Abstract
Binding of the chemotaxis response regulator CheY-P promotes switching between rotational states in flagellar motors of the bacterium Escherichia coli. Here, we induced switching in the absence of CheY-P by introducing copies of a mutant FliG locked in the clockwise (CW) conformation (FliGCW). The composition of the mixed FliG ring was estimated via fluorescence imaging, and the probability of CW rotation (CWbias) was determined from the rotation of tethered cells. The results were interpreted in the framework of a 1D Ising model. The data could be fit by assuming that mutant subunits are more stable in the CW conformation than in the counterclockwise conformation. We found that CWbias varies depending on the spatial arrangement of the assembled subunits in the FliG ring. This offers a possible explanation for a previous observation of hysteresis in the switch function in analogous mixed FliM motors—in motors containing identical fractions of mutant FliMCW in otherwise wild-type motors, the CWbias differed depending on whether mutant subunits were expressed in strains with native motors or native subunits were expressed in strains with mutant motors.
Introduction
The bacterial flagellar motor contains a switch that is responsible for changes in the direction of rotation, and consists of rings of three types of proteins: FliG, FliM, and FliN. The FliG ring, with 26 subunits, interfaces with the stator. Each FliG subunit, and hence the FliG ring, prefers to be in the lower-energy counterclockwise (CCW) conformation, causing the motor to rotate in a default CCW direction. The FliM ring acts as a receptor for the ligand CheY-P, and CheY-P binding stabilizes thermally driven transitions of the switch to the CW state (1). The switch responds ultrasensitively to chemotactic signals, causing the probability of CW rotation (CWbias) to increase in a sigmoidal fashion over a narrow range of CheY-P concentrations (2–4).
In strains lacking the cheY gene, motors rotate exclusively CCW, and cells swim smoothly. Switching can be induced in such strains either via changes in temperature (5) or by creating motors that contain mixtures of wild-type (WT) and mutant subunits predisposed to be in the CW conformation. Minamino et al. (6) studied WT strains containing various amounts of FliGCW, a CW-locked mutant of Salmonella (ΔPAA) found by Togashi et al. (7), and concluded that only a small fraction of FliG has to be in the ΔPAA form to affect CW-CCW switching. In cheY-deleted strains, as more FliGCW was expressed, tethered cells went from exclusively CCW to partially CW with pauses, and then to exclusively CW. Bren and Eisenbach (8) studied strains containing various amounts of a CW-biased mutant of FliM, and found hysteresis. For the same numbers of mutant versus WT subunits in the motor, the CWbias differed depending on whether mutant subunits were expressed in strains with native motors or native subunits were expressed in strains with mutant motors.
Here, we generated switching in Escherichia coli strains lacking the cheY gene by expressing mixtures of FliGCW (9) and FliGWT. We employed total internal reflection fluorescence microscopy (TIRFM) to determine the makeup of the mixed motors. We developed a 1D Ising model to quantitatively describe the induction of switching as a function of the switch composition. We assumed that the FliGCW subunits were more stable in the CW conformation than in the CCW conformation, in contrast to the FliGWT subunits, which were more stable in the CCW conformation. We obtained the best fits to experimental data when the free-energy difference between the two conformations of the mutant (Em) was approximately twice that of the free-energy difference (E) between the two conformations of the WT subunit. Further theoretical analysis showed that for a fixed number of mutant switch subunits in otherwise WT rings, the CWbias varied depending on the arrangement of the mutants in the rings. Based on this, we quantitatively explained the observed hysteresis in switch function in mixed FliM motors by assuming that nearest-neighbor interactions between individual FliM subunits influence FliM assembly. Hysteresis in switch function was not observed for mixed FliG rings. Therefore, it is unlikely that nearest-neighbor FliG-FliG interactions (which depend on the conformations of neighboring FliG subunits) contribute significantly to FliG assembly.
