Key Points
PAX5-JAK2 is the first nuclear DNA-binding JAK2 fusion protein with kinase activity.
JAK2 inhibitors block the kinase activity of PAX5-JAK2.
Abstract
PAX5-JAK2 has recently been identified as a novel recurrent fusion gene in B-cell precursor acute lymphoblastic leukemia, but the function of the encoded chimeric protein has not yet been characterized in detail. Herein we show that the PAX5-JAK2 chimera, which consists of the DNA-binding paired domain of PAX5 and the active kinase domain of JAK2, is a nuclear protein that has the ability to bind to wild-type PAX5 target loci. Moreover, our data provide compelling evidence that PAX5-JAK2 functions as a nuclear catalytically active kinase that autophosphorylates and in turn phosphorylates and activates downstream signal transducers and activators of transcription (STATs) in an apparently noncanonical mode. The chimeric protein also enables cytokine-independent growth of Ba/F3 cells and therefore possesses transforming potential. Importantly, the kinase activity of PAX5-JAK2 can be efficiently blocked by JAK2 inhibitors, rendering it a potential target for therapeutic intervention. Together, our data show that PAX5-JAK2 simultaneously deregulates the PAX5 downstream transcriptional program and activates the Janus kinase-STAT signaling cascade and thus, by interfering with these two important pathways, may promote leukemogenesis.
Introduction
The fusion protein PAX5-JAK2 has been recurrently detected in B-cell precursor acute lymphoblastic leukemia (BCP-ALL).1-4 Both fusion partner proteins play key roles in hematopoiesis, and somatic mutations in their encoding genes have been found in different hematologic neoplasms.5-7
The paired box transcription factor PAX5, a master regulator of B-cell commitment and maintenance,6 is a frequent target of genetic alterations in BCP-ALL.5,8 In ∼2% to 3% of the cases, structural rearrangements result in the expression of PAX5 in-frame fusion genes.1,2,4,5,8-10 PAX5 fusion partners comprise a heterogeneous group of genes encoding transcription factors, structural proteins, kinases, and genes with thus far unknown functions.1,2,8,9,11-13 Regardless of the functional and structural diversity of the fusion partners, a unique feature of PAX5 fusions is the retention of the PAX5 DNA-binding domain, conferring nuclear localization and the ability to occupy PAX5 target sites.14
Generally, it is hypothesized that PAX5 fusions act as aberrant transcription factors antagonizing wild-type PAX5 function in a dominant negative mode.1,8,9,11,15-18 However, in a recent study, we have shown that a subset of the PAX5 fusion proteins may have a cellular context-dependent activation potential, indicating that some PAX5 fusions may also activate target genes, thus arguing against their simplified trans-dominant negative function.14
Janus kinase 2 (JAK2) belongs to a family of nonreceptor tyrosine kinases and is involved in signal transduction from many cytokine and growth hormone receptors regulating hematopoiesis, immunity, growth, and development.19 Besides gain-of-function mutations, mainly found in or near the pseudokinase (JH2) domain, JAK2 also participates in gene rearrangements resulting in the expression of in-frame fusion transcripts encoding chimeric proteins.4,7,20-28 A common feature of all JAK2 fusions is the retention of the catalytically active kinase (JH1) domain, and for several of them, constitutive activation has been demonstrated.3,4,23,28-30 Moreover, BCR-JAK2, ETV6-JAK2, and SEC31A-JAK2, which are all localized in the cytoplasm, are activated upon dimerization via domains provided by the partner proteins, which in turn leads to the activation of the JAK-signal transducer and activator of transcription (STAT) signaling cascade.23,28-30
Because the activation of the JAK-STAT pathway plays a pivotal role in leukemogenesis, a number of small molecule inhibitors is currently under clinical investigation.7,31,32 In this respect, PAX5-JAK2+ leukemia has been found within the recently identified BCR-ABL1-like (Ph-like) BCP-ALL subtype, which is, at least in part, characterized by genetic alterations resulting in constitutive kinase and cytokine receptor signaling, and it has been suggested that some of these patients might benefit from targeted therapies.3,4
We herein demonstrate that PAX5-JAK2 represents the first nuclear JAK2 fusion protein that not only displays DNA-binding capacity and deregulates PAX5 target genes but also possesses an active kinase domain and constitutively activates the JAK-STAT signaling pathway. By showing that JAK2 inhibitors efficiently block hyperactivation of the kinase, we substantiate the notion that PAX5-JAK2 represents a potential druggable target for therapeutic intervention.
Material and methods
Patients
This study includes pediatric patients enrolled in the ALL-Berlin-Frankfurt-Münster (BFM) 2000 and the Dutch Childhood Oncology Group (DCOG) ALL-8, ALL-9, ALL-10, and Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL) 97/03 clinical trials. Informed consent was obtained from the patients, their parents, or their legal guardians in accordance with the Declaration of Helsinki. A detailed description of the patients analyzed by gene expression profiling is provided in supplemental Tables 1-3, available on the Blood Web site.
Constructs and transposon vectors
The coding regions of PAX5-JAK2, PAX5, and JAK2 were polymerase chain reaction (PCR)-amplified with Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes) according to the manufacturer’s instructions using cDNA of the patients or NALM-6 cells. N-terminal V5- or tandem hemagglutinin (HA)-tagged versions were cloned into the following vectors: pcDNA3 (Invitrogen), pIRES-EGFP (Clontech), and the inducible sleeping beauty construct pITR-TCE-Ins-UTR33,34 (supplemental Figure 1G) (kindly provided by E. Kowarz and R. Marschalek, Johann Wolfgang Goethe-University, Frankfurt/Main, Germany). Mutations within the kinase and the DNA-binding domain were introduced using the QuikChange Site-Directed Mutagenesis Kit (Agilent). Details concerning the mutant forms of PAX5 and PAX5-JAK2 and the sequences of the oligonucleotides used for the generation of the constructs are provided in the supplemental Methods and supplemental Table 4.
Cell culture, Ba/F3 starvation, drug treatment, and apoptosis staining
HEK293, HeLa, and JAK2-deficient γ2a fibrosarcoma cells35 (kindly provided by B. Strobl, University of Veterinary Medicine, Vienna, Austria) were maintained in Dulbecco’s modified Eagle medium high glucose (4.5 g/L) with stable glutamine (PAA, Laboratories) containing 10% fetal bovine serum (PAA, Laboratories) and antibiotics. NALM-6 and interleukin 3 (IL3) secreting WEHI-3B cells were maintained in RPMI 1640 with GlutaMAX (Gibco) containing 10% fetal bovine serum and antibiotics. Ba/F3 cells (kindly provided by M. Busslinger, Research Institute of Molecular Pathology, Vienna, Austria) were grown in the latter medium supplemented with 10% IL3 containing WEHI-3B supernatant. Selection of Ba/F3 and NALM-6 cells and induction of protein expression were carried out using 2 µg/mL puromycin and 250 ng/mL doxycycline, respectively. For starvation experiments, cells were washed 3 times with medium without IL3; 106 Ba/F3 cells were seeded and cultured for 7 days in the presence or absence of IL3. Viable cell counts were determined every 24 hours using trypan blue exclusion staining.
