Abstract
Scorpions represent an iconic lineage of arthropods, historically renowned for their unique bauplan, ancient fossil record and venom potency. Yet, higher level relationships of scorpions, based exclusively on morphology, remain virtually untested, and no multilocus molecular phylogeny has been deployed heretofore towards assessing the basal tree topology. We applied a phylogenomic assessment to resolve scorpion phylogeny, for the first time, to our knowledge, sampling extensive molecular sequence data from all superfamilies and examining basal relationships with up to 5025 genes. Analyses of supermatrices as well as species tree approaches converged upon a robust basal topology of scorpions that is entirely at odds with traditional systematics and controverts previous understanding of scorpion evolutionary history. All analyses unanimously support a single origin of katoikogenic development, a form of parental investment wherein embryos are nurtured by direct connections to the parent's digestive system. Based on the phylogeny obtained herein, we propose the following systematic emendations: Caraboctonidae is transferred to Chactoidea new superfamilial assignment; superfamily Bothriuroidea revalidated is resurrected and Bothriuridae transferred therein; and Chaerilida and Pseudochactida are synonymized with Buthida new parvordinal synonymies.
Keywords: Arthropoda, arachnids, missing data, paralogy, relict, transcriptomics
“All scorpions look generally alike.”
Gary Allan Polis (1990)
1. Introduction
The evolutionary origins of scorpions (order Scorpiones), one of the most recognizable and charismatic groups of arthropods, have long been shrouded in mystery and engulfed by dispute. Scorpions first appeared in the Silurian and are represented extensively in the Palaeozoic fossil record [1–3], with ca 2000 described extant species surviving to the present day [4]. The relationship between scorpions and Eurypterida, the extinct group referred to as ‘sea scorpions', has been a matter of historical debate [5–10]. The placement of scorpions within Arachnida was also considered controversial [9–13], but recent phylogenomic studies have favoured the Arachnopulmonata hypothesis: the sister group relationship of scorpions and tetrapulmonates (i.e. spiders and allied orders) [14,15].
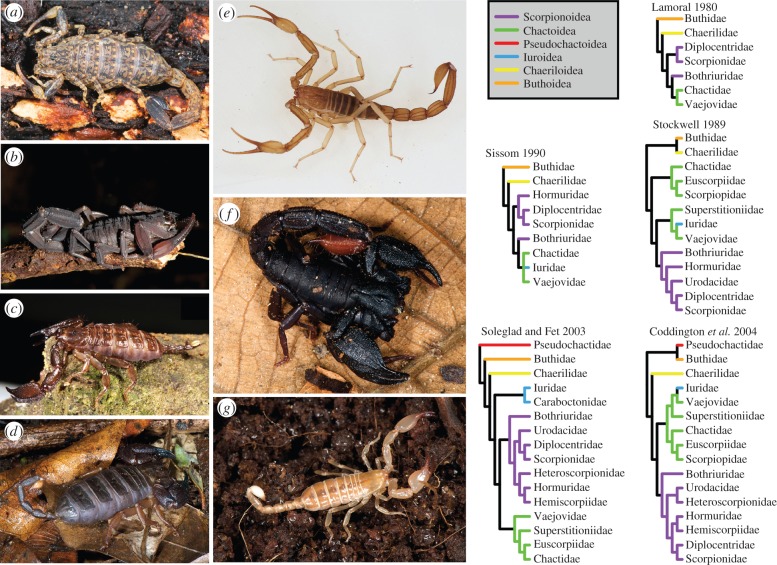
Paralleling the placement of scorpions among the arachnid orders, the internal phylogeny of scorpions has also long been contentious [16–22]. Numerous workers have historically emphasized different morphological characters of scorpions, resulting in variable hypotheses of phylogeny (figure 1), none of which has been generally accepted [21,22]. Traditionally, scorpions are divided into two morphologically distinct groups, the family Buthidae and the non-buthids [16,19,23]. The phylogenetic position of the relictual families Chaerilidae and Pseudochactidae, which share characters with both groups, has been particularly debated [16,19,23,24].
Figure 1.
Left: (a–g) exemplars of scorpion diversity. (a) Isometrus sp. (Buthidae); (b) Tityus obscurus (Buthidae); (c) Liocheles australasiae (Hormuridae); (d) Opisthacanthus cf. asper (Hormuridae); (e) Vietbocap lao (Pseudochactidae); (f) Brotheas granulatus (Chactidae); (g) Belisarius xambeui (Troglotayosicidae). Right: historical hypotheses of scorpion relationships. Colours in tree topologies correspond to superfamilies, as designated by Soleglad & Fet [20]. Photos: G. Giribet. (Online version in colour.)
All previous inferences of scorpion higher level relationships (and ensuing disputes) have been grounded in morphological characters, whose use may be limited in a group that exemplifies morphological stasis [22,24]. Despite widespread use of molecular sequence data for phylogenetic reconstruction, a scorpion phylogeny based on molecular data has yet to be proposed. Use of molecular sequence data from one to three loci is currently limited to analyses of individual families [25,26], or a subset of families in the absence of outgroup taxa [20]. The lack of a reliable phylogeny for scorpions is a major impediment for evolutionary studies of morphology and genomics. For instance, despite significant research efforts on scorpion venoms by means of estimating species trees (EST), transcriptomic or genomic sequencing approaches, conclusions pertaining to the evolution of toxins remain tenuous due to an unknown underlying species tree (e.g. [27–29]). This also holds for investigations of various morphological characters [30–32].
The high gene content exhibited by the first sequenced scorpion genome is suggestive of extensive gene family turnover and duplication events [28], a discovery paralleled by evidence of retention and neofunctionalization of various paralogues early in scorpion evolution [33]. In order to leverage the high gene content of scorpion genomes for testing phylogenetic relationships, we sequenced full, strand-specific transcriptomes from all extant scorpion superfamilies. We present here, to the best of our knowledge, the first complete higher level molecular phylogeny of scorpions.
2. Material and methods
Methods are described in greater detail with full references in the electronic supplementary material.
(a). Species sampling and molecular techniques
Paired-end (150 bp) transcriptomes were generated for 25 scorpion and one pseudoscorpion species. Additional datasets used as outgroups were obtained from a previous study [15] or from GenBank. Collecting locality information, statistics on sequencing yields and accession numbers are provided as the electronic supplementary material, table S1. All extant families of scorpions were sampled, except for Hemiscorpiidae and Heteroscorpionidae (previously considered sister to, or part of, Hormuridae and Urodacidae, respectively [20,34]), and Typhlochactidae (considered part of or sister to Superstitioniidae; [20,35]). Tissue preservation and RNA sequencing are as described by Sharma et al. [15]. All sequenced libraries are accessioned in the Sequence Read Archive. Other materials (described below) are deposited in the Dryad Digital Repository (doi:10.5061/dryad.n0qr5).