Materials and Methods
Strains and plasmids
Strains (all derivatives of the WT strain RP437 (10)) were constructed to allow variable expression of FliGCW or FliGWT in a WT (genomic fliGWT) or mutant (genomic fliGCW) background, respectively. We refer to these strains as forward and reverse strains, respectively. The forward strain (PL14) was constructed by transforming strain PL4 (ΔcheY, sticky fliC (11)) with pTrc99A- fliGCW. The reverse strain (PL99) was constructed by transforming strain PL34 (ΔcheY, sticky fliC, fliGCW (11)) with ptrc99A-fliGWT. The fliGCW mutation was generated in E. coli by introducing a ΔPAA deletion (7) in the WT fliGWT allele. Strain PL34 was transformed with pHL31 (eYFP-fliGWT, ptrc99A (12)) to create strain PL86. Strains were grown in tryptone broth at 33°C to OD600 = 0.5 in the presence of ampicillin (100 μg/l). Isopropyl β-D-1-thiogalactopyranoside (IPTG; 0–15 μM) was added at the start of the growth culture to control the level of expression of the pTrc99A constructs. To create motors with eYFP-FliGWT subunits, strain DFB225 (a fliG null strain (13)) was transformed with pHL31 and pKAF131 (sticky fliC, pACYC184-fliCsticky (1)). An IPTG concentration of 10 μM resulted in tethered motor speeds similar to those of WT tethered motors, and these motors were imaged via TIRFM to estimate , the average intensity of motors whose FliG content was entirely eYFP-FliGWT. Cells were washed twice with motility buffer (0.01 M potassium phosphate (pH 7.0), 10−4 M EDTA, 0.067 M NaCl) and sheared before tethering.
Image analysis and cell selection
Tethered cell rotation was recorded using a digital camera (DCC1240M; Thorlabs) at 67 fps. Rotational speeds and switching statistics were determined using custom-written codes in MATLAB (The MathWorks, Natick, MA) (11). Ellipsoidal fitting to contrast-enhanced or binary images of the cells yielded the change in cell orientation with time. A fraction of tethered cells (<10%) continuously jiggled back and forth without completing full rotations, which were interpreted as indefinite pausing. These cells were not analyzed. Among the cells that rotated and switched, only those with absolute motor speeds >1 Hz were retained for further analysis (9). This selection criterion was applied uniformly to all of the data collected for TIRF-based estimation of the fraction of FliG in the FliGCW form, or the phase-contrast-based estimation of CWbias. To calculate CWbias, motor switching traces were converted to binary traces by assigning all positive (negative) speeds a value of 1 (−1). CWbias was then calculated as the fraction of the time motors spent rotating CW over a 20 s running window.
TIRF background correction
Tethered motor intensities were determined from raw TIRF images as previously described (9). These intensities were then corrected for background contributions due to eYFP-FliG subunits that were present in the cell but not associated with motors. To calculate the contributions of nonmotor eYFP-FliG, we expressed eYFP-only subunits in the cytoplasm and measured the average background contributions from TIRF images of tethered motors. The intensities of motors containing eYFP-FliG were corrected for this background at the relevant induction levels. It is reasonable to assume that the cytoplasmic eYFP contribution correctly reflects the contribution of nonmotor FliG-eYFP, since that latter is known to be present in the cytoplasm, e.g., in cells with a FliF null background (12). The same protocol was applied in the determination of .
Results and Discussion
CWbias as a function of FliGCW expression
We constructed the forward strain (PL14) by introducing a vector (ptrc99A) encoding the fliGCW allele into a strain carrying genomic fliGWT but lacking the cheY gene. Expression was controlled via the concentration of the inducer (IPTG). We also carried out the experiment in reverse: the reverse strain (PL99) was constructed by introducing the same vector encoding the fliGWT allele into a strain carrying genomic fliGCW, also lacking the cheY gene. Motors were observed to switch in either strain. The switching traces were converted to binary traces, as discussed in the Materials and Methods. A sample binary version is shown in Fig. 1 A. The CWbias was estimated as a function of the inducer levels for both strains, as shown in Fig. 1 B. The squares represent experiments conducted using the forward strain. The circles represent experiments conducted using the reverse strain. As expected, increasing cellular concentrations of FliGCW in the forward strain led to an increase in the CWbias (squares), and increasing cellular concentrations of FliGWT in the reverse strain led to a decreasing CWbias (circles).
Figure 1.
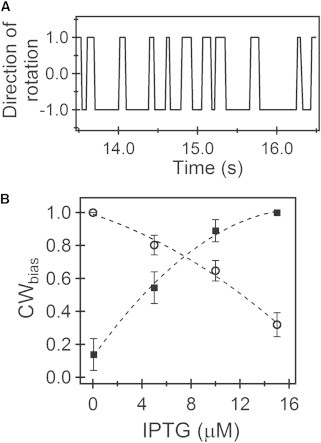
Mixed-motor switching. (A) Sample binary switching trace of a tethered motor consisting of a mix of CW-only and WT FliG subunits. Positive (negative) values indicate CCW (CW) rotation. (B) Measurements of average CWbias calculated from switching traces as a function of inducer (IPTG) concentrations. Sample sizes ranged from n = 18 motors to n = 42 motors. Error bars are standard errors and the dotted curves are a guide to the eye. Squares indicate experiments in the forward strain (fliGWT background with a vector expressing fliGCW) and circles indicate experiments in the reverse strain (fliGCW background with a vector expressing fliGWT).