The drug treatment was conducted with ruxolitinib (INCB18424), fedratinib (SAR302503), or dasatinib (all from Selleckchem) using the indicated concentrations. For phospho-STAT5 and gene expression analysis, Ba/F3 cells were washed 2 times with RPMI without supplements and treated for 1 and 8 hours with inhibitor, respectively. For apoptosis staining, a no-wash procedure36 was performed 24 and 48 hours after treatment. Cells were stained with Annexin-V-allophycocyanin (eBioscience) and 4,6 diamidino-2-phenylindole (DAPI) for 15 minutes at room temperature and immediately analyzed. Data were acquired on a fluorescence-activated cell sorter (FACS) Calibur or FACS Fortessa cytometer (Becton Dickinson) and analyzed using FlowJo (Tree Star).
Transfection and electroporation
Transient transfection of HEK293, HeLa, and γ2a cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Electroporation of Ba/F3 and NALM-6 cells was performed using the Amaxa Cell Line Nucleofector Kit V or T (Lonza) according to the manufacturer’s instructions.
Indirect immunofluorescence
HeLa cells grown on coverslips were transiently transfected with the respective HA-tag constructs. Twenty-four hours after transfection, cells were fixed using 1% formaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton-X100 in PBS, and blocked with 3% bovine serum albumin and 0.2% Triton-X100 in PBS. Washed coverslips were incubated with primary and secondary antibodies (supplemental Table 7), followed by nuclei counterstaining with DAPI solution and mounting in Mowiol 4-88 (Sigma-Aldrich).
Western blotting and reverse phase protein arrays
Cells were lyzed in high salt buffer (10 mM Tris-HCl, pH 7.5, 400 mM NaCl, 0.5% NP40, and 0.3% Triton X-100) with 125 µM ortho-pervanadate (Sigma-Aldrich) and protease inhibitor cocktail (Roche). Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and blocked using blocking reagent (Roche). Western blotting was performed with appropriate dilutions of primary antibodies followed by incubation with IRDye secondary antibodies (Pierce) and detected using the Odyssey system (LI-COR Biosciences). The antibodies used are listed in supplemental Table 7.
Reverse phase protein arrays were performed as described previously37-40 in collaboration with E. Petricoin (George Mason University). The experimental procedure and analysis are described in supplemental Methods.
Chromatin immunoprecipitation and quantitative real-time PCR
Chromatin immunoprecipitation (ChIP) was performed as described elsewhere14 using 2 × 107 cells and 2 μg mouse V5 (Invitrogen) or normal IgG antibody (sc-2025; Santa Cruz). Quantitative real-time PCR was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) using iQ-SYBR-Green-Supermix (Bio-Rad) and primers specific for known PAX5 target loci or a negative control region (supplemental Table 5). Analysis and normalization were performed using the 7500 Software v2.0.6 (Applied Biosystems), and signals were normalized to the individual input DNA using the 2−∆Ct method (percent input in ChIP).
For mRNA quantification, RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using Moloney monkey leukemia virus reverse transcriptase (Promega), oligo(dT)18, and random hexamers. Target gene expression was normalized to Abl1 and Gusb reference gene expression. Primers are listed in supplemental Table 6.
Luciferase assays
HEK293 cells were transiently cotransfected with 200 ng firefly luciferase vector pGL4-CD79A-TSS14 or luc-CD19 (kindly provided by M. Busslinger),41 and 600 ng expression vector containing either wild-type PAX5 or a PAX5 fusion and 20 ng pRL-null (Promega) for normalization purposes. Luciferase activity was measured on an Enspire plate reader (Perkin Elmer) 48 hours after transfection using the Dual-Glo Luciferase Assay System (Promega). Firefly to Renilla luciferase count ratios were further normalized to empty expression vector and empty luciferase vector controls. Results represent the means ± standard deviation (SD) of ≥3 independent experiments, each conducted in triplicate. Statistical analysis was performed using an unpaired 2-tailed Student t test comparing the mean fold activation values of 3 independent experiments with the hypothetical value of 1.
Intracellular phosphoprotein analysis
For intracellular phosphoprotein staining,42,43 cells were harvested and fixed with 2% methanol-free formaldehyde (Polysciences) for 15 minutes at room temperature. Cells were centrifuged and permeabilized by the addition of ice-cold methanol while vortexing and immediately transferred to −20°C for 1 to 3 hours. After centrifugation, the cells were resuspended in PBS supplemented with 0.02% bovine serum albumin and stained for 1 hour at room temperature using the appropriate antibody (supplemental Table 7). For staining of phospho-JAK2, cells were further incubated with a labeled secondary antibody (Jackson ImmunoResearch).
Gene expression analysis by microarray technology
Gene expression profiling of primary leukemia samples was conducted using HG-U133-PLUS2 arrays (Affymetrix). RNA was isolated using Trizol (Invitrogen), and the quality was checked on a Bioanalyzer (Agilent). cRNA target synthesis and GeneChip processing were performed according to standard protocols (Affymetrix). Microarray data analyses were performed in compliance with Minimum Information About a Microarray Experiment (MIAME) guidelines. All further analyses were carried out in the R statistical environment using Bioconductor packages.44 Affymetrix CEL files were preprocessed using frma45 (using the “robust_weighted_average” method). Batch effects were removed using combat.46 Probe sets with z-scores <2 (using the barcode function in the frma package) in all samples and probe sets with an SD <0.5 were excluded. Finally, 1 probe set (the most variable across all samples) was chosen for each gene. Together, this filtering resulted in a matrix with 6217 probe sets.
Four groups were defined according to their PAX5 status: PAX5-JAK2 (n = 5; 4 primary bone marrow samples collected at diagnoses and 1 matched relapse sample), PAX5-ETV6 (n = 4), PAX5-C20orf112 (n = 6), and BCP-ALL cases with no PAX5 alteration named PAX5-WT (n = 17). A detailed description of the patients and the selection criteria is provided in supplemental Tables 1 to 3. Differentially expressed genes were determined using a moderated t test in the R package “limma.”47 All P values were corrected for multiple testing using the Benjamini-Hochberg correction. Genes with a log2 expression ratio (logFC) >1 and P values <.05 were considered significantly different. For data visualization, gene expression relative to the mean expression of PAX5-WT was calculated. Additional details are described in supplemental Methods. Microarray data are available at Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo; accession number GSE56449).