(b). Sequence assembly and orthology assignment
Quality filtering, trimming of reads and strand-specific transcriptomic assemblies were conducted as described in the electronic supplementary material. Predicted open reading frames (ORFs) were assigned to orthologous groups using the Orthologous MAtrix (OMA) algorithm, (OMA stand-alone v.0.99u; [36,37], which has been shown to outperform alternative approaches towards identification of true orthologues and to minimize type I error in orthology assignment [38]. Additional scorpion taxa not sequenced by us (electronic supplementary material, table S1) were obtained from GenBank. For Sanger-sequenced EST and 454 libraries, redundancy reduction was done with CD-HIT as described in the electronic supplementary material. Owing to the small size of additional datasets and/or the quality of the genome of Mesobuthus martensii, predicted ORFs were assigned to orthologous groups using OMA in two separate runs, one for Buthidae (Iurus, Chaerilus and the pseudochactids used as outgroups; taxon occupancy criterion set to representation in at least 19 taxa) and a second for ‘Chactoidea’ and Scorpionoidea sensu stricto (Iurus and Bothriurus used as outgroups; taxon occupancy criterion set to representation in at least 16 taxa). This was done for computational expediency as well as to ensure representation of the smallest libraries in supermatrices.
(c). Phylogenomic analyses of supermatrices
In order to discern the potential effects of several confounding factors in phylogenomic reconstruction, several supermatrices were constructed: (i) according to gene occupancy (Matrices 1–4), (ii) retaining only orthogroups with demonstrable compositional homogeneity (Matrix 5), (iii) with algorithmic matrix reduction ([39], see also [40]; http://mare.zfmk.de) (Matrices 6–7), (iv) by tertiles of per cent pairwise similarity (Matrices 8–10), and (v) by retaining only verified single-copy orthologues common to Arthropoda (Matrices 11–14).
In order to explore the trade-off between number of genes and matrix completeness, four supermatrices were constructed by varying gene occupancy threshold (Matrices 1–4 containing 136, 599, 1557 and 5025 genes, respectively; figure 2a). Alignment and masking of ambiguously aligned positions were conducted as described in the electronic supplementary material.
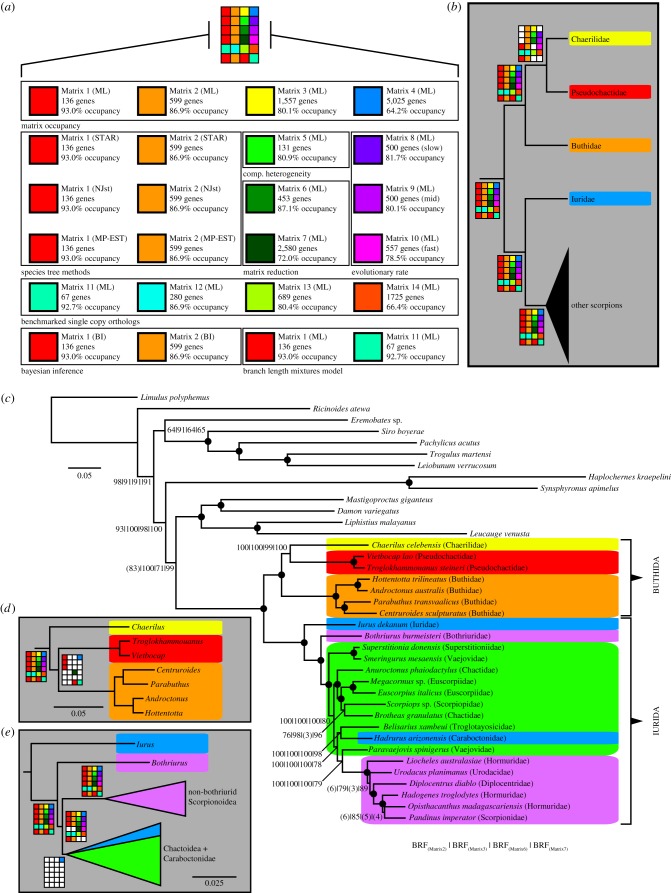
Figure 2.
Phylogenomic analyses of scorpions. (a) Supermatrix composition with numbers of genes and gene occupancies, and accompanying methods. (b) Basal topology of Scorpiones. Shaded squares in Navajo plots indicate recovery of a given node in the corresponding analysis. (c) Tree topology of Scorpiones inferred from ML analysis of 1557 gene matrix. Numbers on nodes indicate bootstrap resampling frequencies from ML analyses of Matrices 2, 3, 6 and 7. Black circles indicate maximal nodal support frequencies for all four matrices. (d) Alternative topology at the base of Buthida obtained in seven analyses. (e) Alternative topology within Iurida obtained in one analysis. Shading corresponds to superfamilies (as in figure 1). (Online version in colour.)
To assess compositional heterogeneity, we analysed each orthogroup in the 1557 gene dataset using BaCoCa v. 1.1 [41]. A supermatrix was constructed by retaining only the 131 most compositionally homogeneous orthogroups, defined as having a relative composition frequency variability value below 0.05. Biases stemming from compositional heterogeneity were thus minimized in this 131 gene supermatrix (Matrix 5).
To implement an algorithmic approach to matrix reduction, we used the MAtrix REduction (MARE) method, which estimates informativeness of every orthogroup based on weighted geometry quartette mapping [39]. Reduction of the 599 gene matrix (Matrix 2) resulted in the retention of 453 orthogroups (Matrix 6), and reduction of the 5025 gene matrix (Matrix 4) in the retention of 2580 orthogroups (Matrix 7).
To assess the possibility that evolutionary rate may be conflated with phylogenetic signal, we created three additional supermatrices by dividing orthogroups of the 1557 gene supermatrix (chosen for balance between dataset size and matrix occupancy) approximately into tertiles, using per cent pairwise identity as a proxy for evolutionary rate (Matrix 8 containing the 500 slowest evolving genes, Matrix 9 the 500 genes of intermediate rate and Matrix 10 the 557 fastest evolving genes). In addition, we trialled tree topologies from supermatrices upon culling fast-evolving sites using TIGER v. 1.02 [42], but we observed major loss of phylogenetic signal upon removing sites ranked in one or more of the fastest evolving bins (of 10 equally sized bins), yielding a basal polytomy for two different matrices and the non-monophyly of scorpions. Those analyses are not included in this study, but are available upon request.
To assess the possibility of incorrect topologies stemming from inadvertent inclusion of paralogues, we identified in Matrices 1–4 all tick (Ixodes scapularis) orthologues that were found to occur in single-copy across Arthropoda, as identified in the BUSCO-Ar database of OrthoDB [43]. The intersection of BUSCO-Ar orthologues and Matrices 1–4 constituted the basis for Matrices 11–14, respectively (figure 2a).
