Abstract
Lipid composition of the membrane and rhodopsin packing density strongly modulate the early steps of the visual response of photoreceptor membranes. In this study, lipid-order and bovine rhodopsin function in proteoliposomes composed of the sn-1 chain perdeuterated lipids 14:0d27-14:1-PC, 16:0d31-16:1-PC, 18:0d35-18:1-PC, or 20:0d39-20:1-PC at rhodopsin/lipid molar ratios from 1:70 to 1:1000 (mol/mol) were investigated. Clear evidence for matching of hydrophobic regions on rhodopsin transmembrane helices and hydrophobic thickness of lipid bilayers was observed from 2H nuclear magnetic resonance order parameter measurements at low rhodopsin concentrations. Thin bilayers stretched to match the length of transmembrane helices observed as increase of sn-1 chain order, while thicker bilayers were compressed near the protein. A quantitative analysis of lipid-order parameter changes suggested that the protein adjusts its conformation to bilayer hydrophobic thickness as well, which confirmed our earlier circular-dichroism measurements. Changes in lipid order parameters upon rhodopsin incorporation vanished for bilayers with a hydrophobic thickness of 27 ± 1 Å, suggesting that this is the bilayer thickness at which rhodopsin packs in bilayers at the lowest membrane perturbation. The lipid-order parameter studies also indicated that a hydrophobic mismatch between rhodopsin and lipids triggers rhodopsin oligomerization with increasing rhodopsin concentrations. Both hydrophobic mismatch and rhodopsin oligomerization result in substantial shifts of the equilibrium between the photointermediates metarhodopsin I and metarhodopsin II; increasing bilayer thickness favors formation of metarhodopsin II while oligomerization favors metarhodopsin I. The results highlight the importance of hydrophobic matching for rhodopsin structure, oligomerization, and function.
Introduction
Rhodopsin, the mammalian dim-light photoreceptor, is the best-characterized G protein-coupled receptor. It is also the only one, as of this writing, for which several high-resolution structures are available in both the dark-adapted and photoactivated states (1–3). Absorption of light leads to the formation of an active metarhodopsin II state (MII) in equilibrium with an inactive metarhodopsin I state (MI) (4). MII binds and activates the heterotrimeric G-protein transducin (Gt), setting off a biochemical amplification cascade that results in a drop of cGMP concentration (5) which, in turn, leads to hyperpolarization of the plasma membrane and the signaling of second-order neurons. In vivo, the reaction ends with rhodopsin phosphorylation by rhodopsin kinases that allow arrestin binding, which prevents further activation of transducin (6).
The lipid bilayer is a powerful allosteric modulator of rhodopsin function. We, and others, have shown that changes in membrane elastic properties as well as specific molecular properties of lipids in the first layer of lipids surrounding rhodopsin, are key determinants of the MI-MII equilibrium (7–11). The influence of hydrophobic mismatch on rhodopsin oligomerization and interfacial lipid-protein interactions was studied by electron paramagnetic resonance (EPR) (12), freeze fracture electron microscopy (13), and more recently by nuclear magnetic resonance (NMR) (14), circular dichroism (14), fluorescence energy transfer (15), and coarse-grained, molecular dynamics simulations (CG-MD) (16). NMR, circular dichroism, and CG-MD showed that mismatch reduction occurred by small adaptations of the bilayer thickness together with rhodopsin conformational changes (14,16). EPR (17,18), electron microscopy (13), and CG-MD (16) reported that rhodopsin oligomerization is bilayer-thickness-dependent. Experiments were generally undertaken at lipid/protein molar ratios (P/L) of 100 or lower. A concentration-dependent oligomerization was observed by fluorescence energy transfer in 16:0-18:1-PC membranes (15). As for function, Botelho et al. (15) reported that the amount of MII formed after photoactivation peaked in membranes with monounsaturated acyl chains of 20 carbons.
In isolated disk membranes of rod photoreceptors, high-resolution atomic force microscopy images revealed rhodopsin to be heavily oligomerized (19), in contradiction to earlier reports (20). However, the functional role of rhodopsin oligomerization is unknown. Indeed, rhodopsin was found to efficiently activate Gt and to promote normal rhodopsin kinase phosphorylation and normal arrestin-1 binding as a monomer (21). In model membranes, increased packing density that may result in formation of rhodopsin oligomers, reduced rhodopsin activation (22).
The data suggest that there are at least three ways of relieving the energetic constraints imposed by a mismatch between the hydrophobic length of a G protein-coupled receptor and bilayer thickness:
-
1.
lipids can modulate the membrane hydrophobic thickness by stretching or disordering their acyl chains at the lipid-protein interface;
-
2.
the protein might adjust its conformation to the surrounding lipid matrix; or
-
3.
it might oligomerize.
