Abstract
AIM: To explore the microRNA (miRNA) profiles and to determine the key miRNAs within the side population (SP) cells of the gastric cancer cell line MKN-45.
METHODS: We used fluorescence-activated cell sorting and Hoechst 33342 labeling to obtain SP cells from the human gastric carcinoma cell line MKN-45. The miRNA expression profiles of the SP and major population (MP) cells were examined using a miRNA gene chip, and key miRNAs were obtained according to aberrant expression and the miRNAs’ possible targets as predicted by bioinformatics.
RESULTS: Using a significance criterion of a 1.5-fold or greater difference in expression level, we observed an increase in the expression of 34 miRNAs and a decrease in the expression of 34 miRNAs when comparing SP to MP cells. Using quantitative real-time reverse transcription-polymerase chain reaction to test for differentially expressed miRNAs combined with bioinformatics results, we found that the downregulated miRNAs, such as hsa-miR-3175 and hsa-miR-203, and the upregulated miRNAs, including hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star, may be important in maintaining and regulating the characteristics of SP cells.
CONCLUSION: There are key miRNAs expressed within the SP cells of the gastric cancer cell line MKN-45, and include hsa-miR-3175, hsa-miR-203, hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star.
Keywords: ATP-binding cassette transporters, Side population cells, Benzimidazoles (Hoe 33342), Stomach neoplasm, Stem cells, MircoRNA
Core tip: MicroRNAs (miRNAs) act as one mechanism by which coding genes are regulated, and they are involved in many biological and pathological processes including carcinogenesis. However, identifying specific miRNAs is difficult, because many miRNAs possess more than one target and many target genes are regulated by more than one miRNA. We used fluorescence-activated cell sorting and Hoechst 33342 labeling to obtain side population (SP) cells from the human gastric carcinoma cell line MKN-45. The miRNA expression profiles of SP and major population cells were examined by miRNA gene chip analysis, and key miRNAs were identified according to aberrant expression and their possible targets as predicted by bioinformatics.
INTRODUCTION
Gastric cancer, with an estimated 986600 new cases worldwide in 2008, is the fourth leading form of cancer and accounts for more than 8% of total cancers[1]. It is one of the most common and fatal malignancies in East Asia, despite a decreased incidence in the West[2]. The most effective and specific methods available to treat the disease include local resection, chemotherapy and radiotherapy. However, the question remains regarding whether there are other, more effective and specific methods to treat gastric cancer.
The high mortality rate for gastric cancer is due to increased cases of relapse and metastasis. The cancer stem cell (CSC) hypothesis has received increased attention recently for its convincing explanation for the initiation of relapse and metastasis in several types of carcinomas, including gastric carcinoma[3-5]. From a large amount of research, several malignant tumor tissues and cell lines have been discovered that possess CSCs, and include acute lymphoblastic leukemia and several solid tumors such as breast, colon, prostate, and gastric cancers[6-9].This finding has strengthened the hypothesis that the initiation of relapse and metastasis may be caused by CSCs. SP cells are a subpopulation of many normal tissues, cancer tissues and cell lines that possess CSC-like phenotypes. However, the exact nature of these cells has yet to be elucidated, particularly regarding the profiles of key miRNAs and their target genes. Our study used fluorescence-activated cell sorting (FACS) to sort side population (SP) cells from gastric cancer cell lines, and then microRNA (miRNA) gene chip analyses were used to examine the miRNA expression profiles of SP and major population (MP) cells. Finally, key miRNAs were obtained according to aberrant expression, and their possible targets were predicted by bioinformatics.
MATERIALS AND METHODS
Cell culture
The human gastric adenocarcinoma cell line MKN-45 was obtained from the Cancer Institute, Chinese Academy of Medical Science and was separately maintained at the Royal Park Memorial Institute (RPMI) in 1640 medium (Invitrogen, United States) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin G, and 100 μg/mL streptomycin. The cells were maintained at 37 °C in a humidified 5% CO2 incubator.