To account for rate heterogeneity, particularly in pseudoscorpion outgroups [15], analyses of all supermatrices incorporated mixture models (CAT + LG4XF or CAT + GTR; [44,45]) in both maximum-likelihood (ML) and Bayesian inference (BI) analyses, as detailed in the electronic supplementary material. RAxML v. 7.7.5 [46] and PhyloBayes MPI v. 1.4f [47] were used for ML and BI analyses, respectively.
To account for heterotachy, we implemented ML analyses with mixed branch length models [48] using PhyML + M3L [48–50] for our most complete matrices (Matrix 1 and Matrix 11; sequence occupancy more than 90%), with four branch length mixtures. Heuristic details are provided in the electronic supplementary material.
(d). Phylogenomic analyses of gene trees
As concatenation methods can mask phylogenetic conflict when strong gene tree incongruence is incident, we conducted species tree approaches on best-scoring ML gene tree topologies of constituent orthogroups of the most complete datasets, Matrix 1 (136 genes; 93.0% occupancy) and Matrix 2 (599 genes; 86.9% occupancy). To examine incongruence of constituent genes, we inferred best-scoring ML gene trees for all orthogroups included in these supermatrices. Species trees were estimated from partial gene trees using three partially parametric methods: STAR [51], MP-EST [52] and NJst [53].
To quantify levels of gene tree incongruence, we calculated for every node in Matrices 3 and 4, the available number of potentially informative gene trees (i.e. trees containing at least one member of each descendant branch and two distinct outgroups) and the number of gene trees congruent with those nodes [54]. We mapped these quantities both for the concatenated ML topology recovered by Matrix 3, as well as for alternative topological hypotheses corresponding to traditional systematic relationships.
Following a supernetwork approach, gene trees were decomposed into quartettes using SuperQ v. 1.1 [55], and a supernetwork assigning edge lengths based on quartette frequencies was inferred selecting the ‘balanced’ edge-weight optimization function.
3. Results
(a). Supermatrix approaches
Our analyses of multiple data matrices (figure 2a) result in a well-supported tree topology of scorpions that is greatly incongruent with traditional morphological hypotheses. All analyses yielded a grouping of Buthidae with Chaerilidae (the putative sister group of the remaining Iurida (iuroids, scorpionoids and chactoids) [19,20,23]) and Pseudochactidae (the putative sister group of all other scorpions [20]) (figure 2b,c). To facilitate discourse, we henceforth refer to this trio of families as the revised Buthida and synonymize parvorders Chaerilida and Pseudochactida with Buthida new synonymies (table 1). The family Chaerilidae was either recovered as sister group to the clade (Pseudochactidae + Buthidae) (figure 2d) or, in the majority of analyses, as sister group to Pseudochactidae (Matrices 2 and 3; figure 2b), indicating some discordance at the base of Buthida.
Table 1.
Revised higher level classification of extant scorpions. (Taxa of questionable monophyly are indicated with asterisks. Taxa of untested phylogenetic placement using molecular sequence data are indicated with question marks.)
| Order Scorpiones Koch, 1837 |
| Suborder Neoscorpionina Thorell & Lindström, 1885 |
| Infraorder Orthosterni Pocock, 1911 |
| Parvorder Buthida Soleglad & Fet 2003 |
| Superfamily Buthoidea Koch, 1837 Family Buthidae Koch, 1837 |
| Superfamily Chaeriloidea Pocock, 1893 new parvordinal assignment Family Chaerilidae Pocock, 1893 |
| Superfamily Pseudochactoidea Gromov, 1998 new parvordinal assignment Family Pseudochactidae Gromov, 1998 |
| Parvorder Iurida Soleglad & Fet 2003 |
| Superfamily Iuroidea Thorell, 1876 Family Iuridae Thorell, 1876 |
| Superfamily Bothriuroidea Simon, 1880 revalidated Family Bothriuridae Simon, 1880 |
| Superfamily *Chactoidea Pocock, 1893 Family Caraboctonidae Kraepelin, 1905 new superfamilial assignment Family *Chactidae Pocock, 1893 Family Euscorpiidae Laurie, 1896 Family Scorpiopidae Kraepelin, 1905 Family Superstitioniidae Stahnke, 1940 Family Troglotayosicidae Lourenço, 1998 Family ?Typhlochactidae Mitchell, 1971 Family *Vaejovidae Thorell, 1876 |
| Superfamily Scorpionoidea Latreille, 1802 Family Diplocentridae Karsch, 1880 Family ?Hemiscorpiidae Pocock, 1893 Family ?Heteroscorpionidae Kraepelin, 1905 Family *Hormuridae Laurie, 1896 Family *Scorpionidae Latreille, 1802 Family Urodacidae Pocock, 1893 |
Every analysis obtained a basal split between Buthida and Iurida (Iuridae + the remaining scorpions). All analyses refuted the monophyly of Iuroidea (diphyletic), Scorpionoidea (diphyletic) and Chactoidea (paraphyletic or polyphyletic), as well as the monophyly of three diverse families—Chactidae, Hormuridae and Vaejovidae—with maximal nodal support (figure 2b,c; electronic supplementary material, figures S1–S20). A single analysis (Matrix 4; 5025 genes) nearly recovered the monophyly of Chactoidea, albeit with a nested inclusion of Caraboctonidae (a member of the superfamily Iuroidea) (figure 2e; electronic supplementary material, figure S4). This anomalous tree topology is discussed in detail below.
The placement of Iurus dekanum and Bothriurus burmeisteri in a grade at the base of the remaining Scorpionoidea and Chactoidea (including Caraboctonidae) was nearly invariable (figure 2b,e). For this reason, we transfer Caraboctonidae to Chactoidea new superfamilial assignment, and the family Bothriuridae to the resurrected superfamily Bothriuroidea revalidated (table 1).
The uniformity of these results indicates that the trade-off between missing data and number of genes analysed does not have a major effect on basal phylogenetic resolution in this case. Accordingly, the distribution of gene representation was nearly uniform for all supermatrices analysed, particularly for ingroup terminals (electronic supplementary material, table S2). The ingroup basal topology yielded by the smaller matrix (Matrix 1) was similar to that of the 1557 gene supermatrix (Matrix 3; electronic supplementary material, figure S3), with minor topological differences among the chactoid lineages and within Buthida.
The basal topology yielded by the 131 gene matrix comprising the most compositionally homogeneous orthogroups (Matrix 5) was identical to that of the 1557 gene supermatrix (electronic supplementary material, figure S7); only a pair of topological differences was observed within the chactoid lineages. An algorithmic approach to maximization of matrix informativeness (MARE) similarly indicated consistency in tree topologies. ML analysis of both reduced supermatrices (Matrices 6 and 7) resulted in the same topology as their respective precursors (Matrices 2 and 4, respectively; figure 2; electronic supplementary material, figures S8 and S9), indicating that discordance at the base of Buthida and elsewhere is not the result of differential gene informativeness in various supermatrices.