Depending on membrane composition, all of these mechanisms might contribute to relieving stresses in lipid-protein interactions.
The aim of this study was to quantitatively follow stresses in membranes from rhodopsin incorporation as function of bilayer thickness from 14 to 20 carbons per lipid hydrocarbon chain by 2H NMR order parameter measurements. Rhodopsin oligomerization was assessed by measuring lipid-order parameter changes as a function of rhodopsin concentration ranging from rhodopsin/lipid molar ratios of 1:1000–1:70. Oligomerization was detected as a fractional decrease of rhodopsin’s influence on lipid order. Function of rhodopsin was followed by measurement of the MI/MII equilibrium. The data suggest that hydrophobic mismatch is dealt with via a combination of changes including an adjustment of bilayer thickness, a change of rhodopsin conformation, and rhodopsin oligomerization.
Materials and Methods
Preparation of reconstituted membranes
Sample preparation was carried out in complete darkness. The phospholipids 14:0d27-14:1-PC (1-perdeuterio- myristoyl-2-myristoleoyl-sn-glycero-3-phosphocholine), 16:0d31-16:1-PC (1-perdeuterio-palmitoyl-2-palmitoleoyl-sn-glycero- 3-phosphocholine), 18:0d35-18:1-PC (1-perdeuterio-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine), and 20:0d39-20:1-PC (1-perdeuterioarachidoyl-2-eicosanoyl-sn-glycero-3-phosphocholine) were synthesized by Avanti Polar Lipids (Alabaster, AL). The lipids are chromatographically pure (>99%), with a low level of acyl-chain migration (<3%).
Rhodopsin was purified from bovine retinas using procedures that were developed in the Litman laboratory (23). Rhodopsin fractions in 3 wt % octylglucoside (OG), which gave an ultraviolet-visible absorption intensity ratio at 280:500 nm of 1.8 or lower, were used. To eliminate the possibility that minor differences in rhodopsin purity may influence results, experiments were conducted using one batch of rhodopsin for reconstitution of the entire series of lipids.
For rhodopsin reconstitution, a glass round-bottom flask was coated with phospholipids by slow rotation and removal of solvent in a stream of pure nitrogen gas. The rhodopsin-OG solution was added to lipid-OG mixed micelles such that the OG/lipid molar ratio was 10:1 and the rhodopsin/phospholipid molar ratio was 1:70, 1:125, 1:250, 1:500, and 1:1000. The sample was vortexed to complete solubilization of the lipid and then equilibrated for 12 h under argon. Subsequently, this rhodopsin/lipid micellar solution was added dropwise at a rate of ∼400 μL/min to deoxygenated PIPES buffer (10 mM PIPES, 100 mM NaCl, 50 μM DTPA, pH = 7.0) under rapid stirring, resulting in formation of unilamellar proteoliposomes. Typically, the final OG concentration was 7.2 mM, which is well below the critical micelle concentration (24).
The proteoliposome dispersion (2.5–3 mL) was then dialyzed against 1 L of PIPES buffer (Slide-A-Lyzer membrane, 10 kDa cutoff; Pierce, Rockford, IL). The buffer was exchanged three times over 16–24 h. The final rhodopsin concentration in the samples was measured by light absorption at 500 nm assuming a molar extinction coefficient ε500 = 40,600 M−1 cm−1 (25). The concentration of residual OG in the lipid bilayers was lower than 0.4 mol % of the lipid concentration as determined by solution 1H NMR on a small aliquot of sample dissolved in deuterated methanol.
For NMR experiments on dark-adapted rhodopsin, the proteoliposomes were pelleted at 500,000×g and 4°C for 12 h in a TLX-Optima centrifuge (Beckmann-Coulter, Fullerton, CA). The pellet was then extruded through porous anodic aluminum oxide (AAO) Anopore filters (Whatman, GE Healthcare Bio-Sciences, Pittsburgh, PA) with a nominal pore diameter of 0.2 μM and a thickness of 60 μM, which resulted in formation of tubular, rhodopsin-containing bilayers supported by the pore walls of AAO as described previously (26). Extrusion was performed at ambient temperature, which is well above the main phase transition temperature of 14:0d27-14:1-PC (Tm < 0°C), 16:0d31-16:1-PC (Tm < 0°C), and 18:0d35-18:1PC (Tm = 4°C), and at 35°C in a heated glovebox for the 20:0d39-20:1-PC (Tm = 17.5°C) membranes.
