Background: Even though ceramide selectively induces apoptosis in multiple cancer models, the mechanisms by which ceramide limits cancer metastasis are unknown.
Results: The ceramide nanoliposome (CNL) inhibits cancer cell metastasis by inducing anoikis and inhibiting extravasation via lysosomal degradation of CD44.
Conclusion: Ceramide reduces the metastatic potential of multiple cancer cell models.
Significance: CNL may have therapeutic utility in preventing and treating metastatic cancers.
Keywords: Anoikis, Breast Cancer, CD44, Ceramide, Metastasis, Carcinoma, Extravasation
Abstract
The ceramide nanoliposome (CNL) has shown promise in being able to treat a variety of primary tumors. However, its potential for treating metastatic cancer remains unknown. In this study, we demonstrate that CNL increases anoikis while preventing cancer cell extravasation under both static and physiological fluid flow conditions. Mechanistically, CNL limits metastases by decreasing CD44 protein levels in human breast and pancreatic cancer cells via lysosomal degradation of CD44, independent of palmitoylation or proteasome targeting. siRNA down-regulation of CD44 mimics CNL-induced anoikis and diminished extravasation of cancer cells. Taken together, our data indicate that ceramide limits CD44-dependent cancer cell migration, suggesting that CNL could be used to prevent and treat solid tumor metastasis.
Introduction
200,000 new cases of breast cancer are diagnosed in the United States each year (1). In such cases, the primary tumors can typically be surgically resected, but once metastasis to other organs occurs, the disease becomes incurable. Current treatments for metastatic breast cancer include drugs that target receptors that are overexpressed in breast cancer, including the estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2. However, effective treatments to limit the metastatic potential of triple negative breast cancers are presently not available.
Cancer metastasis has been described as a series of events termed the “metastatic cascade” (2). The first step of the metastatic cascade involves cell detachment from the tumor. Metastatic cells then secrete proteases to degrade the extracellular matrix and migrate toward blood vessels, which tend to be leaky near tumors. Metastatic cells then invade between endothelial cells to enter the bloodstream. To survive in the bloodstream, metastatic cancer cells have to become resistant to anoikis, which is programmed cell death caused by the detachment of epithelial cells from the extracellular matrix. Even if tumor cells are able to overcome anoikis and survive in the bloodstream, they still have to exit the bloodstream via extravasation before they can form a secondary tumor. The process of extravasation also involves protease degradation of endothelial cell-cell junctions. Finally, tumor cells must survive in a foreign microenvironment and proliferate to form a secondary tumor. In this paper, we will examine the ability of ceramide to regulate the metastasis-related processes of anoikis and extravasation.
CD44 is a transmembrane adhesion molecule that has been implicated in metastasis. It is found at high levels in breast cancer (3), melanoma, and pancreatic cancer (4). Extracellularly, CD44 can bind endothelial cell surface molecules, such as E-selectin (5), as well as components of the extracellular matrix, most notably hyaluronic acid (3). Intracellularly, CD44 binds ezrin, facilitating cancer cell migration and metastasis (6).
Ceramide, a sphingolipid that plays a structural role in cell membranes and acts as a second messenger, induces apoptosis in a wide variety of cell types, although on an equimolar basisless ceramide is needed to induce apoptosis in cancer cells (7, 8). We use a short chain version of ceramide (C6 ceramide) that has been encapsulated in a liposome to effectively deliver such a hydrophobic entity to cells both in vitro and in vivo. We call this nanoscale drug the ceramide nanoliposome (CNL).2 Previous studies using the MDA-MB-231 cell line used herein demonstrated that 5–50 μm CNL inhibited breast cancer cell viability while having no effect on normal mammary epithelial cells (9). In addition, intravenous injection of 36 mg/kg CNL every other day inhibited tumor growth in mouse models of breast cancer (9). Furthermore, CNL blocked neurotensin-induced breast cancer cell migration and matrix metallopeptidase secretion (10). Given the ability of CNL to induce apoptosis in cancer cells and to inhibit neurotensin-induced breast cancer cell migration, we hypothesized that CNL would be able to induce anoikis and inhibit extravasation in metastatic carcinoma cells, including breast cancer, pancreatic cancer, and melanoma cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Treatments
MDA-MB-231 human breast cancer cells were cultured in DMEM (Cellgro) containing 10% FBS (Atlanta Biologicals), 1% GlutaMAX (Gibco), and 1% antibiotic-antimycotic (Invitrogen) at 37 °C and 5% CO2. MDA-MB-468 human breast cancer cells were cultured in DMEM/F12 (Cellgro) containing 10% FBS and 1% antibiotic-antimycotic at 37 °C and 5% CO2. T47D human breast cancer cells, Panc-1 human pancreatic cancer cells, and B16 murine melanoma cells were cultured in RPMI 1640 (Cellgro) containing 10% FBS (Atlanta Biologicals) and 1% antibiotic-antimycotic (Invitrogen) at 37 °C and 5% CO2. MDA-MB-231 (human breast adenocarcinoma) cells were obtained from American Type Culture Collection. EI cells are fibroblast L-cells that overexpress ICAM-1 and E-selectin. They were provided by Dr. Scott Simon (UC Davis, Davis, CA). Ceramide and all other lipids were purchased from Avanti Polar Lipids. The PBS was from Cellgro, and the Z-VAD-fmk caspase inhibitor and pepstatin A were from Enzo Life Sciences. Scrambled siRNA and validated CD44 siRNA were from Invitrogen. 2-Bromopalmitate, chloroquine, E64D, ammonium chloride, and trypan blue were purchased from Sigma-Aldrich.
Liposome Formulation and Extrusion
Lipids, dissolved in chloroform (CHCl3), were combined according to the following molar ratios: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine:1,2-distearyl-sn-glycero-3-phosphocholine:C8 mPEG 750 ceramide:1,2distearyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethelene glycol)-2000] (ammonium salt):C6 ceramide (1.75:3.75:0.75:0.75:3). Ceramide-free liposomes (ghost) containing the same ratio of lipid components were used as a negative lipid control. Lipid mixtures were dried under a stream of nitrogen gas, hydrated with PBS, and then heated above lipid transition temperatures. The resulting solution underwent sonication for 1 min followed by 11 extrusions through a 100-nm polycarbonate membrane. The amount of total lipids in C6 ceramide and ghost liposomal formulations determined by tandem mass spectrometry remained constant in all experiments.
MTS Cell Viability Assay
Cytotoxicity of nanoliposomal ceramide in breast cancer cells was measured by plating 5,000 (MDA-MB-468) or 10,000 (MDA-MB-231) cells into 96-well plates followed by growth for 24 h in a humidified 37 °C cell culture incubator. Next, cells were treated with ghost liposomes or CNL for 24 h in 10% serum medium. After 24 h, cytotoxicity was measured using the CellTiter 96 aqueous nonradioactive cell proliferation assay (Promega).
Anoikis Assay
10,000 cells/well were plated into either cell culture-treated or ultra low adherent 96-well plates (Corning) and incubated at 37 °C and 5% CO2 overnight. Cells were then treated with ghost liposomes and CNL for 24 h, after which time they were subjected to the Apo-ONE caspase 3/7 assay (Promega) per the manufacturer's protocol to measure apoptosis.