To estimate the makeup of the FliG ring in these mixed motors, we expressed an eYFP-FliGWT construct from the same ptrc99A expression system (pHL31 (12)) in strains carrying genomic fliGCW (PL86), and imaged tethered motors via TIRFM. The intensities of eYFP-FliGWT were calculated as described in Lele et al. (9) and further detailed in Materials and Methods. Such measurements were repeated for different induction levels. A representative population measurement at 5 μM IPTG is shown in Fig. 2 A. To determine the fraction of eYFP-FliGWT in the FliG rings, we normalized the average intensity values (obtained from population measurements at each induction level) by the average intensity () of motors whose FliG was entirely eYFP-FliGWT. To estimate , we expressed eYFP-FliGWT in a fliG null strain (DFB225) and measured intensities in functional tethered motors. The variation of the fraction of eYFP-FliGWT as a function of inducer concentration is shown in Fig. 2 B. These measurements were limited to inducer concentrations of 0–10 μM because at higher inducer concentrations (15 μM or more), the cellular eYFP-FliGWT signals were strong enough to make motor intensity profiles difficult to separate from the background. Hence, we estimated the value of the eYFP-FliGWT fraction at 15 μM IPTG by extrapolating the curve in Fig. 2 B using a simple fit . It is possible that the incorporation of eYFP-FliGWT subunits is different from FliG subunits, which would be a limitation of our study. However, the speeds of eYFP-FliGWT motors were essentially the same as those of FliGWT motors (data not shown), indicating that motor assembly was not adversely affected by the fusion product.
Figure 2.
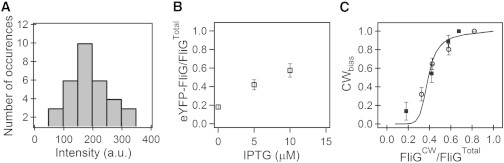
(A) TIRFM measurements of the intensities of YFP-FliGWT subunits in otherwise CW-only tethered motors (in strains with fliGCW on the genome and an IPTG-inducible vector carrying eyfp-fliGWT). This particular measurement (n = 33 motors) was obtained at an IPTG concentration of 5 μM. (B) Mean YFP-FliG/FliGTotal fractions (from distributions of the sort shown in A) of YFP-FliG subunits incorporated in motors, shown as a function of inducer concentration. (C) CWbias as a function of the fraction of FliGCW in otherwise WT motors. Squares (circles) indicate experiments in which strains carried fliGWT (fliGCW) on the genome and a vector expressing fliGCW (fliGWT). No hysteresis in switch function is seen. The black curve is the model prediction for N = 26, E = 0.25, Em = 0.45, and J = 1.9.
As noted above, hysteresis in the switch function was observed in motors made from mixtures of FliMCW and FliMWT subunits (8). Specifically, for the same ratio of FliMCW and FliMWT subunits, a higher CWbias was observed in strains in which FliMWT was expressed from a plasmid compared with strains in which FliMWT was expressed genomically. A comparison of whole-cell extracts with basal-body extracts showed that whole-cell FliMCW/FliMWT ratios were a good approximation for motor FliMCW/FliMWT ratios. The reasons for the hysteresis were not clear.
To test whether a similar hysteresis is observable in our data, we plotted CWbias, shown in Fig. 1 B, as a function of the fraction of FliGTotal that is FliGCW, inferred from Fig. 2 B. The results are shown in Fig. 2 C. The squares represent data over varying FliGCW expression levels in the forward strain, and the circles represent data over varying FliGWT expression levels in the reverse strain. In the former case, we calculated the fractions of FliGCW from the data in Fig. 2 B by making two assumptions. First, the expression levels of either kind of FliG subunit from ptrc99A (or from the genome) are similar given their comparable sizes (fliGWT allele = 996 bp; fliGCW allele = 987 bp). This is a reasonable assumption considering that the promoters are the same, and the cloning sites/plasmid copy numbers/induction levels (for expression from the vector) remain the same. Second, as found for FliM mixed motors, the intracellular FliGCW/FliGWT ratios are a good approximation of the motor FliGCW/FliGWT ratios. This assumption is valid for the following reasons: Consider the ratio of the expression level of the mutant at two different induction levels (). It is straightforward to show that if the latter assumption is correct, then
We estimated the ratios in the right-hand side from the data in Fig. 2 B and independently determined the ratio in the left-hand side by directly measuring the average whole-cell brightness via TIRFM. The right-hand ratios were 0.3 (IPTG1 = 0 μM, IPTG2 = 5 μM) and 0.52 (IPTG1 = 5 μM, IPTG2 = 10 μM). These compared favorably with the left-hand ratios, 0.38 (IPTG1 = 0 μM, IPTG2 = 5 μM) and 0.54 (IPTG1 = 5 μM, IPTG2 = 10 μM), lending support to the assumption. The data in Fig. 2 C suggest that mixed FliG motors do not exhibit the same kind of hysteresis in switching that was observed in mixed FliM motors.
