Abstract
Polygenic risk scores, based on risk variants identified in genome-wide-association-studies (GWAS), explain a considerable portion of the heritability for schizophrenia (SZ) and bipolar disorder (BD). However, little is known about the combined effects of these variants, although polygenic neuroimaging has developed into a powerful tool of translational neuroscience. In this study, we used genome wide significant SZ risk variants to test the predictive capacity of the polygenic model and explored potential associations with white matter volume, a key candidate in imaging phenotype for psychotic disorders.
By calculating the combined additive schizophrenia risk of seven SNPs (significant hits from a recent schizophrenia GWAS study), we show that increased additive genetic risk for SZ was associated with reduced white matter volume in a group of participants (n = 94) consisting of healthy individuals, SZ first-degree relatives, SZ patients and BD patients. This effect was also seen in a second independent sample of healthy individuals (n = 89). We suggest that a moderate portion of variance (~4%) of white matter volume can be explained by the seven hits from the recent schizophrenia GWAS.
These results provide evidence for associations between cumulative genetic risk for schizophrenia and intermediate neuroimaging phenotypes in models of psychosis. Our work contributes to a growing body of literature suggesting that polygenic risk may help to explain white matter alterations associated with familial risk for psychosis.
Highlights
-
•
We demonstrate that compared to controls, white matter volume is reduced in SZ relatives, SZ patients and BD patients.
-
•
Seven schizophrenia risk SNPs are associated with reduced white matter volume across the psychosis spectrum.
-
•
Increased schizophrenia risk is associated with reduced white matter volume
1. Introduction
Neurobiological models of schizophrenia (SZ) and bipolar disorder (BD) implicate altered interregional connectivity and suggest that some of the changes in white matter structure may be heritable (Kaymaz and van Os, 2009; McDonald et al., 2004; McIntosh et al., 2006). This hypothesis is supported by studies that show that decreased white matter volume may be an indicator of genetic liability to both schizophrenia (Francis et al., 2013; Kaymaz and van Os, 2009; McDonald et al., 2004; McIntosh et al., 2006; Oertel-Knochel et al., 2012; van Haren et al., 2012) and bipolar disorder (Emsell and McDonald, 2009; Hasler et al., 2006; Matsuo et al., 2012). Genome wide association studies (GWAS) are identifying an increasing number of single-nucleotide polymorphism (SNPs), significantly associated with risk for schizophrenia and/or bipolar disorder (Cardno and Owen, 2014; GWAS Consortium, 2011; Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Schulze et al., 2014; Smoller et al., 2013), although the functional mechanisms by which these variants confer risk for psychosis remain largely unknown. A recent study reported that a combined measure of schizophrenia risk variant load (polygenic schizophrenia score or risk profile score; RPS) was significantly associated with white and total brain volume, in both healthy and affected individuals (Terwisscha van Scheltinga et al., 2013). The study further suggested that the association was driven by a smaller number of SZ-associated SNPs (n = 186) than the SNP set that best predicted WM volume (n = 2020) or diagnosis (n = 14,751). However, Papiol et al. (2014) failed to replicate associations between polygenic risk score and white matter volume, using the same discovery data set (PGC-SZ1; GWAS Consortium, 2011) and largely the same SNPs (n = 185) hypothesised to be associated with both schizophrenia and WM volume. Papiol et al. (2014) suggested there were only minor differences between study designs, which were unlikely to account for between-study variability (including sample size, ethnicity, genotyping methods and scanner type). Considering that disrupted white matter has been repeatedly observed in relatives of patients with schizophrenia and patients with bipolar disorder (Emsell and McDonald, 2009; Francis et al., 2013; Hasler et al., 2006; Matsuo et al., 2012; McDonald et al., 2004; McIntosh et al., 2006; Oertel-Knochel et al., 2012; van Haren et al., 2012), we suggest that common genetic risk factors should contribute to the neurobiological mechanisms that underlie volumetric white matter disturbances. At present, it is not known which molecular pathways are responsible for modulating white matter at the genetic level, and thus in the present study, we employ an RPS model calculated only using robust, genome-wide significant SNPs for schizophrenia (GWAS Consortium, 2011). We aim to explore the association between additive effects of these seven schizophrenia risk SNPs (7-SNP SZ-RPS) with white matter volume. We additionally base our hypothesis on the findings of Terwisscha van Scheltinga et al. (2013) who reported that at least one of these top-hits from the PGC-SZ1 study (rs17662626 near PCGEM1) was implicated in this prior association, and risk associated with these highly significant SNPs may produce more reliable associations with imaging phenotypes than SNPs added to a polygenic model with less stringent p thresholds and less robust ORs. We hypothesise that larger risk profile scores are associated with reduced white matter volume across the psychosis spectrum.
2. Materials and methods
2.1. Participants
2.1.1. Frankfurt site
A total of n = 94 participants were included and divided into four groups: SZ patients, first-degree relatives, BD patients and healthy controls. The SZ patient group consisted of n = 24 patients of the Department of Psychiatry, Psychosomatic Medicine and Psychotherapy of the University of Frankfurt, Germany (mean age [years]: M = 38.10 [SD = 10.34]) diagnosed according to the DSM-IV criteria (Guze, 1995). At the time of measurement, all patients were treated with atypical antipsychotics. We assessed present psychopathological symptoms in the SZ group using the Positive and Negative Symptom Scale (PANSS) (Kay et al., 1987). The relative group consisted of n = 12 first-degree unaffected relatives of SZ patients (M [years] = 45.12 [12.67]) who were requested to provide a letter from the attending psychiatrist confirming the index patients' diagnosis. We further conducted a semi-structured interview with the relatives (German version of the Premorbid Adjustment Scale (PAS)) (Krauss et al., 2000) in order to further back-up the index patients' diagnosis. A third group included n = 20 patients with bipolar disorder I (M [years] = 39.00 [12.10]) as defined by the DSM-IV criteria (APA, 1994) and diagnosed by a board-certified psychiatrist at the Dept. of Psychiatry and Psychotherapy of Frankfurt University Medical School. Current depressive symptoms were assessed using the German version of the Beck Depression Inventory II (BDI II; Hautzinger et al., 2006), and current mania symptoms were assessed using the German version of the Bech Rafaelsen Mania Scale (BRMAS; Bech, 1981). At the time of the measurements, all patients were in disease remission (see definition in the clinical testing), were without any comorbid axis I or II disorders (including substance abuse or addiction) and were on stable medication at a minimum of 4 weeks prior to measurement (see Table 1). Remitted state in the bipolar patient group was defined by BDI II scores of < 18 (M BD patients = 10.81 [9.56]) and BRMAS scores of < 7 (M BD patients = 0.77 [1.79]) (see Table 1 for further sociodemographic details and Supplementary Table S1 for medication status).