Overexpression of CD44
The retroviral backbones encoding GFP (control) or CD44s and a blasticidin selection marker (obtained from Addgene) were transfected with Lipofectamine 3000 into HEK293 cells growing on 100-mm dishes together with packaging plasmids GAG-Pol and VSV-G. Forty-eight hours later, culture medium was harvested and passed through a 0.45-micron filter to remove cell debris. Viral stocks were used undiluted right away or stored at −80 °C. T47D cells were plated on 60-mm plates at a density of 10,000/cm2. The next day, retroviral stocks were thawed, and Polybrene (Sigma) was added to them at a final concentration of 8 μg/ml. Growth media was removed from the T47D plates and replaced with ∼1.75 ml of retroviral stock with Polybrene per plate. Plates were sealed with Parafilm, placed in a tabletop centrifuge equipped with a backed rotor, and centrifuged at 25 °C for 45 min at 1220 × g. After centrifugation, 5 ml of fresh growth medium was added to the cells. The medium was changed 3 h thereafter or the next morning. 3–4 days after infection, the cells were transferred to growth media containing 10 μg/ml blasticidin for 7 days. Based on our previous experience, all noninfected T47D cells died at this time; however, with virus-infected cells, we saw numerous surviving colonies. These colonies were pooled, and the resulting cells were kept in 5 μg/ml blasticidine for an additional week.
Transfection of siRNA
100 nm siRNA was transfected into MDA-MB-231 cells using Nucleofection (Lonza) following the manufacturer's cell line-specific protocol. 48 h after transfection, CD44 levels were measured, and Transwell migration, extravasation, and anoikis assays were performed.
Mammosphere Assay
5,000 cells/well were grown in 6-well ultra low adherent plates (Corning) in 2 ml of growth medium. Mammospheres were counted using a Nikon Eclipse phase contrast microscope. Trypan blue was used to determine cell viability. Living cells exclude Trypan blue and remain clear, whereas dead cells take up the Trypan blue dye and turn blue.
Transwell Cell Migration Assay
The Transwell cell migration assay was performed using 24-well Transwell inserts (Greiner Bio-one). Cells were serum-starved overnight before the experiment. 200,000 MDA-MB-231 cells in 200 μl of serum-free medium containing 0.002% BSA were added to the upper well of the Transwell chamber. 600 μl of medium with or without chemoattractant (10% FBS) was added to the lower chamber. Cells were allowed to settle for half an hour before being treated. After incubation for 6 or 24 h at 37 °C, all cells were stained with calcein AM (BD Biosciences). Cells in the lower chamber were then detached from the Transwell membrane using trypsin, after which time the cells from the lower chamber (i.e. those that had migrated) were transferred to a black 96-well plate, and fluorescence was measured with an excitation wavelength of 485 nm and an emission wavelength of 520 nm using a fluorescent plate reader. Increased fluorescence corresponds to more cells.
Flow Chamber Extravasation Assay
The in vitro microfluidic device used to simulate cellular extravasation under dynamic conditions consisted of a modified chemotactic Boyden chamber. The bottom piece of the chamber was a 48-well acrylic plate (Neuro Probe, Gaithersburg, MD), and the top piece was an acrylic plate with an inlet and an outlet for flow media. The two plates were separated by a 0.02-inch thick silicone gasket (PharmElast; Trelleborg, Hudson, MA). A 7-cm × 2-cm opening was cut in the center of the gasket to form the flow field. A monolayer of EI cells was grown to confluence on one side of a sterilized polyvinylpyrrolidone-free polycarbonate filter (8 μm pore size; Neuro Probe, Gaithersburg, MD) that was coated with fibronectin (30 μg/ml, 3 h) (BD Discovery Labware, Bedford, MA). The other side of the filter was scraped before placing it in the chamber to remove EI cells that may have grown on the bottom side of the filter. The center 12 wells of the bottom plate were filled with soluble type IV collagen (100 μg/ml in DMEM, 0.1% BSA) (BD Discovery Labware, Bedford, MA). The rest of the wells were filled with medium (DMEM, 0.1% BSA). The cells being studied (untreated, ghost, CNL, siScr, or siCD44) (500,000 cells) were mixed in the flow medium (DMEM, 0.1% BSA), the chamber was placed in an incubator (37 °C, 4 h), and the medium was circulated through the chamber at the desired shear rate (50 or 100 s−1). The volumetric flow rate (Q) was related to the wall shear stress (τW = 6μQ/wh2), where μ is the fluid viscosity, h is height, and w is width of the flow field. The filter was removed from the chamber and stained with HEMA-3 (Fisher Scientific, Pittsburgh, PA). The filter was then attached to a microscope slide, and the top side was scraped to remove the EI cell monolayer. The migrated cells were then quantified in five different locations (mm2) and averaged for each slide. At least three slides were analyzed for each case.
Western Blots
Whole cell lysates were isolated using Nonidet P-40 lysis buffer as previously described (11). Protein quantification was completed using the DC protein assay (Bio-Rad). Protein samples were prepared by heating at 70 °C for 10 min after the addition of denaturing sample buffer. Proteins were separated using SDS-PAGE on a 4–12% gel (Life Technologies) and transferred to a nitrocellulose membrane (General Electric). Antibodies were diluted in 5% BSA in Tris-buffered saline/Tween-20. After 1 h of blocking in 5% BSA, membranes were incubated with the primary antibody, washed, incubated with the horseradish peroxidase-conjugated secondary antibody, and then washed again. Protein bands were visualized using a commercially available chemiluminescence kit (Thermo Scientific). The following antibodies were used: CD44 (catalog no. 3570; Cell Signaling), caveolin-1 (catalog no. sc-7875; Santa Cruz), EGF receptor (catalog no. 4267; Cell Signaling), transferrin receptor (catalog no. 13-6800; Life Technologies), goat anti-mouse (catalog no. sc-2005; Santa Cruz), goat anti-rabbit (catalog no. sc-2004; Santa Cruz), and β-actin (catalog no. A-5441; Sigma).
RT-PCR
Real time PCR was performed using TaqMan gene expression assay (Life Technologies) on an ABI 7900HT qPCR instrument. The PCR consisted of 0.5 μl of 20× TaqMan gene expression assay, 5 μl of 2× TaqMan gene expression master mix, and 5 μl of cDNA template (100 ng) in a total volume of 10.5 μl. The assay included a negative control that lacked any reverse transcriptase, as well as each of the test cDNAs.
Immunofluorescence
The cells were fixed in 4% paraformaldehyde, washed in PBS, permeabilized in 0.1% Triton X-100, and incubated in 0.5% SDS for antigen retrieval. The mouse monoclonal CD44 antibody (catalog no. 3570; Cell Signaling; 1:400) was then applied overnight at 4 °C, followed by an AlexaFluor 546 goat-anti-mouse secondary antibody (catalog no. A-11030; Molecular Probes; 1:500) for 2 h at room temperature. The Nuclei were stained with Hoechst 34580, and the slides were permanently mounted. For negative controls, the primary antibody was omitted. Immunostained cells were then imaged using a confocal microscope (TCS SP8, Leica) with a 63× objective. Images were processed and quantified using Imaris 3D-4D image analysis software (Bitplane). At least three cells were imaged per treatment.
Statistical Analysis
The data are presented as means ± S.E. Statistical analysis between two samples was performed using the Student's t test. Comparisons of more than two groups were performed using one-way analysis of variance with Bonferroni's correction. A p value of p < 0.05 was considered to be statistically significant, and the experiments were repeated three times. To analyze the interaction between CNL and siCD44 on cell migration, a two-way analysis of variance model was set up. This part of the analysis was performed using statistical software SAS version 9.3 (SAS Institute, Cary, NC). The significance level used was 0.05. For the anoikis experiments, a two-way analysis of variance model was used to compare the mean activity levels among the groups (adherent/nonadherent) and treatments (no treatment, ghost, and CNL). Their interaction was examined by an interaction plot. Pair-wise comparisons were performed to examine the interaction between treatment and group using Tukey's test.