Results
PAX5-JAK2 binds to PAX5 target loci and activates these genes
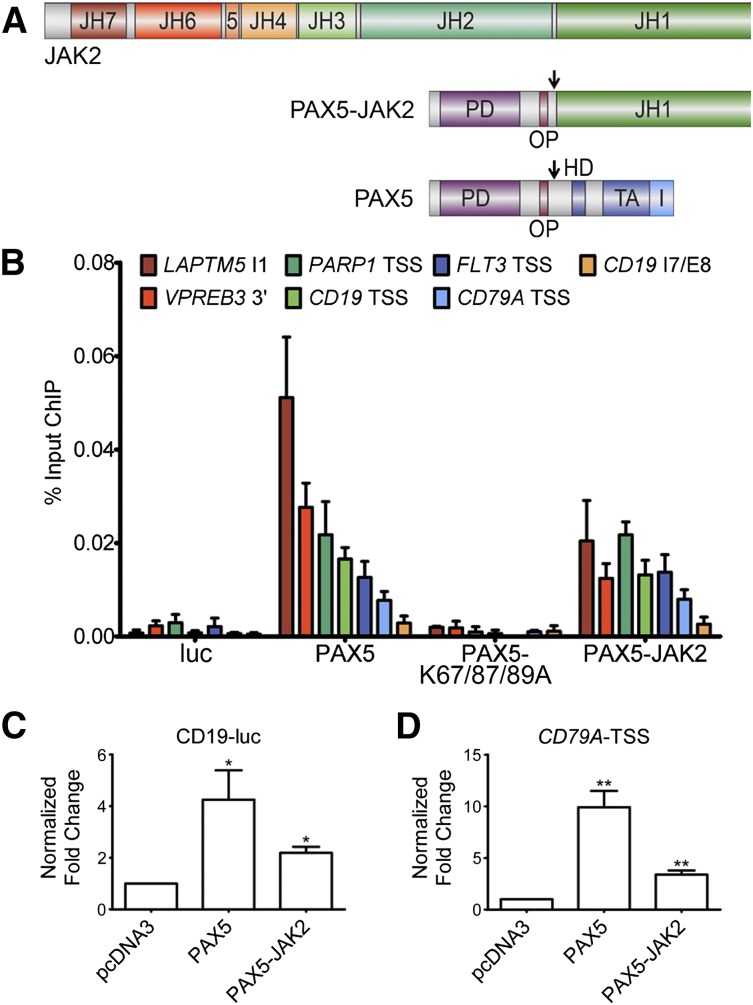
Given the fact that the PAX5 DNA-binding domain is retained in the fusion protein (Figure 1A), we investigated whether PAX5-JAK2 binds to known endogenous PAX5 target loci. ChIP followed by quantitative real-time PCR with PAX5 target site–specific primers was performed in NALM-6 cells inducibly expressing the indicated proteins. Significantly higher levels of specific DNA were precipitated in cells expressing V5-PAX5 and V5-PAX5-JAK2 compared with luciferase or the DNA-binding deficient mutant (V5-PAX5-K67/87/89A)-expressing cells (Figure 1B). No signals above background levels were obtained using primers targeting a negative control region (CD19 intron7/exon8), in uninduced NALM-6 cells, or using an IgG control antibody (supplemental Figure 1A-C). Because the expression level of PAX5-JAK2 is comparable to that of wild-type PAX5 (supplemental Figure 1D), the lower ChIP signal intensities may reflect a decreased binding affinity.
Figure 1.
PAX5-JAK2 is able to bind and regulate PAX5 target genes. (A) Schematic representation of the PAX5-JAK2 fusion protein, PAX5, and JAK2. PD, paired DNA-binding domain; OP, octapeptide; HD, partial homeodomain; TA, transactivation domain; I, inhibitory domain; JH1, kinase domain; JH2, pseudokinase domain; JH3-4, SH2-like domain; JH6-7, FERM domain; arrows indicate nuclear localization signals. (B) Chromatin immunoprecipitation followed by quantitative real-time PCR analysis of NALM-6 cells carrying a luciferase vector (luc) or expressing V5-tagged PAX5, a DNA-binding deficient mutant thereof (V5-PAX5-K67/87/89A), or V5-PAX5-JAK2. The amount of DNA precipitated with V5 antibody was determined by quantitative PCR using PAX5 target loci-specific primers and normalized to input DNA. The results of 1 of 3 representative experiments are shown. 3′, 3-prime region; TSS, transcription start site; I, intron; E, exon. (C-D) Reporter gene assays in HEK293 cells transfected with (C) luc-CD19 or (D) pGL4-CD79A-TSS luciferase reporter and indicated V5-tagged expression vectors. Firefly was normalized to Renilla luciferase activity and to the empty luciferase vector and the expression vector control. The bars represent the means ± SD of ≥3 independent experiments each performed in triplicate. Significance levels were determined using an unpaired 2-tailed Student t test comparing the mean fold activation values of 3 independent experiments with the hypothetical value of 1 (*P < .05, **P < .01, ***P < .001).
As PAX5-JAK2 occupies endogenous wild-type PAX5 target sites (Figure 1B), we next investigated its impact on target gene expression. In reporter gene assays, using either a luciferase vector carrying the endogenous CD79A promoter or the luc-CD19 vector, PAX5-JAK2 activated both constructs (Figure 1C-D). However, the activation capability of PAX5-JAK2 was lower compared with PAX5, and in competition assays, increasing amounts of PAX5-JAK2 had no significant influence on the activation potential of PAX5 (supplemental Figure 1E-F).
The nuclear protein PAX5-JAK2 is phosphorylated and activates STATs
Because the JH1 kinase domain is present in PAX5-JAK2 (Figure 1A), we examined whether the fusion itself is tyrosine phosphorylated and whether it activates downstream STAT proteins.
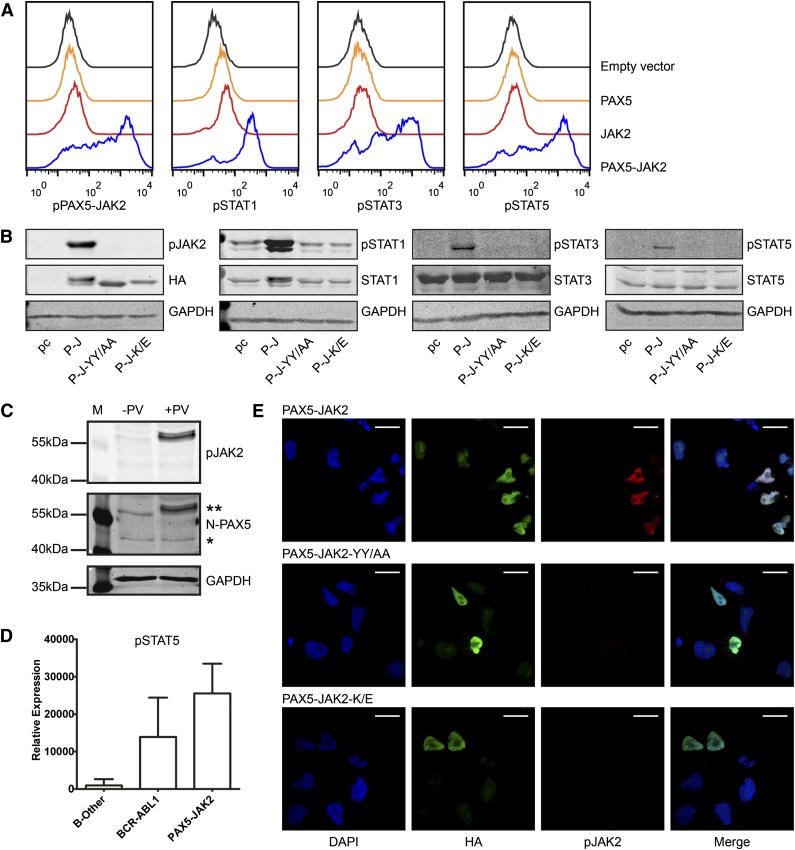
As shown by intracellular phosphoprotein analyses using flow cytometry, transiently transfected, unstimulated JAK2-deficient γ2a cells exhibited a high degree of tyrosine phosphorylation of PAX5-JAK2 (pPAX5-JAK2) (Figure 2A), strongly suggesting cytokine stimulation-independent kinase activity of the fusion protein. Additionally, ectopic expression of PAX5-JAK2 resulted in constitutive phosphorylation of STAT1, 3, and 5 (Figure 2A). Western blotting, including not only a mutant of the tyrosine residues in the activation loop of the kinase (PAX5-JAK2-YY/AA) but also a kinase dead mutant (PAX5-JAK2-K/E), both of which abolished PAX5-JAK2 phosphorylation, confirmed that STAT activation is pPAX5-JAK2 dependent (Figure 2B). Importantly, analysis of the pPAX5-JAK2 status in bone marrow cells obtained from a PAX5-JAK2+ patient confirmed that PAX5-JAK2 is also phosphorylated in primary leukemia cells (Figure 2C). As assessed by reverse phase protein arrays, similar to BCR-ABL1+ samples, two PAX5-JAK2+ cases showed higher levels of pSTAT5 compared with B-other leukemia (Figure 2D). The phosphorylation of other STATs in PAX5-JAK2+ leukemia varied, and, due to the small number of patients, no conclusions could be drawn (supplemental Figure 2A). As for other kinase-activated fusions,48-50 pSTAT5 is essential for transformation and disease maintenance, STAT5 is probably the main downstream STAT target of PAX5-JAK2.
