Summary
Methods to isolate and culture primary prostate epithelial stem/progenitor cells (PESCs) have proven difficult and ineffective. Here, we present a method to grow and expand both murine and human basal PESCs long term in serum- and feeder-free conditions. The method enriches for adherent mouse basal PESCs with a Lin−SCA-1+CD49f+TROP2high phenotype. Progesterone and sodium selenite are additionally required for the growth of human Lin−CD49f+TROP2high PESCs. The gene-expression profiles of expanded basal PESCs show similarities to ESCs, and NF-kB function is critical for epithelial differentiation of sphere-cultured PESCs. When transplanted in combination with urogenital sinus mesenchyme, expanded mouse and human PESCs generate ectopic prostatic tubules, demonstrating their stem cell activity in vivo. This novel method will facilitate the molecular, genomic, and functional characterization of normal and pathologic prostate glands of mouse and human origin.
Highlights
-
•
Basal prostate epithelial stem/progenitor cells (PESCs) are expanded in vitro
-
•
Expanded PESCs can differentiate into glandular structures in vitro and in vivo
-
•
A surface-marker screen identifies marker candidates for prostate basal stem cells
-
•
Gene-expression analysis shows a role of NF-kB signaling in PESC differentiation
Trumpp, Sprick, and colleagues describe a method that enables efficient isolation and in vitro expansion of primary human and mouse prostate epithelial stem/progenitor cells (PESCs). The utility of this method for discovering novel markers and functional pathways important for prostate biology is demonstrated by the identification and functional verification of the involvement of NF-kB signaling during differentiation of PESCs.
Introduction
Several model systems have been developed to understand the pathologically altered pathways observed during benign prostatic enlargement and prostate cancer, the latter being the most common type of cancer in men. It has been suggested that epithelial stem/progenitor cells (PESCs) are critical for the regulation and maintenance of the prostatic gland and that they also play an important role in prostate cancer development (Choi et al., 2012; Goldstein et al., 2010; Lu et al., 2013; Visvader, 2011; Wang et al., 2009). PESCs, like other somatic tissue stem cells, are thought to be rare, with a frequency of 1%–5% (Goldstein et al., 2011; Lukacs et al., 2010). Isolation and ex vivo expansion of PESCs is further complicated by their dependence on poorly understood factors supplied by a prostate stem cell niche composed of smooth muscle cells, fibroblasts, neuroendocrine cells, and differentiating and mature prostate epithelial cells (Goldstein et al., 2010; Morrison and Spradling, 2008; Wang et al., 2009). Although significant progress has been made, current culture techniques allow for only limited expansion of prostate epithelial cells (PrECs), which rapidly cease to proliferate (Chaproniere and McKeehan, 1986; Litvinov et al., 2006; Rhim et al., 2011). Human telomerase reverse transcriptase (hTERT)-mediated immortalization has been used to optimize in vitro cultures of primary PrECs (Kogan et al., 2006). Although hTERT-immortalized cells have prolonged in vitro lifespans, they show significant changes compared with normal PrECs, limiting their value as a model system (Klinger et al., 2006). Culture methods using serum-free media conditions with or without additional murine 3T3 feeder cells to grow murine and human PrECs have been described, but serial passaging is limited and these strategies allow neither significant enrichment nor expansion of the stem/progenitor compartment (Kabalin et al., 1989; Peehl and Stamey, 1986; Robinson et al., 1998). In contrast, growing PrECs in semisolid medium using Matrigel facilitates their growth as prostaspheres that retain PESCs with self-renewal capacity in vitro. However, prostaspheres are difficult to manipulate, and the spheres consist of only few PESCs surrounded by a large number of more differentiated PrECs (Xin et al., 2007). More recently, dissociated murine and human PESCs were isolated by flow cytometry (fluorescence-activated cell sorting [FACS]). However, this method is limited by the low frequency of PESCs in conjunction with the small amount of material obtainable from human biopsies, as well as the lack of a suitable culture systems for maintaining or expanding undifferentiated PESCs (Goldstein et al., 2010, 2011; Lukacs et al., 2010; Miki and Rhim, 2008). Here, we report specific workflows and novel, robust, simple, serum- and feeder-free culture techniques to maintain and expand functional primary basal PESCs of mouse and human origin.
Results
Expansion and Maintenance of Primary Murine Basal PESCs in Serum-free Cultures
To develop conditions that would allow us to maintain and expand ex vivo isolated primary murine PESCs, we used single-cell suspensions obtained from whole murine prostates as the starting material. FACS analysis revealed that these cell mixtures contained 4.5% ± 1.5% of SCA-1+CD49f+TROP2+ cells, a phenotype previously used to define basal PESCs (Figures 1A and S1A; Goldstein et al., 2008, 2011; Lukacs et al., 2010). To identify which of the three markers is most critical for further enrichment of basal PESCs, we performed castration experiments. In response to castration and the associated androgen decay, a basal progenitor hyperplasia is commonly observed (Evans and Chandler, 1987; Wu et al., 2007). As expected, we found that TROP2 was robustly upregulated in the basal progenitor cells of the hyperplastic epithelium of castrated mice, confirming the previous finding that TROP2 is a specific marker for basal PESCs (Stoyanova et al., 2012). In contrast, both testosterone-treated castrated mice and unmanipulated wild-type mice displayed the presence of columnar luminal epithelial cells, with low TROP2 expression in both basal and luminal cells (Figure S1B and data not shown).
Figure 1.
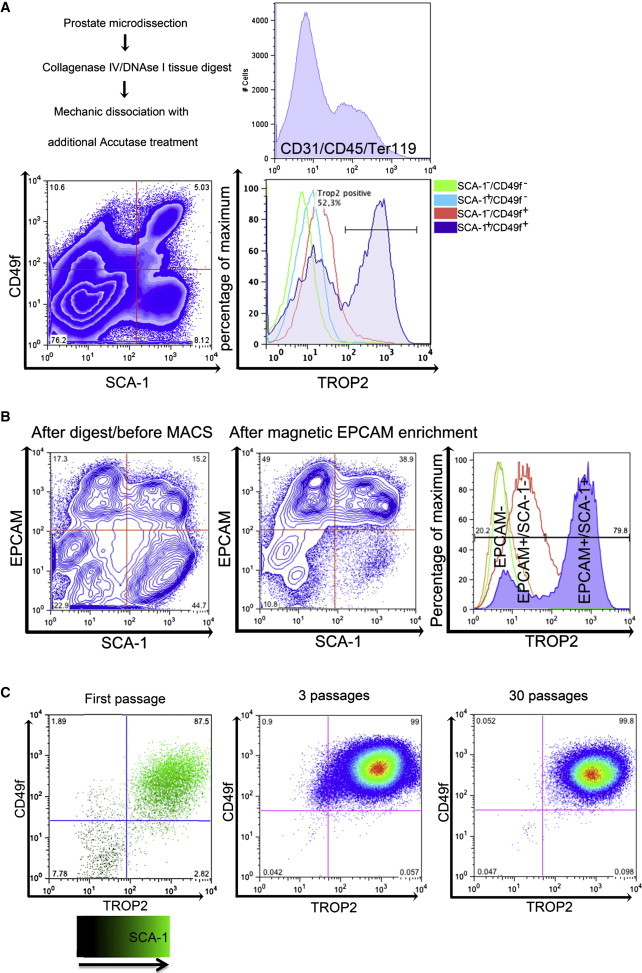
Isolation, Magnetic Separation, and Expansion of Primary Murine Basal PESCs In Vitro
(A) FACS characterization of murine prostate cells after primary dissociation into single cells and staining with SCA-1, CD49f, TROP2, and lin cocktail (Ter119/CD31/CD45). Expression of TROP2 on lin− cells was evaluated in relation to CD49f and SCA-1 expression.