RESULTS
The Ceramide Nanoliposome Induces Anoikis in Carcinoma Cells
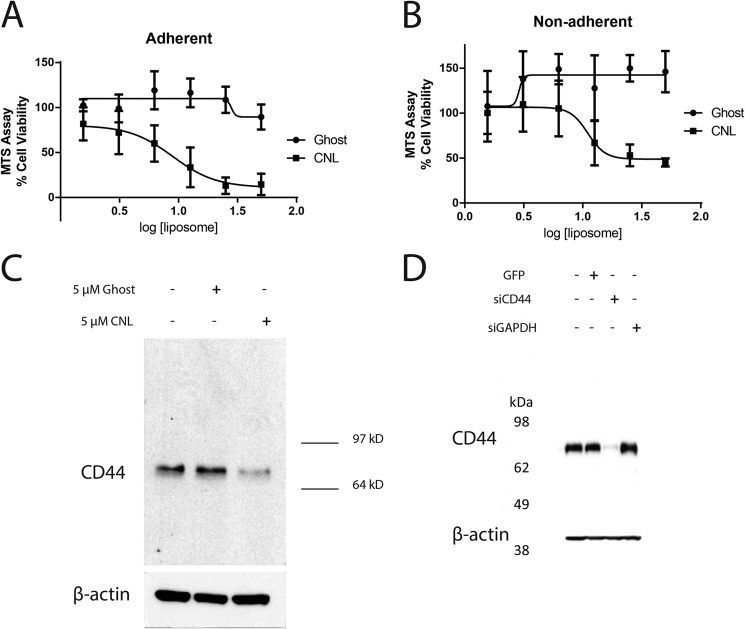
Initially, an MTS assay revealed a cytotoxic IC50 of 8.93 μm (95% CI: 6.12–13.0) for CNL when administered to MDA-MB-231 triple negative breast cancer cells under adherent conditions (Fig. 1A). Under the same conditions, ghost liposomes (negative control) had no effect on cell viability (Fig. 1A). Under nonadherent conditions, CNL inhibited MDA-MB-231 cell viability with an IC50 of 11.0 μm (95% CI: 6.39–19.1) (Fig. 1B). Again, ghost liposomes did not affect cell viability (Fig. 1B). Under nonadherent conditions, cells may have more surface area exposed to drug. However, the IC50 values that we obtained under adherent and nonadherent conditions (8.93 and 11.0 μm, respectively) are not significantly different (p = 0.463) as determined by a four-parameter model. In addition, mass spectrometry now confirms that a similar amount of ceramide is reaching both adherent and nonadherent cells, despite differences in surface area (data not shown). Therefore, we conclude that any difference in surface area between adherent and nonadherent cells does not affect the ability of CNL to target these cells.
FIGURE 1.
Effects of CNL on MDA-MB-231 cells. A, MDA-MB-231 cells were treated with 1.56–50 μm ghost liposomes or CNL for 24 h under adherent conditions, followed by MTS assay for cell viability. The IC50 of CNL was 8.93 μm. n = 4. Bar, S.E. B, MDA-MB-231 cells were treated with 1.56–50 μm ghost liposomes or CNL for 24 h under nonadherent conditions, followed by MTS assay for cell viability. The IC50 of CNL was 11.0 μm. n = 3. Bars, S.E. C, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL for 24 h, followed by Western blot analysis of CD44 and β-actin (loading control). D, siRNA results in 97% knockdown of CD44s in MDA-MB-231 cells. Cells were treated with 100 nm GAPDH siRNA or CD44 siRNA for 48 h, followed by Western blot analysis of CD44.
Subsequently, a lower concentration (5 μm) of CNL was used to investigate the ability of CNL to regulate metastatic processes in MDA-MB-231 cells. Because CD44 levels have been associated with metastasis (12), we attempted to determine whether CNL regulates CD44. Indeed, treatment with 5 μm CNL for 24 h significantly reduced CD44 levels in MDA-MB-231 breast cancer cells (Fig. 1C). Note that there is only one visible band at ∼80 kDa, which is the size of the standard form of CD44 (CD44s). CD44 variants would have a higher molecular weight, whereas cleaved CD44 would have a lower molecular weight. Thus, our data indicate that CNL targets CD44s. We next utilized siRNA against CD44s to mimic the effects of CNL. We were able to achieve 97% knockdown of CD44s using siRNA (Fig. 1D).
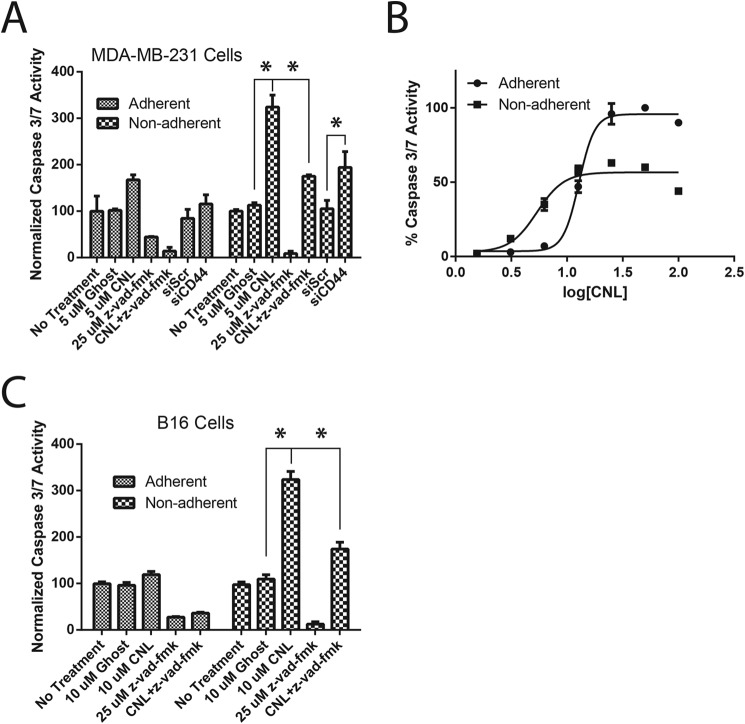
Next, we investigated the ability of CNL to induce anoikis in MDA-MB-231 breast cancer cells. Metastatic cancer cells must develop resistance to anoikis to survive in the bloodstream or lymphatic system. For these studies, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL under adherent and nonadherent conditions and then assayed for caspase 3/7 activity, which indicates apoptosis. Results showed that CNL was able to significantly increase apoptosis under nonadherent conditions, consistent with an increase in anoikis (Fig. 2A). This effect was prevented by Z-VAD-fmk, a pan-caspase inhibitor, indicating that this was indeed caspase-dependent anoikis. We also used siRNA against CD44s to mimic the effects of CNL. Knocking down CD44s using siRNA in MDA-MB-231 cells also significantly increased anoikis (Fig. 2A), suggesting that inhibition of CD44s may mediate the ability of CNL to induce anoikis. We next performed dose-response experiments to more fully characterize the effect of CNL on caspase 3/7 activity under adherent and nonadherent conditions. CNL activated caspase 3/7 with and an EC50 of 12.8 μm (95% CI: 12.4–13.2) under adherent conditions and an EC50 of 5.38 μm (95% CI: 4.53–6.39) under nonadherent conditions (Fig. 2B). This difference in EC50 values is highly significant (p < 0.0001), indicating that CNL activates caspase 3/7 more potently under nonadherent conditions compared with adherent conditions. The ability of CNL to induce anoikis in metastatic carcinoma cells was then confirmed in the B16 murine melanoma cell line (Fig. 2C).
FIGURE 2.