Figure 2.
PAX5-JAK2 is phosphorylated and activates STAT proteins. (A) Intracellular phosphoprotein analyses using flow cytometry were performed in JAK2-deficient γ2a cells transfected with the indicated proteins. One of 3 representative experiments is shown. Black lines, empty vector, pIRES2-EGFP; orange lines, PAX5; red lines, JAK2; blue lines, PAX5-JAK2. (B) Western blotting of whole cell lysates of γ2a cells. pc, pcDNA3 empty vector; P-J, PAX5-JAK2; P-J-YY/AA, tyrosine mutant; P-J-K/E, kinase dead mutant (GAPDH ∼ 37 kDa, HA ∼ 60 kDa, pJAK2 ∼ 60 kDa, STATs/pSTATs ∼ 90 kDa). (C) PAX5-JAK2 phosphorylation detected in whole cell lysate of a bone marrow sample of a PAX5-JAK2+ patient. −PV, without pervanadate; +PV, treated with phosphatase inhibitor pervanadate; **PAX5-JAK2; *PAX5. (D) pSTAT5 levels in bone marrow cells of patients with PAX5-JAK2+ (n = 2), BCR-ABL1+ (n = 33) or B-other (n = 43) leukemia were measured by reverse phase protein arrays, and levels were normalized for total protein levels in each cell lysate. (E) Immunofluorescence was performed in HeLa cells transfected with (top) HA-tagged PAX5-JAK2, as well as with the mutated forms (middle) PAX5-JAK2-YY/AA and (bottom) PAX5-JAK2-K/E using an HA (green) and a p-Y1007/8-JAK2 (red) antibody and DAPI (blue) for DNA counterstaining. Images were acquired on a Leica TCS SP5 equipped with an HCX PL APO CS 63.0×1.40 oil objective in sequential scan mode using identical settings for all conditions. White bars indicate 20 µm.
To further investigate the capability of pPAX5-JAK2 to induce STAT5 activity, we conducted reporter gene assays using a 6x-STAT5-TK-luc reporter construct. PAX5-JAK2 significantly activated the reporter in γ2a cells, whereas its mutant forms or wild-type JAK2 did not (supplemental Figure 2B). Collectively, PAX5-JAK2 itself is phosphorylated and not only phosphorylates downstream STATs but also activates STAT5.
Main signaling functions of wild-type JAK2 are linked to its cytoplasmatic localization.7 In this context, all JAK2 fusion proteins investigated thus far reside in the cytoplasm and therefore can activate the canonical JAK-STAT pathway.7,51 However, a common feature of PAX5 fusions is their mainly nuclear localization.14 To verify that PAX5-JAK2, which retains the nuclear localization signal and the paired domain of PAX5 (Figure 1A), is also a nuclear protein, we ectopically expressed it in cell lines and performed indirect immunofluorescence (Figure 2E; supplemental Figure 2C). In line with its DNA-binding capacity (Figure 1B), pPAX5-JAK2 displayed exclusive nuclear localization (Figure 2E).
The majority of JAK2 fusion proteins possess an oligomerization domain provided by the fusion partner, which facilitates trans-autophosphorylation.7,51 However, PAX5-JAK2 lacks a self-interaction motif, and self-association was ruled out by co-immunoprecipitation experiments (supplemental Figure 2D). Because the kinase dead PAX5-JAK2 mutant was completely unphosphorylated, trans-phosphorylation by another kinase is highly unlikely. Moreover, a DNA-binding–deficient PAX5-JAK2 mutant revealed no impairment of phosphorylation (supplemental Figure 2E), indicating that the autophosphorylation of PAX5-JAK2 is DNA binding independent.
Treatment with JAK2 inhibitors leads to death of PAX5-JAK2–transformed cells
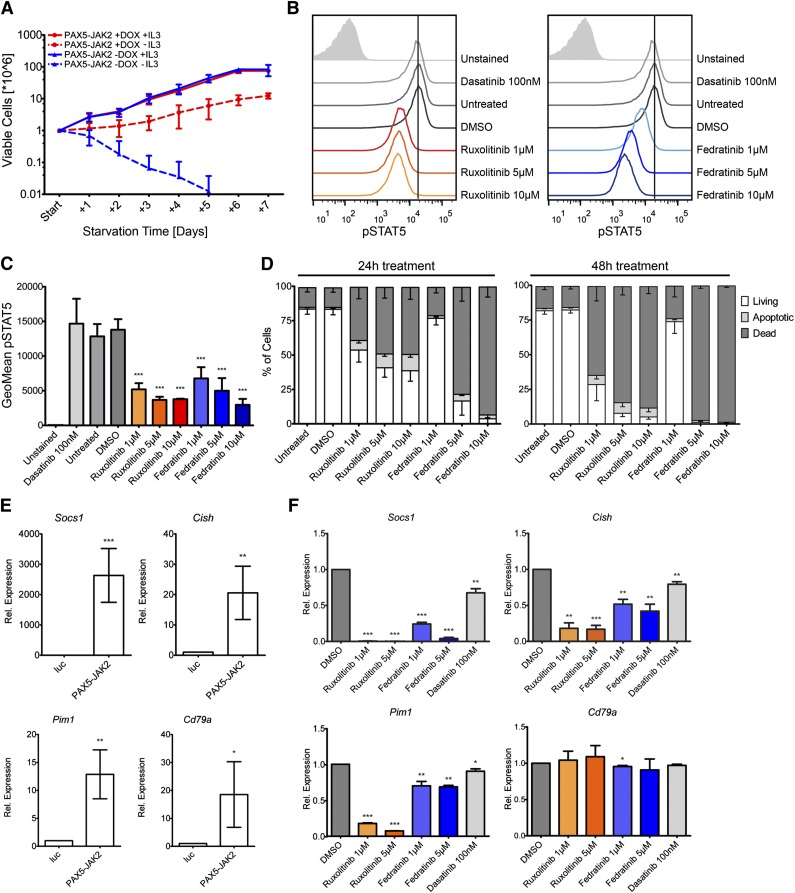
To determine the in vitro transforming potential of PAX5-JAK2, we ectopically expressed it in IL3-dependent Ba/F3 cells using an inducible vector system. Western blotting confirmed that only on administration of doxycycline PAX5-JAK2 was expressed, and consequently, the STATs were phosphorylated (supplemental Figure 3A). Twenty-four hours after PAX5-JAK2 induction, IL3 starvation experiments were started. Although in the presence of IL3, no differences in growth between Ba/F3 cells with or without PAX5-JAK2 induction were observable, following IL3 deprivation, only cells expressing the fusion protein survived (Figure 3A). Although under IL3 starvation, the cells showed a decreased growth rate, PAX5-JAK2 clearly triggered factor-independent proliferation of Ba/F3 cells.