Table 1.
Frankfurt sample: Demographic, clinical and cognitive data for all subject groups. SZ = schizophrenia patients, BD = bipolar patients, REL = first-degree relatives of patients with schizophrenia and CON = controls. RHS = Revised Hallucination Scale and MWT = multiple word choice test. VBM data were available for 94 participants (n = 38 healthy controls, n = 12 relatives of patients with schizophrenia, n = 24 patients with schizophrenia, n = 20 patients with bipolar disorder). The table contains sociodemographic and clinical data for the VBM sample (n = 94). p = significant on an α ≤ 0.05-threshold.
| SZ Mean ± SD | REL Mean ± SD | BD Mean ± SD | CON Mean ± SD | Significance | |
|---|---|---|---|---|---|
| N | 24 | 12 | 20 | 38 | 94 |
| Gender (f/m) | 10/14 | 7/5 | 11/9 | 20/18 | Kruskal–Wallis, p = 0.422 |
| Years of education | 14.13 ± 3.10 | 16.00 ± 2.12 | 15.54 ± 2.08 | 16.43 ± 1.76 CON vs. SZ: t = 2.22, p = 0.006 |
F = 4.76, p = 0.006 |
| Parental education | |||||
| Mother | 13.15 ± 2.65 | 14.11 ± 3.10 | 13.08 ± 3.12 | 13.50 ± 3.19 | F = 0.30, ns |
| Father | 14.08 ± 2.54 | 13.12 ± 3.22 | 14.12 ± 3.45 | 14.80 ± 2.51 | F = 1.43, ns |
| Age | 38.10 (10.34) | 45.12 (12.67) | 39.00 (12.10) | 37.15 (11.14) | F = 2.13, p = 0.114 |
| MWT-B | 105.55 (9.90) | 107.45 (101.43) | 108.13 (7.65) | 112.01 (7.77) | F = 2.42,p = 0.080 |
| Years of illness | 13.65 ± 6.81 | 10.11 ± 7.08 | |||
| Age of onset | 21.10 ± 4.51 | 29.17 ± 11.68 | |||
| Medication (4 weeks stable at minimum) | 27 atypical neuroleptics | 30 mood stabiliser + 12 anti-depressants | |||
| BDI II | 10.81 (9.56) | 1.98 (3.58) | t = 37.79, p < 0.001 | ||
| BRMAS | 0.77 (1.79) | 0.56 (1.01) | z = −0.36, ns | ||
| PANSS sum | 66.41 (16.43) | ||||
| PANSS positive | 17.41 (5.34) | ||||
| PANSS negative | 17.56 (5.90) | ||||
| PANSS general | 33.68 (8.49) |
2.1.1.1. Comparison of the two patient groups' sociodemographics
There were some differences in the sociodemographic variables between the patient groups, although they were matched for age, gender and education. Schizophrenia and bipolar patients differed regarding age of onset (SZ: M [years] = 21.10 [4.51]; BD: M [years] = 29.17 (11.68]; t = 8.37, p = 0.03) and duration of illness (SZ: M [years] = 13.65 [6.81]; BD: M [years] = 10.11 [7.08]; t = 6.54, p = 0.04). This was due to the clinical characteristics of the diseases, as the mean age of onset for both disorders is different (about 21–26 years for schizophrenia, about 30 years for bipolar disorder), and duration of illness depends on the age of onset and the mean age of the samples, and thus will generally differ in age-matched groups with different psychotic disorders.
During the time of the study patients had to be in (at least partial) remission in order for them to comply with the scanning requirements. All patients were on maintenance medication, and the medication doses have been stable for at least 4 weeks upon measurement. We therefore computed the chlorpromazine equivalents for each SZ patient using the formula by Woods (2003), and ‘amitryptilin equivalents’ for each BD patient as described by Ali (1998), and assessed potential influence of the equivalents on the main imaging results.
The control group comprised n = 38 healthy controls (M [years] = 37.15 [11.14]) (see Table 1) who were recruited through local media advertisements. The exclusion criteria for the control subjects and for the relatives were any axis I and axis II disorders according to DSM-IV, left-handedness, current drug-abuse, and neurological diseases.
All subjects underwent the Structured Clinical Interview for DSM-IV (SCID; Wittchen et al., 1996) in order to confirm the clinical diagnosis (in the patient groups), or to ensure that none of the non-patient participants suffered from any psychiatric disorder (relatives, controls). None of the controls had any positive family history of psychosis. All groups were matched for handedness (all right handed; measured with The Edinburgh Handedness Inventory (Oldfield, 1971)) age, gender, education and parental education. SZ patients and controls showed significant differences in the years of education but the years of education in the parents' group was comparable, and the significant differences are due to illness specifications.
We also assessed verbal intelligence using the Multiple Choice Word Comprehension Test (MWT-B; Lehrl, 2005) in order to rule out that potential group differences in these scores may influence the main results. The participants showed no significant group effect in the scores of the MWT-B (verbal intelligence) (F = 2.42, ns) (see Table 1). The participants were provided with a description of the study and gave written informed consent before participation. Experimental procedures were approved by the ethical board of the medical department of the Goethe-University, Frankfurt, Germany.