Fluorescence-activated cell sorting
Nearly confluent MKN-45 cells were harvested by trypsinization with 0.25% Trypsin EDTA (Invitrogen, United States), centrifuged at 1000 r/m for 10 min, washed twice with phosphate buffered saline (PBS), re-suspended at 1 × 106 cells/mL in pre-warmed 37 °C medium of RPMI 1640 with 2% FCS and passed through 40 μm cell strainers (BD Falcon, United States) to obtain single-cell suspensions. The cells were then labeled with Hoechst 33342 (Sigma-Aldrich, United States) at a concentration of 5 μg/mL, and the labeled cells were incubated in the dark for 60-75 min in a 37 °C water bath with intermittent mixing, with or without 75 μmol/L verapamil (Sigma-Aldrich, United States). The cells were suspended in ice-cold PBS containing 2% FBS after staining, and maintained at 4 °C until flow cytometry analysis. Cells were labeled with 1 μg/mL propidium iodide (PI) to assess viability 5 min before examination. The stained cells were analyzed using a FACS Aria II (BD Biosciences, San Jose, CA, United States). The Hoechst dye was excited by an ultraviolet laser at 375 nm, and its fluorescence measured with 450/40 nm (Hoechst blue) and 695/40 LP (long-pass, Hoechst red) optical filters.
miRNA microarray assay and quantitative real-time quantitative reverse transcription-polymerase chain reaction
We used the Affymetrix gene chip miRNA 2.0 array according to the manufacturer’s instructions for our miRNA microarray assay (Affymetrix Int, CA, United States). Each miRNA microarray chip contained 1100 identified human miRNAs probes. Briefly, total RNA (200 ng) was polyadenylated and tagged with biotin HSR ligation, reverse transcribed, and the cDNA was hybridized to a miRNA bead chip for 16 h at 48 °C. The hybridized miRNA bead chip was washed for 1 h in a washing station, and the arrays were scanned on an Affymetrix reader. Microarray data for each sample were normalized to the median, and the processing and analysis was performed with Affymetrix software.
Total RNA, including miRNA, was extracted from MP and SP cells using Trizol reagent according to the manufacturer’s instructions. SYBR green mRNA quantitative real-time polymerase chain reaction (PCR) was performed using the sequence-specific stem-loop primers supplied by RiboBio (Guangzhou, China). Quantitative reverse transcription PCR (RT-PCR) was conducted using a standard SYBR Green PCR kit (QIAGEN) protocol with a Light Cycler 480 real-time instrument (Roche). The relative expression was calculated using the 2-dCT method. The transcription levels of U6 were used as an internal control.
Bioinformatic analysis
In this study, predicted targets of novel miRNA were analyzed and determined using three publicly available algorithms including MiRanda (http://www.microrna.org), PicTar (http://pictar.mdc-berlin.de) and TargetScan (http://www.targetscan.org/). These searchable websites predict biological targets of miRNAs by searching for the presence of conserved 8-mer and 7-mer sites that match the seed region of the miRNA and provide details of the 3’-UTR alignments with predicted sites. To decrease the number of false-positive results, only putative target genes predicted by at least two programs were accepted. In addition, we used a Capital-Bio Molecule Annotation System Version 3.0 to perform gene ontology analysis on the target genes and the specific biological process categories that were enriched.
RESULTS
Gastric cancer cell MKN-45 contain SP cells
Through the use of FACS, we determined that the gastric cancer cell line MKN-45 contained SP cells. We set the sorting gate according to the ability of cells to efflux Hoechst 33342 and their sensitivity to verapamil. The lower left quadrant of the FACS profile, which showed Hoechst blue and could be blocked by verapamil, was defined as the SP. The top right quadrant of the FACS profile, which showed Hoechst red and was not blocked by verapamil, was defined as the MP. We targeted these cells and differentiated them for further study. In the MKN-45 cells, the percentage of SP was 2.0% of the total cells (Figure 1A).
Figure 1.
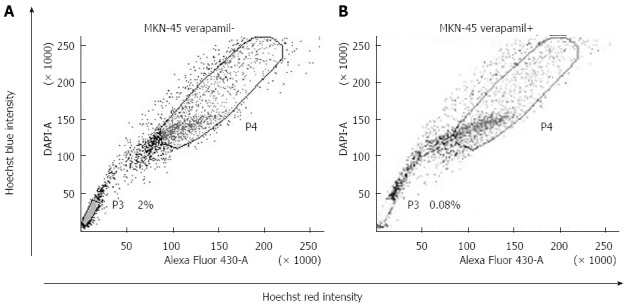
Side population cell analysis. The P3 gate was the side population (SP) cells and the P4 gate was the major population (MP) cells. A: SP ratio in MKN-45 was 2.0%; B: The SP cells obviously decreased with both Hoechst 33342 and verapamil. Figure 1 cited partly from the article of Not All Side Population Cells Contain Cancer Stem-Like Cells in Human Gastric Cancer Cell Lines which published in Dig Dis Sci 2013; 58: 132-139.
miRNA expression profiles and quantitative real-time RT-PCR
To investigate whether miRNAs were differentially expressed in SP and MP cells, we compared their miRNA expression profiles using a miRNA microarray (Figure 2A and B). Using a significance criterion of a 1.5-fold or greater difference in expression level, we observed the increased expression of 34 miRNAs and the decreased expression of 34 miRNAs in SP vs MP cells. The top 15 upregulated and 11 downregulated miRNAs are shown in Table 1. Next, we used quantitative real-time RT-PCR to test the differentially expressed miRNAs, and we found that downregulated miRNAs, such as hsa-miR-3175 and hsa-miR-203, and upregulated miRNA, including hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star, may be important in the maintenance and regulation of SP cell characteristics.
