Abstract
North American tree species, subspecies and genetic varieties have primarily evolved in a landscape of extensive continental ice and restricted temperate climate environments. Here, we reconstruct the refugial history of western North American trees since the last glacial maximum using species distribution models, validated against 3571 palaeoecological records. We investigate how modern subspecies structure and genetic diversity corresponds to modelled glacial refugia, based on a meta-analysis of allelic richness and expected heterozygosity for 473 populations of 22 tree species. We find that species with strong genetic differentiation into subspecies had widespread and large glacial refugia, whereas species with restricted refugia show no differentiation among populations and little genetic diversity, despite being common over a wide range of environments today. In addition, a strong relationship between allelic richness and the size of modelled glacial refugia (r2 = 0.55) suggest that population bottlenecks during glacial periods had a pronounced effect on the presence of rare alleles.
Keywords: species distribution models, allozyme, phylogeography, palaeoecology, quaternary, last glacial maximum
1. Introduction
The current Holocene represents one of many relatively short, warm interglacial periods of the Quaternary, while cold temperatures and extensive continental ice have dominated North America for the 2.4 Myr of the Pleistocene. Evolutionary processes leading to geographical differentiation of tree populations, genetic varieties and in some cases subspecies, have therefore taken place in landscape quite different from today [1]. The evolution of distinct genetic varieties or subspecies is often attributed to presumed disjunct populations during glacial periods, where geographical isolation in combination with different selection pressures or genetic drift would allow subspecies to form [1]. Similarly, low levels of modern genetic diversity and particular signatures in allele frequency distributions are attributed to species being subjected to historical periods of restricted population sizes, referred to as genetic bottlenecks, or to founder effects during recolonization [2].
Important evolutionary events and processes that have shaped species' genetic diversity can therefore be better understood in the light of their historical biogeographies, which have traditionally been inferred by two complimentary research approaches. Dated macrofossil and pollen records, largely from lake cores and packrat (Neotoma) middens, have been used by palaeoecologists to infer glacial refugia and reconstruct post-glacial migration routes (e.g. [3,4]). The second approach, phylogeography, infers historical isolation of populations by analysing present-day geographical patterns of neutral genetic markers. Modern genetic population structure can be screened comprehensively with a moderate research effort, but evolutionary events inferred from genetic markers cannot be linked to a specific time and location in the past. Nevertheless, phylogeographic studies allow formulating and testing hypotheses about refugia and migration routes, summarized in several review papers for North America [5–8].
These reviews highlight a number of recurring patterns in the post-glacial vegetation histories of western North American tree species. Several widespread tree species have genetically distinct varieties or subspecies in climatically distinct coastal and interior regions, e.g. Pinus contorta (lodgepole pine), Pinus ponderosa (ponderosa pine), Pseudotsuga menziesii (Douglas fir). These could have emerged through east–west separation of populations by the coastal Cascade Mountains throughout cycles of glaciations and de-glaciation [8–10]. On the other hand, many temperate tree species of the Pacific northwest with disjunct coastal and interior populations show little or no genetic differentiation, e.g. Tsuga mertensiana (mountain hemlock) and Thuja plicata (western red cedar), suggesting post-glacial recolonization from a single refugium [11,12]. Also, refugia west of the continental ice, a Beringian refugia and nunatak refugia (where mountaintops emerge from the ice) have been proposed (e.g. [13]). There is some fossil and genetic evidence that refugia west of the continental ice contained tree species, allowing for rapid recolonization of northern coastal areas [9,10]. Similarly, white spruce and certain other boreal tree species may have found habitat in ice-free Beringia, allowing southward post-glacial recolonization routes [5,6,14].
Genetic and palaeoecological data have previously been interpreted in conjunction to make biogeographic inferences (e.g. [14]), and in recent years a third approach, species distribution modelling, has been increasingly used to complement information from palaeoecological or phylogeographic studies [15–17]. Each approach has different weaknesses with inescapable trade-offs among generality, precision and realism, as has any model [18]. Biogeographic inferences from palaeoecological data offer high precision and realism, but no generality for their paucity of records. Genetic data offer realism and generality in determining putative refugia, but they lack spatial and temporal precision for biogeographic reconstructions. Habitat models offer generality and are spatially and temporally explicit, but they lack realism for a host of assumptions they make. Combining information from several approaches, however, can allow for a more comprehensive interpretation of the available data (e.g. [17,19]).
Species distribution models are based on statistical approaches that correlate species census data with the environments (often just climate conditions) in which they occur. Their interpretation is sometimes difficult, because of assumptions that we know to be false or at least problematic [20]. This includes the assumption that current species are in equilibrium with their climatic niche, that their climatic tolerances remain constant over time, that climate is the main determinant of species distributions, that biotic interactions remain constant and that there is sufficient time for species to attain new equilibrium conditions (if forecasts or hindcasts are interpreted as actual distributions rather than as potential habitat). It has been argued that these assumptions are least problematic for applications at continental scales and long time frames, where local demographic processes and biological interactions that these models ignore may not have noticeable effects [21]. However, long time frames potentially allow for evolutionary processes that compromise niche constancy [3,22].
In this study, we contribute reconstructions of glacial refugia and post-glacial migration histories for 22 western North American forest trees. We build on two previous methodological investigations where we selected and optimized species distribution modelling techniques specifically for this task, and subsequently tested the accuracy and limitations of the chosen techniques with respect to equilibrium issues, niche constancy and no-analogue climates [22,23]. Here, we use ensemble projections from three selected methods for palaeoclimate reconstructions from 6000, 9000, 11 000, 14 000, 16 000 and 21 000 years before the present from two coupled atmospheric-ocean general circulation models (GCMs). Our primary goal is to use habitat reconstructions to augment phylogenetic and palaeoecological data, and provide a third perspective to evaluate the merit of biogeographic hypotheses regarding the existence of glacial refugia in Beringia and along the Pacific coast, the evolution of subspecies in widespread conifers and the origin of Pacific northwest inland rainforests.
Second, we investigate if genetic diversity in neutral genetic markers corresponds to the number, size and contiguity of modelled glacial refugia as expected from population genetic theory. The analysis serves as an assessment of the role of historical biogeography on modern genetic diversity, and could potentially provide an indirect evaluation of the realism of model-based habitat reconstructions at continental scales. As measures of species-wide genetic diversity, we compiled or calculated the number of alleles per locus (A) and expected heterozygosity (He) for 473 populations of 22 tree species from published allozyme marker studies (electronic supplementary material, table S3). Allozyme markers have been superseded by a range of molecular genetic methods, but for this study they provided a consistent method for all tree species for which we also had pollen and fossil data available. Since it is not possible to accurately track habitat of individual populations over time, we do not attempt to explain within-species differences in genetic diversity, but instead conduct a broad species-level analysis, investigating to what extent number and size of glacial refugia or other life history attributes may explain modern genetic diversity in western North American tree species.
