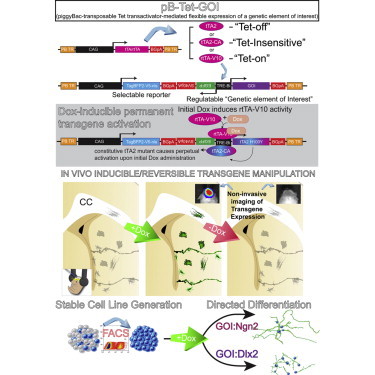
Summary
Precise methods for transgene regulation are important to study signaling pathways and cell lineages in biological systems where gene function is often recycled within and across lineages. We engineered a genetic toolset for flexible transgene regulation in these diverse cellular contexts. Specifically, we created an optimized piggyBac transposon-based system, allowing for the facile generation of stably transduced cell lineages in vivo and in vitro. The system, termed pB-Tet-GOI (piggyBac-transposable tetracycline transactivator-mediated flexible expression of a genetic element of interest), incorporates the latest generation of tetracycline (Tet) transactivator and reverse Tet transactivator variants—along with engineered mutants—in order to provide regulated transgene expression upon addition or removal of doxycycline (dox). Altogether, the flexibility of the system allows for dox-induced, dox-suppressed, dox-resistant (i.e., constitutive), and dox-induced/constitutive regulation of transgenes. This versatile strategy provides reversible temporal regulation of transgenes with robust inducibility and minimal leakiness.
Graphical Abstract
Highlights
-
•
pB-Tet-GOI features the latest generation of Tet transactivators (tTAs) and variants
-
•
piggyBac transposition allows for genomic insertion of pb-Tet-GOI
-
•
pb-Tet-GOI provides flexible control of transgenes in vitro and in vivo
-
•
tTA variants permit reversible, constitutive, or induced constitutive expression
In this article, Breunig and colleagues describe a stable, flexible genetic system for inducible and reversible gene expression. The doxycycline-mediated system, termed pB-Tet-GOI, is non-leaky, robust, and allows for “On→Off→On,” “Off→On→Off” and inducible-constitutive temporal control (“Off→permanently On”) of transgenes in vivo and in vitro, as well as a dox-resistant variant for strong, constitutive expression of transgenes.
Introduction
Complex multicellular organisms contain a wide spatial and temporal diversity of cell types. Gene expression and signaling mechanisms are tightly coordinated processes that are recycled throughout development, maturation, and aging, thus maintaining a smaller overall gene number. This makes the investigation of gene function dependent on the ability to precisely manipulate gene expression in each spatial and temporal context. In the CNS, one such example is the Notch signaling pathway, which is involved in a multitude of cell types from development to late neurodegeneration (Ables et al., 2011). The dynamic cellular contexts and diverse signaling interactions create the need to temporally regulate gene activity in order to precisely study gene function. Moreover, transgene manipulation has emerged as an important technique to study and treat disease. Specifically, directing neuronal subtype differentiation is important for disease modeling, and regulated growth factor secretion is a promising therapy for several neurodegenerative diseases (Behrstock et al., 2006; Marchetto et al., 2011).
Tetracycline (Tet)-regulated systems have been used to temporally and spatially regulate gene expression in various methodologies (Furth et al., 1994; Gossen and Bujard, 1992). Specifically, the bacterial Tet transactivator (tTA) has been optimized to silence gene expression downstream of a Tet-regulated promoter in the presence of doxycycline (dox), a Tet analog. In addition to this “Tet-Off” system, a “Tet-On” system uses a reverse tTA (rtTA) in order to activate transgene expression in the presence of dox (Mansuy and Bujard, 2000). Tet-inducible systems have suffered from “leaky” gene expression and low inducibility of transgene expression over baseline (Mansuy and Bujard, 2000). These drawbacks, along with epigenetic silencing of these elements through endogenous methylation, contribute to the limitations related to CNS transgene regulation (Zhu et al., 2007). While new tTA and rtTA variants help overcome some of these limitations (Zhou et al., 2006), their use in neural stem cell populations is unexplored.
Plasmid electroporation (EP) to the brain has become a commonly used method of in vivo transgenesis in development (Breunig et al., 2007; Saito and Nakatsuji, 2001). However, like most episomal methods of transgenesis, EP suffers from plasmid dilution (Chen and LoTurco, 2012). Thus, studying complex lineage trees or postnatal neuro- and gliogenesis has been limited (Chen and LoTurco, 2012). To mitigate this, the recently characterized piggyBac transposon system permits stable integration of transgenes and can be used with various standard in vitro transfection methods to generate stable cell lines (Chen and LoTurco, 2012; García-Marqués and López-Mascaraque, 2013; Kahlig et al., 2010).