2.1.2. Cardiff site
We recruited an opportunistic sample comprising of n = 89 right-handed Caucasian healthy controls (M [years] = 24.42 [SD = 4.58]). We administered a detailed biographical and sociodemographic questionnaire, and included comparable measures to those used in the Frankfurt sample, in order to ensure that none of the controls had a lifetime history of mental or behavioural disorders (including any axis I or axis II disorder), use of psychotropic medication or neurological disease. After explaining the study we obtained written informed consent. The study was approved by the research ethics committee of the School of Psychology, Cardiff University.
2.2. MRI data acquisition
2.2.1. Frankfurt site
MRI data were acquired using a Trio 3 T scanner (Siemens, Erlangen, Germany) at Frankfurt University with a standard transmit–receive head coil. Anatomical data consisted of one MDEFT (Modified Driven Equilibrium Fourier Transform) sequence (Deichmann et al., 2004) with 1 × 1 × 1 mm3 voxel size and 176 slices.
2.2.2. Cardiff site
MRI data were acquired using a 3 T GT HDx system at the Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University. High resolution T1-weighted three dimensional images were acquired using a fast spoiled gradient echo sequence (FSPGR) with 172 contiguous sagittal slices of 1 mm thickness.
All anatomical MRI scans were reviewed by a neuroradiologist who did not find any pathology, i.e., focal or local atrophy, lacunar infarcts or extensive microangiopathy.
2.3. MRI image processing
2.3.1. Frankfurt & Cardiff sites
For all subjects, we used standard Voxel-based morphometry (VBM) routines including the DARTEL tool to preprocess the anatomical data sets, resulting in non-linear, modulated, smoothed images, defined as ‘absolute volume’. This process was performed with SPM8 (statistical parametric mapping [Wellcome Department of Imaging Neuroscience, London, UK]) running on a current version of the MATLAB software.
First, all white matter images went under rigorous quality control checked by a) visual inspection (for artefacts, structural abnormalities and pathologies) and statistically (for homogeneity of covariance and distribution). Second, customised T1 templates and prior images of grey matter (GM), white matter (WM) and cerebro-spinal fluid (CSF) were created from all participants. For the segmentation, we followed the steps provided by the SPM8 guidelines (light bias regularisation [0.001], 60 mm bias FWHM cut-off, warping regularisation of 4, affine regularisation to the ICBM European brain template (linear registration), sampling distance of 3, manual quality check; for details of the preprocessing steps, see Oertel-Knochel et al., 2012). Finally, the images were smoothed (Ashburner and Friston, 2000) with a Gaussian kernel of 8 × 8 × 8 mm3 (FWHM), whereby the intensity of each voxel was replaced by the weighted average of the surrounding voxels, in essence blurring the segmented image. Considering the results previously reported (Terwisscha van Scheltinga et al., 2013), we only sampled white matter volume, using white matter masks provided by the WFU PickAtlas (http://www.fmri.wfubmc.edu/downloads) toolbox in SPM8 (Maldjian et al., 2003). Absolute white matter volume estimates (measured in cubic decimetres (dm3)) for each white matter mask were extracted using the MarsBaR toolbox (http://marsbar.sourceforge.net/).
2.4. Genotyping, SNP selection and risk profile score calculation
2.4.1. Frankfurt site
Genotyping of the seven candidate SNPs was performed using the Illumina GoldenGate assay (Illumina, Inc., San Diego, CA, USA) using the BeadXpress platform. Assays were designed for the experiment using Illumina's Assay Design Tool (http://support.illumina.com/array/array_software/assay_design_tool.ilmn). In total, 94 blood-derived genomic DNA samples were genotyped to verify the seven SNPs (for SNP sequences see Supplementary Table S2). Nucleic acid quality and concentration were evaluated using PicoGreen assay (Life Technologies, Carlsbad, CA, USA). GoldenGate genotyping was performed according to the manufacturer's protocols. Genotype calling and annotation were performed using GenomeStudio (Illumina, Inc., San Diego, CA, USA).
2.4.2. Cardiff site
Genomic DNA was obtained from saliva using Oragene OG-500 saliva kits. Individuals were genotyped using customised Illumina HumanCoreExome BeadChip SNP genotyping arrays (Illumina, Inc., San Diego, CA). Quality control was implemented in PLINK (Purcell et al., 2007). Individuals were excluded for ambiguous sex, genotyping completeness less than 97%, cryptic relatedness (up to third degree relatives by identity of descent), and non-European ethnicity as detected in iterative EIGENSTRAT analyses of an LD-pruned data set.
2.4.3. Discovery sample
Our risk profile score (7-SNP SZ-RPS) calculation was performed using the method described by Purcell et al. (International Schizophrenia Consortium et al., 2009). Briefly, schizophrenia risk (based on odds ratios; ORs) was estimated from an international GWAS of 9394 schizophrenia cases and 12,462 controls (PGC-schizophrenia stage-one; GWAS Consortium, 2011). Risk profile scores for schizophrenia were calculated by averaging the number of risk alleles for each SNP for each person, weighted by the natural logarithm of its OR, estimated by the schizophrenia GWAS (GWAS Consortium, 2011).
2.4.4. Target sample
We considered SNPs that were significantly associated with schizophrenia risk based on the recent genome scan for schizophrenia (GWAS Consortium, 2011). SNP genotype data were available for seven SNPs that were associated with schizophrenia at a genome-wide level (MIR137 [rs1625579] p = 1.59 × 10−11; CCDC68 [rs12966547] p = 2.60×10−10; CNNM2 [rs7914558] p = 1.82×109; NT5C2 [rs11191580 1] p = 11×10−8; MMP16 [rs7004633] p = 2.75×10−8; CSMD1 [rs10503253] p = 4.14×10−8; PCGEM1 [rs17662626] p = 4.65×10−8). A single risk profile score (7-SNP SZ-RPS) based upon the additive schizophrenia risk of these seven SNPs was created for each individual in the Frankfurt (n = 94) and Cardiff (n = 89) samples.