2. Material and methods
While most species distribution models are built with species presence–absence or community composition data, we used high-resolution maps of ecosystems to characterize the climate space of a dependent class variable. Ecosystem delineations for the continental USA and Canada west of 100° latitude were compiled from various public data sources (as described in [22,23]). Species frequencies and probabilities of presence were calculated for each ecological region based on 55 743 forest inventory plot records within the ecoregion boundaries (as described in [23]). The ecosystem-based modelling approach was chosen for its high model specificity (i.e. low tendency to overpredict), while maintaining reasonable model sensitivity. This was particularly advantageous when approaching climate combinations that are not well represented in model training data [23].
Modern climate data were generated for the 1961–1990 normal period based on a 1 km resolution digital elevation model with the publically available software package ClimateWNA [24]. Ten climate variables which correlated least were selected via principal component analysis as predictor variables: mean annual precipitation, mean temperature of the warmest month, mean temperature of the coldest month, difference between January and July temperature (continentality), May to September precipitation, number of frost-free days, number of growing degree-days above 5°C, and summer and annual dryness indices. Palaeoclimate data were generated with the delta method, overlaying low-resolution palaeoclimatic GCM hindcasts expressed as anomalies on high-resolution current climate grids. We use hindcasts from the Community Climate Model (CCM1) for the periods 6000, 11 000, 14 000, 16 000 and 21 000 years ago and the Geophysical Fluid Dynamics Laboratory model (GFDL) for the periods 6000, 9000, 14 000, 16 000 and 21 000 years ago (as described in [26,27]).
Ecosystem projections were generated with three species distribution modelling methods that permit a categorical dependent variable: (i) Random Forest, an ensemble classification and regression tree technique, (ii) standard discriminant analysis, and (iii) minimum Mahalanobis distance method [22,23]. Ecosystem projections were made for each time period available in each GCM, and subsequently linked with species probabilities of presence. We produced a single ensemble model output for each tree species by averaging the probability of presence from all model runs. Ensemble modelling methods have been shown to produce more accurate model outputs than individual methods alone [23,25].
Probability of presence projections were validated using the area under the curve of the receiver operating characteristic (AUC) statistic. We used standard cross-validation techniques, where present-day plot data were randomly split into a model training dataset and an independent set for model validation. In addition, we used 3571 fossil pollen, macrofossil and packrat midden records from 835 study sites, compiled from various sources (as described in [23]) for completely independent model validation, with no possibilities of autocorrelations or other dependencies among training and validation data. For standard cross-validations, AUC values for ensemble projections used in this study averaged 0.92, which is considered an excellent model fit. For a breakdown of AUC values by species and individual technique, refer to the electronic supplementary material, table S1 (left column). For completely independent validations against fossil and pollen data, average AUC values for ensemble projections were 0.73 and 0.75 for the CCM1 and GFDL palaeoclimate reconstructions, respectively. Lower values are expected due to model limitations, but also because of the nature of the palaeoecological validation data itself. For example, pollen deposits are restricted to certain landscape features and topographic positions, such as bogs or lakes, where the source of pollen are different ecological habitats in the broader surroundings. Other completely independent model validations (e.g. [26,27]) yield validation statistics in line with our results (electronic supplementary material, table S1).
Data on modern genetic diversity of the tree species, measured as allelic richness and expected heterozygosity, were recorded or calculated from allele frequency tables in the published literature. A complete list of genetic data by population is provided in the electronic supplementary material, table S3. Bias adjustments for small sample sizes were not carried out because bias is expected to be small for genetic markers with low polymorphisms and most studies have sample sizes above 15 (95% of all samples) or above 30 (78% of all samples used here). Furthermore, bias owing to investigators' choices of samples per population would only compromise the analysis if confounded with predictor variables such as modelled size and number of refugia or life-history traits, which seems implausible.
As potential explanatory variables for allelic richness and heterozygosity, we calculated total species range area, total perimeter, the number of discrete polygons and contiguity as the ratio of total area to total perimeter. These metrics were calculated for each individual model run with the SDMTools package for the R programming environment [28], and included all grid cells with a predicted species frequency greater than zero. While we add ice coverage to maps of species hindcasts for illustrative purposes, ice-covered habitat projections were not manually excluded when calculating landscape metrics.
In addition to our species distribution metrics, we assigned life-history traits to species based on descriptions in the USDA Silvics of North America manual [29]. We match the life-history categories used by Hamrick et al. [30], who showed in a large meta-analysis that various life-history traits, including general plant type, range size, dispersal mechanism, pollination vector, breeding system and successional syndrome may influence genetic diversity. Here, for perspective, we include two traits: successional status and seed dispersal mechanism. Life-history traits that did not differ among species in this study were omitted (e.g. reproduction or breeding systems) as were those that were already accounted for in the quantitative landscape metrics for the current species distribution.
To investigate potential explanatory variables for expected heterozygosity and allelic richness, we use a regression tree procedure implemented with the mvpart package for the R programming environment [28]. Regression tree analysis is a recursive variance partitioning method that minimizes within-group variance in one or more dependent variables (heterozygosity or allelic richness). A separate set of categorical or continuous predictor variables (life-history traits, modelled modern ranges, and refugia sizes and counts) are used as criteria to split the dependent dataset [31]. All predictor variables are considered for each recursive split and secondary predictors at each level of the regression tree are reported for alternate interpretations.
3. Results
(a). Reconstruction of glacial refugia
A summary of projected range reconstructions for all species suggest a number of regions where different groups of tree species may have found refuge during the last glacial maximum (figure 1 and table 1). Today's subalpine tree species, such as Abies lasiocarpa (subalpine fir), Pinus albicaulis (whitebark pine) and Picea engelmannii (Engelmann spruce), may have found equivalent climate habitat primarily in the northern basins, from the Williamette Valley in the northwest to the New Mexico plateau in the southeast. Boreal species, such as Picea glauca (white spruce), Picea mariana (black spruce), Pinus contorta (lodgepole pine) and Populus tremuloides (trembling aspen), may have found climate habitat equivalent to today's boreal conditions near the eastern limits of the study area, including the High Plains and southwest Tablelands of Colorado and New Mexico. Species that occur in interior lowlands under relatively dry conditions, including Larix occidentalis (western larch), Pinus ponderosa and Pinus edulis (pinyon pine), may have found suitable habitat in the southern mountain ranges. Coastal species of the Pacific northwest were often restricted to relatively small areas along the California coast during the last glacial maximum. For a species-level breakdown of projected habitat corresponding to the regions in figure 1, refer to the electronic supplementary material, table S2 and figure S1A.