All experiments were conducted at a temperature of 37°C, which is well above the main-phase transition temperature of all four lipids. For each experiment, two AAO filters (diameter 25 mm) and one polycarbonate filter (nominal pore size 0.8 μm) were stacked and flushed with several milliliters of PIPES buffer before extruding the dispersion of proteoliposomes. The 1-mL proteoliposome suspension (1 mg lipid/mL) was then extruded 10 times through the stack of filters at a rate of 0.01 mL/s, resulting in entrapment of multilamellar bilayers inside AAO pores. All but a single tubular bilayer covering the inner AAO pore surface were removed by flushing the filters with 5–10 mL of PIPES buffer at a rate of 0.2 mL/s as reported earlier (26).
NMR experiments
Solid-state 2H NMR experiments were carried out on a model DMX500 spectrometer (Bruker, Billerica, MA) equipped with a flat coil 1H,X-probe (Doty Scientific, Columbia, SC) operating at a 2H NMR resonance frequency of 76.8 MHz. Data were acquired at 37°C with a quadrupolar echo pulse sequence, d1-90°x-τ-90°y-τ-acq, at a relaxation delay time d1 = 250 ms, a 5-μs 90° pulse, a delay time τ = 50 μs, and a 200 kHz spectral width. Typically, 150,000 transients were acquired. Order-parameter profiles, mosaic spread of bilayer orientations, and resonance line-width were determined by fitting the spectra with a program written in the software MATHCAD (PTC, Needham, MA). The program reported a smoothed order parameter profile of lipid hydrocarbon chains, the orientational distribution function of bilayer normals (assumed to be Gaussian), and the resonance line-width of all resolved quadrupolar splittings.
Measurement of MII/MI ratio after photoactivation
The equilibrium constant Keq = [MII]/[MI] was determined from rapidly acquired spectra of the MI-MII equilibrium as previously described in Straume et al. (27). Briefly, vesicles were diluted to a rhodopsin concentration of 0.3 mg/mL in pH 7.0 PBS buffer and equilibrated at 37°C in a thermally regulated sample holder. A set of four absorption spectra were collected sequentially in a model No. 8453 diode array spectrophotometer (Agilent, Santa Clara, CA). These included the spectra acquired
-
1.
after the sample was equilibrated in the dark at 37°C;
-
2.
three seconds after the sample was 15–20% bleached by a 520-nm flash;
-
3.
ten minutes after addition of 30 mM hydroxylamine to convert bleached rhodopsin to opsin and retinal oxime; and
-
4.
after complete bleach of the sample.
Individual MI and MII spectra were deconvolved from spectra of their equilibrium mixture, with [MI] and [MII] determined using extinction coefficients at their absorbance maxima of 44,000 and 38,000 cm−1, respectively (28).
Results
Average lipid-order parameters
Dark-adapted rhodopsin was incorporated at rhodopsin/lipid values ranging from 1:1000 to 1:70 into phosphatidylcholine membranes with sn-1 saturated/perdeuterated and sn-2 monounsaturated acyl chains of 14–20 methylene segments in length. The order parameters of the saturated lipid hydrocarbon chains in position sn-1 were measured with a precision of ±0.001. Lipid-order parameters at the protein-lipid interface are altered by lipid-protein interactions, and may differ from lipid order away from the protein. 2H-NMR measurements report order parameters that are averaged on a timescale of 10−4–10−5 s. Also, assuming a typical lateral diffusion rate of lipids of 1 × 10−11 m2 s−1 suggests that lipids may easily move over typical distances between monomeric rhodopsin molecules within microseconds at all investigated rhodopsin concentrations.
The measured 2H NMR spectra of rhodopsin-containing samples (see Fig. 1) show only one set of slightly broadened quadrupolar splittings in the presence of rhodopsin. Therefore, it is reasonable to assume that lipids near rhodopsin and in the bulk of the lipid matrix are in rapid exchange. This is different from EPR measurements that operate on a timescale of 10−8 s, which yields distinct EPR spectra for lipids in the first layer surrounding rhodopsin and away from it. Those measurements indicated that ∼25 lipids form a first shell surrounding rhodopsin (29), which is in reasonable agreement with geometry of a rhodopsin monomer.
Figure 1.

2H NMR spectra of 14:0d27-14:1-PC bilayers containing rhodopsin at a P/L from 0 to 1:70 (only the left half of the spectra are shown). The AAO-supported sample of tubular lipid bilayers was oriented such that the pores are aligned parallel to the static magnetic field of the NMR instrument, resulting in a preferential perpendicular orientation of the bilayer normal to the magnetic field. Resonances are somewhat broadened by mosaic spread of bilayer orientations. Samples displayed only one set of quadrupolar splittings, suggesting that lipids exchange rapidly between the rhodopsin interface and the bulk of the lipid matrix on a timescale of 10−5 s.