(B) Epithelial enrichment using EPCAM-MACS. FACS analyses before and after enrichment; distribution of TROP2 in the EPCAM−, EPCAM+/SCA-1−, and EPCAM+/SCA-1+ populations.
(C) Polychromatic plot of Sca-1/CD49f/TROP2 expression on cultured murine cells after first passage. Comparison of CD49f/TROP2 expression after the third and 30th passages in vitro.
FACS analysis was used to further characterize the PrEC subpopulations. The pan-epithelial marker epithelial cell adhesion molecule (EPCAM) was expressed in both the TROP2low/neg cells and TROP2high basal PESCs, whereas EPCAM− cells did not express TROP2, consistent with a stromal phenotype (Figure S1D and data not shown). Thus, magnetic-activated cell sorting (MACS) purification was used to enrich for EPCAM+ cells to eliminate EPCAM− stromal cells and prevent fast overgrowth of the epithelial cells (Figures 1B, S1C, and S1D). Standard medium for primary PrECs (PrEGM; Lonza) was used with addition of the ROCK inhibitor Y-27632 in order to inhibit dissociation-induced apoptosis of epithelial cells (Liu et al., 2012; Zhang et al., 2011b). To further increase the specific plating efficiency of PESCs, as monitored by expression of CD49f and TROP2, we tested various culture surfaces. Although primary PrECs did not form significant numbers of colonies on standard culture flasks, significantly better attachment to hydrophobic surfaces was observed (Figures S1E and S1F). However, those conditions did not support proliferation, as cell numbers rapidly decreased within 5 days, indicating suboptimal culture conditions. Therefore, we next optimized the culture medium by including supplements that were previously used to expand embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) (Chen et al., 2011). We used combinatorial analyses of stem cell media components to design a novel “mouse prostate medium” (MPM) that supports the growth of primary murine PrECs. This medium contains Advanced Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with additional glutamine, glucose, EGF, bFGF, IGF-I, transferrin, and insulin (Figure S1G; Experimental Procedures).
After digestion of the murine prostate into single cells, enrichment using EPCAM-MACS, and growth of cells on hydrophobic surface flasks using MPM, the PrECs enriched for SCA-1+CD49f+TROP2high basal PESCs and could be stably expanded for more than 30 passages ex vivo (Figure 1C). To examine whether the fast dominance of SCA-1+CD49f+TROP2high cells was caused by their superior survival in these culture conditions, we determined apoptosis by staining for AnnexinV and propidium iodide (PI). Indeed, differentiated TROP2low cells underwent cell death, whereas TROP2high basal epithelial PESCs survived and proliferated (Figure S2A).
In summary, the method presented here is a feeder-free culture method for the in vitro expansion and maintenance of primary murine basal PESCs with an SCA-1+CD49f+TROP2high phenotype.
Differentiation Capacity of Murine Basal PESCs
Most basal PESCs that are highly enriched for SCA-1+CD49f+TROP2high express the prostate basal cell markers tumor protein p63 (TP63) and cytokeratin 5 (CK5), whereas cytokeratin 8 (CK8) and androgen receptor (AR), which are typically found on differentiated luminal cells, are rarely expressed (Figure 2A). As all cells showed an almost uniform SCA-1+CD49f+TROP2high PESC phenotype (Figure 1C), we tested their capacity to differentiate and self-renew in culture. For this purpose, the cells were transferred into semisolid growth conditions containing Matrigel and the formation of spheres was evaluated in comparison with unselected bulk prostate cells. The results show that 27% ± 7.4% of these cells had sphere-forming capacity (Figure 2B). Importantly, sphere formation was accompanied by a morphologic transition into organized epithelial tubule-like structures. The spheres resembled differentiated structures that retained TP63+ basal cells as well as transit-amplifying cells, as indicated by co-expression of CK5 and CK8. In addition, spheres contained cells expressing AR and NKX3-1, consistent with a more differentiated luminal phenotype (Figure 2B). FACS analysis of spheres demonstrated the switch of CD49f+/TROP2high to a CD49f+/TROP2low phenotype, which is associated with the transition from a stem/progenitor to a more differentiated state (Goldstein et al., 2010). To address the capacity of those cells to serially form spheres, we sorted single CD49f+/TROP2high and CD49f+/TROP2low cells out of entire spheres and replated them in semisolid medium. As expected, only the CD49f+/TROP2high cells were able to serially form spheres and thus were the only cells with self-renewal activity (Figure S2B).
Figure 2.
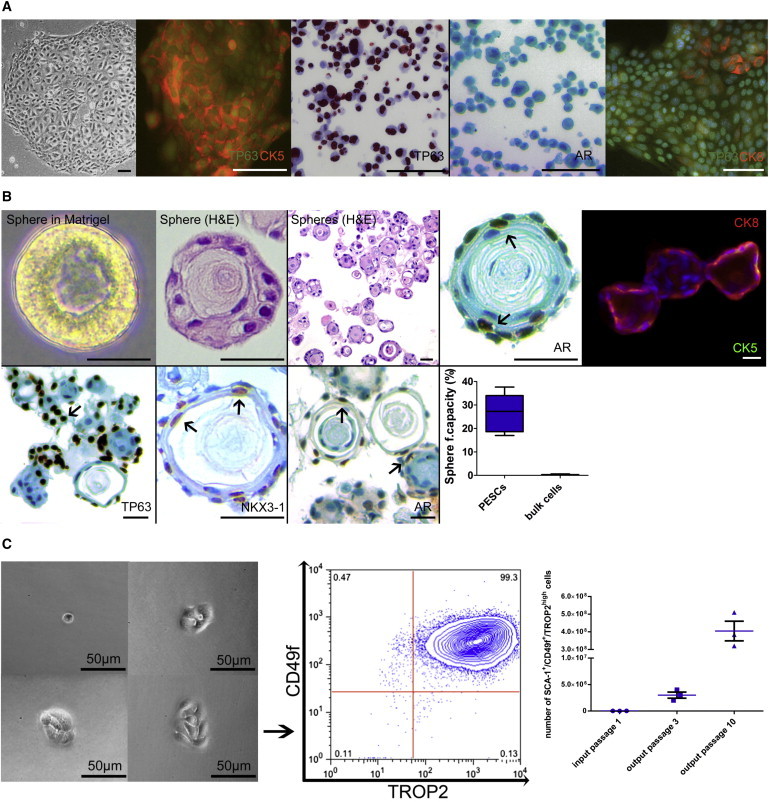
Characterization and Differentiation of Murine Basal PESCs
(A) IHC and immunofluorescence characterization of 2D cultured murine basal PESCs. Scale bar, 100 μm.
(B) Characterization of differentiated murine prostaspheres. Morphology in semisolid Matrigel; immunofluorescence and IHC of prostaspheres. Scale bar, 100 μm. For internal validation of TP63/AR/NKX3-1 antibodies, see Supplemental Experimental Procedures. The sphere-forming capacity of enriched PESCs was compared with bulk digested cells, n = 5 independent PESC preparations, p < 0.01 as determined by Student’s two-tailed t test.
(C) In vitro self-renewal. Colonies derived from single-cell-sorted cultured PESCs retain their SCA-1+/CD49f+/TROP2high phenotype after colony outgrowth. Amplification (cell numbers) of SCA-1+/CD49f+/TROP2high cells using the MPM culture method, n = 3.
To address the self-renewal activity and proliferation of single sorted SCA-1+CD49f+TROP2high cells, we regrew them in adherent cultures and evaluated their phenotype after colony outgrowth was observed. Almost all of the cells retained a SCA-1+/CD49f+/TROP2high PESC phenotype. Additionally, the cells could be expanded to up to 4 × 108 cells in only ten passages (Figure 2C).