Figure 1.
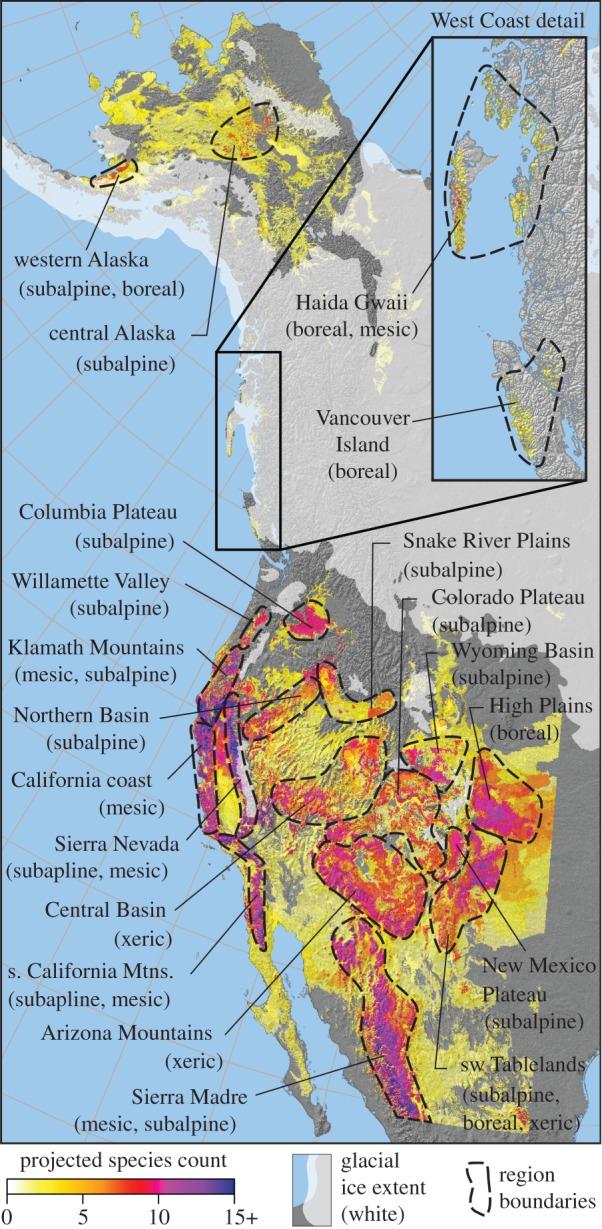
Reconstructed glacial refugia for 22 western North American tree species at the last glacial maximum (21 000 years ago), averaged across two GCM palaeoclimate simulations (GFDL and CCM1). Regions of high species richness have been labelled, and species statistics for each region are provided in the electronic supplementary material, table S2. Table 1 provides equivalent summary statistics by species for broader regions.
Table 1.
List of species included in this study with the representation of individual species in refugia by broad regions to aid interpretation of figure 1. (A detailed breakdown for regions exactly corresponding to figure 1 is provided in the electronic supplementary material, table S2. NC, northern coastal refugia including Alaska, Haida Gwaii and Vancouver Island in figure 1; SC, mesic southern coastal areas including California coast, Klamath Mtns, Sierra Nevada and the s. California Mtns in figure 1; NIB, northern interior basins with subalpine conditions including the Columbia Plateau, Northern Basins, Snake River Plains and Wyoming Basin; SIH, southern interior highlands with xeric conditions including Arizona Mountains, Colorado Plateau, Central Basin, New Mexico Plateau and southwest Tablelands; HP, high plains with boreal conditions; SM, Sierra Madre with mesic and subalpine climate equivalent.)
| location of refugia by species (% of total species refugia) |
|||||||
|---|---|---|---|---|---|---|---|
| no. | species | NC | SC | NIB | SIH | HP | SM |
| 1 | Abies amabilis | 23 | 45 | 26 | 1 | 0 | 5 |
| 2 | Abies lasiocarpa | 12 | 8 | 45 | 27 | 7 | 1 |
| 3 | Abies procera | 0 | 90 | 0 | 3 | 0 | 7 |
| 4 | Acer macrophyllum | 0 | 88 | 0 | 0 | 0 | 12 |
| 5 | Alnus rubra | 0 | 89 | 0 | 3 | 0 | 8 |
| 6 | Calocedrus decurrens | 0 | 76 | 1 | 6 | 0 | 17 |
| 7 | Cupressus nootkatensis | 44 | 45 | 0 | 0 | 0 | 11 |
| 8 | Larix occidentalis | 0 | 29 | 1 | 58 | 0 | 12 |
| 9 | Picea engelmannii | 6 | 4 | 10 | 70 | 8 | 2 |
| 10 | Picea glauca | 33 | 0 | 2 | 18 | 47 | 0 |
| 11 | Picea mariana | 53 | 0 | 1 | 2 | 44 | 0 |
| 12 | Picea sitchensis | 13 | 76 | 1 | 2 | 1 | 7 |
| 13 | Pinus albicaulis | 3 | 25 | 47 | 22 | 3 | 0 |
| 14 | Pinus contorta | 15 | 28 | 17 | 30 | 9 | 1 |
| 15 | Pinus edulis | 0 | 3 | 0 | 46 | 0 | 51 |
| 16 | Pinus monticola | 0 | 80 | 9 | 6 | 0 | 5 |
| 17 | Pinus ponderosa | 0 | 26 | 0 | 56 | 2 | 16 |
| 18 | Populus tremuloides | 5 | 3 | 4 | 64 | 20 | 4 |
| 19 | Pseudotsuga menziesii | 0 | 46 | 5 | 35 | 3 | 11 |
| 20 | Thuja plicata | 13 | 63 | 0 | 15 | 0 | 9 |
| 21 | Tsuga heterophylla | 16 | 65 | 8 | 2 | 0 | 9 |
| 22 | Tsuga mertensiana | 3 | 91 | 1 | 2 | 0 | 3 |
Figure 1 also conveys that much of the land area south of the ice did not serve as refugia for more than a few tree species considered in this study. The low-frequency yellow areas in figure 1 tend to be occupied by species adapted to xeric conditions (e.g. Pinus ponderosa, Pinus edulis and interior populations of Pseudotsuga menziesii). Today's nearest climate equivalents to these regions are open woodlands rather than closed canopy forests. We also find areas of very high species richness (figure 1, purple), where a large majority of all species modelled in this study was predicted to find suitable habitat conditions. This includes the Sierra Nevada, the California Coast Mountains and the Sierra Madre.