Figure 2.
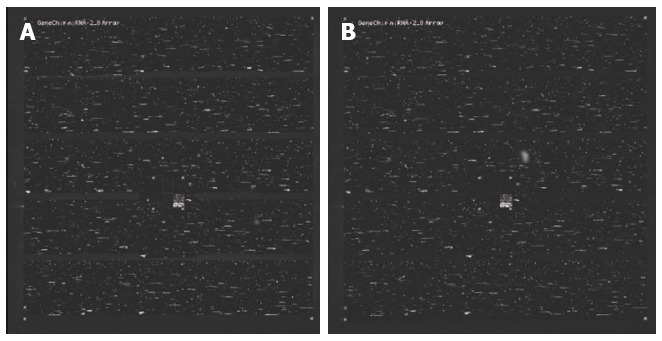
MicroRNA microarray. Affymetrix genechip 2.0 was used to examine microRNA expression profiles of SP and MP cells of MKN-45. A: Hybridized genechip with microRNA of SP cells; B: Hybridized genechip with microRNA of MP cells. SP: Side population; MP: Major population.
Table 1.
MicroRNA microarray and quantitative real-time-polymerase chain reaction analyses results
| Down-regulated genes | Fold change microarray |
ΔCT |
-ΔΔCT |
Upregulated genes | Fold change microarray |
ΔCT |
-ΔΔCT |
| qRT-PCR | qRT-PCR | ||||||
| hsa-miR-3175 | 7.29 | 21.82 | -1.32 | hsa-miR-25-star | 27.35 | 6.63 | 4.47 |
| hsa-miR-1246 | 3.46 | 14.07 | -0.64 | hsa-miR-1275 | 5.13 | 13.77 | 0.48 |
| hsa-miR-663 | 3.12 | 10.69 | -0.13 | hsa-miR-339-5p | 4.03 | 15.63 | 0.89 |
| hsa-miR-1281 | 2.93 | 14.72 | -0.74 | hsa-miR-140-3 p | 3.56 | 13.30 | 0.41 |
| hsa-miR-422a | 2.56 | 14.41 | -0.69 | hsa-miR-197 | 3.36 | 14.08 | 0.63 |
| hsa-miR-1228-star | 2.41 | 16.62 | -0.98 | hsa-miR-362-5p | 3.01 | 15.25 | 0.86 |
| hsa-miR-1975 | 2.10 | 12.84 | -0.38 | hsa-miR-324-5p | 2.75 | 17.85 | 1.06 |
| hsa-miR-1826 | 2.09 | 14.18 | -0.67 | hsa-miR-34a | 2.70 | 23.46 | 2.32 |
| hsa-miR-1915 | 2.08 | 10.83 | -0.14 | hsa-miR-210 | 2.17 | 13.30 | 0.44 |
| hsa-miR-4284 | 2.07 | 14.34 | -0.65 | hsa-miR-500-star | 2.06 | 11.06 | 0.18 |
| hsa-miR-203 | 1.57 | 26.13 | -3.84 | hsa-miR-130a | 1.77 | 22.49 | 2.13 |
| hsa-miR-130b | 1.61 | 17.34 | 0.85 | ||||
| hsa-miR-18a | 1.60 | 13.41 | 0.46 | ||||
| hsa-miR-29a | 1.57 | 15.46 | 0.80 | ||||
| hsa-miR-149 | 1.54 | 11.96 | 0.32 | ||||
Differentially expressed microRNAs (miRNAs) in SP and MP cells of the gastric cancer cell line MKN-45 as determined by miRNA microarray and quantitative real-time-PCR analyses. These results showed that the miRNA profiles of SP cells differed from the miRNA profiles of MP cells. Moreover, downregulated miRNAs, such as hsa-miR-3175 and hsa-miR-203, and upregulated miRNAs, including hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star, showed significant differences in expression between SP and MP cells when tested by quantitative real-time PCR. SP: Side population; MP: Major population; PCR: Polymerase chain reaction.