Human Basal PESCs Require Additional Progesterone and Sodium Selenite
After we established the primary murine PESC culture, we adapted the method to culture and expand PESCs isolated from human prostate. Single-cell suspensions of primary human prostate cells were obtained from patients with benign prostatic hyperplasia (BPH; Figure S3A). Subsequently, EPCAM+ cells were enriched by MACS and cultured as described above for murine cells (MPM conditions). Since human cells did not expand at first, different culture surfaces were evaluated in conjunction with the addition of multiple stem cell media components. In contrast to mouse cells, human PrECs grew exclusively on BD Primaria surfaces, and not on hydrophobic surfaces (Figure 3A). Furthermore, addition of N2 supplement resulted in a significantly higher cell yield as compared with MPM alone (Figure 3B). Since the N2 supplement contains various components, we sought to further define the specific contribution of N2 ingredients. These experiments revealed that the combination of MPM plus progesterone and sodium selenite (termed human prostate medium [HPM]) is optimal for the outgrowth of primary human prostate epithelial colonies (Figure 3C). Cells that were enriched for the described human CD49f+/TROP2high basal PESC phenotype showed a high cloning efficiency (18% ± 2%) and expressed CK5 and TP63 as basal cell markers (Figures 3D and 3F). Similar to what was observed under the murine conditions, human PESCs could be stably expanded for more than 20 passages ex vivo, and cell numbers of up to 2.0 × 108 could be achieved after only eight passages (Figures 3E and S3B). Moreover, by transferring these cells into semisolid Matrigel, we were able to induce differentiation at a defined time point, resulting in a high sphere-forming capacity (20% ± 4%) and also demonstrating the capacity of a subset of cells to differentiate into CD49f+/TROP2low cells (Figures 3F and 3G; Goldstein et al., 2008). Using anti-human EPCAM+ MACS enrichment followed by growth in Primaria flasks and HPM (plus ROCK inhibitor Y-27632), we were able to demonstrate the ex vivo expansion and maintenance of primary human basal PESCs in the absence of serum and feeder cells.
Figure 3.
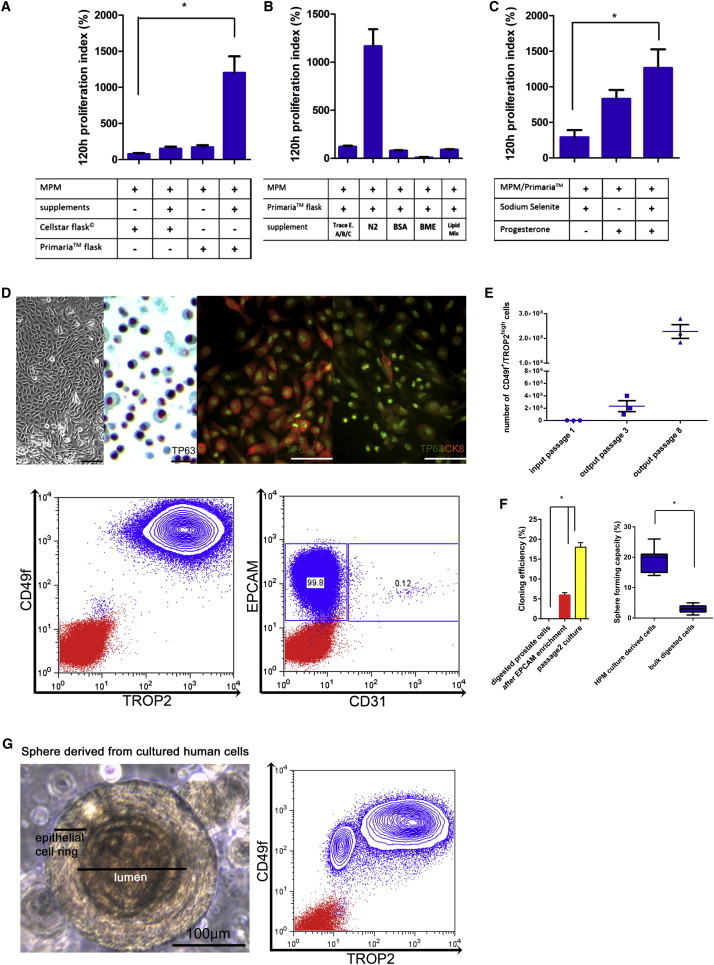
Adherent Enrichment and Expansion of Primary Human Basal PESCs In Vitro
(A–C) Combinatorial testing of factors for optimal expansion and enrichment of human PESCs. Starting with the defined MPM composition, the effect of different factors was determined by changes in the cell numbers after 120 hr (proliferation index 120 hr), n = 8 independent PESC preparations each. Statistical significance was evaluated by one-way ANOVA followed by Bonferroni post hoc tests, p < 0.05.
(A) Initial cell population after EPCAM enrichment and plating with ROCK inhibitor; additional supplementation of trace elements A/B/C, N2 supplement, BSA, BME, and lipid mix.
(B) Addition of N2 supplement to the MPM formulation is the main driver of proliferation for human basal PESCs.
(C) Progesterone and sodium selenite are the N2 constituents necessary for proliferation (HPM conditions).
(D) Micrographs, IHC, and immunofluorescence of cultured human basal PESCs, basal marker TP63, the luminal marker CK8, and the prostate cancer marker AMACR. Scale bars, 20 μm and 100 μM (for the immunofluorescence pictures). Flow-cytometric analyses of human basal PESCs at passage 5 for expression of CD49f, TROP2, EPCAM, and CD31.
(E) Amplification (cell numbers) of CD49f+/TROP2high cells using the HPM culture method; n = 3 independent PESC preparations.
(F) Cloning efficiency as determined by in vitro limiting dilution and Matrigel-sphere-forming capacity of human PESCs cultured under the final optimal conditions (Primaria surface and HPM); n = 5 independent PESC preparations, p < 0.01 as determined by Student’s two-tailed t test.
(G) Morphology and expression of TROP2 and CD49f of differentiated human prostaspheres.
Primary Basal PESC Cultures Are Suitable for Medium- to High-Throughput Assays
To determine the genetic stability of PESC cultures, we performed karyotyping using multiplex fluorescence in situ hybridization (M-FISH) of human PESCs ex vivo. Karyotyping was performed on cultures of three different passage numbers, analyzing 15 individual metaphases each. This analysis confirmed a normal male karyotype (46,XY) (Figure 4A).
Figure 4.
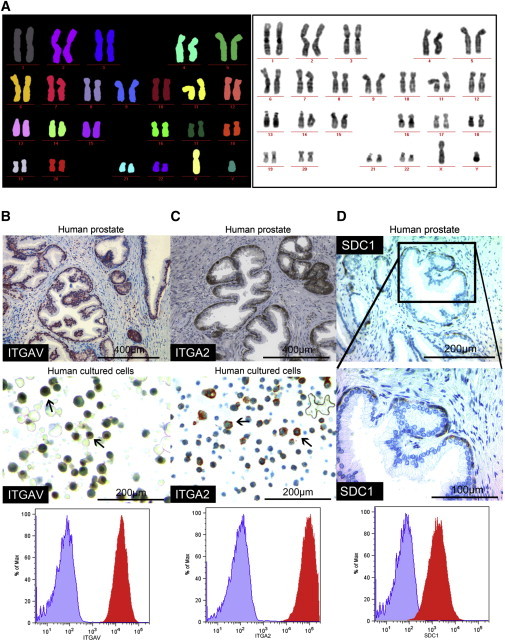
Genetic Stability of PESCs and Histologic Validation of FACS Array Screen-Identified Proteins in Human Prostate Glands
(A) M-FISH analysis of human PESCs in culture demonstrates a normal male karyogram (46,XY) with no signs of chromosomal aberrations; 24-color fluorophore-labeled metaphase chromosomes and their discrimination after visualization and computerized output.