1D Ising model
To explain the sigmoidal nature of CWbias versus the fraction of FliGCW molecules shown in Fig. 2 C, we analyzed switching in the framework of a simple 1D Ising model (14–16) with periodic boundary conditions. We described the Hamiltonian for the system as
| (1) |
where Em (E) is a measure of the free-energy difference between the CW and CCW conformations of an individual FliGCW (FliGWT) subunit, and J is the interaction energy between neighboring subunits, which we assume is the same irrespective of the type of subunits. This is a reasonable assumption considering that biochemical data have shown that FliGCW has a conformation similar to that of FliGWT in the CW state (6). It is this interaction through which the mutant subunits influence the propensity of neighboring WT subunits to adopt the CW conformation (CW propensity). N is the total number of subunits, and m is the number of mutant subunits. The first term on the right in Eq. 1 sums up the interactions between neighboring subunits. The second (third) term represents the free-energy contributions of N-m (m) WT (mutant) FliG subunits. All values are in units of kBT, where kB is Boltzmann’s constant and T is the absolute temperature. Each of the N (=26) FliG subunits can adopt one of two conformations: CCW () or CW (). For a single mutant subunit (m = 1), the partition function can be determined via the transfer matrix approach (17):
| (2) |
We assume typical values of J (∼4/2) and E (= 0.5/2) reported in the literature for the flagellar switch (18,19). For increasing numbers of m, the partition function can be suitably modified to include the configuration or arrangement of the ring. For example, if two mutant subunits (m = 2) are separated by two FliGWT subunits, , whereas if the two mutant subunits are separated by eight FliGWT subunits, then . The CWbias can then be calculated as
| (3) |
The extreme conformations (CW and CCW) are not the only ones adopted by the ring. Entropic factors cause intermediate configurations to have finite probabilities, consistent with the notion of mixed states (18). However, we do not discuss those here, since the CWbias is a very good indicator of the physiologically relevant activity of the complex. To determine the effect of increasing fractions of FliGCW on the CWbias, we first randomly generated all possible arrangements of m FliGCW subunits in a ring consisting of (N – m) FliGWT subunits (5000 samples). Next, for each configuration, the partition function Z was determined and the CWbias was calculated from Eq. 3. The CWbias was then averaged over all 5000 values. In this fashion, the CWbias versus FliGCW fractions (= m/N) was calculated. The predictions (black curve) are shown in Fig. 2 C. The curve, which is a reasonable fit to the experimental data, was calculated by assuming Em ∼2 E.
Effect of assembly on CWbias
Cooperativity plays an important role in the assembly of large complexes in biological systems (20). In the context of the assembly of the FliG ring, cooperativity refers to the preferential association of WT subunits together, minimizing contact with FliGCW subunits to reduce the energy penalty (J) incurred at the boundaries between the two kinds of subunits. A second possibility is that the FliG ring is assembled noncooperatively, such that individual FliGCW subunits are incorporated at random in a ring of otherwise FliGWT subunits. As the number of mutant molecules (m) increases, the number of ways to arrange them in the ring increases as well. Certain ring configurations are entropically favored, and on an average many more motors would have those particular ring configurations. We examined how such FliGCW sequestration, or lack thereof, affected the CWbias.