However, the robustness of the new tTA and rtTA variants and the ability of pBASE to promote stable genetic integration of such complex inducible/reversible genetic systems in stem and progenitor-derived cells have not yet been described. Our results indicate that this genetic system can be an efficient and highly flexible mediator of transgene expression in diverse biological contexts.
Results
We constructed a custom genetic system for stable Tet-regulated flexible transgene expression using piggyBac transposition, termed pB-Tet-GOI (piggyBac-transposable Tet-mediated flexible expression of a genetic element of interest). pB-Tet-GOI consists of three plasmids: (1) a pBase-expressing plasmid for transposon integration, (2) a reverse Tet transactivator (rtTA) plasmid (or transactivator variants [tTA2 and tTA2-CA] discussed below) to allow for dox-regulation of the GOI, and (3) a Tet response GOI plasmid (Figure 1A). This tripartite system provides flexible, “mix and match” use of the different tTA2/rtTA variants with different GOI response plasmids. pBASE itself recognizes the piggyBac terminal repeats (TRs) that flank the rtTA and response plasmids and thereby catalyzes stable genomic insertion of these plasmids to avoid plasmid dilution with cell division (Chen and LoTurco, 2012). Importantly, the pBase plasmid is episomal and dilutes with cell division, preventing the stably inserted rtTA and response plasmids from continuously “hopping” in and out of the genome. The rtTA plasmid yields constitutive expression of rtTA-V10 protein. With dox, the rtTA-V10 binds to the Tet-Bi promoter of the response plasmid and induces bi-directional transcription of two genes of interest (i.e., “Tet-On”). Finally, the Tet response plasmid features a constitutively expressed TagBFP2 reporter with a nuclear localization signal (NLS) and a V5 epitope tag (Figure 1A) for fluorescent and immunocytochemical (ICC) identification of transgenic cells. In addition, this design was employed to minimize spurious Tet-responsive gene expression by using the TagBFP2 cistron as a genetic insulator, minimizing possible upstream promoter/enhancer activity due to insertion in active chromatin and preventing transcriptional interference—including enhancer activity attributed to the upstream piggyBac TR (Shi et al., 2007).
Figure 1.
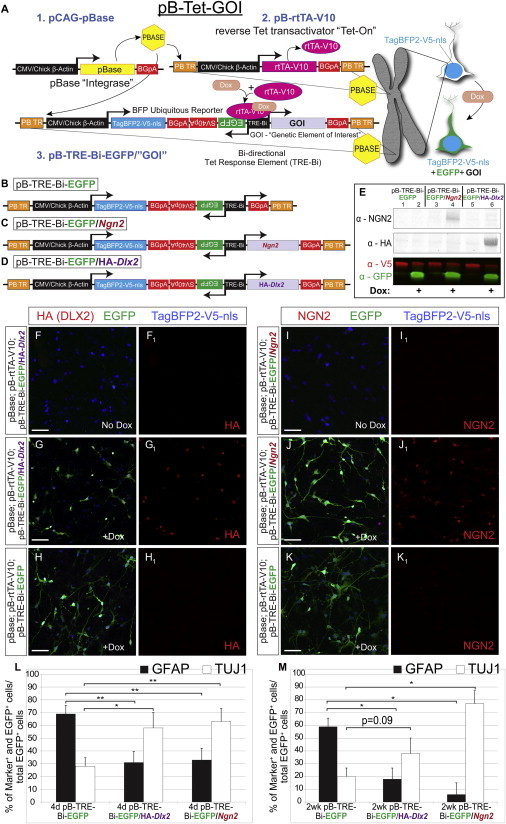
Validating the pB-Tet-GOI System for Transgene Manipulation
(A) Schematic of pB-Tet-GOI system for transposon-mediated integration along with inducible and reversible transgene expression.
(B–D) Response plasmids utilized for directed differentiation experiments.
(E) Western blot analysis of response groups grown with and without dox.
(F–K) Immunocytochemical staining for HA (DLX2 epitope tag), GFP (dox reporter), TagBFP2 (constitutive reporter), and NGN2 in HuNPCs nucleofected with indicated response plasmids.
Scale bar, 100 μm.
(L and M) Quantification of HuNPCs harboring indicated response plasmids and differentiated for 4 days (I) or 2 weeks (J) after dox.
Error bars represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01; n = 3 biological replicates per condition per time point.
See also Figure S1.