With regard to the impact of evolutionary rate on phylogenetic signal, we examined three sub-partitions of a relatively complete matrix (Matrix 3), thereby controlling for the size (i.e. number of sites) and completeness of analysed matrices (note similar dataset occupancies of Matrices 8–10; figure 2a). Barring the placement of Scorpiops sp. and the outgroup species Eremobates sp. in the slowest evolving tertile (Matrix 8, 500 genes), the topologies obtained were identical to the concatenated 1557 gene ML phylogeny (Matrix 3; electronic supplementary material, figures S10–S12). This result contrasts with other studies that have shown a demonstrable effect of evolutionary rate on tree topology [15,56,57], specifically concluding that slowly evolving genes may be more suitable for accurate phylogenetic reconstruction when: (i) long-branch attraction artefacts are incident, and (ii) deep nodes are of interest. But in the present study, we observed relatively uniform patristic distances from the MRCA of Opiliones and Scorpiones upon conducting a Bayesian relative rates test (procedure provided in the electronic supplementary material), suggesting that none of the scorpion species we analysed constitutes a long-branch terminal when inferred with high-occupancy matrices (electronic supplementary material, figure S21). In addition, recent molecular divergence time estimates of arachnids suggest that the diversification of scorpions may not constitute deep (i.e. Palaeozoic) nodes, contrary to previous conjecture [58].
As an external test of OMA's accuracy in predicting single-copy orthologues, the subset of orthologues in Matrices 1–4 that overlapped with benchmarked ‘universal’ single-copy orthologues common to all arthropods was used to construct another family of supermatrices (Matrices 11–14). These similarly yielded the same basal topology as the majority of analyses, even when the number of constituent orthologues was as low as 67 genes (figure 2; electronic supplementary material, figures S15–S18).
Analyses with mixed branch length models of heterotachy recovered a single branch length category for both Matrices 1 and 11. Basal scorpion relationships (i.e. recovery of Buthida, Iurida, etc.) were not affected by the use of the branch length mixture models, suggesting that heterotachy does not strongly affect our dataset (figure 2b; electronic supplementary material, figures S19 and S20).
(b). Species tree and supernetwork approaches
All three semi-parametric species tree methods (MP-EST, STAR and NJst; see Material and methods) applied to genes included in Matrices 1 and 2 recovered the same basal split between the Buthida and Iurida (figure 2b; electronic supplementary material, figures S5 and S6). The STAR and NJst trees derived from gene trees of Matrix 1 recovered the sister relationship of Buthidae and Pseudochactidae (as in the concatenated ML analysis of Matrix 1), but the MP-est method applied to the same dataset recovered the sister relationship of Chaerilidae and Pseudochactidae (as in the concatenated BI analysis of Matrix 1). These data indicate some incongruence at the base of Buthida. Inversely, the MP-est method applied to both datasets recovered a more nested placement of Bothriurus with respect to all other analyses, indicating additional incongruence within ‘Chactoidea’ (including Caraboctonidae).
Upon quantifying gene tree incongruence, we observed large numbers of potentially informative genes for almost every node in Matrix 3 (more than 1000 potentially informative genes per node) and Matrix 4 (more than 2000 potentially informative genes per node) (electronic supplementary material, figures S22 and S23). The fraction of potentially informative genes that was congruent with a given node was generally high for both datasets, except for nodes corresponding to divergences within ‘Chactoidea’. Intriguingly, similar proportions were obtained for the sister group of Pseudochactidae in Matrix 3 (sister to Buthidae: 0.240; sister to Chaerilidae: 0.230) and Matrix 4 (sister to Buthidae: 0.286; sister to Chaerilidae: 0.250), corroborating incongruence at the base of Buthida. Only a small proportion of gene trees was congruent with the alternative placement of Pseudochactidae at the base of the scorpion tree or the placement of Chaerilidae at the base of Iurida in either dataset (less than 0.15). Similarly, the traditionally held monophyly of Iuroidea, Scorpionoidea (including Bothriuridae), and several families was not supported.
All supernetworks (corresponding to Matrices 2–4) indicate consistency with the major result of the supermatrix and species tree approaches, with largely tree-like networks that bear reticulations (indicative of gene conflict) at the base of Buthida and within Iurida (the nodes corresponding to the base of ‘Chactoidea’) (electronic supplementary material, figure S24). Comparatively less gene tree incongruence is observed than in other arthropod datasets (e.g. [15,54]).
4. Discussion
(a). A robust hypothesis of scorpion relationships
This study comprises, to our knowledge, the first comprehensive treatment of scorpion phylogenetic relationships with molecular sequence data sampling all major lineages (superfamilies, sensu [20]). All analyses converged upon a basal tree topology of scorpions that is greatly at odds with traditional hypotheses based on morphology, at every taxonomic level (figure 2c). Unprecedented aspects of our tree topology include the unambiguous inclusion of Buthidae, Chaerilidae and Pseudochactidae in a clade (parvorder Buthida) sister to the remaining scorpions (parvorder Iurida), controverting the overemphasized significance of plesiomorphic anatomy in Pseudochactidae, or the morphological similarities between Iurida and Chaerilidae [20,24]. The non-monophyly of all superfamilies containing multiple constituent families (Chactoidea, Iuroidea and Scorpionoidea), as well as the non-monophyly of several families represented by multiple terminals, indicates pervasive and strong discordance between traditional systematics and molecular phylogenetic signals.
This result is unusual because morphological characters are selected and defined a priori by investigators for their informativeness. In many arthropod clades, clear stepwise gains or losses of morphological characters have historically implied certain basal relationships in groups like insects (e.g. flight; holometaboly), centipedes (e.g. lateral spiracles; number of leg-bearing segments), harvestmen (e.g. direct sperm transfer; median ocelli; paired tarsal claws), and spiders (e.g. unsegmented opisthosoma; venom glands; labidognathous chelicerae), and these evolutionary trends have been robustly validated by phylogenomic data [14,54,59–64]. In the case of scorpions, barring the clear separation of buthids from non-buthid lineages, there has been little agreement as to how scorpion families are related.
The tree topology we obtained across all analyses indicates that most of the character systems commonly used in scorpion systematics are uninformative at the superfamilial level, due to autapomorphic character state distributions with respect to superfamilies or families (electronic supplementary material, figure S25 and table S3) [19,24,65]. A handful of characters unites Iurida and supports the mutual monophyly of Buthidae, Chaerilidae and Pseudochactidae. Almost no characters support interfamilial relationships within Buthida or Iurida, or simply conflict with one another (e.g. within Buthida: cheliceral dentition; hemispermatophore structure; lamellar surface of book lungs). Others still that are variable within these two clades demonstrate homoplasy with respect to the molecular topology (electronic supplementary material, figure S25 and table S3). Our results therefore indicate a need for statistical evaluation of informative discrete morphological character systems (sensu [66]), as well as reassessment of palaeontological systematics of the group [1–3].