Figure 3.
PAX5-JAK2 transforms Ba/F3 cells, and its kinase activity can be blocked by JAK2 inhibitors. (A) Growth rates of Ba/F3 cells in the presence (red) or absence (blue) of doxycycline with (full lines) or without (dashed lines) IL3 were monitored over 7 days by trypan blue exclusion staining. The means ± SD of 3 independent biological replicates are shown. (B) IL3-independent Ba/F3 cells expressing PAX5-JAK2 were treated with the indicated inhibitors for 1 hour. Intracellular pSTAT5 levels were measured by flow cytometry analysis. One of 3 representative experiments is shown. (C) Summary of geometric means ± SD (n = 3) of pSTAT5 levels as determined in B. (D) Cytokine-independent Ba/F3 cells expressing PAX5-JAK2 were treated with the indicated inhibitors for (left) 24 and (right) 48 hours. Staining for apoptotic/dead cells was performed using Annexin-V-allophycocyanin and DAPI, respectively. Means of 3 independent biological replicates are shown. Error bars represent SD. (E) Ba/F3 cells harboring the luc-control vector or PAX5-JAK2 vector were treated with doxycycline for 24 hours. Gene expression was measured using quantitative real-time PCR. Expression values were normalized using Abl1 and Gusb as control genes. Expression relative to luc-control of ≥3 independent experiments is shown. Significance levels were determined using an unpaired 2-tailed Student t test comparing the mean fold activation values of 3 independent experiments with the hypothetical value of 1 (*P < .05, **P < .01, ***P < .001). (F) Cytokine-independent Ba/F3 cells expressing PAX5-JAK2 were treated with indicated inhibitors for 8 hours. Relative gene expression was determined as in E.
To assess the effectiveness of JAK2 inhibitors against the constitutive kinase activity of PAX5-JAK2, we treated IL3-independent Ba/F3 cells expressing PAX5-JAK2 with ruxolitinib and fedratinib.32 Application of both compounds clearly decreased pSTAT5 levels, whereas they remained unaffected when the cells were treated with the Src-family tyrosine kinase inhibitor dasatinib (Figure 3B-C). The effect of the JAK2 inhibitors on pSTAT5 was confirmed in NALM-6-EcoR cells stably expressing PAX5-JAK2 (supplemental Figure 3B). Inhibitory effects of ruxolitinib on PAX5-JAK2 were also observed by Roberts et al.4 Furthermore, we show that drug treatment leads to apoptosis induction and rapid cell death (Figure 3D).
Known JAK-STAT target genes such as Socs1, Pim1, or Cish52 and notably also Cd79a were significantly upregulated in PAX5-JAK2–expressing Ba/F3 cells (Figure 3E), but Cd19, which in Ba/F3 cells is also not activated by wild-type PAX5 (data not shown), was not. Although the expression of JAK-STAT targets was reduced on treatment with JAK inhibitors, Cd79a levels remained unaffected (Figure 3F), clearly showing a correlation between decreasing pSTAT5 levels and target gene expression. A moderate reduction of JAK-STAT target genes (Figure 3F) was observed on dasatinib treatment; however, it neither affected pSTAT5 levels (Figure 3B-C) nor induced apoptosis (data not shown).
Although, due to its nuclear localization, the function of PAX5-JAK2 may be uncoupled from cytoplasmatic interaction-mediated JAK-STAT signaling, JAK2 inhibitors block STAT5 activation, downregulate target gene expression, and cause cell death.
PAX5-JAK2 gene expression profiles reveal deregulation of JAK-STAT and PAX5 target genes
To further elucidate the function of PAX5-JAK2, we performed gene expression profiling of PAX5-JAK2+ primary leukemia samples. The obtained gene expression signature was compared with datasets from B-other BCP-ALL with normal PAX5 status and lacking known sentinel alterations (non–BCR-ABL1-like, PAX5-WT), as well as cases harboring a PAX5-C20orf112 or PAX5-ETV6 fusion.
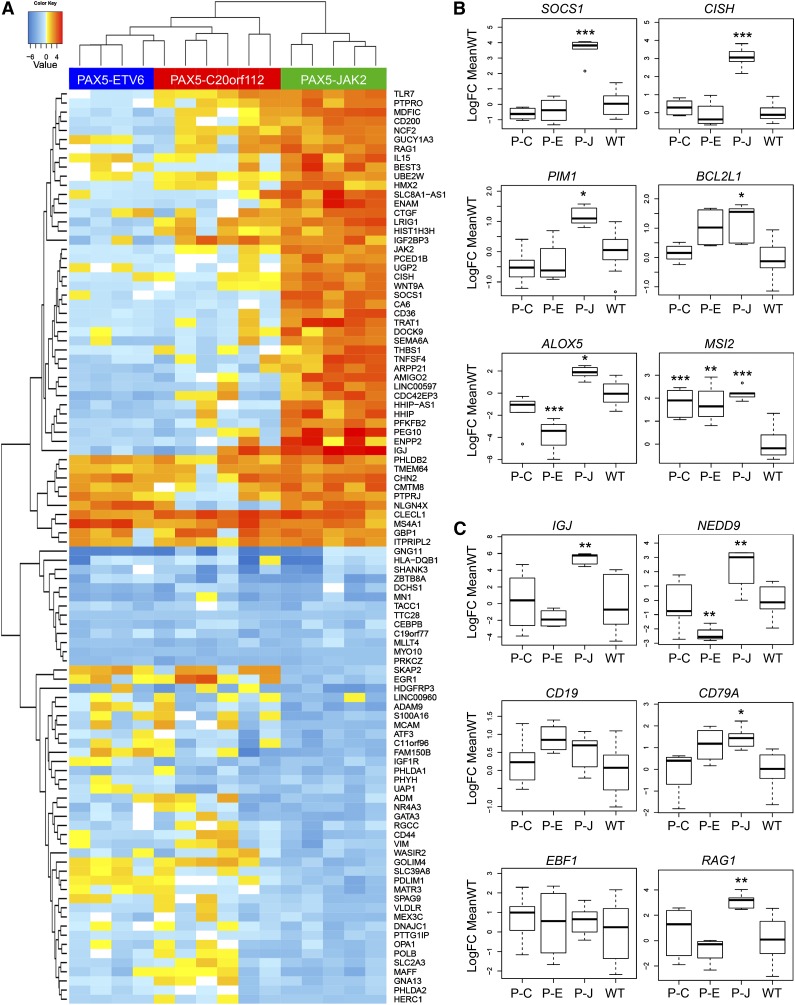
The most salient findings of these analyses are the confirmation of a JAK-STAT signature in PAX5-JAK2+ leukemia and the fact that the most differentially expressed genes show an expression pattern distinct from samples carrying other PAX5 fusions as shown by unsupervised hierarchical clustering and principal component analysis (Figure 4A; supplemental Figure 4A-B; supplemental Table 8).
