the input and unbound fractions (Fig. 3B). Atg30 was immunoprecipitated with WT Pex3, as expected, but pulled down much less Pex3m, even when the difference in Atg30 levels between WT PEX3 and pex3m was accounted for (Fig. 3B).
To address whether Atg30 and Pex3 associate directly, an in vitro interaction assay of His-Atg30 with GST-Pex3ΔN, GST-Pex3mΔN, and GST was performed using purified recombinant proteins. His-Atg30 interacted with GST-Pex3ΔN, but not with GST-Pex3mΔN or GST alone (Fig. 3C). This clearly indicates that this interaction is direct and that the pex3m mutation disrupts the binding with Atg30.
Peroxisomal Localization of Atg30 Is Unaffected in pex3m Cells
In WT cells, Atg30 targets to the peroxisomal membrane, where it interacts with Pex3 (16). As interaction is impaired between Atg30 and Pex3m, Atg30 localization on the peroxisomal membrane was studied by fluorescence microscopy with genomically tagged Atg30-YFP. Atg30 localization was unaffected by pex3m, as seen by co-localization of Atg30-YFP with peroxisomal blue fluorescent protein (BFP)-tagged PTS1 in 48 ± 6% of WT cells and 56 ± 7% of pex3m cells (p is non-significant) (Fig. 3D, white arrowheads). The Pex3-Atg30 interaction is not essential for the peroxisomal localization of Atg30, as the localization of Atg30 was similar in WT and pex3m cells.
Atg30 Is Differentially Phosphorylated in pex3m Cells
The ability of Atg30-YFP to localize to the peroxisomal membrane in pex3m cells as efficiently as in WT cells was an unexpected result. Pex3 has been postulated to be the peroxisomal docking factor for Atg30 not only because of their interaction, but also because of the altered localization of Atg30 in Δpex3 cells (16). Atg30 is massively phosphorylated by an unknown kinase as a trigger for peroxisome degradation. It is differentially phosphorylated at Ser-71 to regulate Atg8 binding and at Ser-112 to modulate its interaction with Atg11 (15). It is also known that Atg30 is hypophosphorylated in Δpex3 cells (16); however, the rational explanation for this phenotype was the mislocalization of Atg30. Interestingly, in pex3m cells, where Atg30 is properly localized, it was also hypophosphorylated (Fig. 4A). These results suggest that efficient Pex3-Atg30 interaction is required to induce the correct phosphorylation of Atg30, which is necessary for pexophagy.
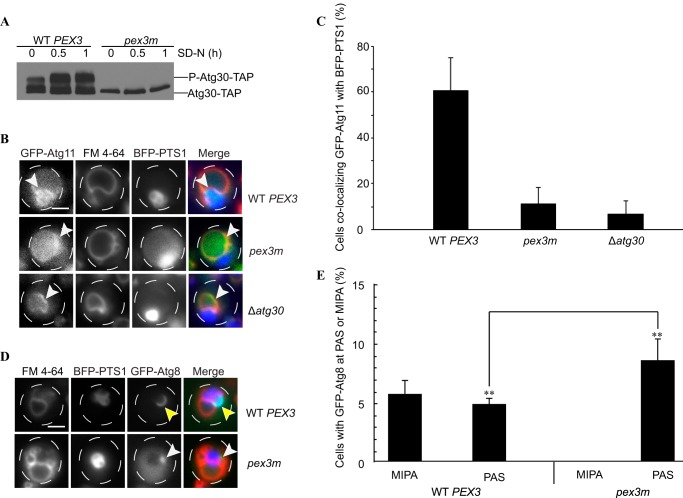
FIGURE 4.
pex3m affects Atg30 phosphorylation, Atg11 localization, and phagophore formation. A, Δypt7 (WT PEX3-HYGROR or pex3m-HYGROR) cells containing Atg30-TAP were followed after peroxisome induction to monitor the phosphorylation status of Atg30 upon switching the medium to SD−N. P-Atg30-TAP denotes the phosphorylated Atg30 species. B, WT PEX3-HYGROR, pex3m-HYGROR, and Δatg30 cells containing GFP-Atg11, BFP-PTS1, and FM 4-64 were visualized after switching to SD−N (1 h). White arrowheads indicate GFP-Atg11 localization. Scale bar = 2 μm. C, cells from B were quantified for GFP-Atg11 accumulation in the interface of the vacuole (FM 4-64) and peroxisomes (BFP-PTS1) (mean % of three experiments ± S.E.). D, Δpex3 cells were transformed with plasmids containing either WT PEX3 or pex3m along with GFP-Atg8 and BFP-PTS1 to monitor phagophore (MIPA) formation (yellow arrowheads) or PAS organization (white arrowheads) upon induction of pexophagy. Scale bar = 2 μm. E, MIPA and PAS structures from D were quantified (mean % of three experiments ± S.E.). **, p < 0.001.