Consequently, a 2H NMR experiment will report lipid-order averaged over lipid properties near the protein and in the bulk of the lipid matrix according to the equations
| (1) |
| (2) |
| (3) |
where 〈Slav〉 is the average order parameter of lipids, 〈Slf〉 is the order parameter of free lipids away from rhodopsin, 〈Slb〉 is the order parameter of lipids in the boundary layer surrounding rhodopsin, nlf is the number of free lipids away from rhodopsin, nlt is the total number of lipids, alb is the number of boundary lipids surrounding rhodopsin, and xr/1 is the rhodopsin/lipid. For simplicity, it was assumed that the boundary layer consists of 25 lipid molecules surrounding every rhodopsin molecule as a first layer.
Equation 3 predicts that without protein oligomerization or overlap of lipid boundary layers surrounding the protein, the average lipid-order 〈Slav〉 changes linearly with the rhodopsin/lipid value, xr/1. In Fig. 2, the corresponding plots are shown. Indeed, the curves for all four lipids at low rhodopsin content seem to change linearly with xr/1. Statistically significant deviations from linearity are observed for 14:0d27-14:1-PC at molar ratios larger than 1:250, for 16:0d31-16:1-PC larger than 1:250, for 18:0d35-18:1-PC larger than 1:125, and for 20:0d39-20:1-PC larger than 1:500. Most likely, those deviations from linearity are the result of rhodopsin oligomerization that occur at higher rhodopsin concentrations (see Fig. 3).
Figure 2.
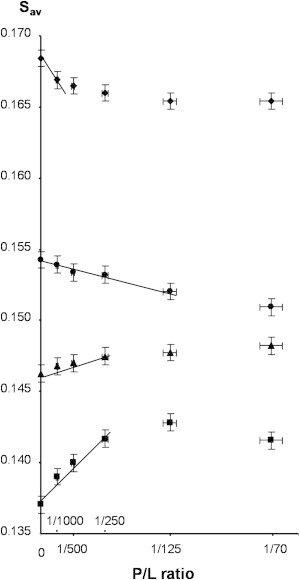
Average sn-1 chain order parameters, Sav, of 14:0d27-14:1-PC (squares), 16:0d31-16:1-PC (triangles), 18:0d35-18:1-PC (circles), and 20:0d39-20:1-PC (diamonds) bilayers containing dark-adapted rhodopsin at P/L from 0 to 1:70. Experiments were conducted at a temperature of 37°C. The average order parameters of lipid bilayers increases continuously with increasing hydrocarbon chain length. Incorporation of rhodopsin increases order of lipids with a chain length of 14 and 16 carbon atoms per chain and decreases order of lipids with 18 and 20 carbon atoms per chain. (Straight lines through the data points) Protein concentration ranges over which lipid-order changes can be approximated by a linear function.
Figure 3.
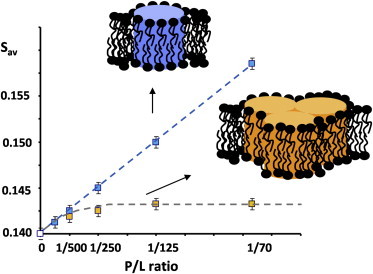
Predicted dependence of changes in average lipid order, Sav, as a function of rhodopsin concentration reported as P/L: (blue points) monomeric rhodopsin; (yellow points) rhodopsin with increasing oligomerization. To see this figure in color, go online.
Rhodopsin MI/MII equilibrium
The ratio of concentrations of photointermediates, Keq = [MII]/[MI], was measured by ultraviolet-visible spectroscopy as outlined in Materials and Methods. Assuming a Boltzmann distribution, the logarithm of Keq is proportional to the difference in free energies between the photointermediates MII and MI, ΔΔG ∝ ln Keq. The experimental results are reported in Fig. 4. For convenience, data are plotted such that ln Keq = 0 for rhodopsin in 14:0d27-14:1-PC is extrapolated to infinite dilution. At all investigated rhodopsin/lipid ratios, the amount of MII formed increased with increasing bilayer thickness. At the lowest rhodopsin content, this increase saturated for 18:0d35-18:1-PC and 20:0d39-20:1-PC. With increasing rhodopsin content, a steady decrease of the amount of MII formed was observed for 14:0d27-14:1-PC, 16:0d31-16:1-PC, and 18:0d35-18:1-PC. For bilayers of 20:0d39-20:1-PC, the amount of MII peaks at a rhodopsin/lipid of 1:250 and decreases at higher rhodopsin content as well.
Figure 4.