We first generated a Tet response plasmid termed pB-TRE-Bi-EGFP (Figure 1B) expressing enhanced green fluorescent protein (EGFP). Next, we incorporated the transcription factor NEUROGENIN 2 (NEUROG2) that promotes glutamatergic-like neuron generation during cortical development (Zhang et al., 2013) and the DLX2 homeobox protein that promotes interneuron generation (Petryniak et al., 2007) into separate response plasmids to functionally test the ability of our system to inducibly direct differentiation. These two plasmids were defined as pB-TRE-Bi-EGFP/Ngn2 (i.e., Neurog2) and pB-TRE-Bi-EGFP/HA-Dlx2 (Dlx2 with a hemagglutinin [HA] epitope tag) (Figures 1C and 1D). For initial tests of the system, we transfected mouse N2a cells and a human cortical progenitor cell line (HuNPC) (Svendsen et al., 1998) with the three different response plasmids, along with rtTA-V10 and pBase. After culture for 4 days with or without dox, western blot analysis revealed robust GFP protein levels only in N2a cells grown with dox and no substantive GFP production without dox (Figure 1E). These collective results highlight both the inducibility and non-leakiness of the system. V5 epitope levels (from the TagBFP2-NLS in the response plasmid) demonstrated equivalent transfection and protein loading (Figure 1E). Moreover, NGN2 and HA (from the Dlx2 construct) were detected only in their respective groups when ICC analysis showed that HuNPCs nucleofected with pB-TRE-Bi-EGFP/HA-Dlx2 and grown with dox produced TagBFP2, GFP, and HA. In contrast, cells grown without dox produced only TagBFP2 (Figures 1F–1G1). Similarly, HuNPCs nucleofected with pB-TRE-Bi-EGFP constitutively expressed nuclear TagBFP2 but expressed GFP only with dox (Figures 1H and 1H1). Finally, NGN2 and GFP were induced only with dox in the pB-TRE-Bi-EGFP/Ngn2 group (Figures 1I–1J1). Interestingly, even though HuNPCs nucleofected with pB-TRE-Bi-EGFP are cortically derived, they did not express detectable NGN2 (Figures 1I1 and 1K1). Taken together, pB-Tet-GOI was readily inducible, non-leaky, and could be used to bidirectionally express a reporter and GOI simultaneously in vitro.
To analyze whether the pB-Tet-GOI system expresses proteins that are biologically active and at sufficient levels to direct specific neuronal differentiation, HuNPCs were nucleofected with each response plasmid and differentiated for up to 6 weeks. Live cells were imaged daily for GFP and TagBFP2 autofluorescence. Notably, each group displayed divergent morphological changes and growth patterns over time (Figures S1A–S1C). Sibling cells from these three groups were fixed at 4 and 14 days for ICC examination of differentiation into TUJ1+ neurons and glial fibrillary acidic protein (GFAP)+ astroglia using confocal microscopy (Figures S1D–S1F2). Induced expression markedly increased neurogenesis at the expense of astrogliogenesis as early as 4 days and to a lesser extent at 14 days in both the pB-TRE-Bi-EGFP/HA-Dlx2 and pB-TRE-Bi-EGFP/Ngn2 groups (Figures 1L and 1M). The divergent morphologies were most clearly seen by computer-assisted cell tracing of the GFP+ cells at this time point (Figures S1G–S1I).
To assess the ability of our system to generate pure populations of stable undifferentiated human cell lines, we nucleofected three groups of HuNPCs as mentioned above, expanded them in culture without dox, and then sorted cells to isolate BFP+ cells (Figures S1J–S1L). All groups, expanded as undifferentiated neurospheres without dox, did not express GFP but did express the neural progenitor marker, SOX2 (Figures S1M–S1M2 and data not shown). Following dox addition, pB-TRE-Bi-EGFP/Ngn2 gained GFP expression while SOX2 expression was lost, suggesting that the immature cells can differentiate on demand by dox addition (Figures S1N–S1N2 and S1O–S1O2).
In addition to rtTA-V10 (Tet-On), we engineered tTA2 (“Tet-Off”) and tTA2-CA (“Tet-insensitive”) plasmids (Figure 2A). tTA2-CA has a H100Y mutation, making it dox insensitive (Hecht et al., 1993) to provide ubiquitous GOI expression without dox. Finally, we incorporated tTA2-CA into the response plasmid (instead of the transactivator plasmid) to permit temporally inducible but subsequently permanent expression of genes under the Tet-Bi promoter, yielding pB-TRE-Bi-EGFP/i-tTA2-CA). More precisely, dox induces rtTA-V10 activation, leading to i-tTA2-CA expression and continual TRE-Bi self-activation (Figure 2A).
Figure 2.