Since individual FliGCW subunits prefer to be in the CW conformation, the energetic penalty incurred by introducing a single FliGCW in an otherwise WT ring is 4J. Adding more FliGCW subunits adjacent to the single subunit does not result in additional penalties since no new boundaries (interfaces between a mutant and neighboring WT subunits) are created, as shown in the upper schematic in Fig. 3 A. However, for m nonsequestered subunits (bottom), there are 2m boundaries, resulting in an energetic penalty of m × 4J that increases the overall propensity of the ring to adopt the CW conformation. The increase in propensity also occurs in part due to entropic factors that ultimately reduce the partition function with increasing R-1. The effect of assembly on the CWbias as a function of the number of FliGWT subunits between pairs of mutants is shown on the right in Fig. 3 A. For simplicity, we calculated the plots by assuming that R-1 is the same between distinct pairs. The ordinate is CWbias normalized by the expected value for two adjacent mutant subunits (R-1 = 0), CW§bias. As can be observed in the figure, CWbias/CW§bias is the lowest for the adjacent pair and progressively increases to a final value as the number of FliGWT subunits separating the pair increases. As noted before, this effect is amplified by an increasing number of mutants. This dependence of CWbias on the configuration is present at finite coupling energies between subunits (18,19). Such a dependence would vanish under the limiting and unrealistic case of infinite coupling between subunits (J→ ∞), as specified by the concerted model of allostery (i.e., the Monod-Wyman-Changeux model (21)).
Figure 3.
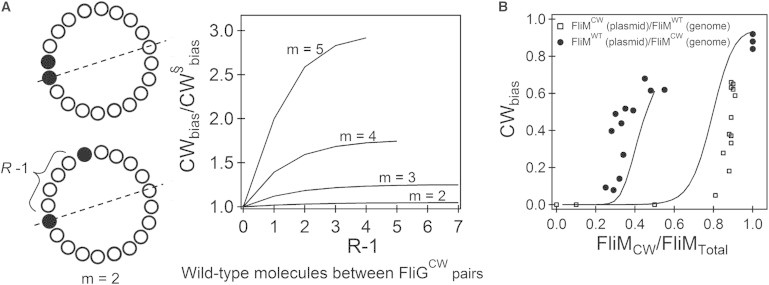
(A) The schematic on the left illustrates the differences in the configuration for sequestered (top) and nonsequestered (bottom) mutants. Black circles indicate FliGCW subunits. The dotted line indicates the line of symmetry (for m = 2), which limits the number of unique ways to arrange the mutants. On the right is CWbias plotted as a function of the gap (R-1) for varying m (FliGCW subunits). The ordinate is a ratio of the CWbias (calculated for increasing R-1 WT subunits between m-1 pairs) to the CW§bias expected when all m mutants are sequestered during ring assembly (R-1 = 0). E = 0.25, Em = 0.45, J = 1.45. (B) Role of assembly in generating hysteresis in switch function (symbols indicate experimental data from Bren and Eisenbach (8)). Squares (circles) indicate experiments in which strains carried fliMWT (fliMCW) on the genome and a vector expressing fliMCW (fliMWT). The black curve on the left was calculated for rings assembled by maximizing the possible separations between all m-1 mutants. The black curve on the right was calculated by sequestering all mutant FliM subunits during ring assembly. N = 34, E = 0.25, Em = 0.45, J = 1.45.
Possible origin of hysteresis in switch function
Fig. 3 B shows the previously observed hysteresis in switch function (8). The squares represent experiments in a strain in which a FliMCW mutant was expressed via an inducible plasmid in cells (strain A) carrying fliMWT on the genome. The circles represent experiments in cells (strain B) in which FliMWT was expressed via an inducible plasmid in a strain carrying the mutant fliMCW on the genome. As can be seen, the switch function (CWbias) exhibits a hysteresis depending on the type of genomic fliM. One explanation for the hysteresis effect could be the dependence of CWbias on the arrangement of mutant subunits in an otherwise WT ring. If there are differences in the way the ring assembles in the two types of strains, then it is possible to use the mixed-ring model to explain the observed hysteresis. We assumed that FliMCW subunits are sequestered in strain A (R-1 = 0), but are not sequestered in strain B. The two curves in Fig. 3 B represent model predictions based on this simple assumption and explain the observed data reasonably well. As expected, sequestration leads to reduced CWbias until the numbers of FliMCW subunits in motors exceed a particular value.