Bioinformatics analysis
There are 33036 miRNA-target relationships predicted by at least two of the three database software programs MiRanda, TargetScan and Pictar[10-12]. However, there are 469 miRNA-target relationships after experimental verification using the Tarbase database. There are 9 microRNAs with predicted or experimentally verified targets among 68 differentially expressed microRNAs, including 8 upregulated microRNAs and one downregulated microRNA. Hsa-miR-130a shows the largest number of target genes, 406, followed by hsa-miR-29a, at 402. These 9 microRNAs have 2006 targets that have been predicted or experimentally validated (Table 2). The targets are often regulated by more than one miRNA (Table 3). Finally, we can construct a global microRNA regulatory network by integrating the human protein-protein interaction data with the predicted microRNA-target relationships. This network is closely linked with the module function analysis[13,14] (Figure 3).
Table 2.
Differentially expressed microRNAs and their target number and known targets
| microRNA | Sp/Mp (FC) | Target number | Validated target number | Known targets |
| hsa-miR-130a | 1.77 | 406 | 3 | MEOX2; TAC1; HOXA5 |
| hsa-miR-29a | 1.57 | 402 | 3 | DNMT3A; BACE1; DNMT3B |
| hsa-miR-130b | 1.61 | 392 | 0 | |
| hsa-miR-203 | 0.64 | 275 | 0 | |
| hsa-miR-34a | 2. 70 | 260 | 3 | E2F3; CCND1; CDK6 |
| hsa-miR-149 | 1.54 | 129 | 0 | |
| hsa-miR-18a | 1.60 | 104 | 0 | |
| hsa-miR-324-5p | 2.75 | 33 | 0 | |
| hsa-miR-210 | 2.17 | 5 | 1 | EFNA3 |
Bioinformatic analysis of differentially expressed microRNAs indicated that differentially expressed microRNAs had predicted or experimentally verified target genes. Moreover, there are multiple targets for many miRNAs, but most of these targets have not been studied or validated.
Table 3.
Genes that are regulated by more than 4 microRNAs
| Gene ID | Gene name | microRNA number | All microRNAs |
| 80218 | NAA50 | 5 | hsa-miR-203; hsa-miR-18a; hsa-miR-34a; hsa-miR-130a; hsa-miR-130b |
| 164 | AP1G1 | 5 | hsa-miR-324-5p; hsa-miR-130b; hsa-miR-29a; hsa-miR-130a; hsa-miR-203 |
| 7803 | PTP4A1 | 4 | hsa-miR-203; hsa-miR-130a; hsa-miR-130b; hsa-miR-29a |
| 90355 | C5orf30 | 4 | hsa-miR-130b; hsa-miR-18a; hsa-miR-130a; hsa-miR-203 |
| 80829 | ZFP91 | 4 | hsa-miR-130b; hsa-miR-130a; hsa-miR-324-5p; hsa-miR-29a |
| 28514 | DLL1 | 4 | hsa-miR-149; hsa-miR-130a; hsa-miR-130b; hsa-miR-34a |
| 23013 | SPEN | 4 | hsa-miR-203;hsa-miR-130a; hsa-miR-29a; hsa-miR-130b |
| 57659 | ZBTB4 | 4 | hsa-miR-18a; hsa-miR-149; hsa-miR-130a; hsa-miR-130b |
| 23261 | CAMTA1 | 4 | hsa-miR-34a; hsa-miR-203; hsa-miR-130b; hsa-miR-130a |
| 9444 | QKI | 4 | hsa-miR-18a; hsa-miR-130b; hsa-miR-29a; hsa-miR-130a |
| 166336 | PRICKLE2 | 4 | hsa-miR-130b; hsa-miR-29a; hsa-miR-130a; hsa-miR-203 |
| 5156 | PDGFRA | 4 | hsa-miR-130a; hsa-miR-149; hsa-miR-130b; hsa-miR-34a |
Predicted target genes of key microRNAs. Some microRNAs, such as hsa-miR-324-5p, hsa-miR-130b, hsa-miR-29a, hsa-miR-130a, and hsa-miR-203, modulate multiple genes; these results indicate that differentially expressed microRNAs and their targets have a reticular network.
Figure 3.
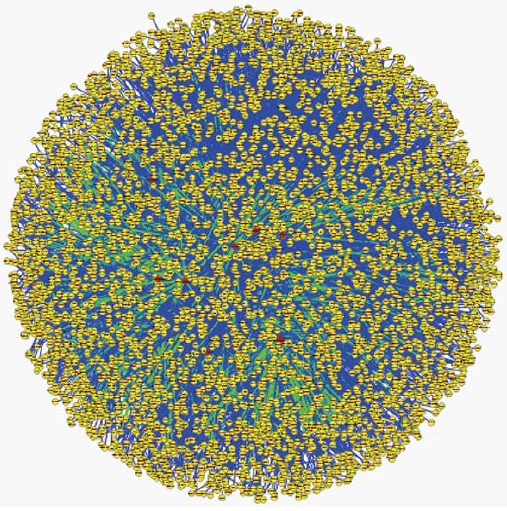
Visualization of the global microRNA regulated network. Red nodes represent microRNA, yellow nodes represent coding genes. Green edge represents the microRNA-target relationship, blue edge represents the protein-protein interaction.