2.5. Statistical analysis
All statistics were performed in IBM SPSS Statistics v21. Initial group differences were explored with a univariate general linear model with group/diagnoses and gender as between subject factors and age/ICV residuals (ICV residuals were created by regressing out WM volume: ICVRES) as covariates. In order to explore the putative effects of our 7-SNP SZ-RPS on WM volume above and beyond the potential confounding effects of age, group ICVRES and diagnoses/group, we performed in a two-step hierarchal regression analyses. Hierarchal regression analyses were performed on white matter volume for each sample (Frankfurt site, Cardiff site) separately. In the first step of the model, covariates of interest were added (Frankfurt: age, gender, ICV & group/diagnoses; Cardiff: age, gender, ICV). We followed up any group differences and/or associations in the first step of the hierarchal regression with one-way ANOVAs/independent sample t-test and bivariate correlations, respectively. In the second step of the model, we used 7-SNP SZ-RPS for SZ as a potential predictor of WM above and beyond the potential effects of demographic confounds. We then proceeded to run two independent partial bivariate correlations between WM volume and 7-SNP SZ-RPS, controlling for potential confounds such as age, gender, intracranial volume (ICV residuals; regressing out WM) and group/diagnosis.
3. Results
3.1. Imaging effects
3.1.1. Frankfurt Site
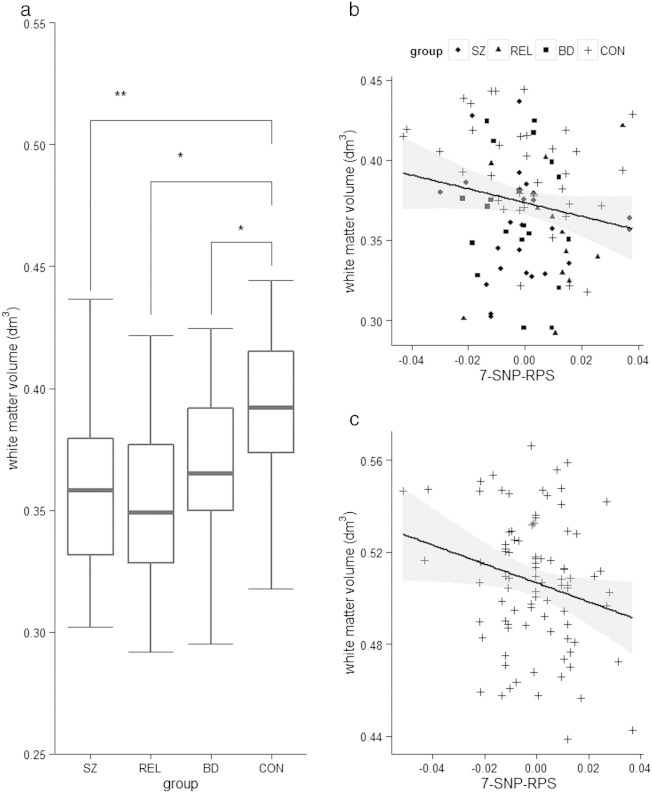
Hierarchical regression revealed that group/diagnosis significantly predicted white matter volume (β = −0.349, p = 0.001). Univariate analysis (adjusted for age, gender and ICVRES) confirmed a significant effect of group/diagnosis (F3,84 = 5.998, p = 0.0009), with corrected pairwise comparisons revealing that the group effect was driven by larger WM volume in healthy controls compared to: schizophrenia patients (pCORRECTED = 0.004), relatives (pCORRECTED = 0.034) and bipolar patients (pCORRECTED = 0.037). Post-hoc independent samples t-test analysis confirmed that the group differences in WM volume between healthy controls and first-degree relatives (t(48) = 3.452, p = 0.001), between the controls and schizophrenia patients (t(60) = 3.944, p ≤ 0.001) and between controls and bipolar patients (t(56) = 2.572, p = 0.008) with higher values for healthy controls in all the cases (see Fig. 1a). There were no significant differences between relatives of schizophrenia patients, schizophrenia patients and bipolar patients (in all cases; p > 0.50). Critically, the polygenic model predicted WM volume above and beyond potentially confounding effects of age, gender, ICV and group/diagnosis (ΔR2 = 0.044, β = −0.211, p = 0.033). Post-hoc partial correlations (controlling for age, gender and ICVRES) revealed that this association was significant in the healthy control group (r = –0.364, p = 0.031) and at the trend level in the bipolar disorder group (r = –0.452, p = 0.068), but not significant in the relatives (r = 0.288, p = 0.497) or schizophrenia patients (r = –0.121, p = 0.600).
Fig. 1.
Frankfurt sample: a. White matter volumes (dm3) across groups, b. correlation between white matter volumes (dm3) and 7-SNP-RPS scores individually for each group in the Frankfurt sample, c. correlation between white matter volumes (dm3) and 7-SNP-RPS scores across the Cardiff sample. Post-hoc analysis revealed that compared to healthy controls, white matter volume was reduced in relatives of schizophrenia patients (p = 0.004), schizophrenia patient relatives (p = 0.034) and bipolar patients (p = 0.037). p-Values are adjusted for multiple comparisons (Bonferroni corrected). Bivariate correlations confirm a negative association between WM volume and 7-SNP SZ-RPS, controlling for age, gender, ICVRES (and diagnosis in the Frankfurt sample) (Frankfurt: r = −0.266, p = 0.033; Cardiff: r = −0.226, p = 0.037). SZ = schizophrenia patients, BD = bipolar patients, REL = first-degree relatives of patients with schizophrenia, and CON = controls. VBM data were available for 94 participants (n = 38 healthy controls, n = 12 relatives of patients with schizophrenia, n = 24 patients with schizophrenia, n = 20 patients with bipolar disorder). p = significant on an α < 0.05-threshold. ** denotes p < 0.01, * denotes p < 0.05. NB: there were no statistical outliers in WM volume in either sample. Shaded grey bars represent 95% confidence intervals.