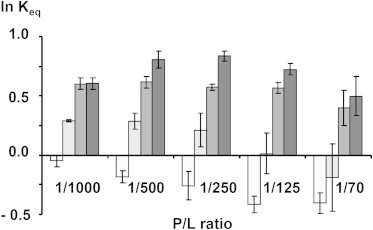
Natural logarithm of the ratio of concentrations of photointermediates MII and MI, ln Keq = ln[[MII]/[MI]] as a function of the P/L and hydrocarbon chain length of phosphatidylcholines. (Lightest to darkest shading) 14:0d27-14:1-PC, 16:0d31-16:1-PC, 18:0d35-18:1-PC, and 20:0d39-20:1-PC.
Discussion
Hydrophobic length of rhodopsin transmembrane helices
It was shown that average order parameters of saturated lipid hydrocarbon chains are related to an effective hydrophobic thickness of lipid bilayers (see Petrache et al. (30) and references therein). For consistency, we used the same definitions for the conversion as in our previous publication (14).
The hydrophobic thickness of rhodopsin-free bilayers changes continuously from 21.2 Å for 14:0d27-14:1-PC to 33.2 Å for 20:0d39-20:1-PC, as shown on the x axis in Fig. 5. The y axis reports the hydrophobic thickness of the first layer of lipids surrounding monomeric rhodopsin. In the absence of rhodopsin, x and y values are identical (solid line). For rhodopsin-containing samples, the lipid-order parameters measured at P/L values of 1:1000 and 1:500 (Fig. 2) were used to calculate an increase or decrease in lipid order of boundary lipids. For simplicity it was assumed that the entire change in order is taking place in the first layer of 25 lipids, while the remainder of lipids is unperturbed. Furthermore, it was assumed that lipids in the boundary layer are in rapid exchange with the bulk such that on the NMR timescale of 10 μs all lipids are equal as observed experimentally. The increase or decrease in order of boundary lipids was then converted to a change in boundary lipid hydrophobic thicknesses that yielded the dashed line in Fig. 5. The solid and dashed lines intersect at a hydrophobic thickness of 27 ± 1 Å. This is the hydrophobic thickness of bilayers at which order-parameter changes, resulting from incorporation of rhodopsin, vanish. It is now the hydrophobic thickness that matches the length of hydrophobic sections on transmembrane helices of rhodopsin.
Figure 5.
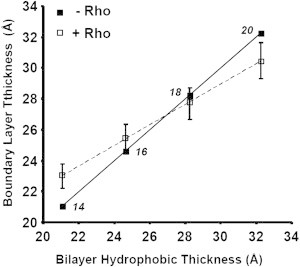
Plot of bilayer hydrophobic thickness-versus-rhodopsin lipid boundary layer thickness of 14:0d27-14:1-PC, 16:0d31-16:1-PC, 18:0d35-18:1-PC, and 20:0d39-20:1-PC. (Solid line) Bilayer thickness in the absence of rhodopsin. The numbers near the data points report the number of carbon atoms per hydrocarbon chain of lipids. (Dashed line) Calculated bilayer thickness in a hypothetical first layer of 25 lipids surrounding monomeric rhodopsin (see text for details); both curves intersect at a hydrophobic thickness of 27 Å. (The finite slope of the dashed line indicates that adjustment of lipid order to rhodopsin is partial.)
The value of 27 Å is in good agreement with data from our earlier experiments on incorporation of rhodopsin into bilayers of 18:0d35-22:6-PC. At the physiological temperature of 36°C, those membranes have a hydrophobic thickness of ∼27 Å and lipid order is almost unperturbed by rhodopsin incorporation (26). A similar value for an optimal bilayer thickness was proposed by Periole et al. (16), using CG-MD.
The assumption that the entire adjustment of order parameters from rhodopsin incorporation occurs in a first layer of 25 lipids surrounding the protein is certainly an oversimplification. In reality, such elastic bilayer deformations decay continuously over several layers of lipid surrounding the protein (31), which would reduce any changes of average length of lipid hydrocarbon chains near the protein that are reported as the dashed line in Fig. 5. Nevertheless, an important conclusion can be drawn from the results. Because the slope of the dashed line is positive, adjustment of hydrophobic lengths of lipids to the protein is only partial. Because any significant increase of exposure of hydrophobic regions on rhodopsin to water is energetically prohibitive (32), an important second conclusion can be drawn from Fig. 5. The structure of rhodopsin adjusts to the hydrophobic thickness of lipid bilayers such that exposure of hydrophobic segments on rhodopsin to water is reduced. Indeed, we had previously observed that helicity of rhodopsin adjusts continuously to the hydrophobic thickness of lipid bilayers (14). It is assumed that rhodopsin adjusts length and tilt of its seven transmembrane helices to match hydrophobic thickness of the lipid matrix. A length adjustment of helices most likely occurs at their ends. Helices are stabilized by intrahelical hydrogen bonding. The turns at either end of helices are stabilized by fewer hydrogen bonds, giving helices freedom to adjust their length to environmental changes.