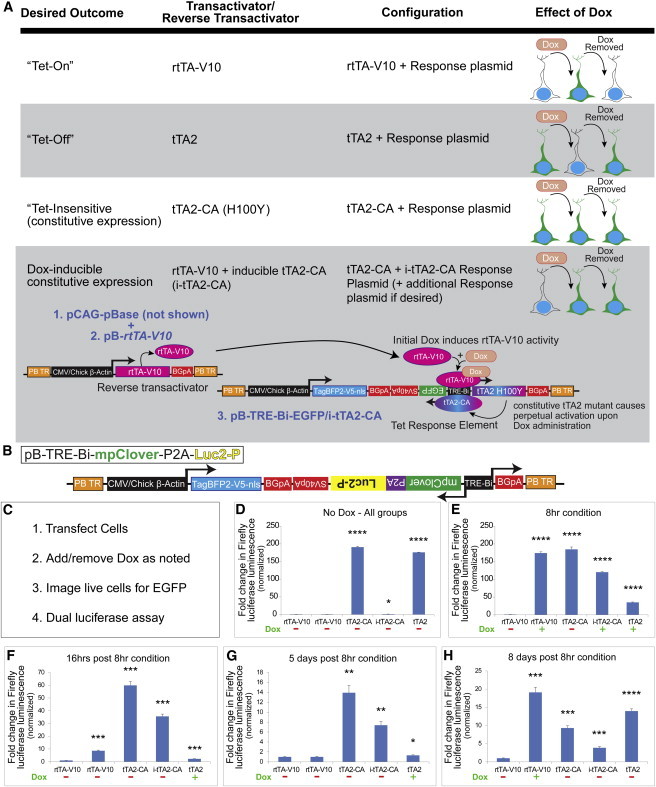
Transactivator Variants Allow for Flexible Manipulation of Transgene Expression
(A) Chart of transactivator variants and expected properties.
(B) Schematic of plasmid encoding pB-TRE-Bi-mpClover-P2A-Luc2P.
(C) Design of in vitro assay for transactivator variant validation.
(D–H) Luciferase assay reveals changes in firefly luminescence in the presence and absence of dox (as noted in panels) for the respective transactivator variants. n = 4 biological replicates per condition per time point. Samples normalized to Renilla.
Error bars represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S2.
In order to rapidly and quantitatively assay these transactivator variants, we incorporated a destabilized firefly luciferase (Luc2P) gene (Figures 2B and 2C). Further, Luc2P was co-expressed using a P2A element with myristolated/palmitolated-Clover (mpClover; Figure 2B), which is an EGFP variant (Lam et al., 2012) tagged to membranes. pB-TRE-Bi-mpClover-P2A-Luc2P-transduced cells did not exhibit mpCLOVER expression or LUC2P activity without dox (Figures 2D–2H; Figures S2A–S2E and SA1). With dox, rtTA-V10 induced mpCLOVER at 8 hr, which was detectable up to 16 hr after dox removal (Figures S2B1–S2C1). However, LUC2P displayed more rapid kinetics in that luminescence was noticeably diminished at 16 hr (Figure 2F). This may reflect the longer half-life of mpCLOVER when compared with destabilized LUC2P. Five days after dox removal, neither mpCLOVER expression nor LUC2P activity was detectable (Figure 2G; Figure S2D1). Dox was subsequently re-added to the culture, and mpCLOVER expression and LUC2P activity was detected (Figure 2H; Figure S2E1). This demonstrates that the system can be induced, reversed, and subsequently induced (i.e., On→Off→On).
As expected, cells expressing the pB-TRE-Bi-EGFP/tTA2-CA (Tet-insensitive) plasmid displayed constitutive mpCLOVER expression and luminescence without dox (Figures 2D–2H; Figures S2A2–S2E2). In contrast, cells expressing the pB-TRE-Bi-EGFP/i-tTA2-CA combined plasmid did not exhibit basal mpCLOVER expression or luminescence without dox (Figure 2D; Figure S2A3). However, after a single dox administration, both were constitutively expressed—even after dox removal (Figures 2E–2H; Figures S2B3–S2E3).
tTA2 (Tet-Off) exhibited high basal luciferase activity and mpCLOVER expression, similar to that of dox-induced rtTA-V10 (Figure 2D; Figure S2A4). mpCLOVER expression decreased with dox addition until it was undetectable at 5 days, and it rebounded 3 days after dox removal (Figures S2B4–S2D4). Luminescence decreased more rapidly (again, presumably due to the attached PEST destabilization domain), approaching baseline at 16 hr after dox addition, though rebounding (along with mpCLOVER fluorescence) after dox removal (Figures 2E–2H; Figure S2E4).