Figure 4.
Gene expression profile of PAX5-JAK2+ patients. (A) Heatmap showing the 50 most up-/downregulated genes in cases with PAX5-JAK2+ leukemia (green bar) relative to the mean expression of the genes in B-cell precursor acute lymphoblastic leukemia without known sentinel alterations and with normal PAX5 status (PAX5-WT). Additionally, PAX5-ETV6+ (blue bar) and PAX5-C20orf112+ (red bar) samples are shown. (B-C) Boxplots depicting log2 fold changes of selected genes relative to the mean expression in PAX5-WT cases; the black line represents the median. Tukey-style whiskers extending to a maximum of 1.5 of the interquartile range beyond the box are depicted. Outliers beyond the range are individually plotted as circles. P values according to “limma” comparing PAX5-WT to the indicated PAX5 fusions are depicted (*P < .05, **P < .01, ***P < .001). P-C, PAX5-C20orf112; P-E, PAX5-ETV6; P-J, PAX5-JAK2; WT, PAX5 wild-type.
In line with its kinase activity, there are typical JAK-STAT targets among the most upregulated genes in PAX5-JAK2+ leukemia compared with PAX5-WT: for instance, SOCS1, CISH, and PIM1 (Figure 4B; supplemental Figure 4C). Although PIM1 enhances tumor growth and confers protection against drug-induced apoptosis,53 it is interesting to note that two other upregulated genes, MSI2 and ALOX5 (Figure 4B), are implicated in leukemic stem cell functions.54-56 Together with the moderate upregulation of the antiapoptotic gene BCL2L1, it appears that PAX5-JAK2 activates the JAK-STAT signaling pathway (supplemental Figure 4C), affects survival, and induces a more stem cell-like phenotype.
In accordance with the function of PAX5-JAK2 as an aberrant transcription factor, not only JAK-STAT, but also PAX5, targets, such as NEDD9 or CD79A, showed differential expression (Figure 4C). Although CD79A displayed the highest expression levels in PAX5-JAK2+ leukemia, no significant differences in CD19 expression were observed (Figure 4C). Furthermore, although IGJ is usually repressed by PAX5,57 it is highly upregulated in PAX5-JAK2+ but not in PAX5-ETV6+ and PAX5-C20orf112+ leukemia. Notably, upregulation of IGJ has been detected in BCR-ABL1–like leukemia,58 suggesting an association with the kinase-activated signature. Moreover, although all leukemia subgroups, independent of their PAX5 status, expressed equal levels of EBF1, an upstream regulator of PAX5, in PAX5-JAK2+ leukemia, RAG1 (Figure 4C) was significantly upregulated, possibly reflecting a slightly different developmental arrest.
To further evaluate the influence of PAX5 fusions on PAX5-responsive targets and genes regulated during B-lymphoid development, we performed gene set enrichment analysis using previously published datasets.57,59-63 This analysis indicates that, at least in the examined subtypes with different PAX5 fusions, the altered gene sets show common and distinct patterns.
The most significant changes between PAX5-JAK2+ and PAX5-WT samples were seen in the previously delineated top regulated gene set differentially expressed in human ETV6-RUNX1+ leukemia with and without PAX5 mutations (supplemental Figure 4D; supplemental Table 9).59 Remarkably, this gene set was also deregulated in leukemia with other PAX5 fusions, indicating that a common subset of genes is affected by PAX5 alterations.
Gene sets representing murine early B-lymphoid development–associated PAX5-activated targets were predominantly expressed at higher levels in PAX5-JAK2+ and PAX5-ETV6+ compared with PAX5-WT leukemia, suggesting that these PAX5 fusions might not generally antagonize wild-type PAX5 function. The lower expression levels of genes activated in mature B cells supposedly reflect the developmental stage of the leukemia rather than a repressive function of PAX5 fusions. Particular gene sets, as, for example, those activated in murine pro-B-cells, were downregulated in PAX5-C20orf112+ leukemia, indicating a potential repressive capacity of this fusion protein. Although some subsets of PAX5-repressed genes appeared significantly upregulated in all PAX5 fusion+ samples, additional distinct subsets seem to be uniquely induced in PAX5-ETV6+ leukemia.
All in all, our gene expression profiling analysis of leukemia samples supports the notion that PAX5-JAK2 has an impact on both the JAK2 and the PAX5 downstream networks and that different PAX5 fusions may result in distinct expression signatures.
Discussion
In this study, we assessed the functional consequences of PAX5-JAK2, a recurrent genetic lesion, which has recently been identified in BCP-ALL.1-3 The PAX5-JAK2 chimeric protein consists of the PAX5 DNA-binding domain and the catalytically active kinase domain of JAK2, suggesting that it may have an impact on both the B-cell transcription program and the activation of the JAK-STAT signaling pathway.
Intriguingly, in stark contrast to the cytoplasmatic localization of all other JAK2 fusion proteins analyzed to date,23,28-30 PAX5-JAK2 localizes exclusively to the nucleus. The nuclear localization of PAX5-JAK2 is most likely driven by the nuclear localization signal and the paired domain of PAX5, the latter enabling the chimera to bind to endogenous PAX5 loci.