(b). A single origin of katoikogenic development
Convergent evolution induced by adaptations to substrate type is prevalent in many lineages of scorpions, possibly driving homoplasy in many characters drawn from external morphology [67,68]. Internal morphology may be less prone to homoplasy that stems from adaptation to substrate, and thus may be more informative at the higher taxonomic levels, as exemplified by mode of embryonic development (electronic supplementary material, figure S25 and table S3). While all scorpions are viviparous, most have large, yolky eggs, with embryonic development occurring in the oviduct, and embryos surrounded by embryonic membranes (apoikogenic development). Only a handful of lineages bears small eggs with no yolk or embryonic membranes; development of the embryos occurs in modified outgrowths of the ovariuterus that enable trophic exchange from the adult female's hepatopancreas to the embryos, via the embryonic chelicerae (katoikogenic development). This unique developmental process unites the non-bothriurid Scorpionoidea, which are invariably recovered as a clade nested within Iurida (figure 2). Concordantly, Bothriuridae, the only putative member of Scorpionoidea that lacks katoikogenic development, was excluded from this clade in all phylogenomic analyses.
The disposition of the digestive glands is distributed in a comparable manner to katoikogenic development (electronic supplementary material, figure S25). All scorpions bear compact digestive glands, excepting the non-bothriurid scorpionoids, which bear digitiform digestive glands [65]. However, the two characters are strongly correlated, probably owing to the physiological and/or physical requirements of katoikogenic development.
(c). Increments to taxonomic sampling reveal additional non-monophyletic groups
The limited taxonomic sampling in this study precludes rigorous investigation of derived relationships, although our analyses surprisingly did suggest the non-monophyly of some families (e.g. Hormuridae, Vaejovidae). Owing to the paucity of genomic resources available for Iurida, few existing datasets can presently be added to our supermatrices, and at considerable expense of matrix occupancy (figure 3; electronic supplementary material, table S2). Adding four small datasets to our analyses (Scorpio maurus palmatus, Heterometrus petersii and two species of Scorpiops) indicated the monophyly of the genus Scorpiops with maximal nodal support, even though few genes (23–35) are available for the Scorpiops species; other relationships within ‘Chactoidea’ were not affected (figure 3a). By contrast, addition of two scorpionids rendered the family Scorpionidae diphyletic in the best-scoring ML topology, with Scorpio maurus palmatus (represented by seven genes) nesting within the non-Liocheles hormurids (bootstrap resampling frequency of 69%; figure 3a).
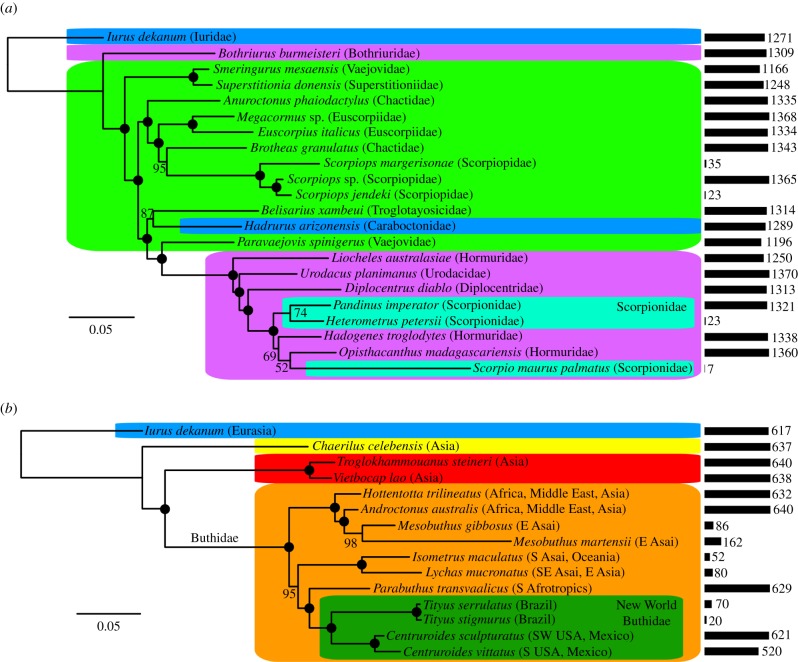
Figure 3.
(a) Phylogeny of Iurida based on ML analysis of 1405 genes, including libraries not sequenced in this study. Bars at right with accompanying numbers indicate gene representation for each terminal. Numbers on nodes indicate bootstrap resampling frequencies. (b) Phylogeny of Buthidae based on ML analysis of 649 genes, including libraries not sequenced in this study. Shading corresponds to superfamilies (as in figure 1) and to distinguish Scorpionidae and New World Buthidae. (Online version in colour.)
The paucity of gene representation for the genera Heterometrus and Scorpio from our dataset, together with the absence of the fourth scorpionid genus, Opistophthalmus, renders the diphyly of Scorpionidae dubious at present. But given robustly supported non-monophyly of Hormuridae and Scorpionoidea sensu lato, the monophyly of Scorpionidae must now be regarded with guarded skepticism as well.
Comparatively more genomic resources are available for Buthidae, the most species-rich family of scorpions, which includes nearly all medicinally significant species. Inclusion of the genome of the buthid M. martensii in tandem with several smaller datasets previously sequenced revealed a robust internal phylogeny of available Buthidae (figure 3b), with the New World buthids (represented here by Centruroides and Tityus) definitively nested within Old World counterparts. This result is somewhat consistent with an earlier inference based on analysis of 16S rRNA sequences [25]; both results controvert the previously hypothesized basal split between Palaeotropical and Neotropical buthids [23].
(d). Topological incongruence of sparse supermatrices is attributable to non-random distribution of missing data
While the supermatrices we analysed constitute simultaneously some of the largest and most complete in arthropod phylogenomic literature [15,59,60], we obtained the aberrant result of a single analysis somewhat consistent with previous morphology-based systematics: the ML analysis of the 5025 gene supermatrix (figure 2e; electronic supplementary material, figure S4). The tree topology recovered by this analysis yields the monophyly of ‘Chactoidea’ (including Caraboctonidae, previously in superfamily Iuroidea) and renders Scorpionoidea paraphyletic instead of polyphyletic.
A corollary of its size, the distinguishing feature of Matrix 4 is its amount of missing data, which exceeds that of all other supermatrices we analysed. While this matrix still contains a formidable degree of completeness (64.2% occupancy), the pernicious effects of missing data have been previously elucidated by Roure et al. [69], among others. Deleterious and misleading effects of missing data in phylogenomic analyses include model misspecification and exacerbation of long-branch artefacts [69], wherefore nearly all recent phylogenomic studies have emphasized maximizing matrix occupancy (e.g. [59–64]). We therefore focused on identifying whether clade-specific absences of data (i.e. non-random distribution of missing cells in the matrix) could be driving support for spurious nodes.