Impaired Recruitment of Atg11 to the RPC in pex3m Cells
In the absence of Atg11, the recruitment of the core autophagy machinery is impeded, as Atg11 is required for the organization of the PAS for selective autophagy pathways (16, 37, 38). Phosphorylation of Atg30 at Ser-112 is required for its interaction with Atg11 (15, 16), and as Atg30 is hypophosphorylated in pex3m cells, we postulated that Atg30-Atg11 interaction might be impaired in the pex3m background. When either WT or pex3m lysates coexpressing Atg30-TAP and FLAG-Atg11 were used for co-immunoprecipitation studies, we observed no interaction between Atg30-TAP and FLAG-Atg11 in pex3m cells, whereas WT cells maintained a strong interaction, even when the difference in Atg30 levels between WT PEX3 and pex3m was accounted for (Fig. 3B).
Atg11 is normally found at the PAS and vacuole membrane in close proximity to the targeted peroxisome after re-localization from a diffuse location around the vacuole prior to activation of pexophagy (42). In pex3m cells, GFP-Atg11 maintained solely vacuole membrane fluorescence, similar to that observed at the onset of pexophagy in Δatg30 cells, and did not re-localize to the vacuole-sequestering membranes or accumulate at the base of the targeted peroxisome, as often as seen in WT cells (61 ± 14% in WT PEX3, 11 ± 7% in pex3m, and 7 ± 6% in Δatg30) (Fig. 4, B and C). Therefore, Pex3 could play an indirect role in the recruitment of Atg11 to the pexophagic RPC.
Phagophore Formation Is Affected in pex3m Cells
We also wondered how the mislocalization of Atg11 in pex3m cells affects the downstream process of phagophore formation. In WT cells, GFP-Atg8 accumulates at the PAS, which is a punctate structure at the vacuole rim in proximity to the peroxisome. During pexophagy, the transient PAS extends into the phagophore (also known as MIPA). Fluorescence microscopy images revealed that pex3m cells expressing BFP-PTS1 and GFP-Atg8 had GFP-Atg8 localized properly at the pexophagy-specific PAS (Fig. 4D). However, the MIPA did not surround targeted peroxisomes in pex3m cells, as opposed to WT cells. The MIPA and PAS were each present in ∼5% of WT cells, whereas pex3m cells accumulated only the PAS (8.5% of cells) as determined by Z-stack analysis (Fig. 4E), with p < 0.001. Therefore, the lack of Atg11 at the RPC in pex3m cells likely leads to defective phagophore formation.
DISCUSSION
We have reported the identification of a distinct Atg30-binding domain in Pex3 that modulates the phosphorylation of Atg30 and the recruitment of Atg11 by Atg30 to the RPC in P. pastoris cells. We highlighted that Pex3 has signaling roles beyond just docking of the pexophagy receptor at the peroxisomal membrane.
Previous studies concluded that in the absence of Pex3, Atg30 would be mislocalized (16). However, those studies shed no light on the nature or physiological role of the interaction of Atg30 with Pex3. Our findings reveal a specific interaction domain of Pex3 that binds directly to Atg30. Additionally, we have shown that post-translational modifications of Atg30 that trigger the onset of pexophagy in yeast can be disrupted by mutations in this interaction domain of Pex3. This is evident when Atg30, which is hypophosphorylated in pex3m cells, fails in the subsequent pexophagy-related interaction with Atg11.
Atg30-YFP properly localized to the peroxisomal membrane in pex3m cells, despite the lack of interaction between Atg30 and Pex3m as judged by co-immunoprecipitation, in vitro binding with recombinant proteins, and yeast two-hybrid studies. This shows that Pex3 is not the sole protein on the peroxisomal membrane that docks Atg30. Our previous study showed that Atg30 also interacts with the peroxisomal integral membrane protein Atg37 (17). Because Pex3 is needed for peroxisome biogenesis, previous studies using Δpex3 cells might have missed additional PMPs, like Atg37, that recruit Atg30 to the peroxisomes.
Pex3 is positioned to be a signaling platform for the peroxisome, as it is a PMP that interacts with many proteins involved in peroxisome biogenesis and dynamics. Atg30 is highly phosphorylated during pexophagy, but is likely kept in an inactive state during peroxisome proliferation. During pexophagy, activation of an unknown factor or signal that depends on Pex3-Atg30 interaction leads to the phosphorylation of the residues on Atg30 responsible for Atg11 interaction.