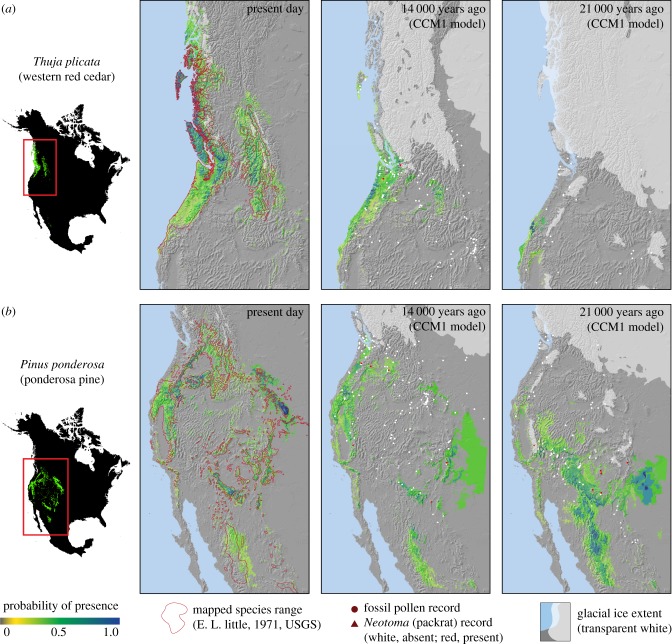
With the exception of Pinus albicaulis, all species had more restricted climate habitats at the last glacial maximum than at present. In particular, coastal species appear to be very restricted compared to their current ranges, as exemplified by Thuja plicata, shown in figure 2a (and in the electronic supplementary material, figure S1T). This pattern is also evident for other species, including Abies amabilis, Abies procera, Alnus rubra, Cupressus nootkatensis and Picea sitchensis (sitka spruce; electronic supplementary material, figures S1A, S1C, S1E, S1G and S1L). Some coastal species also had small simulated refugia west of the continental ice (figure 1, Haida Gwaii and Vancouver Island). Boreal and sub-boreal species were generally least restricted during the last ice age and also have simulated glacial refugia along the northern Pacific coast and in Beringia (figure 1 and table 1). This is exemplified by Picea glauca (electronic supplementary material, figure S1J) but is also visible in glacial ranges of Picea mariana, Pinus contorta and Populus tremuloides (electronic supplementary material, figures S1K, S1N, S1R).
Figure 2.
Maps of projected probability of presence for the present day (1961–1990 normal period) and for the CCM1 climate reconstructions for 14 000 and 21 000 years ago (the last glacial maximum) for (a) Thuja plicata (western red cedar), a mesic species with coastal and interior present-day range and (b) Pinus ponderosa (ponderosa pine), a widespread species in the present day with genetically distinct subspecies. Model projections of all species in all time periods are provided in the electronic supplementary material, figures S1A–S1V.
(b). Species genetic diversity
For species with strong modern genetic differentiation into subspecies and ecotypes, we generally find widespread and diverse refugia. Pinus ponderosa for example, which consists of several subspecies today, had widespread coastal and interior refugia at the last glacial maximum, including the California coast, Arizona Mountains, southwest Tablelands of New Mexico and Colorado, and the Sierra Madre (figure 2b, electronic supplementary material, figure S1Q). For species that today have strong genetic differentiation between coastal and interior populations such as Pinus contorta (var. contorta versus var. latifolia) and Pseudotsuga menziesii (var. menziesii versus var. glauca), we find large glacial refugia east and west of the ice-covered Sierra Nevada (electronic supplementary material, figures S1N and S1S). By contrast, species that today share similar dual interior and coastal distributions but that do not have distinct varieties or subspecies showed only very restricted or no continuous interior habitat. This pattern is exemplified in Thuja plicata, as shown in figure 2a (electronic supplementary material, figure S1T), but can also be seen in Alnus rubra, Cupressus nootkatensis, Pinus monticola, Tsuga heterophylla and Tsuga mertensiana (electronic supplementary material, figures S1E, S1G, S1P, S1U, S1V).
Modern genetic diversity, measured as allelic richness in neutral genetic markers, reveals strong associations with modelled species ranges and number of refugia (the number of discrete areas of contiguous range at the last glacial maximum). In a regression tree analysis, between all the considered life-history traits and landscape metrics, the best explanatory variable for allelic richness is the total area of the projected species range at the last glacial maximum (figure 3a). This can also be visualized with a simple linear regression (figure 4), where Picea glauca and Picea mariana were excluded because their glacial ranges extended much farther east than the area covered by this study (thus underestimating their glacial range size). Additional explanatory variables for allelic richness in the regression tree were contiguity of glacial refugia (a measure of landscape fragmentation) and seed dispersal mechanism. For species with small total refugial area, fragmentation of those refugia was associated with low allelic richness. For species with large glacial refugia, wind-dispersed species had higher allelic richness (figure 3a).
Figure 3.