Three-dimensional reconstructions from electron cryomicrographs of the C-ring in strains carrying the fliGCW gene used in this work have revealed that the most recurring symmetry is 34-fold, indicating that there are likely to be 34 FliM/FliN complexes in CW-only motors (22). However, a recent hypothesis suggests that the FliM assembly in such FliGCW motors carries as few as 26 FliM/FliN complexes, and the ring assembly remains incomplete (23). In presenting the explanation for hysteresis in the FliM ring, we made the assumption that FliM subunits assemble into a complete ring of 34 subunits. However, this is not a necessary assumption for explaining the data, since ultimately the CWbias is determined by the FliG ring conformation. Irrespective of whether or not FliM forms a ring, FliM subunits must interact with FliG subunits (possibly mediated by FliN subunits) to effect a conformational change. Thus, it is possible to reproduce the analysis shown in Fig. 3 B by assuming that the FliGWT subunits in contact with the mutant FliM subunits incorporated in the C-ring have a higher propensity to adopt the CW conformation.
Hysteresis in assembly/disassembly of biological macromolecules is known to occur (24,25). If our explanation of hysteresis in the switch function due to FliM assembly differences is correct, it points to differences in the assembly mechanism of the FliM and FliG rings. FliG assembles before the FliM ring and interacts strongly with the FliF ring (26). It is possible that during the assembly, an energetic penalty is incurred when a FliGCW subunit is positioned next to a FliGWT subunit, influenced by the coupling interactions between the pair. However, these interactions are rendered unimportant by the strong FliF-FliG interactions that likely drive the assembly. As a result, hysteresis is not observed. On the other hand, a significant part of the motor FliM binds through weak interactions in the C-ring, and it is possible that the energetic penalties incurred (influenced by the interactions between a WT and a mutant FliM subunit) make a significant contribution to the assembly. Sequestration is likely achieved through FliM exchange. How such sequestration depends on the FliM properties (e.g., native FliM versus mutant FliM) and how it can be experimentally determined will be a subject of future work.
Author Contributions
P.P.L. and H.C.B. designed the work and wrote the article. P.P.L. performed the research and analyzed the data.
Acknowledgments
We thank Victor Sourjik for sharing the plasmid pHL31 used in this work.
This work was supported by National Institutes of Health grant AI016478.
References
- 1.Scharf B.E., Fahrner K.A., Berg H.C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon U., Camarena L., Stock J.B. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cluzel P., Surette M., Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J., Berg H.C. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 2013;425:1760–1764. doi: 10.1016/j.jmb.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner L., Samuel A.D.T., Berg H.C. Temperature dependence of switching of the bacterial flagellar motor by the protein CheY(13DK106YW) Biophys. J. 1999;77:597–603. doi: 10.1016/S0006-3495(99)76916-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minamino T., Imada K., Namba K. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 2011;9:e1000616. doi: 10.1371/journal.pbio.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Togashi F., Yamaguchi S., Macnab R.M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 1997;179:2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bren A., Eisenbach M. Changing the direction of flagellar rotation in bacteria by modulating the ratio between the rotational states of the switch protein FliM. J. Mol. Biol. 2001;312:699–709. doi: 10.1006/jmbi.2001.4992. [DOI] [PubMed] [Google Scholar]
- 9.Lele P.P., Branch R.W., Berg H.C. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc. Natl. Acad. Sci. USA. 2012;109:20018–20022. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson J.S., Houts S.E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lele P.P., Hosu B.G., Berg H.C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Sourjik V. Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 2011;80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd S.A., Tang H., Blair D.F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson C.J. Models for hemoglobin and allosteric enzymes. Biopolymers. 1968;6:1101–1118. doi: 10.1002/bip.1968.360060806. [DOI] [PubMed] [Google Scholar]
- 15.Duke T.A., Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc. Natl. Acad. Sci. USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Duke T. Cooperative model of bacterial sensing. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1998;58:6399–6406. [Google Scholar]
- 17.Wannier G.H. J. Wiley; New York: 1966. Statistical Physics. [Google Scholar]
- 18.Duke T.A., Le Novère N., Bray D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J., Fahrner K.A., Berg H.C. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 2009;390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson J.R. Cooperativity in macromolecular assembly. Nat. Chem. Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 21.Monod J., Wyman J., Changeux J.P. On the nature of allosteric transitions—a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 22.Thomas D.R., Francis N.R., DeRosier D.J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delalez N.J., Berry R.M., Armitage J.P. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. MBio. 2014;5 doi: 10.1128/mBio.01216-14. e01216-e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasshauer D., Antonin W., Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 25.Singh S., Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 2003;278:18249–18255. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 26.Levenson R., Zhou H., Dahlquist F.W. Structural insights into the interaction between the bacterial flagellar motor proteins FliF and FliG. Biochemistry. 2012;51:5052–5060. doi: 10.1021/bi3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]