(B and C) Staining for ITGAV and ITGA2 in primary human prostate and cultured human PESCs by IHC; surface expression on cultured human PESCs as determined by FACS.
(D) Exclusive expression of SDC1 in the human prostate basal epithelial compartment in vivo as determined by IHC. FACS surface staining of SDC1 in cultured human PESCs. The FACS plot shows the staining intensity for the isotype control (purple) and the specific antibody (red).
See also Figure S4.
The establishment of primary basal PESC cultures grown as 2D adherent cells allowed us to evaluate whether the culture model is suitable for medium- to high-throughput assays. We thus expanded murine and human PESCs to 150 × 106 cells and performed 96-well-based screens to identify cell-surface markers for basal PESCs. Expression of 176 murine and 242 human cell-surface markers was tested on PESCs by flow cytometry (BD Lyoplate; data not shown). The results demonstrated the expression of previously described markers for basal PESCs, including CD29+ and CD49f+ (Goldstein et al., 2011). Moreover, all murine and human basal PESCs expressed high levels of CD24 (Figure S4). Expression of integrin alpha V (ITGAV) and integrin alpha-2 (ITGA2, CD49b) was validated by immunohistochemistry (IHC) on normal prostates, revealing a higher expression within the human prostate basal epithelial layer (Figures 4B and 4C; Collins et al., 2001; Liu and True, 2002). The screen also identified Syndecan-1 (SDC1) as a protein that is exclusively expressed in the human basal prostate compartment and not in differentiated luminal cells (Figure 4D). The identified cell-surface proteins may serve as a basis for future studies on basal PESCs and confirm that our culture methods are suitable for medium- to high-throughput assays.
The Transcriptome of Murine and Human Basal PESCs Is Similar to That of ESCs
To uncover specific differences between basal PESCs and differentiated luminal cells, we compared the gene-expression profiles of mouse and human PESCs with profiles obtained from differentiated sphere cells. As the cultured cells were almost completely comprised of cells with an epithelial phenotype, we were able to establish pure expression profiles without contamination of profiles derived from other, non-epithelial cells of the prostate microenvironment (Figure 5A). Next, we performed gene set enrichment analyses (GSEAs) focusing primarily on gene sets enriched in both human and mouse basal PESCs with a false-discovery rate (FDR) < 0.001 (Mootha et al., 2003; Subramanian et al., 2005). This revealed distinct enrichment of specific gene sets in PESCs that are representative of immature pluripotent cells, especially ESC profiles (Figure 5B; Kesanakurti et al., 2013; Müller et al., 2008; Wong et al., 2008). Important regulators of stem cells and organogenesis, such as SOX2, PRDX1, LMNB1, and PAK1, showed a significantly higher mRNA expression in the undifferentiated PESC cultures (data not shown) (Kim et al., 2011; Yan et al., 2009; Zhang et al., 2011a; Zhu et al., 2009). We tested the levels of expression of human PRDX1 and PAK1 protein by IHC, which demonstrated that both proteins are expressed in the basal epithelial progenitor compartment (Figure S5A). Additionally, we identified LMNB1 (Lamin B1) as a putative new marker for PESCs, as we specifically detected expression within the basal compartment of the normal prostate as well as in cultured PESCs. In contrast, differentiated luminal cells in the prostate as well as in differentiated spheres did not express LMNB1 (Figure S5B).
Figure 5.
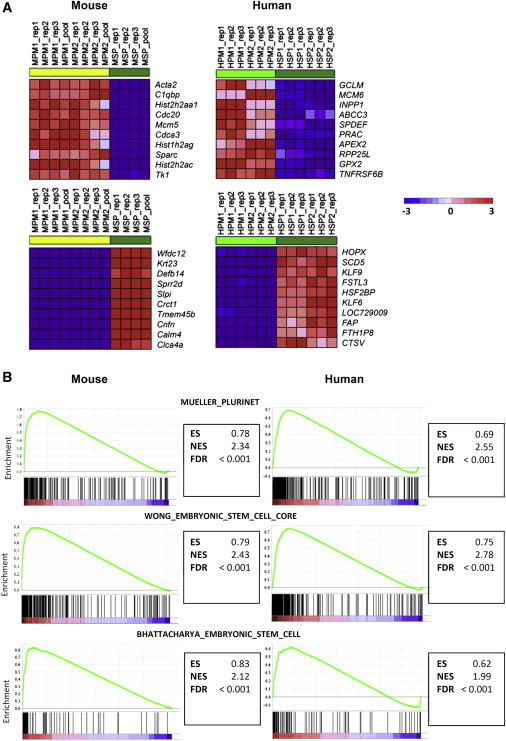
Cultured and Enriched PESCs Demonstrate Similarities to ESCs
(A) The top ten most differentially regulated genes in comparison with basal PESCs and the more differentiated sphere cells. Shown are the top ten upregulated genes in PESCs that are downregulated in spheres (upper two), as well as the top ten downregulated genes in PESCs that are upregulated in spheres (lower two). MPM, murine PESCs; MSP, murine spheres; HPM, human PESCs; HSP, human spheres.
(B) GSEA demonstrates enrichment of ESC gene signatures in undifferentiated cultured basal PESCs (Kesanakurti et al., 2013; Müller et al., 2008; Wong et al., 2008).
See also Figure S5.
GSEAs in Murine and Human PESCs Indicate Regulatory Roles of c-Myc and the TNFα/NF-kB Pathway
To identify potential signaling networks that maintain the undifferentiated versus differentiated state of basal PESCs, we performed GSEA focusing on gene sets that predict distinct transcription factor activities. This revealed upregulation of the MYC gene and multiple MYC targets in undifferentiated basal PESCs compared with differentiated sphere cells (Figure S6A). Moreover, during the process of differentiation into spheres, the PrECs showed a significant enrichment for TNFα- and NF-kB-mediated signaling cascades (Figure 6A). Enrichment plots of MYC targets and TNFα and NF-kB signaling suggest a possible regulatory network between the undifferentiated prostate basal stem cell state and more luminal differentiated spheres (Figure S6B). These results further reveal the upregulation of various signaling pathways in PESCs as compared with the more-differentiated sphere cells (Table S3).
Figure 6.
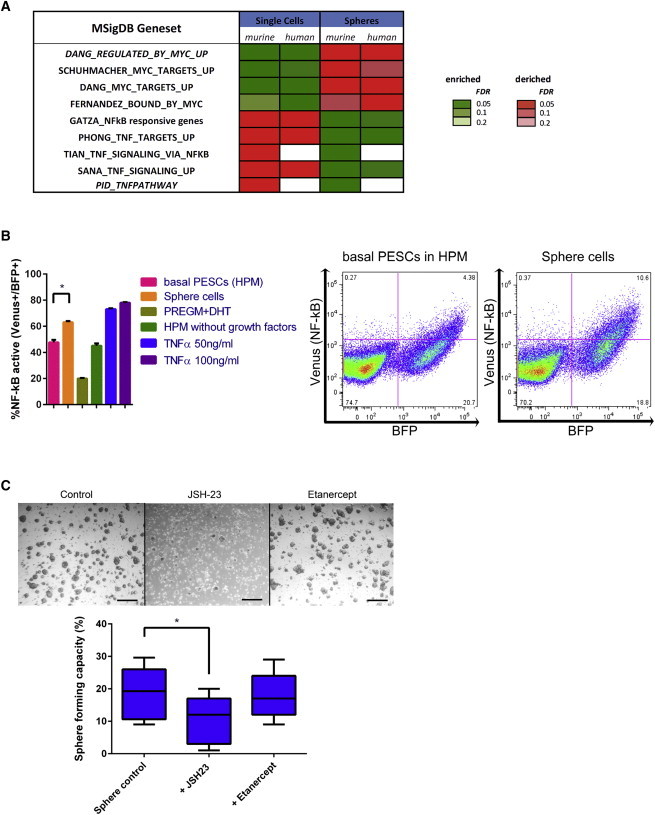
GSEAs Indicate a Functional Role of NF-kB/TNFα in Undifferentiated and Differentiated States of Basal PESCs
(A) Significantly changed gene sets (GSEA) in undifferentiated basal PESCs as compared with differentiated prostasphere cells (Subramanian et al., 2005; Zutter and Santoro, 1990).