To confirm transposition, we nucleofected neural stem cells with our pB-Tet-GOI system, alternate piggyBac expression vectors encoding fluorescent proteins and episomal expression vectors expressing analogous fluorescent proteins in various combinations—with and without pBase. In the presence of pBase (and dox), pB-Tet-Bi-EGFP and pB-mCherry demonstrated negligible loss of co-expression and retained high levels of fluorescence when assayed by vital imaging at 3 and 6 days post-transduction (group 1; Figures S2F and S2G). Without pBase, the identical combination of plasmids (with dox) displayed relatively similar co-expression at 3 days, but both GFP and mCHERRY expression were almost undetectable by 6 days (group 2; Figures S2F and S2G). Mixing different piggyBac or episomal vectors in the presence of pBase demonstrated that the episomal plasmid expression was lost at 3–6 days (groups 3 and 4; Figures S2F and S2G). Conversely, the piggyBac transposed fluorescent proteins were perdurant, arguing against the possibility that non-specific toxicity caused loss of fluorescent protein expression rather than plasmid dilution (groups 3 and 4; Figures S2F and S2G).
tTA systems have shown promise in cell culture, but efficacy in the CNS has been inconsistent. To test the robust, non-leaky, inducible, and reversible characteristics of our pB-Tet-GOI system in vivo, pB-TRE-Bi-EGFP was introduced into ventricular zone (VZ) stem and progenitor cells using postnatal EP, a technique that allows for rapid expression of plasmid-based transgenes in these cells (Boutin et al., 2008; Breunig et al., 2012; Chesler et al., 2008). In the postnatal VZ, radial glia stem and progenitor cells differentiate into the mature cells of the striatum and provide neurons to the olfactory bulb (OB) (Alvarez-Buylla and Garcia-Verdugo, 2002; Carleton et al., 2003; Fernández et al., 2011). Five days after EP of pB-TRE-Bi-EGFP, half of the mice received a single dose of dox followed by brain removal at 1 hr, 1 day, 4 days, 6 days, 10 days, 2 weeks, 3 weeks, and 4 weeks (Figure 3A; Figure S3). The naive (un-stained) fluorescence of TagBFP2 and GFP was imaged to avoid artifactual amplification of these signals (Figure 3B). Coronal brain sections showed nuclear TagBFP2 expression in the subventricular zone, striatum, and OB of all animals (Figures 3C–3L). Six days after treatment, robust GFP expression was observed in groups that received a single dox dose, but not in groups without dox (Figures 3C–3F). This robust expression persisted up to 14 days, decreasing at 3 weeks (Figures 3J and 3J1; Figure S3). Cells at 4 weeks after dox were indistinguishable from cells in animals without dox (Figures 3C–3E, 3K, and 3L), demonstrating the reversibility of transgene induction. Cell counts at the OB core of dox-treated animals revealed a decrease in the GFP+ cell number commencing after 14 days, while TagBFP2+ cell counts remained the same, indicating that the cells were still present but that their GFP expression turned off (Figure 3M). Analysis of the mean fluorescence intensity after the single dox dose revealed the most robust GFP expression at around 6 days, before declining to baseline at approximately 4 weeks (Figure 3N). In total, these data indicate that the pB-Tet-GOI system is a robust, inducible, and reversible method for transgene manipulation in vivo.
Figure 3.
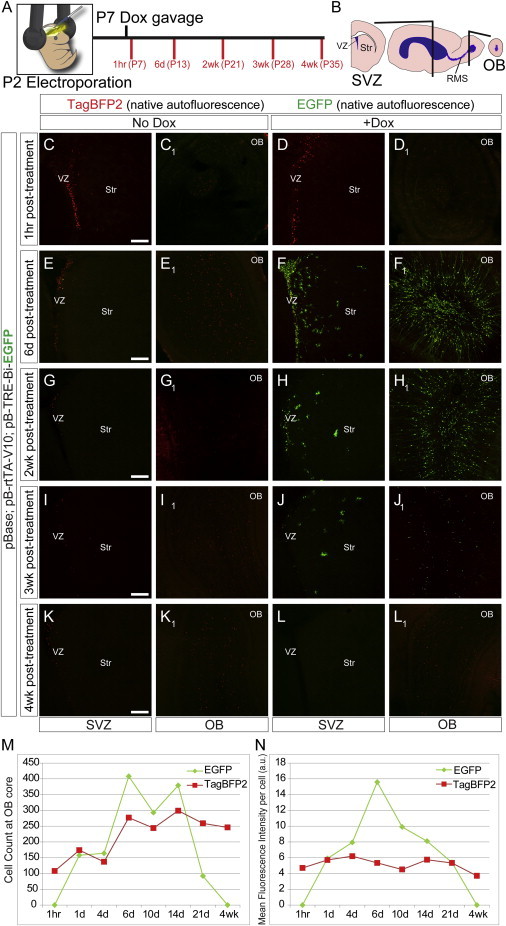
pB-Tet Is Non-leaky, Inducible, and Reversible in the Postnatal Mouse Brain
(A) Experimental timeline of dox administration after electroporation. Control mice received no dox administration.