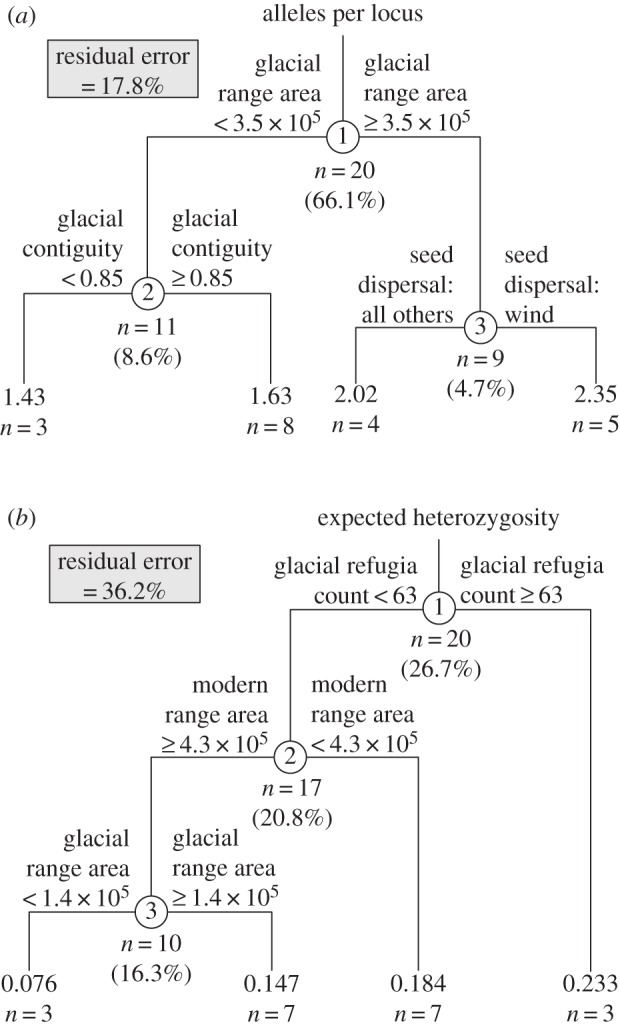
Regression tree analyses of species genetic diversity, as measured by (a) allelic richness and (b) expected heterozygosity. Predictor variables include modern and glacial landscape metrics: total habitat area, contiguity (ratio of perimeter/area) and the number of habitat patches. Also included are two categorical life-history traits: successional stage and seed dispersal mechanism. The number of species in each group (n), and the group mean values for allelic richness or expected heterozygosity are given at terminal nodes. The variance explained by each split is given in parentheses. Alternative criteria for each split are listed in the electronic supplementary material, table S4.
Figure 4.
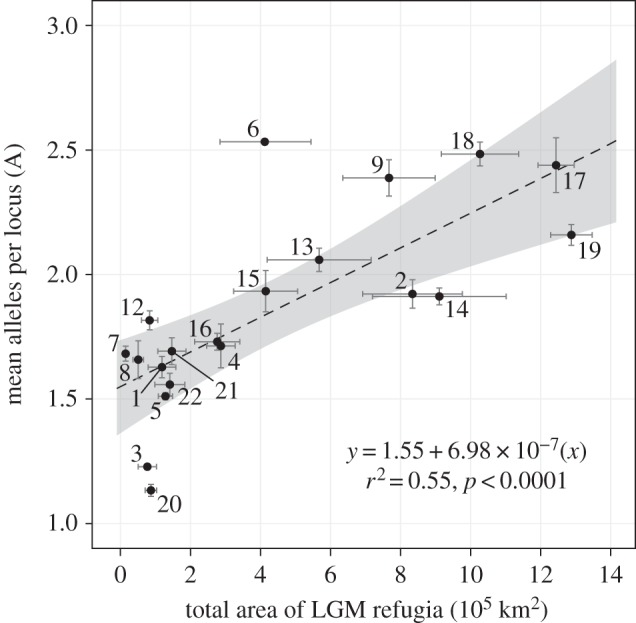
Allelic richness as a function of average modelled species range size at the last glacial maximum. Points represent the average modelled value of six projections based on two GCM-based models of palaeoclimate (CCM1 and GFDL) and three modelling methods. Error bars represent standard errors and the shaded area shows the 95% confidence interval of the regression. Species are annotated by numbers corresponding to table 1.
Landscape metrics and life-history traits accounted for a smaller portion of the variance in expected heterozygosity (figure 3b). The best explanatory variable was the number of glacial refugia (the number of discrete range areas of contiguous range at the last glacial maximum), accounting for 27% of the variance in allozyme heterozygosity in a regression tree analysis. The size of modern and projected glacial range size accounted for another 21% and 16%, respectively. It should be noted that results from regression tree analysis, like all correlative analytical techniques, must consider autocorrelations among predictor variables. Therefore, a comprehensive list of alternative splits for both allelic richness and expected heterozygosity regression trees are provided in the electronic supplementary material, table S4. For the regression tree explaining expected heterozygosity, an almost equally strong explanatory variable in the first split was contiguity of refugia during the last glacial maximum, accounting for 25%, and thus very close to the first choice (27%). The highest expected heterozygosities were therefore associated with high numbers of refugia, greater fragmentation of refugia or both.
4. Discussion
(a). Vegetation history of temperate inland rainforests
Many phylogeographic hypotheses for western North America have focused on the temperate rainforest flora of the Pacific northwest (e.g. [6–8,32]), typically interpreting observations of a north–south genetic division centred in Oregon and inferring the origin of disjunct temperate rainforest communities of northern Idaho and the British Columbia interior. To explain these patterns, three hypotheses were proposed by Brunsfeld et al. [32]. First, the ancient vicariance hypothesis postulates that species were split into coastal and interior refugia, divided by the dry Columbia Basin throughout the Pleistocene. Second, the inland dispersal from the north hypothesis proposes that today's interior ranges were populated post-glacially by migration from refugia in the Cascades to the west. Third, a similar inland dispersal from the south hypothesis posits recolonization of interior distributions from glacial refugia in central Oregon.
Model hindcasts from this study generally support the inland dispersal hypotheses, implying late colonization of inland rainforest communities owing to the lack of stable habitat away from coastal areas. Species with modern disjunct coastal and inland rainforest distributions, e.g. Alnus rubra, Cupressus nootkatensis, Pinus monticola, Thuja plicata, Tsuga heterophylla and Tsuga mertensiana, showed none or very sporadic appearances of suitable interior habitat towards the last glacial maximum (electronic supplementary material, figures S1E, S1G, S1P, S1T, S1U, S1V). This is further supported by fossil data for Tsuga heterophylla and Tsuga mertensiana, which appears in the interior dating back only 3500–4500 years [33,34]. Further, published data suggest little genetic differentiation between coastal and interior tree populations [12]. Plausible paths of recolonization differ among species, with Alnus rubra and Cupressus nootkatensis requiring long-distance dispersal events to reach inland habitats (electronic supplementary material, figure S1E, S1G), Pinus monticola, Thuja plicata, showing connectivity of habitat inland in northern Washington or southern British Columbia (electronic supplementary material, figures S1P, S1T). This path is also supported by the genetic similarity of inland and Washington populations [11]. Finally, Tsuga heterophylla and Tsuga mertensiana show a possible alternative path through northern British Columbia for recolonization of inland refugia by the GFDL model as early as 9000 years ago (electronic supplementary material, figures S1U and S1V).