(B) FACS analyses of NF-kB transcriptional reporter activity in human cells (left, n = 5 independent PESC preparations, statistical significance was evaluated by one-way ANOVA followed by Bonferroni post hoc tests, p < 0.05), % NF-kB active = % Venus/BFP positivity in FITC/Pacific blue cytometer channels and two corresponding FACS plots, demonstrating NF-kB activity in basal PESCs in HPM conditions as compared with increased NF-kB activity in differentiated sphere cells (right).
(C) Inhibition of NF-kB leads to impaired differentiation of human PESCs. Micrographs and corresponding sphere-forming-capacity results of PESCs seeded into regular sphere conditions (PrEGM/Matrigel = sphere control) in comparison with sphere conditions with the addition of NF-kB inhibitor JSH-23 or Etanercept to block soluble TNFα-mediated TNFR binding (n = 5 independent PESC preparations each; statistical significance was evaluated by one-way ANOVA followed by Bonferroni post hoc tests, p < 0.05).
Inhibition of NF-kB, but Not TNFα, Leads to Impaired Differentiation of Human PESCs
To test whether our culture platform is suitable for functional biological analyses, we focused on the postulated regulation of PESC differentiation into spheres by NF-kB or TNFα (Figure 6A). We transfected human PESCs of the CD49f+/TROP2high phenotype with a reporter construct to monitor the transcription factor NF-kB by expression of Venus (pV2b-NF-kB). Compared with the PESCs, more of the sphere cells expressed Venus. Moreover, Venus expression in human PESCs (HPM conditions) increased in response to stimulation by TNF-α (Figure 6B). This increase in NF-kB activity cannot be explained by the use of PrEGM and dihydrotestosterone (DHT), because PrEGM/DHT caused no increase of NF-kB activity alone (Figure 6B). Together, these results suggest that the observed NF-kB activation during the morphological transition into spheres is most likely due to the effect of differentiation itself and is consistent with the transcriptional activation of the NF-kB pathway during differentiation (Figures 6A and 6B). To determine whether the NF-kB pathway is important for differentiation into spheres, we blocked the pathway by the small-molecule inhibitor JSH-23, which inhibits NF-kB nuclear translocation, and assessed the sphere-forming capacity (Shin et al., 2004). JSH-23-treated cultures showed a significantly reduced sphere-forming capacity, whereas blocking of TNFα using the TNF-R2-Fc fusion protein Etanercept had no effect (Figure 6C). These data suggest that a TNFα-independent intrinsic or extrinsic mode of NF-kB activation is critically involved during sphere formation.
Cultured Basal PESCs Demonstrate Stem Cell Function In Vivo
In vivo transplantation assays are among the most commonly used methods for demonstrating stem cell activity. Such methods test the capacity of transplanted stem cells to generate and maintain entire tissue structures comprised of various differentiated cell types. Xin et al. (2003) previously demonstrated the regenerative capacity of the adult prostate epithelium in classical sandwich grafting experiments by co-transplanting prostate epithelium with fetal urogenital sinus mesenchyme (UGSM). These assays demonstrated that signals derived from the UGSM are required for the epithelial cells to generate prostate-gland-like structures in a transplant setting (Cunha and Lung, 1978; Goldstein et al., 2011; Lukacs et al., 2010). To test whether cultured cells retain functional stem cell activity, we lentivirally marked murine and human PESCs by concurrently introducing two fluorescent proteins, tdTomato and Venus (Weber et al., 2008, 2011; Figure S7A). Subsequent subcutaneous (s.c.) transplantation of LeGO-V2/T2 marked SCA-1+CD49f+TROP2high mouse PESCs, which were mixed with unmarked E16 UGSM and revealed prostatic tubules after 10–12 weeks. Their PESC origin was confirmed by immunofluorescence imaging and anti-GFP IHC (Venus). As few as 104 mouse SCA-1+CD49f+TROP2high PESCs were sufficient to induce growth of prostatic tubules when transplanted subcutaneously. Although no engraftment was observed when cells were transplanted intraprostatically (without UGSM), intermediate results were obtained by transplanting cells under the kidney capsule (with UGSM) (Figures 7A and 7B; Table S4). Importantly, new prostatic tubules derived from cultured mouse basal PESCs preserved TP63+ basal cells and demonstrated differentiated AR+-expressing cells encircling the lumina of formed acini, confirming their typical 3D cellular structure at the molecular level.
Figure 7.
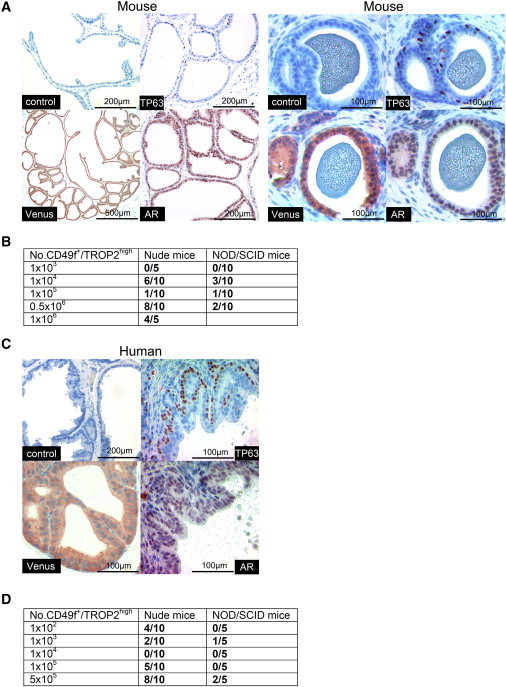
Cultured and Enriched Basal PESCs Preserve Functional Adult Stem Cell Capacity In Vivo
(A) Formation of prostate gland-like acini after s.c. transplantation of cultured Venus+ murine PESCs together with unmarked E16 UGSM. Immunohistochemical staining for TP63 and AR. Venus was detected with an anti-GFP antibody.
(B) Frequency of murine prostate acini regeneration in a limiting-dilution in vivo transplantation assay.
(C) Prostate-gland-like acini formation after s.c. transplantation of cultured Venus+ human PESCs together with unmarked E16 UGSM. Immunohistochemical staining for TP63, AR, and Venus (anti-GFP).
(D) Frequency of human prostate acini regeneration in a limiting-dilution in vivo transplantation assay.
As few as 100 cultured human basal PESCs were able to regenerate prostate acini in nude mice (Figures 7D and S7B; Table S4). For both human and mouse PESCs, nude mice represented the more efficient recipients compared with the more immune-compromised NOD/SCID mice. Regenerated human prostate acini were built up of a single TP63+ basal cell layer and single or multiple layers of differentiated AR+ luminal cells, closely resembling the microscopic anatomy of normal human prostate epithelium (Figures 7C and S7C). In summary, our results demonstrate that cultured murine as well as human primary PESCs are able to regenerate entire prostatic acini, demonstrating that these cells have adult prostate stem cell activity in vivo.