Rhodopsin oligomerization
A mismatch between hydrophobic thicknesses of lipid bilayers and rhodopsin can also be reduced by limiting the fraction of rhodopsin surfaces that are exposed to lipids. This is achieved by oligomerization of rhodopsin molecules. If oligomerization occurs, the number of lipid molecules, alb, that interact with rhodopsin is reduced. According to Eq. 3, this reduces slopes of changes in lipid-order parameters 〈Slav〉 as a function of the rhodopsin/lipid as plotted in Fig. 3. Indeed, such reductions of slope are observed at rhodopsin/lipid as low as 1:500. Changes in slope are larger and tend to occur at lower rhodopsin concentrations for lipid bilayers with hydrophobic thicknesses that deviate the most from the optimal value of 27 Å. The data strongly suggest that rhodopsin oligomerizes in response to increasing elastic stresses in the lipid matrix from rhodopsin incorporation.
The occurrence of rhodopsin clustering in response to the energetic constraints of a hydrophobic mismatch between lipids and protein was reported previously. A chain-length dependence of protein aggregation was observed experimentally by Ryba and Marsh (18), Kusumi et al. (33), and Botelho et al. (15), and with CG-MD by Periole et al. (16).
Functional consequences of hydrophobic mismatch
As seen in Fig. 4, hydrophobic mismatch between lipids and rhodopsin has profound consequences on the MI/MII equilibrium. At all rhodopsin concentrations, there is a trend toward favoring formation of the MII photointermediate with increasing bilayer thickness. Those changes are qualitatively in agreement with expectations from monolayer elastic deformations near the protein (Fig. 6). We and others reported earlier that the MII state of rhodopsin is favored by membranes whose lipids are under negative curvature elastic stress, suggesting that the MII state has a somewhat longer hydrophobic transmembrane region and is more hourglass-shaped (7–11). Curvature elastic stress not only originates from lipid monolayers such as phosphatidylethanolamines with unsaturated hydrocarbon chains that have a tendency to curl like in an inverse hexagonal phase (34), but also from lipid monolayers that have curvature due to a hydrophobic mismatch between lipids and protein (31). Assuming an hourglass-shaped MII state, thicker bilayers relieve stresses upon MII formation in contrast to thinner bilayers for which stresses increase (see Fig. 6). We did not attempt to quantitatively describe the drive toward MII formation via lipid bilayer elasticity, because it is likely that the structural changes in rhodopsin with changing bilayer thickness contribute to the MI/MII energetics as well.
Figure 6.
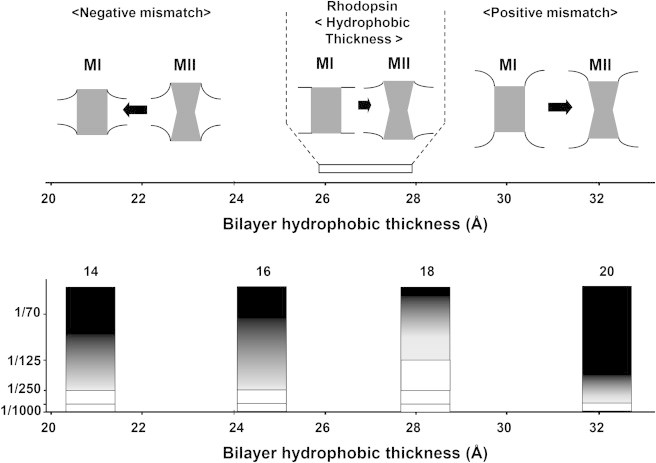
(Top) Cartoon demonstrating adjustment of lipid bilayers to the shape of the MI and MII photointermediates of rhodopsin. Bilayers that are thinner than rhodopsin have negative monolayer curvature, which increases upon MII formation, therefore, favoring formation of MI. Bilayers with a hydrophobic thickness of 27 Å are matched to MI and MII, while thicker bilayers have monolayers with positive curvature near MI and MII. (Bottom) In the case of hydrophobic mismatch between lipids and protein, curvature stress is also reduced by rhodopsin oligomerization that reduces exposure of rhodopsin to lipids; (open) no oligomerization; and (lightest to darkest shading) increasing levels of oligomerization.
Furthermore, the functional data indicate that rhodopsin oligomerization triggered by hydrophobic mismatch between lipids and protein disfavors formation of the MII photointermediate. During our functional experiments, only 15–20% of dark-adapted rhodopsin molecules are bleached. Therefore, upon oligomerization, most of the MI or MII photointermediates have dark-adapted rhodopsin molecules as next-neighbors. Obviously, the free energy of MI photointermediates packed with dark-adapted rhodopsin is lower than the energy of MII photointermediates. Indeed, the larger tilt of transmembrane helices in MII, in particular of helix VI (35), may result in less favorable packing of protein aggregates.