CNL induces anoikis/apoptosis in breast cancer cells via inhibition of CD44s. A, MDA-MB-231 cells were treated with 5 μm ghost, 5 μm CNL, or 25 μm Z-VAD-fmk pan-caspase inhibitor under either adherent or nonadherent conditions for 24 h, followed by an assay for caspase 3/7 activity. n = 3. Bars, S.E. *, p < 0.05. The results show that the treatment (p < 0.0001), group (p = 0.003), and their interaction (p = 0.0166) are all significant. B, MDA-MB-231 cells were treated with 1.56–100 μm CNL for 24 h, followed by an assay for caspase 3/7 activity. The EC50 was 12.8 μm under adherent conditions and 5.38 μm under nonadherent conditions. n = 5. C, B16 cells were treated with 10 μm ghost, 10 μm CNL, or 25 μm Z-VAD-fmk under either adherent or nonadherent conditions for 24 h, followed by an assay for caspase 3/7 activity. n = 5. Bars, S.E. *, p < 0.05. The results show that the treatment (p < 0.0001), group (p < 0.0001), and their interaction (p < 0.0001) are all significant.
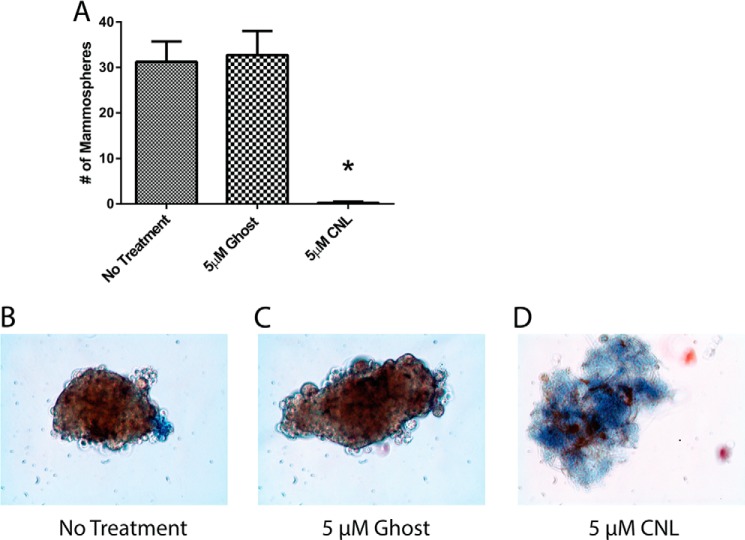
Given the ability of CNL to increase anoikis, studies were performed to investigate whether CNL could prevent mammosphere formation under nonadherent conditions. Mammospheres are thought to arise from cancer stem cells and tend to be highly resistant to anoikis (13). Similar to the caspase 3/7 assay, it was found that CNL significantly inhibited MDA-MB-231 mammosphere formation over the course of 9 days (Fig. 3A). The trypan blue dye exclusion technique was used to confirm mammosphere cell viability, with live cells excluding the blue dye (Fig. 3, B–D). These data support the hypothesis that CNL induces anoikis in breast cancer cells.
FIGURE 3.
CNL decreases breast cancer mammosphere formation. MDA-MB-231 cells were treated with 5 μm ghost or CNL for 9 days, followed by mammosphere analysis using a phase contrast microscope. Blue dye, trypan blue (dead cells). n = 3. Bars, S.E. *, p = 0.0009.
The Ceramide Nanoliposome Inhibits Human Breast Cancer Cell Migration under Static and Physiological Fluid Flow Conditions
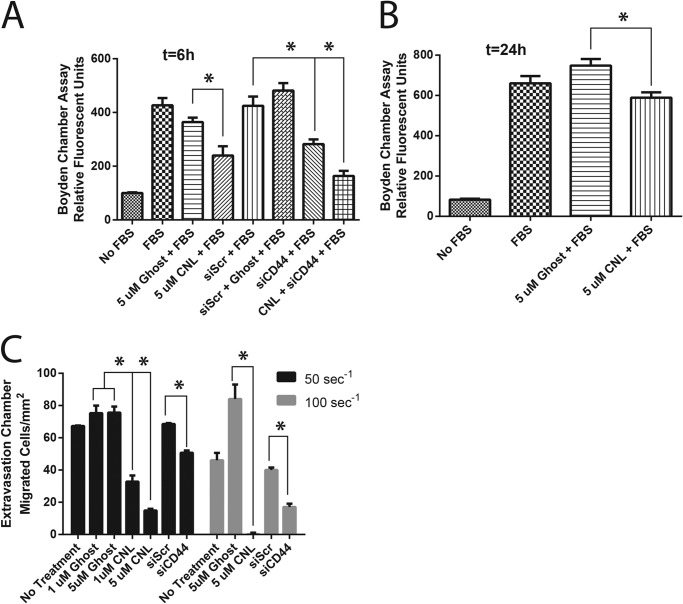
We next investigated whether CNL could block MDA-MB-231 cell migration when FBS was used as a chemoattractant under static conditions (i.e. in the absence of fluid flow). In fact, CNL inhibited cell migration in this cell line at a concentration that does not induce apoptosis after both 6 and 24 h of treatment (Fig. 4, A and B). Again, because CD44 expression is associated with cancer cell migration and extravasation (14, 15), we assessed whether siRNA knockdown of CD44s could mimic the effect of CNL to reduce cancer cell migration. Compared with CNL, similar results were obtained by knocking down CD44s using siRNA (siCD44) (Fig. 4A).
FIGURE 4.
CNL inhibits breast cancer cell migration under static and physiological fluid flow conditions. A and B, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL for 6 h (A) or 24 h (B) in a Transwell cell migration assay. n = 4. Bars, S.E. *, p < 0.05. C, MDA-MB-231 cells were treated with 1–5 μm CNL for 0.5 h or 100 nm scrambled (Scr) or CD44 (siCD44) siRNA for 48 h, followed by an assay for extravasation under fluid flow shear rates of 50 s−1 or 100 s−1. n = 3. Bars, S.E. *, p < 0.05.
To measure cell migration under more physiologically relevant conditions, we used a flow migration chamber to measure the ability of MDA-MB-231 breast cancer cells to migrate through EI endothelial cells under physiological fluid shear rates. EI cells are fibroblast L-cells that overexpress ICAM-1 and E-selectin, thus mimicking endothelial cells. Ligands to ICAM-1 and E-selectin include αvβ3 integrin and CD44; however, only CD44 is expressed in MDA-MB-231 cells (data not shown). The flow migration assay requires both adhesion to endothelial cells and cell migration and thus serves as a model of extravasation (the exit of metastatic cancer cells from the bloodstream). Our previous work demonstrated that shear rate has a much greater effect on adhesion and migration than shear stress (16–18). Thus, we varied the shear rate in our flow migration experiments. Similar to our Transwell results, CNL blocked the ability of breast cancer cells to migrate under flow conditions (Fig. 4C). This effect was increased by increasing concentrations of CNL (1–5 μm) and increasing shear rate (50–100 s−1). In addition, inhibiting CD44s using siRNA had an effect similar to that of CNL. Down-regulation of CD44s decreased extravasation, an effect that was enhanced by increasing the shear rate (Fig. 4C). These results indicate that CNL and CD44s siRNA block breast cancer cell migration through endothelial cells under physiological fluid flow conditions.