3.1.2. Cardiff Site
The hierarchical regression revealed that 7-SNP SZ-RPS predicted WM volume (ΔR2 = 0.051, β = −0.227, p = 0.037). Bivariate correlations confirmed a negative association between WM volume and 7-SNP SZ-RPS, controlling for age and group/diagnosis using the PGZ-SZ-one (Frankfurt: r = –0.266, p = 0.033; Cardiff: r = –0.226, p = 0.037) (See Fig. 1b-c). A regression performed on the combined sample (Stonnington et al., 2008) (independent sample, z-transformed WM volume) confirmed the significant negative association between WM volume and 7-SNP SZ-RPS using the PGC-SZ-one ORs (ΔR2 = 0.042, β = −0.207, p = 0.005). We then performed a post-hoc regression to explore the individual effects of each of the seven SNPs on white matter volume across the whole sample. The seven SNPs (individual effects; rather than polygenic model) also predicted WM volume (controlling for age, gender, ICV and group/diagnoses) (FΔ = 2.976 p = 0.006). We observed that four of the seven SNPs were nominally associated with white matter volume (above and beyond the effects of age and group/diagnoses) including rs10503253 in CSMD1 (β = −0.242, p = 0.001), rs12966547 in CCDC68 (β = −0.168, p = 0.026), rs1625579 in MIR137 (β = −0.161, p = 0.034) and rs17662626 near PCGEM1 (β = −0.141, p = 0.05).
3.2. Demographics & 7-SNP SZ-RPS (both sites)
The 7-SNP SZ-RPS did not differ between the diagnostic groups (F3,93 = 1.617, p = 0.191) within the Frankfurt sample. There was no correlation between 7-SNP SZ-RPS and age in either the Frankfurt (r = 0.036, p = 0.731) or the Cardiff (r = 0.004, p = 0.986) sample, suggesting that the potential associations between 7-SNP SZ-RPS and WM were not a) confounded by age-related reductions in WM and b) not due to shared variance of 7-SNP SZ-RPS and age on WM. In the first step of the model for each sample, age was not a significant predictor of WM volume (Frankfurt: β = −0.144, p = 0.146; Cardiff: β = 0.027, p = 0.799).
3.3. Potential influence of sociodemographics or medication on main imaging results
There were no differences in 7-SNP SZ-RPS between genders at the Cardiff site (F1,88 = 0.196, p = 0.659), in the Frankfurt site (F1,92 = 0.470, p = 0.495) or in the combined sample (F1,182 = 0.604, p = 0.438). We also did additional bivariate correlation analyses (2-tailed) in order to control for potential influence of age of onset and duration of illness on the main imaging results. Here, results indicate that neither age of onset nor duration of illness had any significant association with the VBM parameters in any of the patient groups (p > 0.05). We computed the chlorpromazine equivalents for each SZ patient using the formula by Woods (2003), and ‘amitryptilin equivalents’ (Ali, 1998) for each BD patient. These scores were used to compute correlation analyses between the main imaging results and the intensity of medication. We did not find any influence of these scores on the main results (p > 0.05).
4. Discussion
A growing number of studies are adopting a polygenic approach when exploring neuropsychiatric clinical phenotypes (Dudbridge, 2013). Recent research suggests that up to 32% of the variance in risk for schizophrenia may be captured by the cumulative effects of SNPs (Lee et al., 2012; Ripke et al., 2013). Furthermore, several studies have found associations between increased psychiatric polygenic risk (RPS) and both brain structure (Holmes et al., 2012; Papiol et al., 2014; Whalley et al., 2013) and brain function (Walton et al., 2013b; Walton et al., 2014; Whalley et al., 2012). In the present study, we found that both the clinical phenotype of schizophrenia and familial genetic risk were associated with reduced white matter volume, as indicated by the differences in white matter volume between the healthy controls and SZ first-degree relatives, and between healthy controls and schizophrenia patients. This result supports numerous findings implicating white matter differences in the familial genetic risk associated with schizophrenia (Francis et al., 2013; Oertel-Knochel et al., 2012; van Haren et al., 2012). Moreover, we additionally included a group of BD patients, as they also show reduction in WM volumes, are part of the psychosis spectrum and add more power to the study. Therefore, the initial genetic difference between HC > REL = SZ is our ‘working intermediate phenotype’. We show that this is a ‘genetic’ difference before observing associations with our 7-SNP SZ-RPS.
Crucially, the present study also demonstrated that reduced whole brain white matter was associated with 7-SNP SZ-RPS as measured by the cumulative effect of seven SZ risk loci, which was selectively significant in both healthy control groups. In contrast to previous polygenic imaging studies, this is the first study to use a targeted set of schizophrenia -associated genome wide significant SNPs rather than arbitrary p-threshold cut-offs. It is noteworthy that the estimates for the variance in brain structure explained by the polygenic effects of in our study (R2 = .042) are comparable to the R2 estimates (R2 = .054) originally found by Terwisscha van Scheltinga et al. (2013), which is striking considering that our estimate is only based on a set of seven SNPs.
5. Limitations
Although the study was moderately well-powered for an imaging sample, the 7-SNP SZ-RPS did not delineate our participants based on diagnosis. This suggests that our 7-SNP SZ-RPS model may be underpowered to detect clinical phenotypes. However, as the 7-SNP-RPS was associated with WM volume across our sample sites, we feel that it may reflect a small but significant component of psychosis-related pathophysiology. We did not see an effect of 7-SNP SZ-RPS in the relative or SZ group, but these subsamples may be underpowered or affected by shared environmental confounds. Although we are unable to directly compare our results with those of Terwisscha van Scheltinga et al. (2013) and Papiol et al. (2014) because we did not have genome-wide data for a proportion of our participants, our additional evidence suggests firstly that the genetic susceptibility for schizophrenia is associated with variance in white matter volume and secondly, that this effect may be mediated through a small set of relatively robust risk variants.