A word of caution needs to be added regarding oligomerization and orientation of rhodopsin molecules in reconstituted proteoliposomes. While rhodopsin molecules in the membranes of rod outer segments (ROS) have a uniform orientation, in reconstituted membranes orientation is likely to be random. It is not known whether a random orientation of rhodopsin molecules favors or disfavors rhodopsin oligomerization and whether oligomers of rhodopsin molecules with equal orientation have the same functional properties as molecules with opposite orientations. Also, molecular simulations clearly suggested that the length of hydrophobic segments on rhodopsin is heterogeneous about the circumference of the molecule, which has consequences for the energetics of lipid-protein interaction and rhodopsin oligomerization (31).
The results then raise the question of whether rhodopsin oligomerization takes place in ROS as well. Lipid composition of ROS is complex, and includes a variety of lipid headgroups (36) and lipid hydrocarbon chains (37) as well as variation of cholesterol content depending on the age of the ROS (37,38). Most remarkable is the high content of docosahexaenoic acid chains (22:6n-3) in all major lipid classes. Niu and Mitchell (39) investigated the effect of rhodopsin/18:0-22:6-PC molar ratios on rhodopsin activation in the range of 1/422–1/40 and observed reduced MII formation with increasing rhodopsin concentration as well. However, it was concluded that the presence of polyunsaturated chains attenuates the magnitude of changes.
Although our experimental data do not address oligomerization of rhodopsin in ROS membranes, the trends of 18:0-22:6-PC to yield a hydrophobic thickness of 27 Å at physiological temperature (26) and to attenuate changes in MII formation both suggest that polyunsaturated lipids tend to suppress rhodopsin oligomerization.
Conclusions
The order-parameter measurements show convincingly that a hydrophobic mismatch between lipids and rhodopsin results in an elastic deformation in the lipid bilayer near the protein to adjust membrane thickness to the hydrophobic length of rhodopsin’s transmembrane helices. However, the adjustment of the thickness of bilayers near the protein is only partial. Rhodopsin responds to thickness changes by structural adjustments as we reported earlier (14). Furthermore, rhodopsin reduces unfavorable energy from hydrophobic mismatch between lipids and protein by formation of rhodopsin oligomers to reduce exposure of rhodopsin molecules to lipid.
Hydrophobic mismatch between lipids and rhodopsin has a profound influence on the equilibrium of MI/MII photointermediates. Thicker bilayers clearly favor formation of MII while rhodopsin oligomerization drives the equilibrium toward MI. More experiments need to be conducted to better understand rhodopsin oligomerization. From the results it seems obvious that at the physiological rhodopsin/lipid molar ratio of 1:70, as in membranes of ROS, rhodopsin molecules are likely to influence each other functionally, either directly through oligomerization or indirectly through a superposition of lipid layers that are perturbed by lipid-protein interaction.
References
- 1.Palczewski K., Kumasaka T., Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 2.Choe H.W., Kim Y.J., Ernst O.P. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 3.Scheerer P., Park J.H., Ernst O.P. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 4.Yoshizawa T., Wald G. Pre-lumirhodopsin and the bleaching of visual pigments. Nature. 1963;197:1279–1286. doi: 10.1038/1971279a0. [DOI] [PubMed] [Google Scholar]
- 5.Hargrave P.A., Hamm H.E., Hofmann K.P. Interaction of rhodopsin with the G-protein, transducin. BioEssays. 1993;15:43–50. doi: 10.1002/bies.950150107. [DOI] [PubMed] [Google Scholar]
- 6.Wilden U., Hall S.W., Kühn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl. Acad. Sci. USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soubias O., Teague W.E., Jr., Gawrisch K. Contribution of membrane elastic energy to rhodopsin function. Biophys. J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teague W.E., Jr., Soubias O., Gawrisch K. Elastic properties of polyunsaturated phosphatidylethanolamines influence rhodopsin function. Faraday Discuss. 2013;161:383–395. doi: 10.1039/c2fd20095c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson N.J., Brown M.F. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 10.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 11.Botelho A.V., Gibson N.J., Brown M.F. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 12.Davoust J., Bienvenue A., Devaux P.F. Boundary lipids and protein mobility in rhodopsin-phosphatidylcholine vesicles. Effect of lipid phase transitions. Biochim. Biophys. Acta. 1980;596:28–42. doi: 10.1016/0005-2736(80)90168-6. [DOI] [PubMed] [Google Scholar]