(B) Schematic illustrating coronal planes of SVZ and OB imaged for panels (C)–(L1).
(C–L) Native unstained GFP and TagBFP2 (pseudocolored red) fluorescence was analyzed in the VZ and OB. Significant GFP is observed only with dox.
Scale bar, 200 μm.
(M) TagBFP2+ (all electroporated cells) and GFP+ (dox-responsive) cell counts in the OB core.
(N) Analysis of the mean fluorescence intensity of TagBFP2+ and GFP+ cells in the OB.
SVZ, subventricular zone; Str, striatum; OB, olfactory bulb. Data points represent readings from a single litter used to minimize experimental variation that might result with comparing data across litters. Results are consistent with more than three independent experiments assessing dox activation kinetics in independent litters.
See also Figure S3.
Non-invasively validating transgene expression could confirm appropriate delivery/expression of genetic elements and dox administration. We used a non-invasive imaging method with our well-characterized inducible Luc2P element (Figure 2B) in order to monitor dox induction of the pB-Tet-GOI system. Mice electroporated with a pB-TRE-Bi-mpClover-P2A-Luc2P-containing plasmid, along with pBase and rtTA-V10, were analyzed using in vivo luminescence imaging, which showed a significant induction of luminescence 1 week after dox administration (Figures 4A and 4B; see Figure S4A for timeline). After the initial imaging, several animals were used for histological examination, which demonstrated GFP+ cells in the dox-treated animals and no GFP+ expression in untreated animals (data not shown). One dox-treated animal not receiving a further dose subsequently displayed the basal luminescence reading of untreated littermates, while another littermate receiving another dose exhibited continued luminescence (Figure S4B). Finally, we electroporated littermates with a pB-TRE-Bi-mpClover-P2A-Luc2-P-containing plasmid along with rtTA-V10 (two mice) or tTA2-CA (three mice). As expected, luminescence was observed at similar levels in all animals, with the exception of the littermate that did not receive dox (Figures 4C and 4D). In summary, these data demonstrate the ability to monitor expression of the elements of our genetic system in a non-invasive manner.
Figure 4.
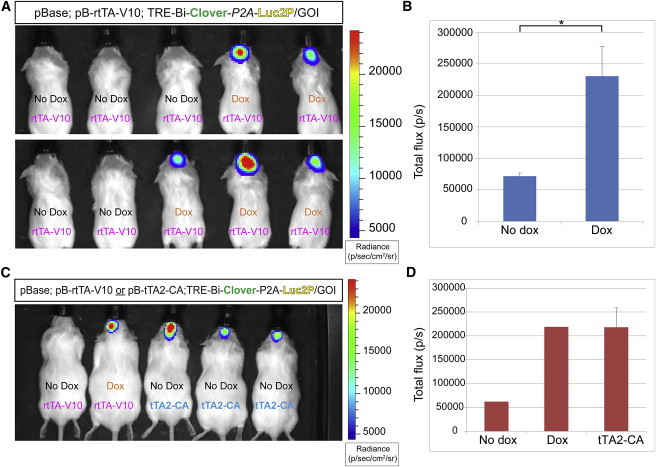
Addition of Luciferase into pB-Tet-GOI Allows Non-invasive Bioluminescence Imaging of Transgene Expression
(A) Bioluminescence analysis revealed firefly activity in mice that received dox (n = 5 biological replicates per condition).
(B) Quantification of total flux (p/s) in mice from (A).
(C) Bioluminescence analysis of tTA2-CA alongside rtTA-V10 (n = 3, all littermates).
(D) Quantification of total flux (p/s) in mice from (C).
Error bars represent mean ± SEM. ∗p < 0.05. See also Figure S4.
Discussion
Here, we characterize pB-Tet-GOI, a customizable, flexible genetic system that achieves stable transgene integration, along with inducible and reversible dox-mediated transgene expression in transduced cells and their resulting lineages without the need for viral packaging and generation of high viral titers. Specifically, we demonstrate the robust dox-mediated regulation of the latest-generation rtTA and tTA2 variants and promoter elements, allowing for transgene expression in vitro and in vivo. Additionally, we provide tTA2-CA and i-tTA2-CA as a constitutive and inducible constitutive activator, respectively, of TRE-Bi for dox-insensitive expression of transgenes.