DISCUSSION
The increased mortality rate for gastric cancer is due to increased cases of relapse and metastasis. Increasing numbers of studies have shown that CSCs are involved in tumor progression and metastasis and are associated with increased aggressiveness and metastasis in vivo but not in vitro[15]. Several published reports have demonstrated that SP cells are being increasingly used as an effective method to obtain and identify stem cells or putative CSCs[6-9]. In our study, we obtained gastric CSCs from MKN-45 cells through sorting by FACS technology, and characterized their CSC properties[16]. Thus, we used SP cells as a gastric CSC model to elucidate their miRNA expression profiles, predict miRNA targets, and analyze possible miRNA modulating mechanisms.
We determined the miRNA expression profiles of SP and MP cells sorted from the MKN-45 cell line using a miRNA microarray, which showed that the miRNA profiles were significantly different between SP and MP cells; the total number of differentially expressed miRNAs was 68. Moreover, we tested the differentially expressed miRNAs using quantitative real-time RT-PCR, and found that downregulated miRNAs, such as hsa-miR-3175 and hsa-miR-203, and upregulated miRNAs, including hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star, showed significant differences between SP and MP cells. In general, miRNAs upregulated in SP cells act as oncogenic miRNAs, while downregulated miRNAs act as tumor suppressors. As a tumor suppressor, hsa-miR-3175 appears to be a novel biomarker; there have been limited studies of this miRNA, and further research to validate whether it is involved in the maintenance of cancer cell stemness is warranted. Conversely, hsa-miR-203 appears to be involved in maintaining the stemness of cancer cells rather than in tumorigenicity, and it plays a crucial role in the progression of human carcinoma. Decreased expression of miR-203 was significantly related to poor differentiation, advanced clinical stage, T3-4 tumor grade, lymph node metastasis, and decreased 5-year overall survival. Furthermore, miR-203 may regulate the expression of E-cadherin mesenchymal transition, and CD44, a marker of CSCs[17], suggesting that miR-203 could be involved in the regulation of CSCs. Interestingly, another study had found that miR-203 was downregulated and Bmi-1 was upregulated in the SP cells of the esophageal squamous cell carcinoma cell line EC9706. MiR-203 over-expressing cells also showed a significant reduction in colony formation that was resistant to chemotherapeutic drug treatment, and tumorigenicity in nude mice. These results indicate that the stem renewal factor Bmi-1 is a direct target of miR-203. The regulation of Bmi-1 by miR-203 may play an important role in controlling the proliferation and self-renewal of esophageal cancer stem-like cells[18]. Our study on miR-203 in SP cells which showed downregulated expression, is in agreement with the above results. However, there is a lack of consensus in the literature about this finding. Stánitz et al[19] reported contradictory findings, in which gastric adenocarcinoma patients with regular alcohol consumption showed an upregulation of miR-203, miR-205, and miR-223[19]. Therefore, we cannot state conclusively that miR-203 acts as a tumor suppressor while maintaining stemness, such as proliferation, self-renewal, and tumorigenicity, of gastric CSCs. In the future, we will continue to study the specific mechanism of action of miR-203 in gastric cancer. As for other oncogenic miRNAs identified in our study, hsa-miR-130a, hsa-miR-324-5p, hsa-miR-34a, and hsa-miR-25-star, are known not to play roles in many biological processes aside from maintaining stemness of CSCs. The results of a previous study show that the low miR-34a expression levels in many cells together with their colony forming efficiency, a characteristic property of CSCs, can be inhibited by miR-34a replacement or synthetic miR-34a in vivo. This finding is in contrast with our study results on miR-34a in vitro. Furthermore, the expression of miR-34a antagomirs in these cells promoted tumor development, indicating that miR-34a is a negative regulator of the tumorigenic properties of cancer cells and the stemness characteristics of CSCs[20,21]. A further study has found that there is a novel regulatory network composed of p53, CD95, let-7, and miR-34a that affects cancer cell survival, differentiation, and sensitivity to apoptotic signals[22]. Moreover, the treatment of pancreatic CSCs with chromatin-modulating agents results in the inhibition of Bcl-2, CDK6 and SIRT1, which are the putative targets of miR-34a. MiR-34a upregulation by these agents also induced acetylated p53, p21 (WAF1), p27 (KIP1) and PUMA (BCL2 binding component 3) in pancreatic CSCs[23]. From these results, we can conclude that miR-34a acts as a major tumor suppressor in maintaining the stemness of CSCs, but the putative targets, potential functions and mechanism of miR-34a in gastric cancer SP cells remain to be explored and detailed in further studies. Another oncogenic miRNA is miR-130a, which showed high levels of expression in some cancer cells[24], especially in some drug-resistant cell lines[25], and low levels in other cancer cells[26,27]. Interestingly, our results on miR-130a showed high levels in SP cells. More information about miR-130a and CSCs, the putative targets regulated by miR-130a, and the mechanism of miR-130a in gastric CSCs requires further study. miR-324-5p and miR-25-star are 2 additional oncogenic miRNAs that were identified in our study. miR-324-5p can inhibit the proliferation of glioma cells via the targeted regulation of glioma-associated oncogene 1[28]. It is also involved in the development of cervical cancer as a result of human papillomavirus infection, which might regulate the oncogene E5. Whether miR-324-5p participates in the regulation of gastric CSCs has yet to be studied. MiR-25-star may be a new player in the behavior of cancer and CSCs, but there is nothing in the literature to date.