The Ceramide Nanoliposome Targets CD44s to the Lysosome in Human Breast Cancer Cells
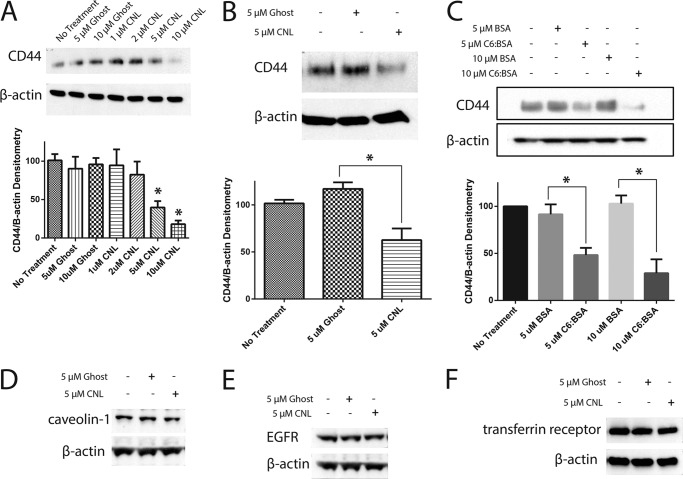
Given that overexpression of CD44s was able to decrease the effects of CNL on breast cancer cells, we next attempted to determine the mechanism by which CNL inhibits CD44s. CD44s expression decreased in a dose-dependent manner after treatment with CNL for 24 h, as shown by Western blot analysis (Fig. 5A). Such a decrease in CD44s would prevent breast cancer cells from binding to EI endothelial cells and extravasating, thus mediating the ability of CNL to prevent extravasation in this system. CD44s protein levels also decreased after a shorter 6-h treatment with CNL compared with untreated or ghost-treated cells (Fig. 5B). To see whether more traditional formulations of ceramide would have the same effect, we conjugated C6 ceramide to BSA (C6:BSA). We then treated MDA-MB-231 cells with 5 or 10 μm C6:BSA, followed by Western blot analysis of CD44s. Our results indicated that, similar to CNL, C6:BSA reduces CD44s levels in a dose-dependent manner (Fig. 5C). We next treated MDA-MB-231 cells with 20 μg/ml cycloheximide or 5 μm CNL in combination with cycloheximide for 0, 1, 2, 3, 14, and 24 h, followed by Western blot analysis of CD44s to determine the effect of CNL on the half-life of CD44s in MDA-MB-231 cells. We found that CD44s normally has a half-life of 8 h 41 min, but that CNL treatment reduces this to 24 min (data not shown). To determine whether the effect of CNL on CD44s was specific or not, we analyzed CNL-treated cells for other plasma membrane proteins, including caveolin-1, EGF receptor, and the transferrin receptor. Our results show that levels of these proteins do not change after treatment with CNL (Fig. 5, D–F), indicating that CNL specifically affects CD44s. CD44 inhibition was further confirmed by immunofluorescence on MDA-MB-231 cells that had been treated with 5 μm CNL for 24 h (Fig. 6A). Although untreated and ghost-treated cells had high levels of CD44 that was found mainly at the plasma membrane, quantification of the images revealed that CNL-treated cells had significantly lower expression of CD44 (Fig. 6, A and B).
FIGURE 5.
CNL inhibits CD44s in MDA-MB-231 cells. A, MDA-MB-231 cells were treated with increasing concentrations of ghost liposomes or CNL for 24 h, followed by Western blot analysis for CD44. n = 3. Bars, S.E. *, p < 0.05. B, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL for 6 h, followed by Western blot analysis for CD44. n = 3. Bars, S.E. *, p < 0.05. C, MDA-MB-231 cells were treated with 5–10 μm BSA or C6 ceramide conjugated to BSA for 24 h, followed by Western blot analysis of CD44. n = 3. Bars, S.E. *, p < 0.0001. D–F, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL for 24 h, followed by Western blot analysis of caveolin-1 (D), EGF receptor (E, EGFR), and the transferrin receptor (F).
FIGURE 6.
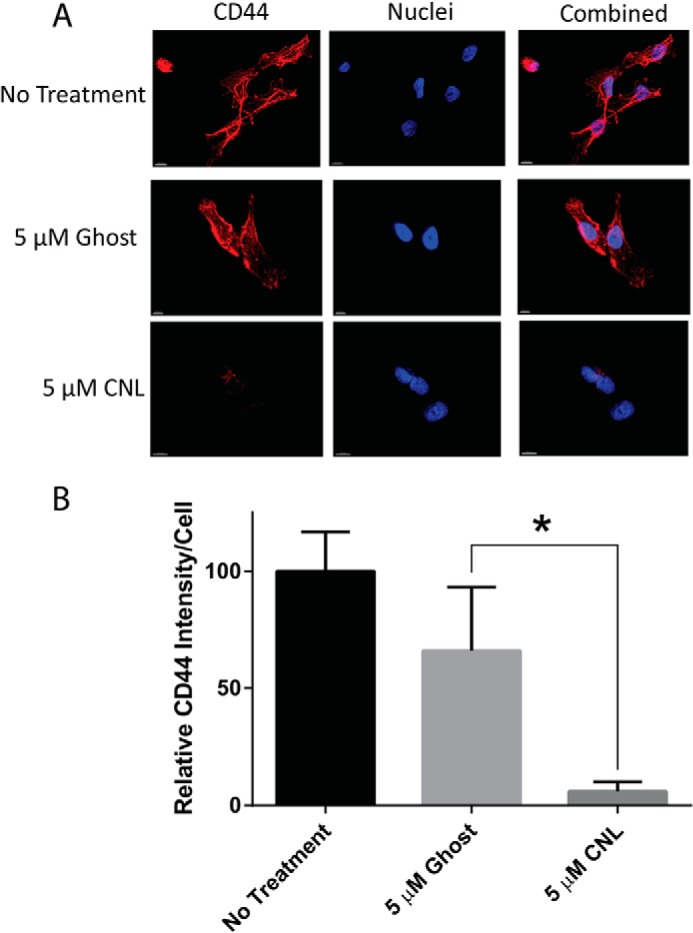
CNL decreases CD44 levels. A, MDA-MB-231 cells were treated with 5 μm ghost liposomes or CNL for 24 h, followed by immunofluorescence of CD44. Red, CD44; blue, nuclei. B, the images shown in A were quantified using Imaris imaging software. n = 3. Bars, S.E. *, p < 0.05.
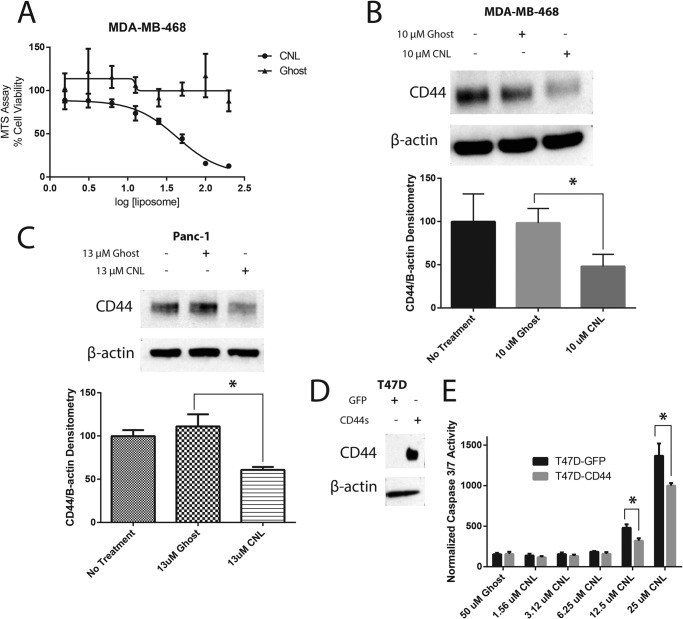
Having established the ability of CNL to inhibit CD44s in the MDA-MB-231 cell line, we next sought to extend this finding to other carcinoma cell lines. We first examined the MDA-MB-468 cell line. Like the MDA-MB-231 cell line, the MDA-MB-468 cell line is a triple negative human breast cancer cell line that expresses high levels of CD44. An MTS cell viability assay indicated that CNL inhibits cell viability in MDA-MB-468 cells with an IC50 of 43 μm, whereas ghost liposomes had a negligible effect on cell viability up to 200 μm (Fig. 7A). In our subsequent experiments, we used a nontoxic concentration of 10 μm CNL and found that CNL inhibited CD44s (Fig. 7B). This effect was further confirmed in Panc-1 human pancreatic cancer cells via Western blot (Fig. 7C). Panc-1 cells were treated with 13 μm CNL for 48 h because the IC50 of CNL on Panc-1 cell viability was previously determined to be 26 μm for a 48-h treatment (19).