Using a permutation-based approach to identify genes with non-random distribution of absences and presences (procedure provided in the electronic supplementary material, Methods section and figure S26a,b; see also [60]), we observed that the number of genes for which missing data distribution is significantly different from random (at α = 0.05) increases disproportionally as the taxon occupancy threshold decreases. Whereas the proportion of constituent genes with non-random distribution of missing data is less than 10% for Matrices 1–3, this proportion increases to 20.2% for the 5025 gene matrix (electronic supplementary material, figure S26c). To test whether non-random distribution of missing data contributed to support for spurious nodes recovered by Matrix 4, we ran a separate ML analysis of only the 4008 genes wherein missing data were randomly distributed. In this analysis, we discovered that nodes corresponding to basal relationships within a putatively monophyletic ‘Chactoidea’ + Caraboctonidae are all unsupported (bootstrap resampling frequencies of 30–36%). The majority of bootstrap replicates support the topology recovered by other analyses, i.e. paraphyly of ‘Chactoidea’ + Caraboctonidae (figure 2c; electronic supplementary material, figure S26d). These results are consistent with a pervasive effect of non-randomly distributed missing data in sparse supermatrices in inflating nodal support frequencies for spurious relationships [69]. We therefore treat the tree topology of Matrix 4 with scepticism and favour instead the basal tree topology depicted in figure 2, which was robustly supported by all other more complete matrices.
5. Conclusion
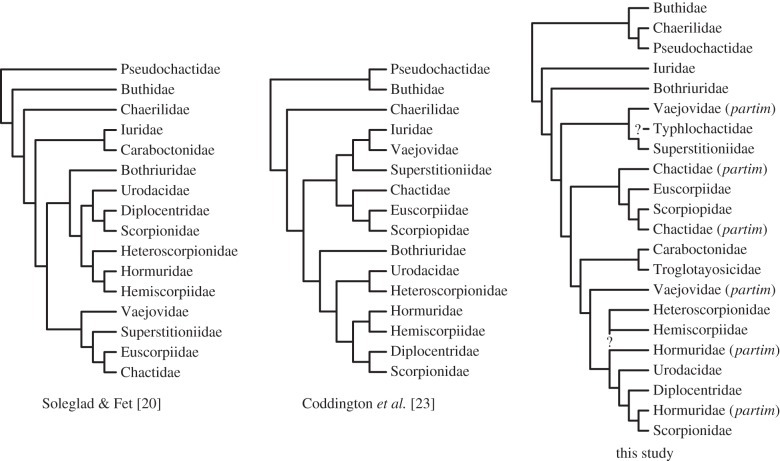
We executed multidimensional analyses of some of the largest and most complete datasets in arthropod phylogenomics to resolve for the first time, to our knowledge, the phylogeny of scorpions, one of the most iconic arthropod groups. In accordance with our results, and to simplify the state of scorpion higher level systematics, we provide herein a revised classification of the group (table 1). The basal topology revealed by our analyses (figure 4), and particularly the placement of Pseudochactidae and Chaerilidae, is anticipated to transform the design of forthcoming studies investigating the early evolution of scorpion venoms, placement of fossils and molecular dating.
Figure 4.
Comparison of historical hypotheses of scorpion relationships (left and middle) with a majority rule consensus all tree topologies obtained in this study. Note non-monophyly of multiple families. Question marks indicate three non-sampled lineages whose putative placement (based on morphology) is now also suspect.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Gonzalo Giribet for use of laboratory and tissue storage facilities for this work. Comments from Gonzalo Giribet and Ward C. Wheeler refined some of the ideas presented herein. Mike Rix (University of Adelaide), Mark Harvey (Western Australian Museum), and Andy Austin and Steve Cooper (South Australian Museum) generously contributed the library of Urodacus planimanus for inclusion in our analyses. Vilaivan Vorlachak and Vianney Catteau provided invaluable assistance to P.P.S. during fieldwork in Laos. Marc M. Santiago assisted with Python-to-R script translation. Sebastian Kvist and Alejandro Oceguera Figueroa kindly facilitated permitting for specimens collected by E.G.S. Comments from Philip Donoghue and two anonymous reviewers greatly improved a previous manuscript draft.
Author contributions
The authors jointly conceived of the ideas presented in this study. Fieldwork was conducted in Mexico by E.G.S., in Europe by L.M., and in Southeast Asia by P.P.S. R.F. conducted molecular work. P.P.S. conducted phylogenomic analyses. All authors contributed to the writing of the manuscript.
Funding statement
Fieldwork was additionally supported by internal funds from the Muséum d'Histoire Naturelle de la Ville de Genève to L.M., and by UNAM-DGAPA-PAPIIT IN213612 to Fernando Álvarez Padilla. This material is based on work supported by the National Science Foundation Postdoctoral Research Fellowship in Biology under grant no. DBI-1202751 to P.P.S.