We were however, able to replicate previous findings suggesting an involvement of rs17662626 (near PCGEM1) in the volume of white matter (Terwisscha van Scheltinga et al., 2013). Other studies looking at individual risk alleles used in the model (such as MIR137, CSMD1, CNNM2) and other psychosis risk SNPs have shown no association with white matter volume (Cousijn et al., 2014; Rose et al., 2013; Rose et al., 2014; Tesli et al., 2013). We suggest that the effect sizes of common risk alleles for SZ are small, so additive models like our 7-SNP SZ-RPS may help to elucidate the combined effects of multiple loci on SZ intermediate phenotypes. Our results support the hypothesis that a subset of schizophrenia risk variants are related to the maturation of white matter volume, suggesting that common variation in these risk loci may increase schizophrenia susceptibility through perturbation of brain connectivity (Voineskos, 2015). Future work should explore biologically informed polygenic models in order to disentangle the molecular pathways that confer susceptibility for deficits in white matter structure.
Conflicts of interest
None of the authors have any conflict of interest to report.
Acknowledgements
This study was supported by the National Centre for Mental Health at Cardiff University with funds from the National Institute for Social Care and Health Research (NISCHR) and the Medical Research Council (grant number; MR/K013041/1). We are grateful to all professionals, patients and volunteers involved with the National Centre for Mental Health (NCMH). NCMH is funded by the National Institute for Social Care and Health Research (NISCHR), Welsh Government, Wales (Grant No. BR09). Polygenic analysis was supported by Medical Research Council (MRC) grant “Behavioural and neurophysiological effects of schizophrenia risk genes: a multi-locus, pathway based approach” (MR/K004360/1).
Footnotes
Supplementary material associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.nicl.2015.03.005.
Appendix A. Supplementary data
Supplementary material
References
- Ali I.M. Long-term treatment with antidepressants in primary care: are sub-therapeutic doses still being used? Psychiatr. Bull. 1998;22(1):15–19. [Google Scholar]
- APA . (4th edition) American Psychiatric Association; Washington, D.C.: 1994. Diagnostic and statistical manual of mental disorders. [Google Scholar]
- Ashburner J., Friston K.J. Voxel-based morphometry — the methods. Neuroimage. 2000;11(6 1):805–821. doi: 10.1006/nimg.2000.0582. 10860804 [DOI] [PubMed] [Google Scholar]
- Bech P. Rating scales for affective disorders: Their validity and consistency. Acta Psychiatr Scand Suppl. 1981;295:1–101. 7044046 [PubMed] [Google Scholar]
- Cardno A.G., Owen M.J. Genetic relationships between schizophrenia, bipolar disorder, and schizoaffective disorder. Schizophr. Bull. 2014;40:504–515. doi: 10.1093/schbul/sbu016. 24567502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousijn H., Eissing M., Fernández G., Fisher S.E., Franke B., Zwiers M., Harrison P.J., Arias-Vásquez A. No effect of schizophrenia risk genes MIR137, TCF4, and ZNF804A on macroscopic brain structure. Schizophr. Res. 2014;159:329–332. doi: 10.1016/j.schres.2014.08.007. 25217366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann R., Schwarzbauer C., Turner R. Optimisation of the 3D MDEFT sequence for anatomical brain imaging: technical implications at 1.5 and 3 T. Neuroimage. 2004;21(2):757–767. doi: 10.1016/j.neuroimage.2003.09.062. 14980579 [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. P.L.O.S. Genet. 2013;9(3) doi: 10.1371/journal.pgen.1003348. 23555274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsell L., McDonald C. The structural neuroimaging of bipolar disorder. Int. Rev. Psychiatry. 2009;21(4):297–313. doi: 10.1080/09540260902962081. 20374145 [DOI] [PubMed] [Google Scholar]
- Francis A.N., Bhojraj T.S., Prasad K.M., Montrose D., Eack S.M., Rajarethinam R., van Elst L.T., Keshavan M.S. Alterations in the cerebral white matter of genetic high risk offspring of patients with schizophrenia spectrum disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;40:187–192. doi: 10.1016/j.pnpbp.2012.08.003. 22910323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guze, S.B., (1995). Diagnostic and Statistical Manual of Mental-Disorders, fourth edition (Dsm-Iv) - Amer-Psychiat-Assoc. American Journal of Psychiatry 152, 1228-1228. [DOI] [PubMed]
- GWAS Consortium Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43(10):969–976. doi: 10.1038/ng.940. 21926974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler G., Drevets W.C., Gould T.D., Gottesman I.I., Manji H.K. Toward constructing an endophenotype strategy for bipolar disorders. Biol. Psychiatry. 2006;60(2):93–105. doi: 10.1016/j.biopsych.2005.11.006. 16406007 [DOI] [PubMed] [Google Scholar]
- Hautzinger M., Keller F., Kühner C. Das Beck Depressionsinventar II. Deutsche Bearbeitung und Handbuch zum BDI II. [!(sb:name)!]; Frankfurt a. M.: 2006. [Google Scholar]
- Holmes A.J., Lee P.H., Hollinshead M.O., Bakst L., Roffman J.L., Smoller J.W., Buckner R.L. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J. Neurosci. 2012;32(50):18087–18100. doi: 10.1523/JNEUROSCI.2531-12.2012. 23238724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. 19571811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S.R., Fiszbein A., Opler L.A. The Positive and Negative Syndrome Scale (PANSS) for schizophrenia. Schizophr. Bull. 1987;13(2):261–276. doi: 10.1093/schbul/13.2.261. 3616518 [DOI] [PubMed] [Google Scholar]