FIGURE 7.
CNL inhibits CD44s in breast and pancreatic cancer cells. A, MDA-MB-468 cells were treated with 1.56–200 μm ghost liposomes or CNL for 24 h, followed by MTS assay for cell viability. The IC50 was 43 μm. n = 3. B, MDA-MB-468 cells were treated with 10 μm ghost liposomes or CNL for 24 h, followed by Western blot analysis of CD44. n = 5. Bars, S.E. p = 0.0014. C, Panc-1 cells were treated with 13 μm CNL for 48 h, followed by Western blot analysis of CD44. n = 3. Bars, S.E. *, p = 0.0099. D, GFP and CD44s were overexpressed in T47D cells, followed by Western blot analysis of CD44 to confirm overexpression. E, T47D-GFP and T47D-CD44 cells were treated with 50 μm ghost or 1.56–25 μm CNL for 24 h, followed by an assay for caspase 3/7 activity. n = 5. Bars, S.E. *, p < 0.05.
Having found that CNL is able to reduce CD44s levels in multiple carcinoma cell lines, we decided to see whether overexpression of CD44s would be able to rescue the effect of CNL. To do this, we overexpressed GFP or CD44s in T47D human breast cancer cells, which do not normally express CD44 (Fig. 7D). We refer to these cells as T47D-GFP and T47D-CD44, respectively. We then performed a caspase 3/7 activity assay in T47D cells expressing GFP or CD44s using increasing concentrations of CNL. Our results indicated that at 12.5 and 25 μm CNL, CD44s overexpression reduced CNL-induced caspase 3/7 activity (Fig. 7E), suggesting that additional mechanisms underlie the apoptotic actions of CNL in cell lines that overexpress CD44s. These data suggest that in cells that do not express CD44, ceramide acts through traditional mechanisms (e.g. activation of SAPK/JNK or MAPK or inhibition of Akt) (20–22). However, when CD44s is overexpressed, these cells are slightly more resistant to CNL, because CD44s expression at the plasma membrane must be reduced for apoptosis/anoikis to occur. Taken together, these data suggest that in cells that overexpress CD44s, CNL in part reduces surface expression of CD44s to induce apoptosis or anoikis. Thus, there are likely CD44-dependent and CD44-independent mechanisms of CNL.
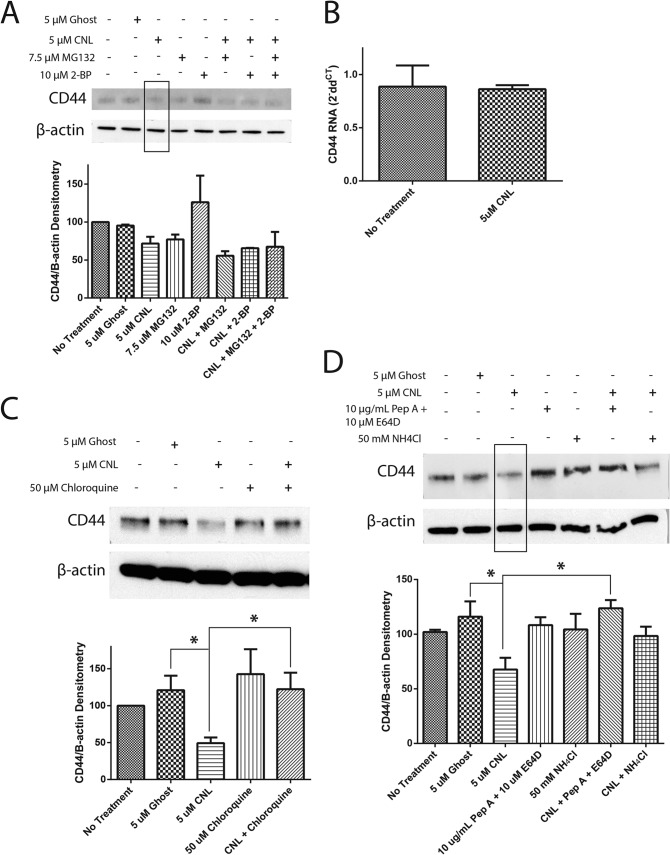
There is evidence that CD44 can be palmitoylated and targeted to lipid rafts (23). To test whether CNL induces palmitoylation of CD44s as an early event in its degradation, we used 2-bromopalmitate, which irreversibly inhibits palmitoyl acyltransferases. Although 2-bromopalmitate by itself actually increased CD44s levels, it was unable to rescue the effect of CNL, indicating that the ability of CNL to decrease CD44s levels is not dependent on palmitoylation of CD44 (Fig. 8A). Similarly, the proteasome inhibitor MG132 was also unable to rescue the effect of CNL on CD44s, indicating that CNL does not target CD44s to the proteasome (Fig. 8A). In addition, the effect of CNL on CD44s was not seen at the RNA level, as shown by RT-PCR (Fig. 8B). Treatment of MDA-MB-231 cells with 5 μm CNL for 24 h had no effect on CD44s RNA levels.
FIGURE 8.
CNL targets CD44s to the lysosome. A, MDA-MB-231 cells were pretreated with 7.5 μm MG132 (proteasome inhibitor) or 10 μm 2-bromopalmitate (2-BP; palmitoyl transferase inhibitor) for 0.5 h, followed by treatment with 5 μm ghost liposomes or CNL for 14 h. Western blot analysis for CD44 was then performed. n = 3. Bars, S.E. B, MDA-MB-231 cells were treated with 5 μm CNL for 24 h, followed by RT-PCR analysis of CD44, using β-actin as an endogenous control. n = 9. Bars, S.E. C, MDA-MB-231 cells were pretreated with 50 μm chloroquine for 1 h, followed by treatment with 5 μm ghost liposomes or CNL for 6 h. Western blot analysis of CD44 was then performed. n = 3. Bars, S.E. *, p < 0.05. D, MDA-MB-231 cells were pretreated with 10 μg/ml pepstatin A in combination with 10 μm E64D or 50 mm NH4Cl for 1 h, followed by treatment with 5 μm ghost liposomes or CNL for 6 h. Western blot analysis for CD44 was then performed. n = 3. Bars, S.E. *, p = 0.05.
To further understand the effect of CNL on CD44s expression, we used chloroquine, which inhibits lysosomal activity by preventing endosomal and lysosomal acidification. Western blot protein analysis revealed that chloroquine was able to inhibit the loss of CD44s caused by CNL, suggesting that CNL targets CD44s to the lysosome for degradation (Fig. 8C). We confirmed the role of the lysosome by using other lysosomal inhibitors, namely pepstatin A in combination with E64D, as well as ammonium chloride. Similar to the results obtained with chloroquine, inhibition of the lysosome with these agents prevented CNL from inhibiting CD44s (Fig. 8D), indicating that CNL targets CD44s to the lysosome for degradation.