- 13.Pearson L.T., Chan S.I., Engelman D.M. Pair distribution functions of bacteriorhodopsin and rhodopsin in model bilayers. Biophys. J. 1983;43:167–174. doi: 10.1016/S0006-3495(83)84337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soubias O., Niu S.L., Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botelho A.V., Huber T., Brown M.F. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Periole X., Huber T., Sakmar T.P. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 17.Davoust J., Schoot B.M., Devaux P.F. Physical modifications of rhodopsin boundary lipids in lecithin-rhodopsin complexes: a spin-label study. Proc. Natl. Acad. Sci. USA. 1979;76:2755–2759. doi: 10.1073/pnas.76.6.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryba N.J.P., Marsh D. Protein rotational diffusion and lipid/protein interactions in recombinants of bovine rhodopsin with saturated diacylphosphatidylcholines of different chain lengths studied by conventional and saturation-transfer electron spin resonance. Biochemistry. 1992;31:7511–7518. doi: 10.1021/bi00148a011. [DOI] [PubMed] [Google Scholar]
- 19.Fotiadis D., Liang Y., Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 20.Chabre M., Cone R., Saibil H. Biophysics: is rhodopsin dimeric in native retinal rods? Nature. 2003;426:30–31. doi: 10.1038/426030b. discussion 31. [DOI] [PubMed] [Google Scholar]
- 21.Bayburt T.H., Vishnivetskiy S.A., Gurevich V.V. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu S.L., Doctrow B., Mitchell D.C. Rhodopsin activity varies in proteoliposomes prepared by different techniques. Biochemistry. 2009;48:156–163. doi: 10.1021/bi801835s. [DOI] [PubMed] [Google Scholar]
- 23.Litman B.J. Purification of rhodopsin by concanavalin A affinity chromatography. Methods Enzymol. 1982;81:150–153. doi: 10.1016/s0076-6879(82)81025-2. [DOI] [PubMed] [Google Scholar]
- 24.Chattopadhyay A., London E. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 25.Wald G., Brown P.K. The molar extinction of rhodopsin. J. Gen. Physiol. 1953;37:189–200. doi: 10.1085/jgp.37.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soubias O., Polozov I.V., Gawrisch K. Functional reconstitution of rhodopsin into tubular lipid bilayers supported by nanoporous media. Biochemistry. 2006;45:15583–15590. doi: 10.1021/bi061416d. [DOI] [PubMed] [Google Scholar]
- 27.Straume M., Mitchell D.C., Litman B.J. Interconversion of metarhodopsins I and II: a branched photointermediate decay model. Biochemistry. 1990;29:9135–9142. doi: 10.1021/bi00491a006. [DOI] [PubMed] [Google Scholar]
- 28.Applebury M.L. Dynamic processes of visual transduction. Vision Res. 1984;24:1445–1454. doi: 10.1016/0042-6989(84)90305-5. [DOI] [PubMed] [Google Scholar]
- 29.Watts A., Volotovski I.D., Marsh D. Rhodopsin-lipid associations in bovine rod outer segment membranes. Identification of immobilized lipid by spin-labels. Biochemistry. 1979;18:5006–5013. doi: 10.1021/bi00589a031. [DOI] [PubMed] [Google Scholar]
- 30.Petrache H.I., Dodd S.W., Brown M.F. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys. J. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mondal S., Khelashvili G., Weinstein H. Quantitative modeling of membrane deformations by multihelical membrane proteins: application to G-protein coupled receptors. Biophys. J. 2011;101:2092–2101. doi: 10.1016/j.bpj.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusumi A., Subczynski W.K., Merkle H. Spin-label studies on phosphatidylcholine-cholesterol membranes: effects of alkyl chain length and unsaturation in the fluid phase. Biochim. Biophys. Acta. 1986;854:307–317. doi: 10.1016/0005-2736(86)90124-0. [DOI] [PubMed] [Google Scholar]
- 34.Gruner S.M. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proc. Natl. Acad. Sci. USA. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altenbach C., Kusnetzow A.K., Hubbell W.L. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boesze-Battaglia K., Albert A.D. Phospholipid distribution among bovine rod outer segment plasma membrane and disk membranes. Exp. Eye Res. 1992;54:821–823. doi: 10.1016/0014-4835(92)90040-y. [DOI] [PubMed] [Google Scholar]
- 37.Boesze-Battaglia K., Hennessey T., Albert A.D. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J. Biol. Chem. 1989;264:8151–8155. [PMC free article] [PubMed] [Google Scholar]
- 38.Boesze-Battaglia K., Albert A.D. Cholesterol modulation of photoreceptor function in bovine retinal rod outer segments. J. Biol. Chem. 1990;265:20727–20730. [PubMed] [Google Scholar]
- 39.Niu S.L., Mitchell D.C. Effect of packing density on rhodopsin stability and function in polyunsaturated membranes. Biophys. J. 2005;89:1833–1840. doi: 10.1529/biophysj.105.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]