A further bioinformatic analysis found that hsa-miR-130a, hsa-miR-29a, hsa-miR-210, and hsa-miR-34a each had more than one predicted or experimentally verified targets, while other miRNAs, such as hsa-miR-203 and hsa-miR-324-5p, had no known predicted or experimentally verified targets (Table 2). However, the latter miRNAs often regulated a common target at the same time (Table 3), such as putative transcription factors, including Dll1 (Delta-like 1), ZBTB4, (the Zinc fingers, C2H2 and BTB domain containing (ZBTB) family member), CAMTA1 (calmodulin-binding transcription activator 1), QKI (the RNA-binding protein Quaking) and PDGFRα (platelet-derived growth factor receptor alpha) among others. The transcription factor Dll1, which is one of the Notch signaling pathway ligands, is involved in the maintenance of stem cells during embryogenesis and in self-renewing tissues of the adult. The epigenetic regulation of the Notch ligand DLL1 controls Notch1 signaling activation in gastric cancer, and Notch1 inhibition is associated with the diffuse type of gastric cancer[29]. CAMTA1 is a putative transcription factor in glioblastoma stem cells that acts as a tumor suppressor gene and is a target of miR-9 and miR-17[30]. QKI is a newly identified tumor suppressor found in multiple cancers whose expression is significantly decreased in most GC tissues; the reduced QKI expression correlates well with poor differentiation status, depth of invasion, gastric lymph node metastasis, distant metastasis, advanced TNM stage, and poor survival[31]. In mouse and human CRC cells, miR-574-5p has been shown to regulate QKI isoforms post-transcriptionally and to cause altered β-catenin and p27 (Kip1) expression, increased proliferation, migration and invasion and decreased differentiation and cell cycle exit[32]. PDGFRα is involved in the resistance of the stem cell factor kit to imatinib mesylate in gastrointestinal stromal tumors[33]. Deficiencies in the downregulation of PDGFRα have been identified as a candidate mechanism for tumor cell proliferation, and the knockdown of PDGFRα by siRNA results in a reduction in cell growth[34]. Other targets, such as E2F3, CCND1 and CDK6, are also important genes that may be involved in tumorigenicity and the maintenance of stemness in CSCs. Therefore, according to our results, differentially expressed microRNAs and their targets have a reticular network. In fact, the maintenance and modulation of stemness in CSCs may be the result of regulation by a double shift network consisting of 2 groups of miRNAs and their target genes working with opposing functions. This work on miRNAs and their possible targets lays the groundwork for future studies in this area.
In summary, we elucidated the expression profiles of microRNAs using SP cells as a gastric CSC model and predicted the key microRNA targets by bioinformatics analysis. These findings will pave the way for further studies on the molecular modulation of cancer mechanisms by microRNA.
ACKNOWLEDGMENTS
The authors thank Jun-Ying Jia and Chun-Chun Liu (Bio-Physics Institute of Chinese Academy) for their help in cell sorting.
COMMENTS
Background
Gastric cancer remains the most common cancer worldwide. The most effective and specific methods available to address the disease include local resection, chemotherapy, and radiotherapy. However, the question remains as to whether there are any other more effective and specific methods to treat gastric cancer. The cancer stem cell (CSC) hypothesis has garnered more attention in recent times for its convincing explanation for the initiation of relapse and metastasis of cancers including gastric carcinoma. Side population (SP) cells are a subpopulation found in many normal tissues, cancer tissues, and cell lines that possess CSC-like phenotypes. However, the exact nature of these cells has yet to be elucidated, especially regarding key microRNA (miRNA) profiles and the target genes regulated by these miRNAs.