The inherent limitations of climate reconstructions and additional uncertainties arising from species distribution modelling should make it obvious that modelled inland migration paths should not be over-interpreted at fine scales. They offer an additional continental-scale perspective to build working hypotheses that could be tested by means of palaeoecological or genetic studies. At local or even intermediate scales, where genetic discontinuities among different mountain ranges are discussed (e.g. [35]), or alternate migration paths are hypothesized (e.g. [7]), we find it difficult to make meaningful contributions for lack of temporal resolution and realism in our habitat reconstructions at fine scales.
(b). Widespread trees with subspecies structure
Both Pinus ponderosa and Pseudotsuga menziesii feature present-day coastal and interior subspecies, divided by topographic barriers such as the Sierra Nevada and Cascade ranges [4,36–38]. Their palaeoecological and phylogeographic data also suggest a common history: disjunct southern coastal and interior refugia at the last glacial maximum and allopatric post-glacial migration northward [4,36]. Their vegetation histories as modelled in this study support separate and persistent interior and coastal populations, with the ice-covered Sierra Nevada serving as a topographic barrier during glacial periods (figure 2a, electronic supplementary material, figure S1Q and S1S). Modelled refugia for both species appear in the Sierra Nevada and Klamath Mountains on the coast and through the Arizona Mountains and southwest Tablelands in the interior, locations which are also confirmed by palaeoecological data [4,36]. Neither Pinus ponderosa nor Pseudotsuga menziesii projections suggest stable habitat north or west of the continental ice.
The phylogeography of interior and coastal subspecies of Pinus contorta is somewhat more complex, with genetic data suggesting additional refugia either in Beringia or along the Pacific coast, possibly in the area of Haida Gwaii [9,10]. The potential for refugia west or north of the continental ice is supported in our GFDL model projections in particular, which show extensive Pinus contorta habitat along the Pacific Coast and in Beringia, the most stable of which appears in Haida Gwaii and western Alaska (electronic supplementary material, figure S1N). A northern refugium for Pinus contorta in the Yukon has been suggested by Wheeler et al. [39] but since questioned [9,40]. Notably, present-day Yukon populations exhibit rare alleles shared by both adjacent interior and coastal populations, suggesting dual recolonization paths from the south interior [40] as well as inland dispersal from the west coast to the Yukon via deep fjords and river valleys [9], a pattern supported by our model reconstructions.
(c). Refugia west and north of the ice
Glacial refugia in Beringia and Haida Gwaii may also explain very high calculated post-glacial migration rates, a phenomenon known as ‘Reid's Paradox’ [41]. For example, the expansion of Picea glauca into northern Canada and Alaska evident from the fossil record requires migration rates of 1.5–2.0 km yr−1, necessitating mechanisms such as repeated long-distance dispersal events [14]. However, the existence of a glacial refugium for Picea glauca and Picea mariana in Alaska, for which there is moderate genetic and palaeoecological evidence [14,35,42], reduces these required migration rates. Our hindcasts based on the GFDL model suggest possible refugia in Beringia both for Picea glauca and Picea mariana (electronic supplementary material, figures S1J and S1K). Other species where both GFDL and CCM1 models suggest refugial habitat in Beringia include Populus tremuloides, and Pinus contorta (electronic supplementary material, figures S1R and S1N), as discussed above. Similar northern refugia have been proposed in other high-latitude regions such as central and northern Europe, based on palaeoecological or genetic data (e.g. [43]) supported by modelling efforts similar to those presented here [44].
Because Arctic oceanic currents were blocked from entering the Pacific by the Beringian land bridge at the last glacial maximum, climate conditions along the Pacific coast were relatively mild. Lower relative sea levels may have allowed for refugia west of the continental ice that could have sustained tree populations in areas around Vancouver Island, Haida Gwaii and the Alaskan Panhandle, including areas now submerged, which are not considered in our analysis. Conclusive palaeoecological evidence of glacial tree populations in the Haida Gwaii area is still lacking, with the early macrofossil evidence for Pinus contorta and Picea sitchensis dating to ca 13 000 and 11 400 years ago, respectively [45]. However, allelic richness of modern Haida Gwaii populations of several species is relatively high, suggesting that these populations were not established from repeated long-distance dispersal events [6,39,46].
Our model hindcasts suggest suitable habitat along the Pacific coast, notably in Haida Gwaii, for many temperate rainforest species including Thuja plicata, Tsuga heterophylla, Tsuga mertensiana, Cupressus nootkatensis, Abies amabilis, Picea sitchensis and Alnus rubra. However, the weak representation in the fossil and pollen record of tree species at the last glacial maximum should serve as a note of caution. Furthermore, Abies amabilis, Picea sitchensis, Thuja plicata, Tsuga heterophylla and Tsuga mertensiana lack genetic evidence of cryptic northern refugia [12,47,48].
(d). Drivers of modern genetic diversity
We find surprisingly strong relationships between modern genetic diversity and landscape metrics that describe species distributions during the last glacial maximum, complementing analysis of life-history traits as drivers of genetic diversity by Hamrick et al. [30]. Our observation that allelic richness is better explained than expected heterozygosity by past vegetation history corresponds to previous studies (e.g. [49]) and fits population genetic theory. Rare alleles, which strongly influence measures of allelic richness, are likely to be lost in population bottlenecks [2] that most probably occurred at the last glacial maximum. By contrast, rare alleles should contribute little to expected heterozygosity, which is instead driven by the frequency and evenness of common alleles that are not readily lost even in very small populations [50]. Instead, large numbers of disjunct refugia that have persisted over long periods of time during the Pleistocene should favour genetic differentiation of past populations and high levels of expected heterozygosity in modern populations, as we found in this analysis (figure 3b).
Our observation of modelled refugial history relating to measures of modern genetic diversity as expected by population genetic theory could also be interpreted as an indicator that both GCMs and species distribution models approach a sufficient degree of realism to establish such continental-scale relationships. We must be careful, however, not to over-interpret habitat reconstructions at local scales. For example, fossil and genetic data suggest coastal populations of Pseudotsuga menziesii (electronic supplementary material, figure S1S) were north of the modelled distribution at the last glacial maximum [4,38]. This may reflect that the species has not migrated as far north as its climatic niche space would allow, and hence hind-casting their realized niche would cause under-predictions in this area. It is therefore essential to draw on multiple sources of data for biogeographic reconstructions. In the electronic supplementary material, figures S1A to S1V, we provide mapped species distributions in conjunction with habitat projections for current climate, as well as fossil records overlaid on hindcasts from two GCMs to provide a visual sense of uncertainty in addition to the statistical evaluations in the electronic supplementary material, table S1.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We also thank the anonymous reviewers whose critical feedback improved the quality of this manuscript.