Through pBase-mediated transposition, stable cell lines can be rapidly generated. This technology not only facilitates the investigation of cell biology in vitro but also permits the in vivo induction of requisite transgenes from transplanted cell populations. Therapeutic applications include induced growth factor delivery from transplanted cells (Behrstock et al., 2006). Also, induced, direct differentiation in vivo permits the transplantation of precursor cells, which expand easier in culture and survive better post-transplantation, followed by dox-mediated maturation.
Our pB-Tet-GOI system allows for more precise investigations of gene function and cell lineages using in vivo EP with the ability to stably genetically trace and reversibly manipulate transgene expression. Notably, the combination of rtTA-V10 and i-tTA2 permits permanent cell lineage tracing after a single pulse. Thus, dox can be used as an orthogonal ligand for gene manipulation with other chemically induced genetic systems such as tamoxifen and RU486-induced transgenes. Finally, there is great potential to combine this system with INTRSECT technology, including alternate recombinases to achieve greater control over transgene manipulation (Fenno et al., 2014).
This genetic system is not limited to use in the CNS and is broadly applicable to many fields of biology. Any context requiring temporal control of transgenes may benefit from the use of the pB-Tet-GOI system. Further, we have recently confirmed that mir30 and miR-E-based small hairpin RNAs (Fellmann et al., 2013) can be similarly induced by our system, providing temporally controlled knockdown experiments (data not shown). Taken together, the pB-Tet-GOI tool kit will be useful for the investigation and elucidation of molecular mechanisms of biological systems in a wide variety of contexts. Further, it holds great promise for the fields of reprogramming and gene therapy.
Experimental Procedures
Mice and Electroporation
CD1 mice were used according to the Cedars-Sinai Medical Center institutional animal care and use committee. EPs were performed as described previously (Breunig et al., 2012). Briefly, postnatal day 2 pups were placed on ice for ∼8 min until unresponsive to tail pressure. A total of 1.2 μl of a plasmid cocktail of pBase (0.5 μg/μl [final]), transactivator (1 μg/μl [final]), and response plasmid (1 μg/μl [final]) diluted in Tris-EDTA buffer buffer was injected into the left lateral ventricle. Fast green dye was added (10%v/v) to visualize injections. Platinum Tweezertrodes delivered five pulses of 120 V (50 ms; separated by 950 ms) from the ECM 830 System (Harvard Apparatus). SignaGel was applied to increase conductance. Mice were then warmed with a heat lamp for recovery.
Cell Culture and Nucleofection
HuNPCs (CNS010 cell line) were expanded as described previously (Svendsen et al., 1997, 1998). Briefly, HuNPCs were expanded as neurospheres in Stemline Media (Sigma S3194) with epidermal growth factor (Sigma E9644), leukemia inhibitory factor (Millipore LIF1010), and antibiotic-antimycotic (Life Technologies 15240-062). Spheres were dissociated for nucleofection with pB-Tet plasmids using the Lonza Nucleofector 2b device per the manufacturer’s recommendations. After nucleofection, media contained B-27 (Life Technologies 12587-010) without growth factors.
Doxycycline
Culture media contained dox (final concentration 100 ng/ml, Clontech 631311). Mice were gavaged with dox at 33 μg/g weight, using a 5-mg/ml stock solution.
Acknowledgments
We thank C. Svendsen for providing the HuNPCs and D. Eisenstat, M. Lin, and J. Loturco for providing plasmids. We thank G. Gowing, B. Shelley, V. Mattis, D. Sareen, and L. Garcia for experimental assistance. We thank S. Svendsen for critical review. We acknowledge support from the Samuel Oschin Comprehensive Cancer Institute Cancer Research Forum Award (M.D., J.J.B.), the Board of Governors RMI of Cedars-Sinai (M.D., J.J.B), the Thrasher-Broidy Trinity College Research Fellowship (W.S.S), the Smidt Family Foundation, and the Paul and Vera Guerin Family Foundation.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
References
- Ables J.L., Breunig J.J., Eisch A.J., Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A., Garcia-Verdugo J.M. Neurogenesis in adult subventricular zone. The Journal of neuroscience. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrstock S., Ebert A., McHugh J., Vosberg S., Moore J., Schneider B., Capowski E., Hei D., Kordower J., Aebischer P., Svendsen C.N. Human neural progenitors deliver glial cell line-derived neurotrophic factor to parkinsonian rodents and aged primates. Gene Ther. 2006;13:379–388. doi: 10.1038/sj.gt.3302679. [DOI] [PubMed] [Google Scholar]