How might Pex3 regulate the phosphorylation status of Atg30? During pexophagy, (i) a conformational change in Atg30, mediated by Pex3, could expose phosphorylation sites to the kinase(s) to activate Atg30; or (ii) Pex3 might act directly or indirectly as a docking site for kinase(s) that may phosphorylate Atg30 (Fig. 5). The kinase(s) and phosphatase(s) responsible for coordinated phosphorylation events on Atg30 have not been discovered yet, and we are in active pursuit. We conclude that peroxisomal Pex3 is more than just a docking factor, and it would be interesting to determine whether other selective autophagy receptors are post-translationally regulated by the organelle-specific protein ligands to control the organelle fate.
FIGURE 5.
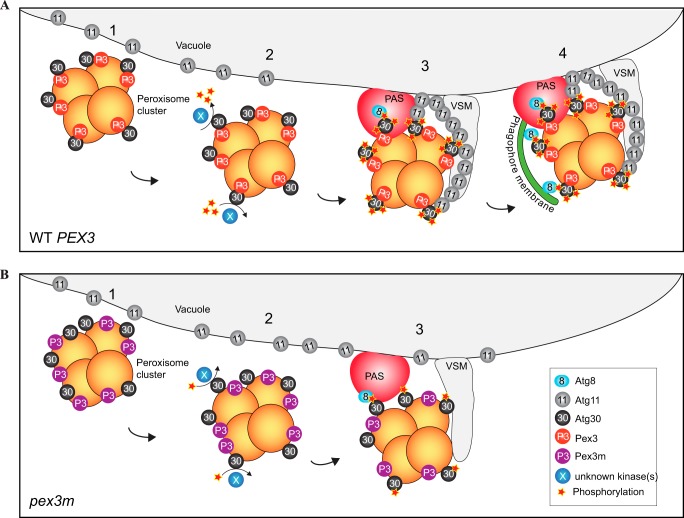
Model for Atg30 activation by Pex3. A, in WT cells, Atg30 and Pex3 localize at the peroxisomal membrane (step 1). After pexophagy induction, Atg30 is phosphorylated by an unknown kinase(s) in a Pex3-Atg30 interaction-dependent manner (step 2). Atg8 and Atg11 interact with phosphorylated Atg30, leading to PAS formation and Atg11 accumulation at the interface between peroxisomes and vacuolar membranes (step 3). The phagophore (decorated by Atg8) extends from the PAS and engulfs the peroxisome cluster (step 4) to allow for fusion with the vacuolar membranes and peroxisome degradation inside the vacuole. B, in pex3m cells, Atg30 is targeted to the peroxisome in a Pex3-independent manner (step 1). Atg30 is not properly phosphorylated (step 2). Despite being hypophosphorylated, Atg8 is still recruited to the PAS, yet Atg11 does not accumulate in the proximity of the peroxisome cluster (step 3).
Acknowledgments
We thank all current laboratory members for critical discussion and review of manuscript and our previous laboratory member Dr. Soojin Lee for generating the pex3 mutants used in Fig. 1. We thanks Gaurav Agrawal for contributions to Fig. 5.
This work was supported, in whole or in part, by National Institutes of Health Grant GM069373 (to S. S.) and Mentored Research Scientist Development Award K01DK094843 (to T. Y. N.).
- PAS
- phagophore assembly site
- RPC
- receptor protein complex
- PMP
- peroxisomal membrane protein
- PTS
- peroxisomal targeting signal
- MIPA
- micropexophagic apparatus
- TAP
- tandem affinity purification
- BFP
- blue fluorescent protein.