Funding statement
Funding for this study was provided by the Canadian Natural Sciences and Engineering Research Council (NSERC) Discovery Grant RGPIN-330527–07 through the Government of Canada and the Alberta Ingenuity grant no. 200500661 through the Government of Alberta. Support for D.R.R. was provided by NSERC and the Alberta Innovates Technology Futures program.
References
- 1.Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. Lond. B 359, 183–195. ( 10.1098/rstb.2003.1388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leberg PL. 1992. Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 46, 477–494. ( 10.2307/2409866) [DOI] [PubMed] [Google Scholar]
- 3.Davis MB, Shaw RG. 2001. Range shifts and adaptive responses to Quaternary climate change. Science 292, 673–679. ( 10.1126/science.292.5517.673) [DOI] [PubMed] [Google Scholar]
- 4.Gugger PF, Sugita S. 2010. Glacial populations and postglacial migration of Douglas-fir based on fossil pollen and macrofossil evidence. Q. Sci. Rev. 29, 2052–2070. ( 10.1016/j.quascirev.2010.04.022) [DOI] [Google Scholar]
- 5.Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bousquet J. 2009. Inferring the past from the present phylogeographic structure of North American forest trees: seeing the forest for the genes. Can. J. Forest Res. 39, 286–307. ( 10.1139/X08-181) [DOI] [Google Scholar]
- 6.Shafer ABA, Cullingham CI, Cote SD, Coltman DW. 2010. Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 19, 4589–4621. ( 10.1111/j.1365-294X.2010.04828.x) [DOI] [PubMed] [Google Scholar]
- 7.Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS. 1997. Chloroplast DNA intraspecific phylogeography of plants from the Pacific northwest of North America. Plant Syst. Evol. 206, 353–373. ( 10.1007/BF00987957) [DOI] [Google Scholar]
- 8.Carstens BC, Brunsfeld SJ, Demboski JR, Good JM, Sullivan J. 2005. Investigating the evolutionary history of the Pacific northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution 59, 1639–1652. ( 10.1554/04-661.1) [DOI] [PubMed] [Google Scholar]
- 9.Fazekas AJ, Yeh FC. 2006. Postglacial colonization and population genetic relationships in the Pinus contorta complex. Can. J. Bot. 84, 223–234. ( 10.1139/B05-150) [DOI] [Google Scholar]
- 10.Godbout J, Fazekas A, Newton CH, Yeh FC, Bousquet J. 2008. Glacial vicariance in the Pacific northwest: evidence from a lodgepole pine mitochondrial DNA minisatellite for multiple genetically distinct and widely separated refugia. Mol. Ecol. 17, 2463–2475. ( 10.1111/j.1365-294X.2008.03761.x) [DOI] [PubMed] [Google Scholar]
- 11.O'Connell LM, Ritland K, Thompson SL. 2008. Patterns of post-glacial colonization by western red cedar (Thuja plicata, Cupressaceae) as revealed by microsatellite markers. Botany 86, 194–203. ( 10.1139/B07-124) [DOI] [Google Scholar]
- 12.Ally D, El-Kassaby YA, Ritland K. 2000. Genetic diversity, differentiation and mating systems in mountain hemlock (Tsuga mertensia) across British Columbia. Forest Genet. 7, 97–108. [Google Scholar]
- 13.Provan J, Bennett KD. 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571. ( 10.1016/j.tree.2008.06.010) [DOI] [PubMed] [Google Scholar]
- 14.Anderson LL, Hu FS, Nelson DM, Petit RJ, Paige KN. 2006. Ice-age endurance: DNA evidence of a white spruce refugium in Alaska. Proc. Natl Acad. Sci. USA 103, 12 447–12 450. ( 10.1073/pnas.0605310103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carstens BC, Richards CL. 2007. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution 61, 1439–1454. ( 10.1111/j.1558-5646.2007.00117.x) [DOI] [PubMed] [Google Scholar]
- 16.Waltari E, Hijmans RJ, Peterson AT, Nyari AS, Perkins SL, Guralnick RP. 2007. Locating Pleistocene refugia: comparing phylogeographic and ecological niche model predictions. PLoS ONE 2, e0000563 ( 10.1371/journal.pone.0000563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metcalf JL, Prost S, Nogués-Bravo D, DeChaine EG, Anderson C, Batra P, Araújo MB, Cooper A, Guralnick RP. 2014. Integrating multiple lines of evidence into historical biogeography hypothesis testing: a Bison bison case study. Proc. R. Soc. B 281, 20132782 ( 10.1098/rspb.2013.2782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levins R. 1966. Strategy of model building in population biology. Am. Sci. 54, 421–431. [Google Scholar]
- 19.Gavin DG, et al. 2014. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytol. 204, 37–54. ( 10.1111/Nph.12929) [DOI] [PubMed] [Google Scholar]
- 20.Araújo MB, Guisan A. 2006. Five (or so) challenges for species distribution modelling. J. Biogeogr. 33, 1677–1688. ( 10.1111/j.1365-2699.2006.01584.x) [DOI] [Google Scholar]
- 21.Pearson RG, Dawson TP. 2003. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 12, 361–371. ( 10.1046/j.1466-822X.2003.00042.x) [DOI] [Google Scholar]
- 22.Roberts DR, Hamann A. 2012. Predicting potential climate change impacts with bioclimate envelope models: a palaeoecological perspective. Glob. Ecol. Biogeogr. 21, 121–133. ( 10.1111/j.1466-8238.2011.00657.x) [DOI] [Google Scholar]
- 23.Roberts DR, Hamann A. 2012. Method selection for species distribution modelling: are temporally or spatially independent evaluations necessary? Ecography 35, 792–802. ( 10.1111/j.1600-0587.2011.07147.x) [DOI] [Google Scholar]
- 24.Wang TL, Hamann A, Spittlehouse DL, Murdock TQ. 2012. ClimateWNA: high-resolution spatial climate data for western North America. J. Appl. Meteorol. Climatol. 51, 16–29. ( 10.1175/Jamc-D-11-043.1) [DOI] [Google Scholar]