Research frontiers
MiRNAs act as one mechanism underlying the regulation coding genes and are involved in many biological and pathological processes including carcinogenesis. Increasingly, aberrant miRNA expression has been found in tumor tissues or cells; however, there has been little research on the miRNAs of CSCs or in SP cells that possess CSC-like characteristics.
Innovations and breakthroughs
This study is the first to use SP cells from the gastric cancer cell line MKN-45 as a CSC model to elucidate the miRNAs profiles, and we discovered key miRNAs within these SP cells. This study provides the groundwork for additional research, and the results of this study provide mechanisms for future miRNA studies.
Applications
Previous studies have investigated the diagnostic and prognostic value of miRNAs in gastric cancer. One such study identified a 7-miRNA signature (miR-10b, miR-21, miR-223, miR-338, let-7a, miR-30a-5p and miR-126) that is an independent predictor for overall survival and relapse-free survival shown by multivariate analysis. However, the most specific and effective miRNAs have yet to be discovered. The association of miRNA deregulation with the pathogenesis and progression of malignant disease illustrates the great potential of utilizing miRNAs as targets for therapeutic interventions. However, the therapeutic potential of miRNA-based treatments in malignant disease remains largely unexplored.
Terminology
Fluorescence activated cell sorting (FACS) is a technique based on nucleic acid dye Hoechst 33342 efflux, which can sort cells into SP cells and MP cells. The CSC hypothesis suggests that tumors consist of tumor-forming, self-renewing CSCs within a large population of non-tumor-forming cancer cells. CSCs resist standard chemotherapy that reduces tumor mass by killing non-stem cells. During remission, CSCs can regenerate all the cell types in the tumor through their stem cell-like behavior, resulting in relapse of the disease. The side population is a subpopulation of many normal tissues, cancer tissues and cell lines that possess CSC-like phenotypes. They can be isolated by fluorescence-activated cell sorting techniques.
Peer-review
This is an interesting manuscript. The study is well designed. In this study, the authors find that the gastric cancer cell line MKN-45 contains SP cells that possess cancer stem-like characteristics, and further bioinformatic analysis found that there are key miRNAs in the cancer stem cell-like SP cells of the gastric cancer cell line MKN-45. However, the specificity and effectiveness of the miRNAs and their diagnostic and prognostic values will require additional studies to test and prove.
Footnotes
Supported by National Natural Science Fund, No. 81000706/H1108; National Key Technology Research and Development Program, No. 2012BA141B01; and the Air Force General Hospital of Chinese PLA Innovation Fund, No. KZ2013035.
Ethics approval: The study was reviewed and approved by the Medical Ethics Committee of PLA Air Force General Hospital.
Conflict-of-interest: There are no conflicts of interest to declare.
Data sharing: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: October 29, 2014
First decision: November 14, 2014
Article in press: January 16, 2015
P- Reviewer: Kwon RS, Lehmann U S- Editor: Yu J L- Editor: Cant MR E- Editor: Zhang DN
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Krejs GJ. Gastric cancer: epidemiology and risk factors. Dig Dis. 2010;28:600–603. doi: 10.1159/000320277. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 5.McDonald SA, Greaves LC, Gutierrez-Gonzalez L, Rodriguez-Justo M, Deheragoda M, Leedham SJ, Taylor RW, Lee CY, Preston SL, Lovell M, et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology. 2008;134:500–510. doi: 10.1053/j.gastro.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Dey D, Saxena M, Paranjape AN, Krishnan V, Giraddi R, Kumar MV, Mukherjee G, Rangarajan A. Phenotypic and functional characterization of human mammary stem/progenitor cells in long term culture. PLoS One. 2009;4:e5329. doi: 10.1371/journal.pone.0005329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Oates JE, Grey BR, Addla SK, Samuel JD, Hart CA, Ramani VA, Brown MD, Clarke NW. Hoechst 33342 side population identification is a conserved and unified mechanism in urological cancers. Stem Cells Dev. 2009;18:1515–1522. doi: 10.1089/scd.2008.0302. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda K, Saikawa Y, Ohashi M, Kumagai K, Kitajima M, Okano H, Matsuzaki Y, Kitagawa Y. Tumor initiating potential of side population cells in human gastric cancer. Int J Oncol. 2009;34:1201–1207. [PubMed] [Google Scholar]