DISCUSSION
This study demonstrates the ability of the ceramide nanoliposome to induce anoikis and inhibit extravasation in metastatic carcinoma cells by inhibiting CD44. Although an abundant body of literature exists on the ability of another sphingolipid, sphingosine-1-phosphate, to regulate cell migration (24), this study demonstrates that ceramide inhibits migration and extravasation in multiple cancer models. Indeed, using sophisticated in vitro models, we have shown that CNL can inhibit multiple steps of the metastatic cascade, including migration, resistance to anoikis, and extravasation.
Under normal conditions, breast epithelial cells are susceptible to anoikis. If these cells were to detach, they would sense a loss of adherence through integrins and would undergo a caspase-dependent programmed cell death termed anoikis (25). For metastatic cancer cells to survive in the lymphatic system or the bloodstream, they must become resistant to anoikis. In this paper, we examined the ability of CNL to increase the caspase-dependent process of anoikis. Although ceramide has previously been shown to induce anoikis in HEK293 (26) and HeLa cells (27), we have now extended this finding to breast cancer and melanoma cells. Hu et al. (27) demonstrated that ceramide-induced anoikis is the result of Golgi fragmentation and inhibition of β1 integrin. In our study, knocking down CD44 had the same effect on anoikis as treating with CNL, and overexpression of CD44 decreased the effect of CNL, supporting the heretofore unknown notion that CNL induces anoikis via down-regulation of CD44. Interestingly, other labs have shown that CD44 mediates anoikis resistance via intracellular signaling (28), including inhibition of the retinoblastoma protein (Rb) (29).
We have shown that CNL induces anoikis. However, even if carcinoma cells evade anoikis, CNL can prevent them from metastasizing by inhibiting extravasation. In this paper, we studied cell migration and extravasation using both static and fluid flow models. Because the static migration and flow migration assays measure different things, it is useful to compare results from both assays, although we would like to remind the reader that there were some differences in the way that the two experiments were carried out. First of all, 10% FBS was used as the chemoattractant in the static migration experiments, whereas soluble collagen IV was used as the chemoattractant in the flow migration experiments. Second, the cells used in the static migration experiments were serum-starved overnight prior to treatment, whereas the cells in the flow migration experiment were not serum-starved. Despite these differences, we feel that insights can still be gained from comparing results from the two assays. Static migration measures only cell motility, whereas flow migration measures both adhesion of cancer cells to the endothelium and cell motility. At a 6-h time point, 5 μm CNL decreased breast cancer cell migration under static conditions by ∼35%, and CD44 siRNA also decreased static migration by ∼35% (Fig. 3A). At the 24-h time point, CNL decreased static migration by 21% (Fig. 3B). Under a physiological flow rate of 50 s−1, 1 μm CNL decreased migration by 56%, 5 μm CNL decreased migration by 80%, and siCD44 decreased migration by 26% (Fig. 4). Under a physiological flow rate of 100 s−1, 5 μm CNL completely abolished cell migration, whereas siCD44 decreased migration by 58% (Fig. 4). Because CNL has a greater effect on flow migration than on static migration, our data indicate that ceramide is affecting both cell motility and adhesion, with the greatest effect seen at the highest shear rate and the highest concentration of CNL. Increased shear forces prevent circulating tumor cells from being able to adhere to endothelial cells, thus preventing extravasation.
CD44, a mediator of breast cancer metastasis, resides on the cell surface of metastatic breast cancer cells and binds to hyaluronan (30) and E-selectin (5). Decreased CD44 levels in breast cancer cells result in decreased adhesion to human bone marrow endothelial cells, indicating a role for CD44 in breast cancer metastasis to bone. In addition, increased CD44 levels lead to increased adhesion to human bone marrow endothelial cells via binding to hyaluronan (31). Furthermore, CD44 increases breast cancer metastasis to the liver in vivo (32).
Here, we have shown for the first time that CNL decreases pro-migratory CD44 protein levels but not RNA levels, demonstrating that the effect of CNL on CD44 is post-transcriptional. In addition, CNL causes CD44 to be targeted to the lysosome. Thankamony and Knudson (23) found that CD44 is localized to lipid rafts before being internalized. This agrees with our findings, because ceramide is found at high concentrations in lipid rafts (21). Thus, the ceramide delivered by CNL likely induces lipid raft formation, leading to internalization of CD44. The decrease in CD44, a pro-metastatic transmembrane protein, helps to explain how CNL can prevent extravasation. In addition, CD44hi/CD24lo mammary epithelial cells have been shown to be resistant to anoikis (33), suggesting that CD44 may help breast cancer cells evade anoikis. Furthermore, CD44 has been described as a cancer stem cell marker (34), and the mammosphere assay used in this paper has been used by some labs as a means of measuring the presence of cancer stem cells (35). Because CNL decreases both CD44 levels and the formation of mammospheres, CNL may be preventing the growth of primary and secondary tumors by inhibiting CD44-expressing cancer stem cells.
Although ceramide has long been known to induce apoptosis (20), we now demonstrate that short chain ceramide nanoliposomes can also modulate the metastasis-related processes of anoikis, cell migration, and extravasation through inhibition of CD44 at concentrations that do not affect cell viability. Ceramide is able to target the metastatic cascade at multiple points by inducing anoikis and inhibiting extravasation. Taken together, our data suggest that ceramide analogs may be useful not only in treating early stage solid tumors (9, 19, 36, 37), but also in preventing and treating late stage, metastatic carcinoma.
Acknowledgments
We thank the Penn State College of Medicine Functional Genomics and Microscopy and Histology Core Facilities.
This work was supported, in whole or in part, by National Institutes of Health Grant CA-125707 (to C. D.). This work was also supported by National Science Foundation Grants CBET-0729091 and CBET-1330633 (to C. D.) and funds from the G. Thomas Passananti Endowment from the Penn State College of Medicine (to M. K.). Penn State Research Foundation has licensed CNL technology to Keystone Nano, Inc. (State College, PA). M. K. is chief medical officer and co-founder of Keystone Nano.