References
- 1.Kjellesvig-Waering EN. 1986. A restudy of the fossil Scorpionida of the world. Palaeontogr. Am. 55, 1–287. [Google Scholar]
- 2.Jeram AJ. 1998. Phylogeny, classifications and evolution of Silurian and Devonian scorpions. In Proc. 17th European Colloquium of Arachnology, Edinburgh, UK (ed. Selden PA.), pp. 17–31. Burnham Beeches, UK: British Arachnological Society. [Google Scholar]
- 3.Dunlop JA. 2010. Geological history and phylogeny of Chelicerata. Arthropod Struct. Dev. 39, 124–142. ( 10.1016/j.asd.2010.01.003) [DOI] [PubMed] [Google Scholar]
- 4.Prendini L. 2011. Order Scorpiones C.L. Koch, 1850. In Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (ed. Zhang Z-Q.), pp. 115–117. Zootaxa 3148 Auckland, New Zealand: Magnolia Press. [Google Scholar]
- 5.Selden PA, Jeram AJ. 1989. Palaeophysiology of terrestrialisation in the Chelicerata. Trans. R. Soc. Edinb. Earth Sci. 80, 303–310. ( 10.1017/S0263593300028741) [DOI] [Google Scholar]
- 6.Dunlop JA. 1998. The origins of tetrapulmonate book lungs and their significance for chelicerate phylogeny. In Proc. 17th European Colloquium of Arachnology, Edinburgh, UK (ed. Selden PA.), pp. 9–16. Burnham Beeches, UK: British Arachnological Society. [Google Scholar]
- 7.Dunlop JA, Braddy SJ. 2001. Scorpions and their sister group relationships. In Scorpions 2001. In Memoriam Gary A. Polis. (eds Fet V, Selden PA.), pp. 1–24. Burnham Beeches, UK: British Arachnological Society. [Google Scholar]
- 8.Scholtz G, Kamenz C. 2006. The book lungs of Scorpiones and Tetrapulmonata (Chelicerata, Arachnida): evidence for homology and a single terrestrialization event of a common arachnid ancestor. Zoology 109, 2–13. ( 10.1016/j.zool.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 9.Giribet G, Edgecombe GD, Wheeler WC, Babbitt C. 2002. Phylogeny and systematic position of Opiliones: a combined analysis of chelicerate relationships using morphological and molecular data. Cladistics 18, 5–70. ( 10.1006/clad.2001.0185) [DOI] [PubMed] [Google Scholar]
- 10.Shultz JW. 2007. A phylogenetic analysis of the arachnid orders based on morphological characters. Zool. J. Linn. Soc. 150, 221–265. ( 10.1111/j.1096-3642.2007.00284.x) [DOI] [Google Scholar]
- 11.Van der Hammen L. 1989. An introduction to comparative arachnology. Leiden, The Netherlands: SPB Academic Publishing. [Google Scholar]
- 12.Weygoldt P, Paulus HF. 1979. Untersuchungen zur Morphologie, Taxonomie und Phylogenie der Chelicerata. Z. Zool. Syst. Evolutionsforschung 17, 85–116, 177–200 ( 10.1111/j.1439-0469.1979.tb00694.x) [DOI] [Google Scholar]
- 13.Wheeler WC, Hayashi CY. 1998. The phylogeny of the extant chelicerate orders. Cladistics 14, 173–192. ( 10.1111/j.1096-0031.1998.tb00331.x) [DOI] [PubMed] [Google Scholar]
- 14.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079–1083. ( 10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 15.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 16.Lamoral BH. 1980. A reappraisal of the suprageneric classification of recent scorpions and their zoogeography. In Verhandlungen. 8. Internationaler Arachnologen-Kongress abgehalten ander Universität für Bodenkultur Wien (ed. Gruber J.), pp. 439–444, 7–12 Juli, 1980 Vienna, Austria: H. Egermann. [Google Scholar]
- 17.Lourenço WR. 1985. Essai d'Interprétation de la Distribution du Genre Opisthacanthus (Arachnida, Scorpiones, Ischnuridae) dans les Régions Néotropicales et Afrotropicale. Étude Taxinomique, Biogéographique, Évolutive et Écologique. University Paris VI; Paris, France: Universit, Pierre et Marie Curie. [Google Scholar]
- 18.Stockwell SA. 1989. Revision of the phylogeny and higher classification of scorpions (Chelicerata), 319 p. PhD thesis, University of Berkeley, Berkeley, California; University Microfilms International, Ann Arbor, MI, USA. [Google Scholar]
- 19.Sissom WD. 1990. Systematics, biogeography, and paleontology. In The biology of scorpions (ed. Polis GA.), pp. 64–160. Stanford, CA: Stanford University Press. [Google Scholar]
- 20.Soleglad ME, Fet V. 2003. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius 11, 1–57. [Google Scholar]
- 21.Fet V, Soleglad ME. 2005. Contributions to scorpion systematics. I. On recent changes in high-level taxonomy. Euscorpius 31, 1–13. [Google Scholar]
- 22.Prendini L, Wheeler WC. 2005. Scorpion higher phylogeny and classification, taxonomic anarchy, and standards for peer review in online publishing. Cladistics 21, 446–494. ( 10.1111/j.1096-0031.2005.00073.x) [DOI] [PubMed] [Google Scholar]
- 23.Coddington JA, Giribet G, Harvey MS, Prendini L, Walter DE. 2004. Arachnida. In Assembling the tree of life (eds Cracraft J, Donoghue MJ.), pp. 296–318. New York, NY: Oxford University Press. [Google Scholar]
- 24.Prendini L, Volschenk ES, Maaliki S, Gromov AV. 2006. A ‘living fossil’ from Central Asia: the morphology of Pseudochactas ovchinnikovi Gromov, 1998 (Scorpiones: Pseudochactidae), with comments on its phylogenetic position. Zool. Anz. 245, 211–248. ( 10.1016/j.jcz.2006.07.001) [DOI] [Google Scholar]
- 25.Fet V, Gantenbein B, Gromov AV, Lowe G, Lourenço WR. 2003. The first molecular phylogeny of Buthidae (Scorpiones). Euscorpius 4, 1–10. [Google Scholar]
- 26.Prendini L, Crowe TM, Wheeler WC. 2003. Systematics and biogeography of Scorpionidae (Chelicerata: Scorpiones), with a discussion on phylogenetic methods. Invert. Syst. 17, 185–259. ( 10.1071/IS02016) [DOI] [Google Scholar]
- 27.Ma Y, Zhao R, Cao Z, Li W. 2010. Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics 10, 2471–2485. ( 10.1002/pmic.200900763) [DOI] [PubMed] [Google Scholar]
- 28.He Y, et al. 2013. Molecular diversity of Chaerilidae venom peptides reveals the dynamic evolution of scorpion venom components from Buthidae to non-Buthidae. J. Proteomics 89, 1–14. ( 10.1016/j.jprot.2013.06.007) [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 4, 2602 ( 10.1038/ncomms3602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Norma-Rashid Y, Lourenço WR, Zhu M. 2013. True lateral eye numbers for extant buthids: a new discovery on an old character. PLoS ONE 8, e55125 ( 10.1371/journal.pone.0055125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kluβmann-Fricke BJ, Prendini L, Wirkner CS. 2012. Evolutionary morphology of the hemolymph vascular system in scorpions: a character analysis. Arthropod Struct. Dev. 41, 545–560. ( 10.1016/j.asd.2012.06.002) [DOI] [PubMed] [Google Scholar]
- 32.Michalik P, Mercati D. 2010. First investigation of the spermatozoa of a species of the superfamily Scorpionoidea (Opistophthalmus penrithorum, Scorpionidae) with a revision of the evolutionary and phylogenetic implications of sperm structures in scorpions (Chelicerata, Scorpiones). J. Zool. Syst. Evol. Res. 48, 89–101. ( 10.1111/j.1439-0469.2009.00540.x) [DOI] [Google Scholar]