- 10.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimeault M, Batra SK. Characterization of nonmalignant and malignant prostatic stem/progenitor cells by Hoechst side population method. Methods Mol Biol. 2009;568:139–149. doi: 10.1007/978-1-59745-280-9_8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Xi H, Cai A, Xia Q, Wang XX, Lu C, Zhang Y, Song Z, Wang H, Li Q, et al. Not all side population cells contain cancer stem-like cells in human gastric cancer cell lines. Dig Dis Sci. 2013;58:132–139. doi: 10.1007/s10620-012-2330-1. [DOI] [PubMed] [Google Scholar]
- 17.Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M, Xiao H. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. Tumour Biol. 2014;35:5953–5963. doi: 10.1007/s13277-014-1790-7. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Jiang X, Li H, Guo L, Jiang W, Lu SH. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014;23:576–585. doi: 10.1089/scd.2013.0308. [DOI] [PubMed] [Google Scholar]
- 19.Stánitz E, Juhász K, Tóth C, Gombos K, Natali PG, Ember I. Evaluation of MicroRNA expression pattern of gastric adenocarcinoma associated with socioeconomic, environmental and lifestyle factors in northwestern Hungary. Anticancer Res. 2013;33:3195–3200. [PubMed] [Google Scholar]
- 20.Basak SK, Veena MS, Oh S, Lai C, Vangala S, Elashoff D, Fishbein MC, Sharma S, Rao NP, Rao D, et al. The CD44(high) tumorigenic subsets in lung cancer biospecimens are enriched for low miR-34a expression. PLoS One. 2013;8:e73195. doi: 10.1371/journal.pone.0073195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One. 2014;9:e90022. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hau A, Ceppi P, Peter ME. CD95 is part of a let-7/p53/miR-34 regulatory network. PLoS One. 2012;7:e49636. doi: 10.1371/journal.pone.0049636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santarpia L, Calin GA, Adam L, Ye L, Fusco A, Giunti S, Thaller C, Paladini L, Zhang X, Jimenez C, et al. A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr Relat Cancer. 2013;20:809–823. doi: 10.1530/ERC-13-0357. [DOI] [PubMed] [Google Scholar]
- 25.Yang LY, Wang HJ, Jia XB, Wang X, Luo J, Zhang XY. [Expression of miR-130a in cisplatin resistant cell lines of ovarian cancer] Sichuan Da Xue Xue Bao Yi Xue Ban. 2012;43:60–64. [PubMed] [Google Scholar]
- 26.Zhu X, Lin Z, Du J, Zhou X, Yang L, Liu G. Studies on microRNAs that are correlated with the cancer stem cells in chronic myeloid leukemia. Mol Cell Biochem. 2014;390:75–84. doi: 10.1007/s11010-013-1958-2. [DOI] [PubMed] [Google Scholar]
- 27.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu HS, Zong HL, Shang M, Ming X, Zhao JP, Ma C, Cao L. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci. 2014;18:828–832. [PubMed] [Google Scholar]
- 29.Piazzi G, Bazzoli F, Ricciardiello L. Epigenetic silencing of Notch signaling in gastrointestinal cancers. Cell Cycle. 2012;11:4323–4327. doi: 10.4161/cc.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schraivogel D, Weinmann L, Beier D, Tabatabai G, Eichner A, Zhu JY, Anton M, Sixt M, Weller M, Beier CP, et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 2011;30:4309–4322. doi: 10.1038/emboj.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bian Y, Wang L, Lu H, Yang G, Zhang Z, Fu H, Lu X, Wei M, Sun J, Zhao Q, et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem Biophys Res Commun. 2012;422:187–193. doi: 10.1016/j.bbrc.2012.04.138. [DOI] [PubMed] [Google Scholar]
- 32.Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, Zhang T, Wang G, Guo Z, Luo Y, et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardsley MR, Horváth VJ, Asuzu DT, Lorincz A, Redelman D, Hayashi Y, Popko LN, Young DL, Lomberk GA, Urrutia RA, et al. Kitlow stem cells cause resistance to Kit/platelet-derived growth factor alpha inhibitors in murine gastrointestinal stromal tumors. Gastroenterology. 2010;139:942–952. doi: 10.1053/j.gastro.2010.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siebzehnrubl FA, Jeske I, Müller D, Buslei R, Coras R, Hahnen E, Huttner HB, Corbeil D, Kaesbauer J, Appl T, et al. Spontaneous in vitro transformation of adult neural precursors into stem-like cancer cells. Brain Pathol. 2009;19:399–408. doi: 10.1111/j.1750-3639.2008.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