- CNL
- ceramide nanoliposome
- CI
- confidence interval
- ICAM-1
- intercellular adhesion molecule 1
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- CD44s
- standard form of CD44
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
REFERENCES
- 1. Siegel R., Naishadham D., Jemal A. (2013) Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 2. Hart I. R. (1989) Immune profile in metastasis. Curr. Opin. Immunol. 1, 900–903 [DOI] [PubMed] [Google Scholar]
- 3. Zöller M. (1995) CD44: physiological expression of distinct isoforms as evidence for organ-specific metastasis formation. J. Mol., Med. 73, 425–438 [DOI] [PubMed] [Google Scholar]
- 4. Lesley J., Hyman R., English N., Catterall J. B., Turner G. A. (1997) CD44 in inflammation and metastasis. Glycoconj. J. 14, 611–622 [DOI] [PubMed] [Google Scholar]
- 5. Dimitroff C. J., Lee J. Y., Rafii S., Fuhlbrigge R. C., Sackstein R. (2001) CD44 is a major E-selectin ligand on human hematopoietic progenitor cells. J. Cell Biol. 153, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin T. A., Harrison G., Mansel R. E., Jiang W. G. (2003) The role of the CD44/ezrin complex in cancer metastasis. Crit. Rev. Oncol. Hematol. 46, 165–186 [DOI] [PubMed] [Google Scholar]
- 7. Stover T., Kester M. (2003) Liposomal delivery enhances short-chain ceramide-induced apoptosis of breast cancer cells. J. Pharmacol. Exp. Ther. 307, 468–475 [DOI] [PubMed] [Google Scholar]
- 8. Liu X., Ryland L., Yang J., Liao A., Aliaga C., Watts R., Tan S. F., Kaiser J., Shanmugavelandy S. S., Rogers A., Loughran K., Petersen B., Yuen J., Meng F., Baab K. T., Jarbadan N. R., Broeg K., Zhang R., Liao J., Sayers T. J., Kester M., Loughran T. P., Jr. (2010) Targeting of survivin by nanoliposomal ceramide induces complete remission in a rat model of NK-LGL leukemia. Blood 116, 4192–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stover T. C., Sharma A., Robertson G. P., Kester M. (2005) Systemic delivery of liposomal short-chain ceramide limits solid tumor growth in murine models of breast adenocarcinoma. Clin. Cancer Res. 11, 3465–3474 [DOI] [PubMed] [Google Scholar]
- 10. Heakal Y., Kester M. (2009) Nanoliposomal short-chain ceramide inhibits agonist-dependent translocation of neurotensin receptor 1 to structured membrane microdomains in breast cancer cells. Mol. Cancer Res. 7, 724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaiser J. M., Imai H., Haakenson J. K., Brucklacher R. M., Fox T. E., Shanmugavelandy S. S., Unrath K. A., Pedersen M. M., Dai P., Freeman W. M., Bronson S. K., Gardner T. W., Kester M. (2013) Nanoliposomal minocycline for ocular drug delivery. Nanomedicine 9, 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lou W., Krill D., Dhir R., Becich M. J., Dong J. T., Frierson H. F., Jr., Isaacs W. B., Isaacs J. T., Gao A. C. (1999) Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 59, 2329–2331 [PubMed] [Google Scholar]
- 13. Liu S., Dontu G., Mantle I. D., Patel S., Ahn N. S., Jackson K. W., Suri P., Wicha M. S. (2006) Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66, 6063–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zohar R., Suzuki N., Suzuki K., Arora P., Glogauer M., McCulloch C. A., Sodek J. (2000) Intracellular osteopontin is an integral component of the CD44-ERM complex involved in cell migration. J. Cell. Physiol. 184, 118–130 [DOI] [PubMed] [Google Scholar]
- 15. Zöller M. (2011) CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 11, 254–267 [DOI] [PubMed] [Google Scholar]
- 16. Slattery M. J., Liang S., Dong C. (2005) Distinct role of hydrodynamic shear in leukocyte-facilitated tumor cell extravasation. Am. J. Physiol. Cell Physiol. 288, C831–C839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang S., Slattery M. J., Dong C. (2005) Shear stress and shear rate differentially affect the multi-step process of leukocyte-facilitated melanoma adhesion. Exp. Cell Res. 310, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang S., Slattery M. J., Wagner D., Simon S. I., Dong C. (2008) Hydrodynamic shear rate regulates melanoma-leukocyte aggregation, melanoma adhesion to the endothelium, and subsequent extravasation. Ann. Biomed. Eng. 36, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Y., DiVittore N. A., Kaiser J. M., Shanmugavelandy S. S., Fritz J. L., Heakal Y., Tagaram H. R., Cheng H., Cabot M. C., Staveley-O'Carroll K. F., Tran M. A., Fox T. E., Barth B. M., Kester M. (2011) Combinatorial therapies improve the therapeutic efficacy of nanoliposomal ceramide for pancreatic cancer. Cancer Biol. Ther. 12, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verheij M., Bose R., Lin X. H., Yao B., Jarvis W. D., Grant S., Birrer M. J., Szabo E., Zon L. I., Kyriakis J. M., Haimovitz-Friedman A., Fuks Z., Kolesnick R. N. (1996) Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature 380, 75–79 [DOI] [PubMed] [Google Scholar]
- 21. Fox T. E., Houck K. L., O'Neill S. M., Nagarajan M., Stover T. C., Pomianowski P. T., Unal O., Yun J. K., Naides S. J., Kester M. (2007) Ceramide recruits and activates protein kinase C zeta (PKC zeta) within structured membrane microdomains. J. Biol. Chem. 282, 12450–12457 [DOI] [PubMed] [Google Scholar]
- 22. Kim H. J., Oh J. E., Kim S. W., Chun Y. J., Kim M. Y. (2008) Ceramide induces p38 MAPK-dependent apoptosis and Bax translocation via inhibition of Akt in HL-60 cells. Cancer Lett. 260, 88–95 [DOI] [PubMed] [Google Scholar]
- 23. Thankamony S. P., Knudson W. (2006) Acylation of CD44 and its association with lipid rafts are required for receptor and hyaluronan endocytosis. J. Biol. Chem. 281, 34601–34609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pyne N. J., Pyne S. (2010) Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 25. Chiarugi P., Giannoni E. (2008) Anoikis: a necessary death program for anchorage-dependent cells. Biochem. Pharmacol. 76, 1352–1364 [DOI] [PubMed] [Google Scholar]
- 26. Widau R. C., Jin Y., Dixon S. A., Wadzinski B. E., Gallagher P. J. (2010) Protein phosphatase 2A (PP2A) holoenzymes regulate death-associated protein kinase (DAPK) in ceramide-induced anoikis. J. Biol. Chem. 285, 13827–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu W., Xu R., Zhang G., Jin J., Szulc Z. M., Bielawski J., Hannun Y. A., Obeid L. M., Mao C. (2005) Golgi fragmentation is associated with ceramide-induced cellular effects. Mol. Biol. Cell 16, 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su Y. J., Lai H. M., Chang Y. W., Chen G. Y., Lee J. L. (2011) Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 30, 3186–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lakshman M., Subramaniam V., Rubenthiran U., Jothy S. (2004) CD44 promotes resistance to apoptosis in human colon cancer cells. Exp. Mol. Pathol. 77, 18–25 [DOI] [PubMed] [Google Scholar]
- 30. Radotra B., McCormick D. (1997) Glioma invasion in vitro is mediated by CD44-hyaluronan interactions. J. Pathol. 181, 434–438 [DOI] [PubMed] [Google Scholar]
- 31. Draffin J. E., McFarlane S., Hill A., Johnston P. G., Waugh D. J. (2004) CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 64, 5702–5711 [DOI] [PubMed] [Google Scholar]
- 32. Ouhtit A., Abd Elmageed Z. Y., Abdraboh M. E., Lioe T. F., Raj M. H. (2007) In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am. J. Pathol. 171, 2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gauger K. J., Hugh J. M., Troester M. A., Schneider S. S. (2009) Down-regulation of sfrp1 in a mammary epithelial cell line promotes the development of a cd44high/cd24low population which is invasive and resistant to anoikis. Cancer Cell Int. 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prince M. E., Sivanandan R., Kaczorowski A., Wolf G. T., Kaplan M. J., Dalerba P., Weissman I. L., Clarke M. F., Ailles L. E. (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 104, 973–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M. A., Daidone M. G. (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511 [DOI] [PubMed] [Google Scholar]
- 36. Adiseshaiah P. P., Clogston J. D., McLeland C. B., Rodriguez J., Potter T. M., Neun B. W., Skoczen S. L., Shanmugavelandy S. S., Kester M., Stern S. T., McNeil S. E. (2013) Synergistic combination therapy with nanoliposomal C6-ceramide and vinblastine is associated with autophagy dysfunction in hepatocarcinoma and colorectal cancer models. Cancer Lett. 337, 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tagaram H. R., Divittore N. A., Barth B. M., Kaiser J. M., Avella D., Kimchi E. T., Jiang Y., Isom H. C., Kester M., Staveley-O'Carroll K. F. (2011) Nanoliposomal ceramide prevents in vivo growth of hepatocellular carcinoma. Gut 60, 695–701 [DOI] [PubMed] [Google Scholar]