- 33.Sharma PP, Schwager EE, Extavour CG, Wheeler WC. 2014. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc. R. Soc. B 281, 20140661 ( 10.1098/rspb.2014.0661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prendini L. 2000. Phylogeny and classification of the superfamily Scorpionoidea Latreille, 1802 (Chelicerata, Scorpiones): an exemplar approach. Cladistics 16, 1–78. ( 10.1111/j.1096-0031.2000.tb00348.x) [DOI] [PubMed] [Google Scholar]
- 35.Prendini L, Francke OF, Vignoli V. 2010. Troglomorphism, trichobothriotaxy and typhlochactid phylogeny (Scorpiones, Chactoidea): more evidence that troglobitism is not an evolutionary dead-end. Cladistics 26, 117–142. ( 10.1111/j.1096-0031.2009.00277.x) [DOI] [PubMed] [Google Scholar]
- 36.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: Orthology inference among 1000 complete genomes. Nucleic Acids Res. 39, D289–D294. ( 10.1093/nar/gkq1238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenhoff AM, Gil M, Gonnet GH, Dessimoz C. 2013. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS ONE 8, e53786 ( 10.1371/journal.pone.0053786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altenhoff AM, Dessimoz C. 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 5, e1000262 ( 10.1371/journal.pcbi.1000262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieselt-Struwe K, von Haeseler A. 2001. Quartet-mapping, a generalization of the likelihood-mapping procedure. Mol. Biol. Evol. 18, 1204–1219. ( 10.1093/oxfordjournals.molbev.a003907) [DOI] [PubMed] [Google Scholar]
- 40.Meusemann K, et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol. Biol. Evol. 27, 2451–2464. ( 10.1093/molbev/msq130) [DOI] [PubMed] [Google Scholar]
- 41.Kück P, Struck TH. 2014. BaCoCa: a heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Mol. Phylogenet. Evol. 70, 94–98. ( 10.1016/j.ympev.2013.09.011) [DOI] [PubMed] [Google Scholar]
- 42.Cummins CA, McInerney JO. 2011. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. Syst. Biol. 60, 833–844. ( 10.1093/sysbio/syr064) [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. 2012. OrthoDB: a hierarchical catalog of animal, fungal, and bacterial orthologs. Nucleic Acids Res. 41, D358–D365. ( 10.1093/nar/gks1116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 45.Le SQ, Dang CC, Gascuel O. 2012. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol. Biol. Evol. 29, 2921–2936. ( 10.1093/molbev/mss112) [DOI] [PubMed] [Google Scholar]
- 46.Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 60, 291–302. ( 10.1093/sysbio/syr010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615. ( 10.1093/sysbio/syt022) [DOI] [PubMed] [Google Scholar]
- 48.Kolaczkowski B, Thornton JW. 2008. A mixed branch length model of heterotachy improves phylogenetic accuracy. Mol. Biol. Evol. 25, 1054–1066. ( 10.1093/molbev/msn042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 50.Hanson-Smith V. 2013. M3L. Google Code Repository See https://code.google.com/p/m3l/.
- 51.Liu L, Yu L, Pearl DK, Edwards SV. 2009. Estimating species phylogenies using coalescence times among sequences. Syst. Biol. 58, 468–477. ( 10.1093/sysbio/syp031) [DOI] [PubMed] [Google Scholar]
- 52.Liu L, Yu L, Edwards SV. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Biol. 10, 302 ( 10.1186/1471-2148-10-302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Yu L. 2011. Estimating species trees from unrooted gene trees. Syst. Biol. 60, 661–667. ( 10.1093/sysbio/syr027) [DOI] [PubMed] [Google Scholar]
- 54.Fernández R, Laumer CE, Vahtera V, Libro S, Kaluziak ST, Sharma PP, Pérez-Porro AR, Edgecombe GD, Giribet G. 2014. Evaluating topological conflict in centipede phylogeny using transcriptomic data sets. Mol. Biol. Evol. 31, 1500–1513. ( 10.1093/molbev/msu108) [DOI] [PubMed] [Google Scholar]
- 55.Grünewald S, Spillner A, Bastkowski S, Bogershausen A, Moulton V. 2013. SuperQ: computing supernetworks from quartets. IEEE/ACM Trans. Comput. Biol. Bioinform. 10, 151–160. ( 10.1109/TCBB.2013.8) [DOI] [PubMed] [Google Scholar]
- 56.Nosenko T, et al. 2013. Deep metazoan phylogeny: When different genes tell different stories. Mol. Phylogenet. Evol. 67, 223–233. ( 10.1016/j.ympev.2013.01.010) [DOI] [PubMed] [Google Scholar]
- 57.Telford MJ, Lowe CJ, Cameron CB, Ortega-Martinez O, Aronowicz J, Oliveri P, Copley RR. 2014. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc. R. Soc. B 281, 20140479 ( 10.1098/rspb.2014.0479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma PP, Wheeler WC. 2014. Cross-bracing uncalibrated nodes in molecular dating improves congruence of fossil and molecular age estimates. Front. Zool. 11, 57 ( 10.1186/s12983-014-0057-x) [DOI] [Google Scholar]
- 59.Simon S, Narechania A, DeSalle R, Hadrys H. 2012. Insect phylogenomics: exploring the source of incongruence using new transcriptomic data. Genome Biol. Evol. 4, 1295–1309. ( 10.1093/gbe/evs104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 61.Sharma PP, Giribet G. 2014. A revised, dated phylogeny of the arachnid order Opiliones. Front. Genet. 5, 255 ( 10.3389/fgene.2014.00255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hedin M, Starett J, Akhter S, Schönhofer AL, Shultz JW. 2012. Phylogenomic resolution of Paleozoic divergences in harvestmen (Arachnida, Opiliones) via analysis of next-generation transcriptome data. PLoS ONE 7, e42888 ( 10.1371/journal.pone.0042888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández R, Hormiga G, Giribet G. 2014. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr. Biol. 24, 1772–1777. ( 10.1016/j.cub.2014.06.035) [DOI] [PubMed] [Google Scholar]
- 64.Bond JE, Garrison NL, Hamilton CA, Godwin RL, Hedin M, Agnarsson I. 2014. Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr. Biol. 24, 1765–1771. ( 10.1016/j.cub.2014.06.034) [DOI] [PubMed] [Google Scholar]
- 65.Volschenk ES, Mattoni CI, Prendini L. 2008. Comparative anatomy of the mesosomal organs of scorpions (Chelicerata, Scorpiones), with implications for the phylogeny of the order. Zool. J. Linn. Soc. 154, 651–675. ( 10.1111/j.1096-3642.2008.00426.x) [DOI] [Google Scholar]
- 66.Blomberg SP, Garland T, Jr, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 67.González-Santillán E, Prendini L. 2013. Redefinition and generic revision of the North American vaejovid scorpion subfamily Syntropinae Kreaepelin, 1905, with descriptions of six new genera. Bull. Am. Mus. Nat. Hist. 382, 1–71. ( 10.1206/830.1) [DOI] [Google Scholar]
- 68.Monod L, Prendini L. 2014. Evidence for Eurogondwana: the roles of dispersal, extinction and vicariance in the evolution and biogeography of Indo-Pacific Hormuridae (Scorpiones: Scorpionoidea). Cladistics 31, 71–111. ( 10.1111/cla.12067) [DOI] [PubMed] [Google Scholar]
- 69.Roure B, Baurain D, Philippe H. 2013. Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Mol. Biol. Evol. 30, 197–214. ( 10.1093/molbev/mss208) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.