REFERENCES
- 1. Tan C. C., Yu J. T., Tan M. S., Jiang T., Zhu X. C., Tan L. (2014) Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol. Aging 35, 941–957 [DOI] [PubMed] [Google Scholar]
- 2. Feng Y., He D., Yao Z., Klionsky D. J. (2014) The machinery of macroautophagy. Cell Res. 24, 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 4. Yang Z., Klionsky D. J. (2009) An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Till A., Lakhani R., Burnett S. F., Subramani S. (2012) Pexophagy: the selective degradation of peroxisomes. Int. J. Cell Biol. 2012, 512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johansen T., Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 10. Motley A. M., Nuttall J. M., Hettema E. H. (2012) Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31, 2852–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki K., Kondo C., Morimoto M., Ohsumi Y. (2010) Selective transport of α-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 285, 30019–30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanki T., Kurihara Y., Jin X., Goda T., Ono Y., Aihara M., Hirota Y., Saigusa T., Aoki Y., Uchiumi T., Kang D. (2013) Casein kinase 2 is essential for mitophagy. EMBO Rep. 14, 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfaffenwimmer T., Reiter W., Brach T., Nogellova V., Papinski D., Schuschnig M., Abert C., Ammerer G., Martens S., Kraft C. (2014) Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 15, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka C., Tan L. J., Mochida K., Kirisako H., Koizumi M., Asai E., Sakoh-Nakatogawa M., Ohsumi Y., Nakatogawa H. (2014) Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 207, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farré J. C., Burkenroad A., Burnett S. F., Subramani S. (2013) Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farré J. C., Manjithaya R., Mathewson R. D., Subramani S. (2008) PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nazarko T. Y., Ozeki K., Till A., Ramakrishnan G., Lotfi P., Yan M., Subramani S. (2014) Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J. Cell Biol. 204, 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma C., Agrawal G., Subramani S. (2011) Peroxisome assembly: matrix and membrane protein biogenesis. J. Cell Biol. 193, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinto M. P., Grou C. P., Alencastre I. S., Oliveira M. E., Sá-Miranda C., Fransen M., Azevedo J. E. (2006) The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J. Biol. Chem. 281, 34492–34502 [DOI] [PubMed] [Google Scholar]
- 20. Snyder W. B., Faber K. N., Wenzel T. J., Koller A., Lüers G. H., Rangell L., Keller G. A., Subramani S. (1999) Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell 10, 1745–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hettema E. H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. (2000) Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang Y., Morrell J. C., Jones J. M., Gould S. J. (2004) PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hattula K., Hirschberg D., Kalkkinen N., Butcher S. J., Ora A. (2014) Association between the intrinsically disordered protein PEX19 and PEX3. PLoS ONE 9, e103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munck J. M., Motley A. M., Nuttall J. M., Hettema E. H. (2009) A dual function for Pex3p in peroxisome formation and inheritance. J. Cell Biol. 187, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R. L., Rachubinski R. A. (2013) An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang J., Mast F. D., Fagarasanu A., Rachubinski D. A., Eitzen G. A., Dacks J. B., Rachubinski R. A. (2009) Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J. Cell Biol. 187, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams C., van der Klei I. J. (2013) Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 438, 395–401 [DOI] [PubMed] [Google Scholar]
- 28. Yamashita S., Abe K., Tatemichi Y., Fujiki Y. (2014) The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato Y., Shibata H., Nakatsu T., Nakano H., Kashiwayama Y., Imanaka T., Kato H. (2010) Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J. 29, 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt F., Treiber N., Zocher G., Bjelic S., Steinmetz M. O., Kalbacher H., Stehle T., Dodt G. (2010) Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J. Biol. Chem. 285, 25410–25417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schueller N., Holton S. J., Fodor K., Milewski M., Konarev P., Stanley W. A., Wolf J., Erdmann R., Schliebs W., Song Y. H., Wilmanns M. (2010) The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. EMBO J 29, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt F., Dietrich D., Eylenstein R., Groemping Y., Stehle T., Dodt G. (2012) The role of conserved PEX3 regions in PEX19-binding and peroxisome biogenesis. Traffic 13, 1244–1260 [DOI] [PubMed] [Google Scholar]
- 33. Vasko R., Ratliff B. B., Bohr S., Nadel E., Chen J., Xavier S., Chander P., Goligorsky M. S. (2013) Endothelial peroxisomal dysfunction and impaired pexophagy promotes oxidative damage in lipopolysaccharide-induced acute kidney injury. Antioxid. Redox Signal 19, 211–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. (2008) Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deosaran E., Larsen K. B., Hua R., Sargent G., Wang Y., Kim S., Lamark T., Jauregui M., Law K., Lippincott-Schwartz J., Brech A., Johansen T., Kim P. K. (2013) NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., Kim J., Alexander A., Cai S., Tripathi D. N., Dere R., Tee A. R., Tait-Mulder J., Di Nardo A., Han J. M., Kwiatkowski E., Dunlop E. A., Dodd K. M., Folkerth R. D., Faust P. L., Kastan M. B., Sahin M., Walker C. L. (2013) A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 15, 1186–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. (2011) Phosphorylation of Ser114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. (2005) Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16, 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. (1992) Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast 8, 613–628 [DOI] [PubMed] [Google Scholar]
- 40. Wiemer E. A., Lüers G. H., Faber K. N., Wenzel T., Veenhuis M., Subramani S. (1996) Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Biol. Chem. 271, 18973–18980 [DOI] [PubMed] [Google Scholar]
- 41. Nazarko T. Y., Farré J. C., Subramani S. (2009) Peroxisome size provides insights into the function of autophagy-related proteins. Mol. Biol. Cell 20, 3828–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr., Klionsky D. J. (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manjithaya R., Jain S., Farré J. C., Subramani S. (2010) A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J. Cell Biol. 189, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]