- Kaymaz N., van Os J. Heritability of structural brain traits: an endophenotype approach to deconstruct schizophrenia. Int. Rev. Neurobiol. 2009;89:85–130. doi: 10.1016/S0074-7742(09)89005-3. 19900617 [DOI] [PubMed] [Google Scholar]
- Krauss H., Marwinski K., Schulze T., Mueller D.J., Held T., Rietschel M., Maier W., Freyberger H.J. Reliability and validity of the German version of the Premorbid Adjustment Scale (PAS) Nervenarzt. 2000;71(3):188–194. doi: 10.1007/s001150050028. 10756527 [DOI] [PubMed] [Google Scholar]
- Lee S.H., DeCandia T.R., Ripke S., Yang J., Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ) International Schizophrenia Consortium (ISC) Molecular Genetics of Schizophrenia Collaboration (MGS) [!(%xInRef|ce:surname)!] P.F., Goddard M.E., Keller M.C., Visscher P.M., Wray N.R. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. 22344220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrl S. Mehrfachwahl-Wortschatz-Intelligenztest MWT-B balingen. Spitta Verlag; unveränderte Auflage: 2005. p. 5. [Google Scholar]
- Maldjian J.A., Laurienti P.J., Kraft R.A., Burdette J.H. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19(3):1233–1239. doi: 10.1016/s1053-8119(03)00169-1. 12880848 [DOI] [PubMed] [Google Scholar]
- Matsuo K., Kopecek M., Nicoletti M.A., Hatch J.P., Watanabe Y., Nery F.G., Zunta-Soares G., Soares J.C. New structural brain imaging endophenotype in bipolar disorder. Mol. Psychiatry. 2012;17(4):412–420. doi: 10.1038/mp.2011.3. 21321565 [DOI] [PubMed] [Google Scholar]
- McDonald C., Bullmore E.T., Sham P.C., Chitnis X., Wickham H., Bramon E., Murray R.M. Association of genetic risks for schizophrenia and bipolar disorder with specific and generic brain structural endophenotypes. Arch. Gen. Psychiatry. 2004;61(10):974–984. doi: 10.1001/archpsyc.61.10.974. 15466670 [DOI] [PubMed] [Google Scholar]
- McIntosh A.M., Job D.E., Moorhead W.J., Harrison L.K., Whalley H.C., Johnstone E.C., Lawrie S.M. Genetic liability to schizophrenia or bipolar disorder and its relationship to brain structure. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B(1):76–83. doi: 10.1002/ajmg.b.30254. 16342281 [DOI] [PubMed] [Google Scholar]
- Oertel-Knöchel V., Knöchel C., Matura S., Rotarska-Jagiela A., Magerkurth J., Prvulovic D., Haenschel C., Hampel H., Linden D.E. Cortical–basal ganglia imbalance in schizophrenia patients and unaffected first-degree relatives. Schizophr. Res. 2012;138(2–3):120–127. doi: 10.1016/j.schres.2012.02.029. 22464726 [DOI] [PubMed] [Google Scholar]
- Oldfield R.C. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. doi: 10.1016/0028-3932(71)90067-4. 5146491 [DOI] [PubMed] [Google Scholar]
- Papiol S., Mitjans M., Assogna F., Piras F., Hammer C., Caltagirone C., Arias B., Ehrenreich H., Spalletta G. Polygenic determinants of white matter volume derived from GWAS lack reproducibility in a replicate sample. Transl. Psychiatry. 2014;4:e362. doi: 10.1038/tp.2013.126. 24548877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. 21926972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. 17701901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Kim Y., Lee S.H., Magnusson P.K., Sanchez N., Stahl E.A., Williams S., Wray N.R., Xia K., Bettella F., Borglum A.D., Bulik-Sullivan B.K., Cormican P., Craddock N., de Leeuw C., Durmishi N., Gill M., Golimbet V., Hamshere M.L., Holmans P., Hougaard D.M., Kendler K.S., Lin K., Morris D.W., Mors O., Mortensen P.B., Neale B.M., O’Neill F.A., Owen M.J., Milovancevic M.P., Posthuma D., Powell J., Richards A.L., Riley B.P., Ruderfer D., Rujescu D., Sigurdsson E., Silagadze T., Smit A.B., Stefansson H., Steinberg S., Suvisaari J., Tosato S., Verhage M., Walters J.T., Multicenter Genetic Studies of Schizophrenia Consortium. Levinson D.F., Gejman P.V., Kendler K.S., Laurent C., Mowry B.J., O’Donovan M.C., Owen M.J., Pulver A.E., Riley B.P., Schwab S.G., Wildenauer D.B., Dudbridge F., Holmans P., Shi J., Albus M., Alexander M., Campion D., Cohen D., Dikeos D., Duan J., Eichhammer P., Godard S., Hansen M., Lerer F.B., Liang K.Y., Maier W., Mallet J., Nertney D.A., Nestadt G., Norton N., O’Neill F.A., Papadimitriou G.N., Ribble R., Sanders A.R., Silverman J.M., Walsh D., Williams N.M., Wormley B., Psychosis Endophenotypes International Consortium. Arranz M.J., Bakker S., Bender S., Bramon E., Collier D., Crespo-Facorro B., Hall J., Iyegbe C., Jablensky A., Kahn R.S., Kalaydjieva L., Lawrie S., Lewis C.M., Lin K., Linszen D.H., Mata I., McIntosh A., Murray R.M., Ophoff R.A., Powell J., Rujescu D., Van Os J., Walshe M., Weisbrod M., Wiersma D., Wellcome Trust Case Control Consortium. Donnelly P., Barroso I., Blackwell J.M., Bramon E., Brown M.A., Casas J.P., Corvin A.P., Deloukas P., Duncanson A., Jankowski J., Markus H.S., Mathew C.G., Palmer C.N., Plomin R., Rautanen A., Sawcer S.J., Trembath R.C., Viswanathan A.C., Wood N.W., Spencer C.C., Band G., Bellenguez C., Freeman C., Hellenthal G., Giannoulatou E., Pirinen M., Pearson R.D., Strange A., Su Z., Vukcevic D., Donnelly P., Langford C., Hunt S.E., Edkins S., Gwilliam R., Blackburn H., Bumpstead S.J., Dronov S., Gillman M., Gray E., Hammond N., Jayakumar A., McCann O.T., Liddle J., Potter S.C., Ravindrarajah R., Ricketts M., Tashakkori-Ghanbaria A., Waller M.J., Weston P., Widaa S., Whittaker P., Barroso I., Deloukas P., Mathew C.G., Blackwell J.M., Brown M.A., Corvin A.P., McCarthy M.I., Spencer C.C., Bramon E., Corvin A.P., O’Donovan M.C., Stefansson K., Scolnick E., Purcell S., McCarroll S.A., Sklar P., Hultman C.M., Sullivan P.F. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. 23974872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose E.J., Hargreaves A., Morris D., Fahey C., Tropea D., Cummings E., Caltagirone C., Bossù P., Chiapponi C., Piras F., Spalletta G., Gill M., Corvin A., Donohoe G. Effects of a novel schizophrenia risk variant rs7914558 at CNNM2 on brain structure and attributional style. Br. J. Psychiatry. 2014;204(2):115–121. doi: 10.1192/bjp.bp.113.131359. 24311551 [DOI] [PubMed] [Google Scholar]
- Rose E.J., Morris D.W., Hargreaves A., Fahey C., Greene C., Garavan H., Gill M., Corvin A., Donohoe G. Neural effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013;162B(6):530–537. doi: 10.1002/ajmg.b.32182. 23839771 [DOI] [PubMed] [Google Scholar]
- Schulze T.G., Akula N., Breuer R., Steele J., Nalls M.A., Singleton A.B., Degenhardt F.A., Nöthen M.M., Cichon S., Rietschel M., Bipolar Genome. McMahon F.J. Molecular genetic overlap in bipolar disorder, schizophrenia, and major depressive disorder. World J. Biol. Psychiatry. 2014;15(3):200–208. doi: 10.3109/15622975.2012.662282. 22404658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington C.M., Tan G., Klöppel S., Chu C., Draganski B., Jack C.R., Jr., Chen K., Ashburner J., Frackowiak R.S. Interpreting scan data acquired from multiple scanners: a study with Alzheimer's disease. Neuroimage. 2008;39(3):1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. 18032068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga A.F., Bakker S.C., van Haren N.E., Derks E.M., Buizer-Voskamp J.E., Boos H.B., Cahn W., Hulshoff Pol H.E., Ripke S., Ophoff R.A., Kahn R.S., Psychiatric Genome-wide Association Study Consortium Genetic schizophrenia risk variants jointly modulate total brain and white matter volume. Biol. Psychiatry. 2013;73(6):525–531. doi: 10.1016/j.biopsych.2012.08.017. 23039932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesli M., Egeland R., Sønderby I.E., Haukvik U.K., Bettella F., Hibar D.P., Thompson P.M., Rimol L.M., Melle I., Agartz I., Djurovic S., Andreassen O.A. No evidence for association between bipolar disorder risk gene variants and brain structural phenotypes. J. Affect. Disord. 2013;151(1):291–297. doi: 10.1016/j.jad.2013.06.008. 23820096 [DOI] [PubMed] [Google Scholar]
- Smoller J.W., Craddock N., Kendler K., Lee P.H., Neale B.M., Nurnberger J.I., Ripke S., Santangelo S., Sullivan P.F., Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. 23453885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haren N.E., Rijsdijk F., Schnack H.G., Picchioni M.M., Toulopoulou T., Weisbrod M., Sauer H., van Erp T.G., Cannon T.D., Huttunen M.O., Boomsma D.I., Hulshoff Pol H.E., Murray R.M., Kahn R.S. The genetic and environmental determinants of the association between brain abnormalities and schizophrenia: the schizophrenia twins and relatives consortium. Biol. Psychiatry. 2012;71(10):915–921. doi: 10.1016/j.biopsych.2012.01.010. 22341827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos A.N. Genetic underpinnings of white matter ‘connectivity’: heritability, risk, and heterogeneity in schizophrenia. Schizophr. Res. 2015;161:50–60. doi: 10.1016/j.schres.2014.03.034. 24893906 [DOI] [PubMed] [Google Scholar]
- Walton E., Geisler D., Lee P.H., Hass J., Turner J.A., Liu J., Sponheim S.R., White T., Wassink T.H., Roessner V., Gollub R.L., Calhoun V.D., Ehrlich S. Prefrontal inefficiency is associated with polygenic risk for schizophrenia. Schizophr. Bull. 2014;40:1263–1271. doi: 10.1093/schbul/sbt174. 24327754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E., Turner J., Gollub R.L., Manoach D.S., Yendiki A., Ho B.C., Sponheim S.R., Calhoun V.D., Ehrlich S. Cumulative genetic risk and prefrontal activity in patients with schizophrenia. Schizophr. Bull. 2013;39(3):703–711. doi: 10.1093/schbul/sbr190. 22267534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley H.C., Papmeyer M., Sprooten E., Romaniuk L., Blackwood D.H., Glahn D.C., Hall J., Lawrie S.M., Sussmann J., McIntosh A.M. The influence of polygenic risk for bipolar disorder on neural activation assessed using fMRI. Transl. Psychiatry. 2012;2:e130. doi: 10.1038/tp.2012.60. 22760554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley H.C., Sprooten E., Hackett S., Hall L., Blackwood D.H., Glahn D.C., Bastin M., Hall J., Lawrie S.M., Sussmann J.E., McIntosh A.M. Polygenic risk and white matter integrity in individuals at high risk of mood disorder. Biol. Psychiatry. 2013;74:280–286. doi: 10.1016/j.biopsych.2013.01.027. 23453289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittchen H.U., Zhao S., Abelson J.M., Abelson J.L., Kessler R.C. Reliability and procedural validity of UM-CIDI DSM-III-R phobic disorders. Psychol. Med. 1996;26(6):1169–1177. doi: 10.1017/s0033291700035893. 8931163 [DOI] [PubMed] [Google Scholar]
- Woods S.W. Chlorpromazine equivalent doses for the newer atypical antipsychotics. J. Clin. Psychiatry. 2003;64(6):663–667. doi: 10.4088/jcp.v64n0607. 12823080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material