- Boutin C., Diestel S., Desoeuvre A., Tiveron M.C., Cremer H. Efficient in vivo electroporation of the postnatal rodent forebrain. PloS one. 2008;3:e1883. doi: 10.1371/journal.pone.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig J.J., Arellano J.I., Macklis J.D., Rakic P. Everything that glitters isn’t gold: a critical review of postnatal neural precursor analyses. Cell Stem Cell. 2007;1:612–627. doi: 10.1016/j.stem.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Breunig J.J., Gate D., Levy R., Rodriguez J., Jr., Kim G.B., Danielpour M., Svendsen C.N., Town T. Rapid genetic targeting of pial surface neural progenitors and immature neurons by neonatal electroporation. Neural Dev. 2012;7:26. doi: 10.1186/1749-8104-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton A., Petreanu L.T., Lansford R., Alvarez-Buylla A., Lledo P.M. Becoming a new neuron in the adult olfactory bulb. Nat. Neurosci. 2003;6:507–518. doi: 10.1038/nn1048. [DOI] [PubMed] [Google Scholar]
- Chen F., LoTurco J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods. 2012;207:172–180. doi: 10.1016/j.jneumeth.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler A.T., Le Pichon C.E., Brann J.H., Araneda R.C., Zou D.J., Firestein S. Selective gene expression by postnatal electroporation during olfactory interneuron nurogenesis. PloS one. 2008;3:e1517. doi: 10.1371/journal.pone.0001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Fenno L.E., Mattis J., Ramakrishnan C., Hyun M., Lee S.Y., He M., Tucciarone J., Selimbeyoglu A., Berndt A., Grosenick L. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández M.E., Croce S., Boutin C., Cremer H., Raineteau O. Targeted electroporation of defined lateral ventricular walls: a novel and rapid method to study fate specification during postnatal forebrain neurogenesis. Neural Dev. 2011;6:13. doi: 10.1186/1749-8104-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth P.A., St Onge L., Böger H., Gruss P., Gossen M., Kistner A., Bujard H., Hennighausen L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Marqués J., López-Mascaraque L. Clonal identity determines astrocyte cortical heterogeneity. Cereb. Cortex. 2013;23:1463–1472. doi: 10.1093/cercor/bhs134. [DOI] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B., Müller G., Hillen W. Noninducible Tet repressor mutations map from the operator binding motif to the C terminus. J. Bacteriol. 1993;175:1206–1210. doi: 10.1128/jb.175.4.1206-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlig K.M., Saridey S.K., Kaja A., Daniels M.A., George A.L., Jr., Wilson M.H. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl. Acad. Sci. USA. 2010;107:1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.J., St-Pierre F., Gong Y., Marshall J.D., Cranfill P.J., Baird M.A., McKeown M.R., Wiedenmann J., Davidson M.W., Schnitzer M.J. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy I.M., Bujard H. Tetracycline-regulated gene expression in the brain. Curr. Opin. Neurobiol. 2000;10:593–596. doi: 10.1016/s0959-4388(00)00127-6. [DOI] [PubMed] [Google Scholar]
- Marchetto M.C., Brennand K.J., Boyer L.F., Gage F.H. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum. Mol. Genet. 2011;20(R2):R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak M.A., Potter G.B., Rowitch D.H., Rubenstein J.L. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Shi X., Harrison R.L., Hollister J.R., Mohammed A., Fraser M.J., Jr., Jarvis D.L. Construction and characterization of new piggyBac vectors for constitutive or inducible expression of heterologous gene pairs and the identification of a previously unrecognized activator sequence in piggyBac. BMC Biotechnol. 2007;7:5. doi: 10.1186/1472-6750-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen C.N., Caldwell M.A., Shen J., ter Borg M.G., Rosser A.E., Tyers P., Karmiol S., Dunnett S.B. Long-term survival of human central nervous system progenitor cells transplanted into a rat model of Parkinson’s disease. Exp. Neurol. 1997;148:135–146. doi: 10.1006/exnr.1997.6634. [DOI] [PubMed] [Google Scholar]
- Svendsen C.N., ter Borg M.G., Armstrong R.J., Rosser A.E., Chandran S., Ostenfeld T., Caldwell M.A. A new method for the rapid and long term growth of human neural precursor cells. J. Neurosci. Methods. 1998;85:141–152. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Vink M., Klaver B., Berkhout B., Das A.T. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 2006;13:1382–1390. doi: 10.1038/sj.gt.3302780. [DOI] [PubMed] [Google Scholar]
- Zhu P., Aller M.I., Baron U., Cambridge S., Bausen M., Herb J., Sawinski J., Cetin A., Osten P., Nelson M.L. Silencing and un-silencing of tetracycline-controlled genes in neurons. PLoS ONE. 2007;2:e533. doi: 10.1371/journal.pone.0000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