Discussion
Here, we provide a novel method to expand and study functional basal PESCs in adherent cultures. Simple serum- and feeder-free conditions were established to grow and expand murine LinnegCD49f+SCA-1+TROP2high and human LinnegCD49f+TROP2high PESCs. The reported method represents a major advance from previous protocols (Goldstein et al., 2011; Lukacs et al., 2010; Rhim et al., 2011; Robinson et al., 1998) and complements the protocol recently proposed by Karthaus et al. (2014) to expand and maintain enriched prostate progenitor cells ex vivo. The method also overcomes the presence of undefined media and culture components such as bovine pituitary extract (Peehl and Stamey, 1986). The ability to significantly expand functional human basal PESCs, in terms of both total number and frequency, will help investigators overcome the bottleneck related to the limited availability of primary prostate patient tissue for cellular, molecular, genomic, and pharmacological analyses.
A key element of the culture conditions is the balanced combination of growth factors and signaling molecules (e.g., EGF, bFGF, IGF, insulin, transferrin, and Rock inhibitor), which apparently generates an artificial androgen-independent PESC microenvironment that promotes the self-renewal and maintenance of 2prostate stem cell fate. Although these factors have been known for a long time in the cell-culture field, it was critical to discover the exact composition of the media in combination with a hydrophobic surface that would allow significant expansion of undifferentiated murine basal PESCs as compared with the widely used standard method (PrEGM). The necessary adaptations to enable human basal PESC amplification included the switch to surface-treated flasks and the addition of sodium selenite and progesterone. Progesterone has also been reported to induce mammary epithelial progenitor cell expansion, indicating that it may promote hormone-controlled epithelial stem cells in general (Joshi et al., 2010). Expanded basal PESCs not only show expression signatures similar to those of pluripotent ESCs and other somatic stem cells but also harbor functional stem cell potential, as demonstrated by their capacity to generate prostatic tubules in vivo. These results are comparable to those obtained in transplantation experiments performed with PESCs isolated from primary prostate biopsies (Goldstein et al., 2010, 2011; Lukacs et al., 2010). The method described here now allows the robust expansion of such primary cells and thus facilitates an in-depth analysis of the molecular programs employed.
One can induce expanded PESCs to differentiate at any desired time point by transferring the cells from adherent conditions into previously described prostasphere culture conditions (Xin et al., 2007). However, the described method cannot be used to study the role of luminal PESCs, which have also been reported to be a self-sustaining lineage (Karthaus et al., 2014). In addition to basal PESCs, luminal PESCs have also been suggested to be the putative cell of origin for prostate cancer (Choi et al., 2012; Goldstein et al., 2010; Wang et al., 2009). Furthermore, in vitro differentiation into spheres can only a serve as a model system and does not resemble the full luminal differentiation program of prostate gland development in vivo. This limitation and the putative presence of transit-amplifying (intermediate) cells have to be considered when using these methods (Ousset et al., 2012; Pastrana et al., 2011). Additionally, the methods we have described for murine cells cannot be used to replace lineage-tracing mouse models—they can only complement the findings from such models. In particular, work by Wang et al. (2013) clearly shows that prostate basal cells develop a substantial plasticity ex vivo when they are removed from their normal environment. In line with this, our experiments confirm the finding that a significant discrepancy exists between the high in vitro sphere-forming capacity of basal PESCs and their capacity to form glands in vivo. In vivo, only a small proportion of basal cells were shown to have a graft-regenerating capacity (Wang et al., 2013). Nevertheless, our methods additionally facilitate the analysis of primary human cells, allowing such cells to be amplified, manipulated, and studied in detail. Clearly, the direct analysis of human cells holds the potential to provide data that are of more relevance to the biology of human development and disease.
The culture method described here creates a novel platform for studying prostate disease etiology and progression. PESCs grown as adherent feeder-free cultures are easy to manipulate (e.g., for transfection and infection) and can be induced to differentiate or transplanted to form prostate tubules in vivo. Thus, this method will provide the basis for various in-depth analyses of epithelial prostate stem cells. First, it provides the basis to selectively expand and study murine basal PESCs isolated from different genetically engineered mice, such as in the PTEN prostate cancer model (Di Cristofano et al., 2001). This may help to identify molecular mechanisms during differentiation and the progression from normal prostate basal stem cells to hyperplastic and possibly even neoplastic epithelium (Carver et al., 2011). However, one has to keep in mind that prostate cancers that arise from basal stem cells may have a different phenotype and clinical outcome compared with those derived from luminal prostate stem cells (Choi et al., 2012; Lu et al., 2013). Second, using co-culture techniques that combine basal PESCs with cellular prostate stromal components (e.g., associated fibroblasts and smooth muscle cells), one can dissect and study important cross regulations between primary PESCs and their corresponding microenvironmental niche to better understand prostate-gland regulation at a more global level. Third, human basal PESCs isolated from patients with BPH can be isolated and studied at the molecular and genomic levels, and subsequently linked to their biologic behavior in vitro and in vivo. An estimated 50% of men show histologic evidence of BPH by the age of 50 years, and 40%–50% of these men become clinically significant, demonstrating the clinical relevance of this novel method. Finally, mouse- or patient-derived and expanded PESCs can be used for high-throughput screens using knockdown or chemical compound libraries. This novel mouse and human method to expand functional PESCs may boost research on normal prostate gland biology and may open up new possibilities for studying the etiology of prostatic diseases.
Experimental Procedures
Prostate Cell Preparation and Identification of Basal/Luminal Prostate Epithelial Cells by Flow Cytometry
Microdissection, enzymatic digestion, and preparation of single cells from male C57Bl/6 mice prostate primary human prostate were performed as described previously (Goldstein et al., 2011; Lukacs et al., 2010). We changed the described enzymatic digestion of the human prostate into a 4 hr routine to obtain higher cell yields (specific steps are provided in Supplemental Experimental Procedures). For isolation of primary human cells from surgical prostate tissues, we obtained informed consent according to the principles of the Declaration of Helsinki. Procedures were approved by the responsible ethics committee of Heidelberg University (permit S-479/2009). For detailed information regarding patient tissues, see Table S2. Identification of basal stem cells by the lineageneg (Ter119−/CD31−/CD45−) SCA-1+CD49f+TROP2high phenotype in the murine prostate, as well as the CD49f+TROP2high phenotype in the human prostate, was performed as described previously (Goldstein et al., 2008, 2010; Lukacs et al., 2010; Xin et al., 2005). CD49f+TROP2low cells have been identified and characterized before as more differentiated epithelial phenotype cells (Goldstein et al., 2008, 2010, 2011). Please see Table S5 for specific antibody information. We began our new enrichment and culture methods after enzymatic digestion of the primary murine or human prostate into single cells.
Adherent Expansion of Primary Murine and Human Basal PESCs
As a combined first step in establishing the murine and human cell cultures, we performed MACS enrichment for EPCAM+ cells after primary preparation of single-cell suspensions from murine and human prostates (Figure 1). For this purpose, we stained digested murine cell suspensions with anti-mouse CD326 (EPCAM)-phycoerythrin (PE) (Clone 8.8; eBioscience) followed by indirect magnetic bead labeling using anti-PE microbeads (Miltenyi Biotec). For human cells, we directly used anti-human EPCAM microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Magnetic enrichment was performed using the autoMACS Pro Separator (Miltenyi Biotec). We altered the tissue culture flask surfaces by comparing negatively charged standard plastic culture flasks (TPP) with hydrophobic (suspension) culture flasks (Cellstar; Greiner Bio-One) or net-negative pretreated surface flasks (Primaria; BD). After evaluating the appropriate culture surface, we tested different combinations of stem cell media components (described in detail in Figures 1, 2, 3, and S1 and Table S1). Cells were plated in either hydrophobic CellStar (Greiner) 24-well plates (murine) or 24-well Primaria (BD) plates (human) with individual combinations of media components (n = 8). We evaluated the proliferation index by evaluating the cell number at a specific time point divided by the number of input cells at time zero. The best media for the expansion of murine prostate basal epithelial progenitor cells, MPM, consists of Advanced DMEM/F12 supplemented with additional glutamine, glucose, EGF, bFGF, LONG R3 IGF-I, holo-transferrin, and insulin. The best media for the expansion of human prostate basal epithelial progenitor cells, HPM, is the MPM formulation plus additional progesterone and sodium selenite.