- 25.Araújo MB, New M. 2007. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47. ( 10.1016/j.tree.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 26.Eskildsen A, le Roux PC, Heikkinen RK, Hoye TT, Kissling WD, Poyry J, Wisz MS, Luoto M. 2013. Testing species distribution models across space and time: high latitude butterflies and recent warming. Glob. Ecol. Biogeogr. 22, 1293–1303. ( 10.1111/Geb.12078) [DOI] [Google Scholar]
- 27.Heikkinen RK, Marmion M, Luoto M. 2012. Does the interpolation accuracy of species distribution models come at the expense of transferability? Ecography 35, 276–288. ( 10.1111/j.1600-0587.2011.06999.x) [DOI] [Google Scholar]
- 28.R Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
- 29.Burns RM, Honkala H, Coords T. 1990. Silvics of North America: 1. Conifers; 2. Hardwoods. In Agricultural Handbook 654, vol. 2, p. 877 Washington, DC: US. Department of Agriculture, Forest Service; See http://www.na.fs.fed.us/spfo/pubs/silvics_manual/table_of_contents.htm. [Google Scholar]
- 30.Hamrick JL, Godt MJW, Shermanbroyles SL. 1992. Factors influencing levels of genetic diversity in woody plant species. Popul. Genet. For. Trees 42, 95–124. ( 10.1007/978-94-011-2815-5_7) [DOI] [Google Scholar]
- 31.De'ath G, Fabricius KE. 2000. Classification and regression trees: a powerful yet simple technique for ecological data analysis. Ecology 81, 3178–3192. ( 10.1890/0012-9658(2000)081[3178:CARTAP]2.0.CO;2) [DOI] [Google Scholar]
- 32.Brunsfeld SJ, Sullivan J, Soltis DE, Soltis PS. 2001. Comparative phylogeography of northwestern North America: a synthesis. In Integrating ecological and evolutionary processes in a spatial context (eds Silvertown J, Antovics J.), pp. 319–339. Oxford, UK: Blackwell Science. [Google Scholar]
- 33.Mehringer PJ. 1996. Columbia River Basin Ecosystems: Late Quaternary environments. In interior Columbia Basin ecosystem management project. Pullman, WA: Department of Anthropology and Geology, Washington State University. [Google Scholar]
- 34.Rosenberg SM, Walker IR, Mathewes RW. 2003. Postglacial spread of hemlock (Tsuga) and vegetation history in Mount Revelstoke National Park, British Columbia, Canada. Can. J. Bot. 81, 139–151. ( 10.1139/b03-015) [DOI] [Google Scholar]
- 35.Jaramillo-Correa JP, Beaulieu J, Bousquet J. 2004. Variation in mitochondrial DNA reveals multiple distant glacial refugia in black spruce (Picea mariana), a transcontinental North American conifer. Mol. Ecol. 13, 2735–2747. ( 10.1111/j.1365-294X.2004.02258.x) [DOI] [PubMed] [Google Scholar]
- 36.Norris JR, Jackson ST, Betancourt JL. 2006. Classification tree and minimum-volume ellipsoid analyses of the distribution of ponderosa pine in the western USA. J. Biogeogr. 33, 342–360. ( 10.1111/j.1365-2699.2005.01396.x) [DOI] [Google Scholar]
- 37.Li P, Adams WT. 1989. Range-wide patterns of allozyme variation in Douglas-fir (Pseudotsuga menziesii). Can. J. Res. 19, 149–161. ( 10.1139/x89-022) [DOI] [Google Scholar]
- 38.Gugger PF, Sugita S, Cavender-Bares J. 2010. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: testing hypotheses from the fossil record. Mol. Ecol. 19, 1877–1897. ( 10.1111/j.1365-294X.2010.04622.x) [DOI] [PubMed] [Google Scholar]
- 39.Wheeler NC, Guries RP. 1982. Population structure, genic diversity, and morphological variation in Pinus contorta Dougl. Can. J. Forest Res. 12, 595–606. ( 10.1139/x82-091) [DOI] [Google Scholar]
- 40.Cwynar LC, Macdonald GM. 1987. Geographical variation of lodgepole pine in relation to population history. Am. Nat. 129, 463–469. ( 10.1086/284651) [DOI] [Google Scholar]
- 41.Clark JS, et al. 1998. Reid's paradox of rapid plant migration: dispersal theory and interpretation of paleoecological records. Bioscience 48, 13–24. ( 10.2307/1313224) [DOI] [Google Scholar]
- 42.Brubaker LB, Anderson PM, Edwards ME, Lozhkin AV. 2005. Beringia as a glacial refugium for boreal trees and shrubs: new perspectives from mapped pollen data. J. Biogeogr. 32, 833–848. ( 10.1111/j.1365-2699.2004.01203.x) [DOI] [Google Scholar]
- 43.Parducci L, et al. 2012. Glacial survival of boreal trees in northern Scandinavia. Science 335, 1083–1086. ( 10.1126/science.1216043) [DOI] [PubMed] [Google Scholar]
- 44.Svenning JC, Normand S, Kageyama M. 2008. Glacial refugia of temperate trees in Europe: insights from species distribution modelling. J. Ecol. 96, 1117–1127. ( 10.1111/j.1365-2745.2008.01422.x) [DOI] [Google Scholar]
- 45.Lacourse T, Mathewes RW, Fedje DW. 2005. Late-glacial vegetation dynamics of the Queen Charlotte Islands and adjacent continental shelf, British Columbia, Canada. Palaeogeogr. Palaeoclim. 226, 36–57. ( 10.1016/j.palaeo.2005.05.003) [DOI] [Google Scholar]
- 46.Wellman HF. 2004. The genetics of selective breeding in western hemlock (Tsuga heterophylla). Vancouver, BC: University of British Columbia. [Google Scholar]
- 47.Yeh FC, Hu XS. 2005. Genetic structure and migration from mainland to island populations in Abies procera Rehd. Genome 48, 461–473. ( 10.1139/g04-127) [DOI] [PubMed] [Google Scholar]
- 48.Holliday JA, Yuen M, Ritland K, Aitken SN. 2010. Postglacial history of a widespread conifer produces inverse clines in selective neutrality tests. Mol. Ecol. 19, 3857–3864. ( 10.1111/j.1365-294X.2010.04767.x) [DOI] [PubMed] [Google Scholar]
- 49.Comps B, Gomory D, Letouzey J, Thiebaut B, Petit RJ. 2001. Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nei M, Maruyama T, Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10. ( 10.2307/2407137) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.