In line with the anticipated dual function of PAX5-JAK2, gene expression profiling of primary leukemia cells showed that typical JAK-STAT and PAX5 targets are among the deregulated genes in PAX5-JAK2+ compared with PAX5-WT leukemia. Furthermore, the expression signature of PAX5-JAK2+ leukemia is clearly distinct from that of other PAX5 chimeras, illustrating for the first time on a genome-wide level that different PAX5 fusion proteins have a distinct impact on gene regulation. The differences in target gene transcription elicited by the fusion proteins may be explained by the lack of the potent C-terminal regulatory elements of PAX5,64 as well as by their distinct DNA affinities and interactions with the wild-type partner proteins.14 Furthermore, transcriptional deregulation may also be influenced by changes in interactions with cofactors, such as chromatin remodeling, histone modifying, basal transcription factor complexes, and E26 transformation-specific factors.65-69 Because nuclear PAX5-JAK2 interacts with chromatin, it may well be involved in chromatin modifications, as recently described for nuclear JAK2.70-72 Together, our data support the concept that PAX5 chimeras have distinct properties conferred by the partner proteins, which are likely to shape their function and impact on leukemia development.14
Our findings regarding the tyrosine phosphorylation in the JH1 kinase domain of PAX5-JAK2 raise the question by which mechanism it may occur. The majority of JAK2 fusion proteins possess an oligomerization domain provided by the fusion partner, which facilitates trans-autophosphorylation.7,51 However, PAX5-JAK2 does not form oligomers. Furthermore, using a kinase dead or DNA-binding–deficient mutant of the chimeric protein, we show that the kinase activation occurs via a mechanism independent of trans-phosphorylation by another kinase and DNA binding. Therefore, we hypothesize that the lack of inhibition by the JH2 domain enhances the basal JH1 kinase activity73 and thereby drives PAX5-JAK2 phosphorylation and activation. Alternatively, conformational changes of PAX5-JAK2 may facilitate its autophosphorylation. Although the underlying mechanism remains unclear, it is tempting to speculate that PAX5-JAK2 activates the JAK-STAT pathway in a manner that is independent from classical cytoplasmatic interactions. Consequently, it is conceivable that PAX5-JAK2 phosphorylates STATs in the nucleus, where they are present also in an unphosphorylated state.74,75
Because of the high frequency of JAK2 mutations in hematologic malignancies, a plethora of small molecule inhibitors has been developed, and several of them are currently under clinical investigation.31,32,76 Although the function of PAX5-JAK2 may be uncoupled from canonical JAK-STAT signaling, JAK2 inhibitors efficiently inhibit its kinase activity, resulting in dephosporylation of its downstream target STAT5, transcriptional downregulation of JAK-STAT target genes, and rapid cell death.
In summary, PAX5-JAK2 represents the first monomeric, autophosphorylating, nuclear, DNA-binding JAK2 fusion protein that, on one hand, has the potential to deregulate PAX5 target genes via its binding to PAX5 target loci and, on the other hand, displays kinase activity and constitutively activates the JAK-STAT signaling cascade in a supposedly noncanonical manner. Importantly, the kinase activity of PAX5-JAK2 can be blocked by JAK2 inhibitors,4 suggesting a potential therapeutic benefit for patients with this specific subtype of leukemia.
Acknowledgments
The authors thank A. Attarbaschi for providing patient material and clinical data; S. Anderl, A. Halfmann, A. M. Husa, W. Mensing, M. König, K. Nebral, A. Schumich, and the FACS core facility of the Children’s Cancer Research Institute for technical assistance; and M. Busslinger, E. Kowarz, R. Marschalek, R. Moriggl, M. Stanulla, B. Strobl, and H. Strobl for providing material.
This work was supported by Austrian Science Fund Grant FWF P21554-B19 (to S.S.), the St. Anna Kinderkrebsforschung e.V., the Tyrolean Cancer Research Institute, the Pediatric Oncology Rotterdam Foundation (J.R.M.M. and M.L.D.B.), and Dutch Cancer Society Program Grant UvA 2008-4265 (to M.L.D.B.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.S. designed and conducted experiments, analyzed data, carried out bioinformatic analyses, and wrote the manuscript; K.F. performed qChIP experiments and revised the manuscript; M.K. carried out and supervised the bioinformatic analyses; J.R.M.M. and M.L.D.B. provided reverse phase protein array and gene expression data; R.K. performed gene expression profiling; S.S. conceived and supervised the study and revised the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sabine Strehl, CCRI, Children’s Cancer Research Institute, St. Anna Kinderkrebsforschung e.V., Zimmermannplatz 10, A-1090 Vienna, Austria; e-mail: sabine.strehl@ccri.at.
References
- 1.Coyaud E, Struski S, Prade N, et al. Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogenetique Hematologique study. Blood. 2010;115(15):3089–3097. doi: 10.1182/blood-2009-07-234229. [DOI] [PubMed] [Google Scholar]
- 2.Nebral K, Denk D, Attarbaschi A, et al. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23(1):134–143. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- 3.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 7.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36(4):529–541. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 9.Kawamata N, Ogawa S, Zimmermann M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci USA. 2008;105(33):11921–11926. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazio G, Daniele G, Cazzaniga V, et al. Three novel fusion transcripts of the paired box 5 gene in B-cell precursor acute lymphoblastic leukemia [published online ahead of print October 10, 2014]. Haematologica. doi: 10.3324/haematol.2014.112193. [DOI] [PMC free article] [PubMed]
- 11.Bousquet M, Broccardo C, Quelen C, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109(8):3417–3423. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- 12.Cazzaniga G, Daniotti M, Tosi S, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61(12):4666–4670. [PubMed] [Google Scholar]
- 13.Nebral K, König M, Harder L, Siebert R, Haas OA, Strehl S. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br J Haematol. 2007;139(2):269–274. doi: 10.1111/j.1365-2141.2007.06731.x. [DOI] [PubMed] [Google Scholar]
- 14.Fortschegger K, Anderl S, Denk D, Strehl S. Functional heterogeneity of PAX5 chimeras reveals insight for leukemia development. Mol Cancer Res. 2014;12(4):595–606. doi: 10.1158/1541-7786.MCR-13-0337. [DOI] [PubMed] [Google Scholar]
- 15.Fazio G, Palmi C, Rolink A, Biondi A, Cazzaniga G. PAX5/TEL acts as a transcriptional repressor causing down-modulation of CD19, enhances migration to CXCL12, and confers survival advantage in pre-BI cells. Cancer Res. 2008;68(1):181–189. doi: 10.1158/0008-5472.CAN-07-2778. [DOI] [PubMed] [Google Scholar]
- 16.Kurahashi S, Hayakawa F, Miyata Y, et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene. 2011;30(15):1822–1830. doi: 10.1038/onc.2010.554. [DOI] [PubMed] [Google Scholar]
- 17.Qiu JJ, Chu H, Lu X, Jiang X, Dong S. The reduced and altered activities of PAX5 are linked to the protein-protein interaction motif (coiled-coil domain) of the PAX5-PML fusion protein in t(9;15)-associated acute lymphocytic leukemia. Oncogene. 2011;30(8):967–977. doi: 10.1038/onc.2010.473. [DOI] [PubMed] [Google Scholar]
- 18.Fazio G, Cazzaniga V, Palmi C, et al. PAX5/ETV6 alters the gene expression profile of precursor B cells with opposite dominant effect on endogenous PAX5. Leukemia. 2013;27(4):992–995. doi: 10.1038/leu.2012.281. [DOI] [PubMed] [Google Scholar]
- 19.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26(47):6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet M, Quelen C, De Mas V, et al. The t(8;9)(p22;p24) translocation in atypical chronic myeloid leukaemia yields a new PCM1-JAK2 fusion gene. Oncogene. 2005;24(48):7248–7252. doi: 10.1038/sj.onc.1208850. [DOI] [PubMed] [Google Scholar]
- 21.Griesinger F, Hennig H, Hillmer F, et al. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes Chromosomes Cancer. 2005;44(3):329–333. doi: 10.1002/gcc.20235. [DOI] [PubMed] [Google Scholar]