Prostasphere Assay and Analysis/Sorting of Sphere-Derived Single Cells
The semisolid prostasphere assay used for in vitro differentiation analyses has been described elsewhere (Xin et al., 2007). Briefly, cultured murine or human prostate basal epithelial cells were resuspended in a 50:50 mixture of Matrigel (BD) and PrEGM (Lonza) and plated around the rim of a well of a 12-well tissue culture plate. The Matrigel mix was allowed to solidify at 37°C and then 800 μl PrEGM was added. To recover the sphere cells for subsequent flow-cytometry analysis/cell sorting, we used Cell Retrieval Solution (BD) followed by sphere digestion into single cells using StemPro-Accutase (GIBCO) in combination with mechanical trituration using a 27-gauge needle and 40-μm filters.
Flow Cytometry and Single-Cell-Sorting Experiments
All cell sortings were performed on BD FACS Aria II or Aria III cell sorters. To minimize loss of cell viability, we performed experiments on fresh cell suspensions out of our culture, prepared shortly before flow cytometry, detaching the cells using StemPro-Accutase (GIBCO). Antibody staining was performed in PBS supplemented with 5 mM EDTA. Please see Table S5 for specific antibody information. Prior to flow cytometry or sorting, cells were filtered using 40-μm filters. The sorting buffer included PBS, 5 mM EDTA and 10 μM Y-27632 ROCK inhibitor (Tocris Bioscience). Forward-scatter height (FSC-H) versus forward-scatter width (FSC-W), and side-scatter height (SSC-H) versus side-scatter width (SSC-W) profiles were used to eliminate cell doublets. Dead cells were eliminated by excluding PI+ cells, whereas contaminating human or mouse Lin+ cells were eliminated by gating on Ter119/CD31/CD45-FITC− for mouse and CD45/CD3-FITC− for human cells. Gates for FACS experiments were determined by using isotype controls for the respective specific antibodies used. Gates were then set to exclude the respective population in the isotype control experiment. In single-cell-sorting experiments, each cell was individually sorted into a different well of a 96-well plate, using a built-in protocol in the FACS Aria II and III software packages, with appropriate adjustments (device: 96-well plate, precision: single-cell). For sorting, we used a 100-μm nozzle. Sorted single cells were additionally evaluated by microscopy. We assessed the true clonogenicity of single basal progenitor cell-derived colonies quantitatively by performing a limiting-dilution analysis in vitro using 96-well plates and L-Calc software (Stem-Cell Technologies) after observing colony outgrowth and confirming a stable SCA-1+/CD49f+/TROP2high phenotype.
Lentiviral Vectors and Lentiviral Gene Transfer
LeGO-V2 (Venus) and LeGO-T2 (tdTomato) were previously described (Weber et al., 2011) and kindly provided by Kristoffer Weber and Boris Fehse. Lentiviral particles were generated as previously described (Kutner et al., 2009). For transduction, prostate cells were cultured in MPM or HPM for 24 hr at a fixed cell number. Target cells were incubated in the presence of 8 μg ml−1 polybrene for 12 hr at 37°C with viral supernatant at a multiplicity of infection (moi) of 50–60 per vector. Transduction efficiency was validated 48–72 hr after transduction using FACS.
Mouse Experiments and Evaluation of In Vivo Stem Cell Capability
All mouse experiments were approved by the animal-protection officers of the German Cancer Research Center (DKFZ) and the state of Baden-Württemberg in accordance with German law (Tierschutzgesetz, G18-12). Male NOD-SCID and nude mice were bred at the animal facility of the DKFZ and maintained under pathogen-free, individual ventilated-cage conditions. E16 UGSM was used for co-injections with culture-derived prostate progenitor cells to provide the necessary growth signals to promote in vivo prostate gland regeneration. Before performing the co-injections, we prepared the UGSM according to previously described protocols (Lukacs et al., 2010). First, we set up timed pregnancies in C57Bl/6 mice and harvested the fetuses at day 16 of pregnancy. We cut the fetus in half below the liver and microdissected the pelvic UGSM under the stereomicroscope while placing the bottom half of the fetus in a supine position and holding the hind legs apart with forceps (Cunha and Lung, 1978). The UGSM was removed and separated intact by gently pulling up on the bladder. To prove the in vivo stem cell capability of our culture-derived cells, we co-injected our LeGO-V2/T2 marked cultured murine or human cells together with E16 UGSM and Matrigel into male nude or NOD/SCID mice subcutaneously into the renal capsule, as well as intraprostatically (without UGSM). For detailed information, see Figure 7 and Table S4. Experimental results were obtained using passage 5 (early) cultured cells, though engraftment could also be observed using late passage numbers (data not shown). To support differentiation, we subcutaneously implanted testosterone pellets (12.5 mg/90-day release; Innovative Research of America). After 10–12 weeks, we harvested the regenerated s.c. grafts for subsequent analyses. Before conducting histological analyses on fixed tissue, we visualized direct Venus fluorescence in freshly dissected s.c. grafts under the fluorescent stereomicroscope.
IHC and Immunofluorescence
For IHC of differentiated spheres, we retrieved prostaspheres from the Matrigel using Cell Retrieval Solution (BD). The spheres were then fixed in 10% buffered formalin and transferred into HistoGel (Thermo Scientific) for subsequent sectioning and staining with various antibodies according to the manufacturer’s instructions. Antibodies for basal epithelial TP63 and differentiated luminal epithelial markers (AR, NKX3-1) were validated on primary prostate tissue before use (for detailed analyses, see Supplemental Experimental Procedures). For immunofluorescence, cells were grown in Cellstar (Greiner Bio-One) or BD Primaria six-well plates, fixed within the wells using BD Cytofix for 30 min at 4°C, and stained overnight with primary antibodies. The next day, staining was done with secondary anti-IgG-Alexa 488 (Invitrogen) and anti-IgG-Alexa 546 (Invitrogen). Finally, cells were counterstained with ProLong antifade reagent (Invitrogen) and visualized with a standard fluorescence microscope. Please see Table S5 for specific IHC and immunofluorescence antibody information. Regenerated s.c. tissue grafts were fixed in 10% buffered formalin and placed in 70% ethanol. Sections (4 μm) were stained with hematoxylin and eosin (H&E) or rabbit polyclonal anti-GFP/Venus antibody (ab290; Abcam). We previously validated the antibody for detection of Venus to prove the in vivo stem cell capability of our culture-derived cells as compared with coinjected untransduced cells of the fetal urogenital sinus.
Author Contributions
T.H. developed the methods and designed the study, performed culture and differentiation experiments in vitro and in vivo, and wrote the manuscript. C.E. helped with lentiviral constructs and performed bioinformatics analyses. C.K. performed experiments and supervised all mouse transplantation experiments. T.R.-W., S.W., and E.N. helped with vector design and experiments. S.M.-G., V.V., and W.W. performed histological analyses and secondary pathologic evaluations. A.J. and B.S. performed M-FISH analyses. I.B., A.S., and S.P. analyzed the data and/or provided intellectual guidance regarding their interpretation. M.R.S. and A.T. designed the study, analyzed and evaluated results, and wrote the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgments
We thank Dr. S. Schmitt and his team at the DKFZ Flow Cytometry Core Facilities for expert technical assistance, and Dr. M. Milsom for discussions and critical reading of the manuscript. We also thank Dr. B. Fehse (University Medical Center Hamburg-Eppendorf) for providing LeGO constructs. This work was supported by the BioRN Spitzencluster “Molecular and Cell Based Medicine,” supported by the German Bundesministerium für Bildung und Forschung (BMBF), the SFB 873 “Maintenance and Differentiation of Stem Cells in Development and Disease” funded by the Deutsche Forschungsgemeinschaft (DFG), and the Dietmar-Hopp Foundation.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Martin R. Sprick, Email: martin.sprick@hi-stem.de.
Andreas Trumpp, Email: a.trumpp@dkfz.de.
Accession Numbers
The expression array data reported in this work have been deposited in the Gene Expression Omnibus under accession number GSE61861.
Supplemental Information
References
- Carver B.S., Chapinski C., Wongvipat J., Hieronymus H., Chen Y., Chandarlapaty S., Arora V.K., Le C., Koutcher J., Scher H. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaproniere D.M., McKeehan W.L. Serial culture of single adult human prostatic epithelial cells in serum-free medium containing low calcium and a new growth factor from bovine brain. Cancer Res. 1986;46:819–824. [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.T., Habib F.K., Maitland N.J., Neal D.E. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J. Cell Sci. 2001;114:3865–3872. doi: 10.1242/jcs.114.21.3865. [DOI] [PubMed] [Google Scholar]
- Cunha G.R., Lung B. The possible influence of temporal factors in androgenic responsiveness of urogenital tissue recombinants from wild-type and androgen-insensitive (Tfm) mice. J. Exp. Zool. 1978;205:181–193. doi: 10.1002/jez.1402050203. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., De Acetis M., Koff A., Cordon-Cardo C., Pandolfi P.P. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- Evans G.S., Chandler J.A. Cell proliferation studies in the rat prostate: II. The effects of castration and androgen-induced regeneration upon basal and secretory cell proliferation. Prostate. 1987;11:339–351. doi: 10.1002/pros.2990110406. [DOI] [PubMed] [Google Scholar]
- Goldstein A.S., Lawson D.A., Cheng D., Sun W., Garraway I.P., Witte O.N. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.S., Huang J., Guo C., Garraway I.P., Witte O.N. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.S., Drake J.M., Burnes D.L., Finley D.S., Zhang H., Reiter R.E., Huang J., Witte O.N. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat. Protoc. 2011;6:656–667. doi: 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- Kabalin J.N., Peehl D.M., Stamey T.A. Clonal growth of human prostatic epithelial cells is stimulated by fibroblasts. Prostate. 1989;14:251–263. doi: 10.1002/pros.2990140306. [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesanakurti D., Chetty C., Rajasekhar Maddirela D., Gujrati M., Rao J.S. Essential role of cooperative NF-κB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene. 2013;32:5144–5155. doi: 10.1038/onc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Sharov A.A., McDole K., Cheng M., Hao H., Fan C.M., Gaiano N., Ko M.S., Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger R.Y., Blum J.L., Hearn B., Lebow B., Niklason L.E. Relevance and safety of telomerase for human tissue engineering. Proc. Natl. Acad. Sci. USA. 2006;103:2500–2505. doi: 10.1073/pnas.0508184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan I., Goldfinger N., Milyavsky M., Cohen M., Shats I., Dobler G., Klocker H., Wasylyk B., Voller M., Aalders T. hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res. 2006;66:3531–3540. doi: 10.1158/0008-5472.CAN-05-2183. [DOI] [PubMed] [Google Scholar]
- Kutner R.H., Zhang X.Y., Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Litvinov I.V., Vander Griend D.J., Xu Y., Antony L., Dalrymple S.L., Isaacs J.T. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66:8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.Y., True L.D. Characterization of prostate cell types by CD cell surface molecules. Am. J. Pathol. 2002;160:37–43. doi: 10.1016/S0002-9440(10)64346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ory V., Chapman S., Yuan H., Albanese C., Kallakury B., Timofeeva O.A., Nealon C., Dakic A., Simic V. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.L., Huang Y.F., You L.R., Chao N.C., Su F.Y., Chang J.L., Chen C.M. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am. J. Pathol. 2013;182:975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Lukacs R.U., Goldstein A.S., Lawson D.A., Cheng D., Witte O.N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki J., Rhim J.S. Prostate cell cultures as in vitro models for the study of normal stem cells and cancer stem cells. Prostate Cancer Prostatic Dis. 2008;11:32–39. doi: 10.1038/sj.pcan.4501018. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Spradling A.C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F.J., Laurent L.C., Kostka D., Ulitsky I., Williams R., Lu C., Park I.H., Rao M.S., Shamir R., Schwartz P.H. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B.D., Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Pastrana E., Silva-Vargas V., Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peehl D.M., Stamey T.A. Growth responses of normal, benign hyperplastic, and malignant human prostatic epithelial cells in vitro to cholera toxin, pituitary extract, and hydrocortisone. Prostate. 1986;8:51–61. doi: 10.1002/pros.2990080107. [DOI] [PubMed] [Google Scholar]
- Rhim J.S., Li H., Furusato B. Novel human prostate epithelial cell culture models for the study of carcinogenesis and of normal stem cells and cancer stem cells. Adv. Exp. Med. Biol. 2011;720:71–80. doi: 10.1007/978-1-4614-0254-1_6. [DOI] [PubMed] [Google Scholar]
- Robinson E.J., Neal D.E., Collins A.T. Basal cells are progenitors of luminal cells in primary cultures of differentiating human prostatic epithelium. Prostate. 1998;37:149–160. doi: 10.1002/(sici)1097-0045(19981101)37:3<149::aid-pros4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Shin H.M., Kim M.H., Kim B.H., Jung S.H., Kim Y.S., Park H.J., Hong J.T., Min K.R., Kim Y. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Stoyanova T., Goldstein A.S., Cai H., Drake J.M., Huang J., Witte O.N. Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via β-catenin signaling. Genes Dev. 2012;26:2271–2285. doi: 10.1101/gad.196451.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H., Halili M.V., Hu Y.P., Price S.M., Abate-Shen C., Shen M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Bartsch U., Stocking C., Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- Weber K., Thomaschewski M., Warlich M., Volz T., Cornils K., Niebuhr B., Täger M., Lütgehetmann M., Pollok J.M., Stocking C. RGB marking facilitates multicolor clonal cell tracking. Nat. Med. 2011;17:504–509. doi: 10.1038/nm.2338. [DOI] [PubMed] [Google Scholar]
- Wong D.J., Liu H., Ridky T.W., Cassarino D., Segal E., Chang H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.T., Altuwaijri S., Ricke W.A., Huang S.P., Yeh S., Zhang C., Niu Y., Tsai M.Y., Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc. Natl. Acad. Sci. USA. 2007;104:12679–12684. doi: 10.1073/pnas.0704940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Ide H., Kim Y., Dubey P., Witte O.N. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lawson D.A., Witte O.N. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc. Natl. Acad. Sci. USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lukacs R.U., Lawson D.A., Cheng D., Witte O.N. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- Yan Y., Sabharwal P., Rao M., Sockanathan S. The antioxidant enzyme Prdx1 controls neuronal differentiation by thiol-redox-dependent activation of GDE2. Cell. 2009;138:1209–1221. doi: 10.1016/j.cell.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Landmann F., Zahreddine H., Rodriguez D., Koch M., Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- Zhang L., Valdez J.M., Zhang B., Wei L., Chang J., Xin L. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS ONE. 2011;6:e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Wurdak H., Wang J., Lyssiotis C.A., Peters E.C., Cho C.Y., Wu X., Schultz P.G. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4:416–426. doi: 10.1016/j.stem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Zutter M.M., Santoro S.A. Widespread histologic distribution of the alpha 2 beta 1 integrin cell-surface collagen receptor. Am. J. Pathol. 1990;137:113–120. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