- 22.Atak ZK, Gianfelici V, Hulselmans G, et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013;9(12):e1003997. doi: 10.1371/journal.pgen.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278(5341):1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 24.Murati A, Gelsi-Boyer V, Adélaïde J, et al. PCM1-JAK2 fusion in myeloproliferative disorders and acute erythroid leukemia with t(8;9) translocation. Leukemia. 2005;19(9):1692–1696. doi: 10.1038/sj.leu.2403879. [DOI] [PubMed] [Google Scholar]
- 25.Peeters P, Raynaud SD, Cools J, et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90(7):2535–2540. [PubMed] [Google Scholar]
- 26.Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, Morton CC. Novel SSBP2-JAK2 fusion gene resulting from a t(5;9)(q14.1;p24.1) in pre-B acute lymphocytic leukemia. Genes Chromosomes Cancer. 2008;47(10):884–889. doi: 10.1002/gcc.20585. [DOI] [PubMed] [Google Scholar]
- 27.Reiter A, Walz C, Watmore A, et al. The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer Res. 2005;65(7):2662–2667. doi: 10.1158/0008-5472.CAN-04-4263. [DOI] [PubMed] [Google Scholar]
- 28.Van Roosbroeck K, Cox L, Tousseyn T, et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood. 2011;117(15):4056–4064. doi: 10.1182/blood-2010-06-291310. [DOI] [PubMed] [Google Scholar]
- 29.Cuesta-Domínguez Á, Ortega M, Ormazábal C, et al. Transforming and tumorigenic activity of JAK2 by fusion to BCR: molecular mechanisms of action of a novel BCR-JAK2 tyrosine-kinase. PLoS ONE. 2012;7(2):e32451. doi: 10.1371/journal.pone.0032451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwaller J, Frantsve J, Aster J, et al. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. EMBO J. 1998;17(18):5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintás-Cardama A, Kantarjian H, Cortes J, Verstovsek S. Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nat Rev Drug Discov. 2011;10(2):127–140. doi: 10.1038/nrd3264. [DOI] [PubMed] [Google Scholar]
- 32.Gotlib J. JAK inhibition in the myeloproliferative neoplasms: lessons learned from the bench and bedside. Hematology Am Soc Hematol Educ Program. 2013;2013:529-537. [DOI] [PubMed]
- 33.Wächter K, Kowarz E, Marschalek R. Functional characterisation of different MLL fusion proteins by using inducible Sleeping Beauty vectors. Cancer Lett. 2014;352(2):196–202. doi: 10.1016/j.canlet.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Emerenciano M, Kowarz E, Karl K, et al. Functional analysis of the two reciprocal fusion genes MLL-NEBL and NEBL-MLL reveal their oncogenic potential. Cancer Lett. 2013;332(1):30–34. doi: 10.1016/j.canlet.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 35.Kohlhuber F, Rogers NC, Watling D, et al. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17(2):695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stemberger J, Witt V, Printz D, Geyeregger R, Fritsch G. Novel single-platform multiparameter FCM analysis of apoptosis: Significant differences between wash and no-wash procedure. Cytometry A. 2010;77(11):1075–1081. doi: 10.1002/cyto.a.20976. [DOI] [PubMed] [Google Scholar]
- 37.Zuurbier L, Homminga I, Calvert V, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24(12):2014–2022. doi: 10.1038/leu.2010.204. [DOI] [PubMed] [Google Scholar]
- 38.Petricoin EF, III, Espina V, Araujo RP, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67(7):3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 39.Paweletz CP, Charboneau L, Bichsel VE, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20(16):1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 40.van den Berk LC, van der Veer A, Willemse ME, et al. Disturbed CXCR4/CXCL12 axis in paediatric precursor B-cell acute lymphoblastic leukaemia. Br J Haematol. 2014;166(2):240–249. doi: 10.1111/bjh.12883. [DOI] [PubMed] [Google Scholar]
- 41.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol Cell Biol. 1995;15(5):2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55(2):61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 43.Schulz KR, Danna EA, Krutzik PO, Nolan GP. Single-cell phospho-protein analysis by flow cytometry. In: Coligan JE, Bierer B, Margulies DH, Shevach EM, Strober W, eds. Brown P, Donovan JC, Kruisbeek AM, guest eds. Protein Analysis by Flow Cytometry. Current Protocols in Immunology. 2012, 96:IV:8.17:8.17.1-8.17.20. [Google Scholar]
- 44.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics. 2010;11(2):242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 47.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20(18):3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 48.Hoelbl A, Schuster C, Kovacic B, et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2(3):98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwaller J, Parganas E, Wang D, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell. 2000;6(3):693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 50.Ye D, Wolff N, Li L, Zhang S, Ilaria RL., Jr STAT5 signaling is required for the efficient induction and maintenance of CML in mice. Blood. 2006;107(12):4917–4925. doi: 10.1182/blood-2005-10-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jatiani SS, Baker SJ, Silverman LR, Reddy EP. Jak/STAT pathways in cytokine signaling and myeloproliferative disorders: approaches for targeted therapies. Genes Cancer. 2010;1(10):979–993. doi: 10.1177/1947601910397187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178(5):2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 53.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013;85(5):629–643. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41(7):783–792. doi: 10.1038/ng.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466(7307):765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kharas MG, Lengner CJ, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24(3):269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 58.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122(15):2622–2629. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45(10):1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27(1):49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 61.Revilla-I-Domingo R, Bilic I, Vilagos B, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31(14):3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pridans C, Holmes ML, Polli M, et al. Identification of Pax5 target genes in early B cell differentiation. J Immunol. 2008;180(3):1719–1728. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- 63.Liu GJ, Cimmino L, Jude JG, et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev. 2014;28(12):1337–1350. doi: 10.1101/gad.240416.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dörfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996;15(8):1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 65.He T, Hong SY, Huang L, et al. Histone acetyltransferase p300 acetylates Pax5 and strongly enhances Pax5-mediated transcriptional activity. J Biol Chem. 2011;286(16):14137–14145. doi: 10.1074/jbc.M110.176289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao H, Lukin K, Ramírez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106(27):11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001;8(6):1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 68.Sigvardsson M, Clark DR, Fitzsimmons D, et al. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22(24):8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McManus S, Ebert A, Salvagiotto G, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30(12):2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dawson MA, Bannister AJ, Göttgens B, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461(7265):819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dawson MA, Bannister AJ, Saunders L, et al. Nuclear JAK2. Blood. 2011;118(26):6987–6988. doi: 10.1182/blood-2011-10-385278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20(10):3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown S, Zeidler MP. Unphosphorylated STATs go nuclear. Curr Opin Genet Dev. 2008;18(5):455–460. doi: 10.1016/j.gde.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 75.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 76.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14(3):504–527. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]